Abstract
The prolamin box (P-box) is a highly conserved 7-bp sequence element (5′-TGTAAAG-3′) found in the promoters of many cereal seed storage protein genes. Nuclear factors from maize endosperm specifically interact with the P-box present in maize prolamin genes (zeins). The presence of the P-box in all zein gene promoters suggests that interactions between endosperm DNA binding proteins and the P-box may play an important role in the coordinate activation of zein gene expression during endosperm development. We have cloned an endosperm-specific maize cDNA, named prolamin-box binding factor (PBF), that encodes a member of the recently described Dof class of plant Cys2-Cys2 zinc-finger DNA binding proteins. When tested in gel shift assays, PBF exhibits the same sequence-specific binding to the P-box as factors present in maize endosperm nuclei. Additionally, PBF interacts in vitro with the basic leucine zipper protein Opaque2, a known transcriptional activator of zein gene expression whose target site lies 20 bp downstream of the P-box in the 22-kDa zein gene promoter. The isolation of the PBF gene provides an essential tool to further investigate the functional role of the highly conserved P-box in regulating cereal storage protein gene expression.
The prolamin seed storage proteins of maize, named zeins, are encoded by five distinct classes of genes that are distinguished by the molecular mass of their protein products (reviewed in ref. 1). Zein gene mRNA and protein expression are limited to the endosperm and coordinately regulated in a temporal fashion, beginning between 8 and 10 days after pollination (DAP), peaking at 16 DAP, and continuing throughout seed development (reviewed in ref. 2). Though cereal seed storage proteins are the primary source of proteins in human diets worldwide and have long been the subject of intense scientific study, our understanding of the molecular mechanisms regulating their expression is limited. Much of what is known about this process in maize is based on the molecular, genetic, and biochemical analysis of the opaque2 locus. This work has demonstrated that the Opaque2 (O2) gene encodes a basic leucine zipper (bZIP) transcription factor (3, 4) that binds to a promoter element in the 22-kDa class of zein genes to activate their expression (5, 6).
The primary effect of the opaque2 mutation is a reduction in the transcription of a specific subset of zein genes (7). This leads to a corresponding decrease in 22-kDa, and to a lesser extent, 15-kDa zein protein compared with wild type. Consistent with these phenotypic effects, only the 22- and 15-kDa zein gene promoters contain O2 binding sites (5, 8). Therefore, additional regulatory factors and promoter elements must mediate the coordinate activation of all classes of zein genes during endosperm development. The prolamin box (P-box) is a good candidate for such a cis-acting regulatory element, because it is present within the promoters of all zein genes as well as many storage protein genes from related cereals (Fig. 1). The P-box was initially identified on the basis of both its highly conserved nucleotide sequence (5′-TGTAAAG-3′) and position (−300 region) relative to the translation start codon of prolamin genes (9, 10). Endosperm nuclear factors have been shown to bind the P-box present in the 19-, 22- and 27-kDa zein gene promoters (11, 12). Further analysis of these protein-DNA interactions indicate that they may be specific to the endosperm (13, 14). Transient expression assays of zein gene promoter activity in maize endosperm suspension culture cells (12, 14) suggest that the P-box plays a positive role in the coordinate activation of zein gene expression during endosperm development. Interestingly, the P-box in the 22-kDa zein gene promoter lies just 20 nucleotides upstream of the binding site for O2, suggesting that O2 may interact with factors binding the P-box to activate 22-kDa zein gene expression (5). Despite the fact that P-box binding factors were one of the first DNA-protein interactions identified in plants (11), the molecular cloning of the gene encoding the protein that binds the P-box has not yet been reported.
Figure 1.

Alignment of cereal seed storage protein gene promoters in the region of the P-box sequence element. The location of each P-box element is given in nucleotides upstream of the translation start codon. GenBank accession numbers for the different promoter sequences are in parentheses: 22-kDa zein (X55722), 19-kDa zein (V01472), 15-kDa zein (M13507), 27-kDa zein (X58197), 10-kDa zein (M23537), α-coixin (X63113), γ-kafirin (X62480), B-hordein (X87232), C-hordein (M36941), γ-hordein (M36378), D-hordein (X84368), α-gliadin (K03076), LMW glutenin (X07747), HMW glutenin (X12929), ω-secalin (X60295), avenin (J05486), and GluB glutelin (X54193).
Recently, a novel class of DNA binding proteins possessing a conserved Cys2-Cys2 zinc-finger motif, named the Dof or MOA domain (15, 16), has been identified from maize, Arabidopsis, pumpkin, and tobacco (17, 18). Three of these proteins have been shown to specifically interact with a 5′-AAAG-3′ or its reverse and complementary 5′-CTTT-3′ sequence motif (16, 18, 19), which is also present in the P-box (Fig. 1). The Arabidopsis Dof protein OBP1 was isolated on the basis of protein–protein interactions with ocs element binding factor proteins (16), bZIP factors that bind ocs element sequences in the cauliflower mosaic virus 35S (CaMV-35S) promoter and related sequences in the Arabidopsis glutathione S-transferase 6 (GST6) promoter (20). In both the CaMV-35S and GST6 promoters, the binding sites for OBP1 and its interacting OBF proteins are closely linked, separated by only 12 bp in the GST6 promoter (20). The binding of a Dof protein to an AAAG DNA sequence motif and its interaction with bZIP factors that bind to neighboring promoter sites suggested that similar interactions between a Dof protein, the P-box, and O2 also may occur on the 22-kDa zein gene promoter. This report describes the molecular cloning of an endosperm-specific maize cDNA that encodes a Dof domain protein. The cloned protein, which we have named prolamin box binding factor or PBF, binds the P-box with the same specificity as the P-box binding activity in maize endosperm nuclear extracts. Additionally, PBF forms protein–protein contacts in vitro with O2, but not with the related maize bZIP protein OHP1. These results indicate that PBF represents the P-box binding activity observed in maize endosperm nuclei, and that specific protein–protein interactions between PBF and O2 are likely to be important in the regulation of 22-kDa zein gene expression. Considering the conserved sequence and position of the P-box among cereal storage protein genes, it is likely that homologous proteins exist in the endosperm of other cereals. The cloning of the maize PBF gene should facilitate the isolation of corresponding genes in other important grain crops, thus promoting comparative studies of their role in cereal storage protein gene expression.
MATERIALS AND METHODS
Cloning of PBF cDNA.
A reverse transcriptase–PCR approach was used to isolate Dof-encoding cDNAs from maize endosperm. An oligo(dT) tag primer [5′-GTCGACTCTAGAGGATCC(T)12-3′] was used to prime first-strand cDNA synthesis from poly(A)-selected 18-DAP maize endosperm mRNA. The tag primer and two primers derived from conserved residues in the Dof domain (see Fig. 3) were subsequently used in nested PCR amplifications of endosperm cDNA. DNA sequencing of the amplified products revealed that the primary product contained a Dof-related sequence. This gene fragment was then used as a probe to screen approximately 1 × 106 plaques of a cDNA library prepared from maize developing endosperm mRNA (21). Using either standard or reduced stringency hybridization conditions, 20 hybridizing plaques were identified. Eight of these were plaque-purified and determined by restriction mapping and DNA sequencing to represent the same PBF cDNA.
Figure 3.
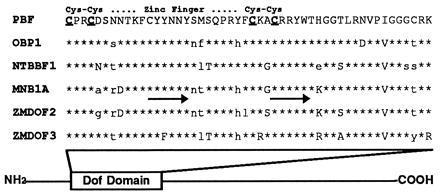
Alignment of Dof domains from maize (PBF, MNB1a [16], ZMDOF2 [14], ZMDOF3 [14]), Arabidopsis (OBP1 [17]), and tobacco (NTBBF1 [18]) genes. ∗ indicate amino acid identity to PBF. Uppercase letters indicate conservative substitutions, whereas lowercase letters denote nonconservative substitutions. The Dof domain is located at the amino terminus for each of these proteins. The regions corresponding to the primer sequences used in the reverse transcriptase–PCR amplification of the PBF cDNA are underlined with arrows.
DNA Binding Assays.
Preparation of nuclear extracts from 16-DAP endosperm (inbred Oh43) and DNA binding assays were performed essentially as described in ref. 5. Three micrograms of protein from endosperm nuclear extracts were incubated in binding buffer at room temperature for 10 min, followed by the addition of 1 × 105 cpm 32P-labeled double-stranded DNA probes and incubation at room temperature 20 min. Bound complexes were resolved on nondenaturing 4% polyacrylamide gels in 0.25× TBE (90 mM Tris/64.6 mM boric acid/2.5 mM EDTA, pH 8.3) at 4°C, dried onto Whatman 3MM paper, and autoradiographed.
For expression of PBF in Escherichia coli, the entire PBF cDNA was first cloned into pBluescript KS. The 1,197-bp NcoI- BamHI fragment from this plasmid, with the NcoI site spanning the start codon, was inserted into NcoI+ BamHI-digested pET-11d (Novagen) and transformed into E. coli strain BL21(D3). Overnight cultures harboring either the recombinant PBF clone or the pET-11d vector without insert were diluted 1:10 in Luria–Bertani medium and grown for 3 hr at 37°C. PBF expression was then induced with 1 mM isopropyl β-d-thiogalactoside from the phage T7 promoter for 1 hr at 30°C. Cells carrying the pET-11d vector with no insert were similarly induced. Bacterial extracts were prepared by pelleting the cells, sonicating in the presence of lysis buffer (10 mM Hepes, pH 7.9/50 mM KCl/1 mM EDTA/1 mM DTT/0.1 mM phenylmethylsulfonyl fluoride/0.5 mg/ml leupeptin/2 mg/ml aprotinin/10% glycerol) and freezing at −70°C. Five micrograms of total protein from the bacterial extracts was incubated in binding buffer with labeled probes in DNA binding assays as described above.
RNA Gel Blot Analysis.
Total RNA was isolated as described (22) from maize wild-type tissues (inbred Oh43) or mutant endosperm homozygous for the o2-R null allele in an Oh43 background (21). RNAs were prepared from 4-day-old seedling roots, 4-day-old seedling shoots, expanding leaves (leaf 10), 1.5-cm immature ears, 1.5-cm immature tassels, 18-DAP embryos, and developing endosperms at 5, 8, 10, 12, 14, 15, 18, 21, and 25 DAP. RNA gel blots were prepared from 10 μg of total RNA, hybridized to random primer-labeled probes, and washed as described in ref. 23. The following gel-purified restriction fragments were used as probes: 720-bp XbaI- SpeI restriction fragment from the cloned PBF cDNA, the entire O2 cDNA (3), and a 653-bp PstI–SacI fragment from the plasmid pSKUBI carrying the maize ubiquitin cDNA (24).
In Vitro Protein–Protein Interaction Assays.
35S-labeled O2, OHP-1 (25), and PBF proteins were produced by in vitro transcription and translation using the TNT system (Promega). Plasmids for the expression of fusion proteins between GST and either the full-length PBF or O2 cDNAs were constructed in the pGEX-2TK vector (Pharmacia). A BamHI site was engineered before the ATG codon of each cDNA clone by PCR and used to generate in-frame fusions to the C terminus of GST, which were subsequently sequenced to ensure fidelity. To produce GST fusion proteins, XL1-Blue E. coli cells harboring either the empty pGEX-2TK vector or the recombinants described above were grown overnight at 37°C, then diluted 1/10 with Luria–Bertani medium and further grown for 1 hr. Protein expression was induced by addition of isopropyl β-d-thiogalactoside to 0.5 mM and cell growth continued for 3 hr at 30°C. Cells were pelleted and resuspended in TNE5 (10 mM Tris, pH 7.5/150 mM NaCl/5 mM EDTA) with 1 mM phenylmethylsulfonyl fluoride and lysed by sonication. Insoluble material was removed by centrifugation after addition of Nonidet P-40 to 1%. GST, or GST fusion proteins were enriched by affinity to glutathione-agarose beads (Sigma) following standard procedures.
The system described by Gyuris et al. (26) was used with minor modifications to test for protein–protein interactions. A typical binding reaction contained 100 μl of a 50% slurry of a particular GST-fusion protein adsorbed to gluthatione-agarose beads equilibrated in NETN buffer (50 mM Tris, pH 7.5/100 mM NaCl/1 mM EDTA/0.1% Nonidet P-40/0.25% gelatin) and 8 μl of TNT-labeled protein. After incubation for 30 min at 20°C the agarose beads were washed four times with 0.01% SDS in NETN and finally resuspended in Laemmli loading buffer. Interacting proteins retained by the resin were resolved by SDS/PAGE in 10% gels and detected by fluorography. Proteins were heated to 37°C for 10 min to promote dissociation of any dimerized bZIP proteins followed by incubation for 30 min at 20°C with gentle agitation (see Fig. 5B). Because bZIP protein homodimer and heterodimer formation has previously been shown to be enhanced in the presence of DNA carrying the bZIP target site sequence (27), the protein–protein complexes (see Fig. 5B) also were allowed to form in the presence of 1 μg of double-stranded oligonucleotide containing an O2 binding site (5).
Figure 5.
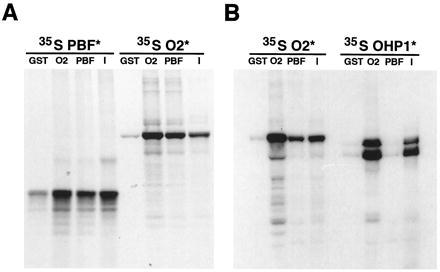
The PBF and O2 proteins interact in vitro. 35S-labeled PBF (predicted molecular mass 30 kDa), O2 (predicted molecular mass 47 kDa), and OHP1 (predicted molecular mass 44 kDa) proteins were incubated with equal amounts of glutathione-agarose beads saturated with either GST alone as a negative control, GST::O2 fusion protein (O2), or GST::PBF fusion protein (PBF). After extensive washes of the beads (see Materials and Methods), the retained proteins were eluted and separated by SDS/PAGE and detected by fluorography. For each experiment a sample of the 35S-labeled protein that was incubated with the GST fusion proteins was run separately as I. (A) The left four lanes show the interaction of GST::O2 with 35S-labeled PBF. The right four lanes show the reciprocal experiment where GST::PBF interacts with 35S-labeled O2. In both experiments the signal recovered by the GST fusion proteins is enhanced compared with that observed with GST alone (negative control). The homodimerization of GST::O2 with 35S-labeled O2 serves as a positive control for interaction. (B) GST::PBF can coprecipitate 35S-labeled O2 but not 35S-labeled OHP1. A selective interaction is reflected by the significant difference in the amounts of 35S-labeled O2 compared with 35S-labeled OHP1 bound by GST::PBF. The amount of 35S-labeled OHP1 signal recovered by GST::PBF is comparable to that observed for the negative control (GST). The homodimerization of O2 and heterodimerization of O2 and OHP1 serve as positive controls for interaction. The two in vitro translated products observed for OHP1 are reproducible (25) and probably represent the full-length OHP1 protein (upper band) and a smaller product (lower band) produced by translation initiation from a methionine 37 residues internal to the start codon.
RESULTS
PBF in Endosperm Nuclei Interacts with the 5′-AAAG-3′ Sequence Motif.
Previous DNaseI footprinting and electrophoretic mobility-shift assays indicated that the P-box binding activity identified in maize endosperm nuclear extracts specifically interacted with the P-box core sequence (12). The introduction of three nucleotide substitutions in the P-box core sequence (from TGTAAAG to TcTAgAc, mutant nucleotides in lowercase letters) dramatically reduced this interaction (12). To further define those nucleotides within the P-box that were recognized by the endosperm nuclear proteins, oligonucleotides that contained a subset of these substitutions (TcTAAAG or TGTAgAc) or a tetramer of the wild-type P-box core sequence (TGTAAAGG) were tested in electrophoretic mobility-shift assays (Fig. 2). A single shifted complex was observed when endosperm nuclear extracts were incubated with a labeled probe carrying the wild-type P-box from the 22Z-4 zein gene promoter (5). This binding activity was greatly enhanced by incubating extracts with a probe containing a tetramer of the P-box core sequence. Consistent with previous reports (12), the TcTAgAc mutated P-box sequence completely eliminated the binding activity present in nuclear extracts. In addition, substitutions specific to the AAAG sequence motif abolished the P-box binding activity present in nuclear extracts, whereas mutations of the TGT sequence alone reduced, but did not eliminate DNA binding. These results indicated that the P-box binding activity present in maize endosperm nuclei interacted primarily with the AAAG sequence motif.
Figure 2.
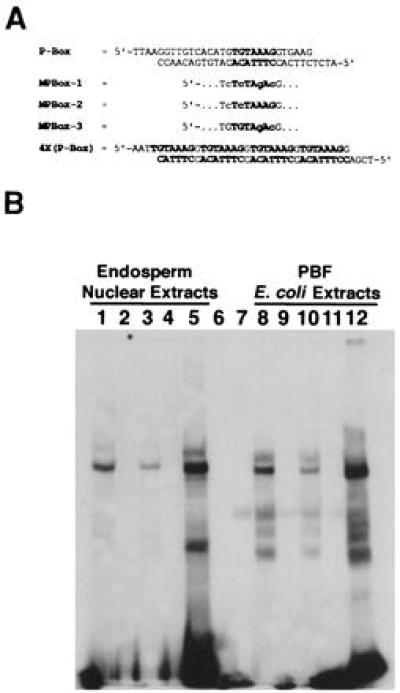
The DNA-binding activities of endosperm nuclear extracts and the PBF cDNA expressed in E. coli. (A) Oligonucleotide sequences of the wild-type P-box from the 22Z-4 promoter (P-box), a tetramer of the P-box core sequence (4XP-Box), and three different mutated derivatives of the P-Box (MPBox-1, MP-Box-2, MPBox-3) used as probes. The MPBox oligonucleotides were identical to the wild-type P-box except for those nucleotide changes in the P-box core sequence indicated in lowercase letters. (B) Electrophoretic mobility-shift assay analysis of the above probes incubated with protein extracts from either maize endosperm nuclei (lanes 1–5), E. coli expressing the PBF cDNA (lanes 8–12), no protein (lane 6), or E. coli harboring the pET-11d vector without insert (lane 7). Lanes 1, 6, 7, and 8 were incubated with the wild-type P-Box probe, lanes 2 and 9 with MPBox-1, lanes 3 and 10 with MPBox-2, lanes 4 and 11 with MP-Box3, and lanes 5 and 12 with the 4X(P-Box) probe. The free probe was partially run off at the bottom of the gel.
Molecular Cloning of a Maize PBF.
Based on the specific interactions between the Arabidopsis OBP1 Dof domain protein with a AAAG DNA sequence motif and its ability to associate with bZIP factors that bind to nearby promoter elements (16), we reasoned that a maize Dof domain protein might interact with the P-box in the 22-kDa zein gene promoter, which lies 20 nucleotides upstream of the binding site for the bZIP protein O2. Using oligonucleotide primers to the conserved Dof domain in reverse transcriptase–PCR assays of maize endosperm RNA, an endosperm cDNA was cloned that encoded a Dof domain protein (Fig. 3). The sequence of this cDNA (deposited in GenBank, accession number U82230) exhibited a high degree of amino acid identity (75–80%) with other Dof domain sequences (17), but is distinct from previously identified maize Dof genes. Outside of the Dof domain, the maize endosperm Dof protein shared no significant amino acid similarity with other Dof proteins. However, the cloned cDNA did show complete sequence identity with a maize EST isolated from an endosperm cDNA library (GenBank accession number T23343).
When the cloned cDNA was expressed in E. coli and bacterial lysates were tested in electrophoretic mobility-shift assays, the cloned Dof protein specifically bound to the P-box in a manner identical to that observed with endosperm nuclear extracts (Fig. 2). The maize Dof protein in the bacterial extracts bound with high affinity to the wild-type P-box probe and demonstrated dramatically increased binding affinity when incubated with the probe containing a tetramer of the P-box core sequence. Similarly, the expressed protein did not bind to the probe where the AAAG sequence motif had been mutated to AgAc. When incubated with the probe containing a mutated TGT sequence, the binding activity of the cloned protein to the probe was reduced in a manner similar to that observed for the endosperm nuclear factor. These results indicate that the cloned maize endosperm Dof cDNA encodes a P-box binding protein that mimics the binding activity of the maize nuclear factor. This protein was therefore named prolamin box binding factor, or PBF.
PBF mRNA Expression Is Endosperm-Specific.
Because both zein gene expression and the interactions between nuclear factors and the P-box appear to be specific to the endosperm (14), the expression of PBF might be expected to be endosperm-specific. The spatial distribution of PBF gene expression was examined among each of the major maize organs (Fig. 4A). PBF mRNA was only detected in RNA samples isolated from endosperm tissue. The temporal accumulation of PBF expression during endosperm development was then investigated (Fig. 4B). PBF expression was first detected at 10 DAP, continued to increase with a peak at 15 DAP, and was maintained throughout the remainder of endosperm development. This profile of expression paralleled that observed for the O2 gene (Fig. 4B) and is consistent with both O2 and PBF being present at significant levels in maize endosperm 1–2 days before the expression of zein gene mRNA, which begins around 12 DAP (1, 7). PBF expression was also observed to be unaffected by null mutations in O2, which demonstrates that PBF expression is not regulated by O2 activity.
Figure 4.
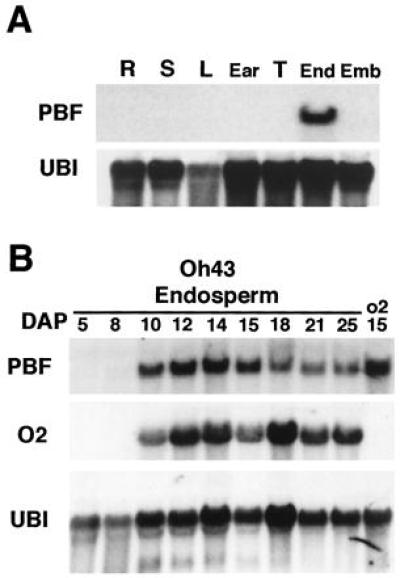
PBF mRNA expression detected by RNA gel blot analysis. (A) Expression of PBF mRNA among different maize organs. R, seedling roots; S, seedling shoots; L, adult leaves; Ear, immature female infloresence; T, immature tassel; End, 15-DAP endosperm; Emb, 18-DAP embryos. The 1.2-kb transcript is only detected in RNA from endosperm. To control for RNA loading, the filter was stripped and reprobed with a maize ubiquitin cDNA fragment that shows expression in all organs. (B) Expression of PBF and O2 mRNA during endosperm development. The expression of both genes is detected beginning 8 DAP and follows a similar profile during the remainder of endosperm development. The o2 lane contains RNA from endosperm at 15 DAP isolated from seeds homozygous for the opaque2-R null allele. The same filter was first probed with PBF, then sequentially stripped and reprobed first with an O2 cDNA fragment followed by a maize ubiquitin cDNA fragment.
PBF Interacts with the O2 Protein in Vitro.
The Dof protein OBP1 was cloned on the basis of protein–protein interactions with OBF bZIP factors (16), demonstrating that members of these two families of DNA-binding proteins can interact. The OBP1 and OBF binding sites are in close proximity within the CaMV-35S and Arabidopsis GST6 promoters, an arrangement that parallels the location of the O2 and PBF binding sites in the 22-kDa zein promoter. Therefore, we tested if PBF can interact with O2. PBF and O2 proteins were produced as fusions to the C terminus of GST and affinity-purified with glutathione beads. The bound fusion proteins were then incubated with 35S-labeled proteins produced in vitro to determine if the labeled proteins would coprecipitate with bound GST fusion proteins (26). The results of reciprocal experiments involving GST::PBF incubated with 35S-O2 and GST::O2 incubated with 35S-PBF are presented in Fig. 5A. Both assays show that the O2 and PBF proteins can recognize one another in vitro, as demonstrated by the increase in labeled proteins recovered after incubation with the GST fusions compared with the nonspecific binding observed for GST alone (negative control). A weaker interaction of GST::PBF with 35S-PBF was also observed. An additional experiment was performed to assess whether PBF forms specific protein–protein contacts with O2, or more generally interacts with the bZIP class of proteins. OHP1 is a bZIP factor that is related to O2 in its bZIP domain, is capable of forming heterodimers with O2, and is also expressed in maize endosperm (25). However, Fig. 5B shows that 35S-OHP1 failed to interact with GST::PBF, whereas protein–protein contacts between O2 and itself, indicative of bZIP homodimerization, as well as heterodimerization between OHP1 and O2, were detected as expected. These results indicate that at least for the O2 and OHP1 endosperm bZIP proteins the association between O2 and PBF is specific.
DISCUSSION
We have cloned a cDNA from maize endosperm that we believe encodes the P-box binding activity present in maize endosperm nuclear extracts. This conclusion is supported by the observation that the PBF cDNA expressed in E. coli produces a protein that binds to the P-box with the same sequence specificity as the factor identified in maize endosperm. We cannot exclude the possibility that some other Dof-domain protein is responsible for the the P-box binding activity observed in endosperm nuclear extracts. However, extensive screening at reduced stringency of our maize endosperm cDNA library with the initial reverse transcriptase–PCR product or the cloned PBF cDNA resulted in the isolation of only PBF cDNAs. This indicates that PBF is at least the most abundant, and probably the only, Dof protein gene expressed at significant levels in maize endosperm.
The mobility in nondenaturing gels of the shifted P-box complexes obtained with cloned PBF showed some variability from experiment to experiment, and differed slightly from the mobility of the P-box binding activity present in endosperm nuclear extracts. This differential migration may indicate that PBF in maize endosperm nuclei is modified posttranslationally (e.g., phosphorylation or glycosylation) in a manner that is not faithfully reproduced in E. coli. Alternatively, PBF protein expressed in E. coli may be sensitive to proteolysis or subject to premature termination of translation.
Endosperm nuclear factors that bind the P-box promoter element were one of the first protein-DNA interactions identified in plants (11). However, extensive screens by our lab and others to identify P-box binding proteins from endosperm cDNA expression libraries with labeled P-box probes were unsuccessful. A probable explanation for the failure to identify PBF by this method is the presence of an in-frame stop codon 18-bp upstream of the ATG initiation codon. The position of this stop codon, coupled with the amino-terminal location of the Dof domain within PBF, would greatly reduce the probability of obtaining a cDNA insert that would produce both an in-frame fusion protein and still contain a functional Dof DNA-binding domain.
The association between PBF and O2 observed in vitro is likely to represent an important functional interaction in vivo. This hypothesis is supported by the fact that O2 and PBF are coexpressed specifically in endosperm tissue and exhibit identical patterns of temporal accumulation during endosperm development. Additionally, PBF failed to interact with OHP1, a bZIP factor that is related to and capable of forming heterodimers with O2 (25), but does not activate 22-kDa zein gene expression (R.L.P. and R.J.S., unpublished work). Further evidence for a functional interaction between PBF and O2 comes from transient expression assays of zein promoter/reporter constructs in maize endosperm suspension culture cells. It previously has been shown that 22-kDa zein promoter activity is enhanced on average 5- to 10-fold in response to O2 expressed from an effector plasmid (28). Recently, we have determined that deletion of the entire P-box element or specific substitutions in the 5′-AAAG-3′ sequence motif (to AgAc) greatly reduced the ability of O2 to transactivate the 22-kDa zein gene promoter (R.L.P., S.P.M., and R.J.S., unpublished work). These results indicate that the binding of an endosperm protein (presumably PBF) to the P-box is required for O2-dependent activation of reporter gene expression. Similar interactions have been proposed for nuclear factors that bind the P-box and a neighboring bZIP consensus sequence from the wheat glutenin and barley C-hordein storage protein genes (29–31).
The protein-interaction assays (Fig. 5) also revealed that PBF interacts with itself in vitro, suggesting that PBF may function as a dimer. Whether such an interaction may be important for DNA binding or some other activity associated with PBF is currently unknown. The GST::PBF self-interaction appears to be weaker than that observed between GST::PBF and O2. This suggests that at least in vitro, the interaction of PBF with O2 is favored over PBF dimerization.
The analysis of both 22-kDa and 27-kDa zein gene promoter activity in transient expression assays with endosperm culture cells (6, 11) suggested that factors binding the P-box participate in the coordinate activation of zein genes during endosperm development. At least three observations regarding PBF and the P-box support this view: (i) PBF mRNA is specific to the endosperm and accumulates immediately before the activation of zein gene expression, (ii) PBF specifically interacts with O2, a known activator of zein gene expression, and (iii) the P-box is required for the O2-dependent trans-activation of the 22-kDa zein gene promoter in transient expression assays. One mechanism for PBF action may be the stimulation of O2 binding to its target sites in 22- and 15-kDa zein gene promoters. The Arabidopsis Dof protein OBP1 significantly stimulates the binding of OBF bZIP proteins to the CaMV-35S and GST6 promoters (16, 20). Because the promoters of the other classes of zein genes lack an O2 binding site, it remains an interesting question whether PBF alone may be sufficient to drive their expression, or if PBF may interact with other transcription factors in addition to O2 to coordinately regulate expression of all zein genes.
The isolation of a cDNA encoding PBF permits the detailed genetic and biochemical analysis of its proposed functional role in coordinately activating zein gene expression. We are currently in the process of obtaining mutations in the PBF gene through a reverse genetics approach (23, 32). Having such mutants will help elucidate the in vivo function of PBF and will provide direct genetic proof that PBF encodes the P-box binding activity present in maize endosperm nuclei. The continued analysis of PBF and its ability to interact with O2 and possibly other endosperm transcription factors will contribute significantly to our understanding of seed storage protein gene expression in maize as well as other cereals. In addition, these studies should offer further insights into the function of this novel class of DNA binding proteins containing the Dof domain.
Acknowledgments
We would like to thank Karam Singh (University of California at Los Angeles) for thoughtful discussion. This work was supported by a grant to R.J.S from the National Institutes of Health (GM41286). J.V.C. was supported by a fellowship and project DGCYT PB94-0404 from the Spanish Ministry of Education. S.P.M. was supported by a National Research Service Award Fellowship from the National Institutes of Health (1 F32 GM18577-01).
ABBREVIATIONS
- PBF
prolamin box binding factor
- O2
Opaque2
- P-box
prolamin box
- DAP
days after pollination
- GST
glutathione S-transferase
- bZIP
basic leucine zipper
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. U82230).
References
- 1.Thompson G A, Larkins B A. BioEssays. 1989;10:108–113. doi: 10.1002/bies.950100404. [DOI] [PubMed] [Google Scholar]
- 2.Aukerman M J, Schmidt R J. In: Results and Problems in Cell Differentiation 20, Plant Promoters and Transcription Factors. Nover L, editor. Berlin: Springer; 1994. pp. 209–233. [Google Scholar]
- 3.Schmidt R J, Burr F A, Aukerman M J, Burr B. Proc Natl Acad Sci USA. 1990;87:46–50. doi: 10.1073/pnas.87.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hartings H, Maddaloni M, Lazzaroni N, Di Fonzo N, Motto M, Salamini F, Thompson R. EMBO J. 1989;8:2795–2801. doi: 10.1002/j.1460-2075.1989.tb08425.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmidt R J, Ketudat M, Aukerman M J, Hoschek G. Plant Cell. 1992;4:689–700. doi: 10.1105/tpc.4.6.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ueda T, Waverezak K, Ward N, Sher M, Ketudat M, Schmidt R J, Messing J. Plant Cell. 1992;4:701–709. doi: 10.1105/tpc.4.6.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kodrzycki R, Boston R S, Larkins B A. Plant Cell. 1989;1:105–114. doi: 10.1105/tpc.1.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cord-Neto G, Yunes J A, da Silva M J, Vettore A L, Arruda P, Leite A. Plant Mol Biol. 1995;27:1015–1029. doi: 10.1007/BF00037028. [DOI] [PubMed] [Google Scholar]
- 9.Forde B G, Heyworth A, Pywell J, Kreis M. Nucleic Acids Res. 1985;13:7327–7339. doi: 10.1093/nar/13.20.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown J W S, Waneldt C, Feix G, Neuhaus G, Schweiger H G. Eur J Cell Biol. 1986;42:161–170. [Google Scholar]
- 11.Maier U-G, Brown J W S, Toloczyki C, Feix G. EMBO J. 1987;6:17–22. doi: 10.1002/j.1460-2075.1987.tb04712.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ueda T, Wang Z, Pham N, Messing J. Mol Cell Biol. 1994;14:4350–4359. doi: 10.1128/mcb.14.7.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maier U G, Grasser K D, Haas M M, Feix G. Mol Gen Genet. 1990;221:164–170. doi: 10.1007/BF00261716. [DOI] [PubMed] [Google Scholar]
- 14.Quayle T, Feix G. Mol Gen Genet. 1992;231:369–374. doi: 10.1007/BF00292705. [DOI] [PubMed] [Google Scholar]
- 15.Yanagisawa S. Nucleic Acids Res. 1995;23:3403–3410. doi: 10.1093/nar/23.17.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang B, Chen W, Foley R C, Buttner M, Singh K. Plant Cell. 1995;7:2241–2252. doi: 10.1105/tpc.7.12.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yanagisawa S. Trends Plant Sci. 1996;1:213–214. [Google Scholar]
- 18.De Paolis A, Sabatini S, De Pascalis L, Costantino P, Capone I. Plant J. 1996;10:215–233. doi: 10.1046/j.1365-313x.1996.10020215.x. [DOI] [PubMed] [Google Scholar]
- 19.Yanagisawa S, Izui K. J Biol Chem. 1993;268:16028–16036. [PubMed] [Google Scholar]
- 20.Chen W, Chao G, Singh K B. Plant J. 1996;10:955–966. doi: 10.1046/j.1365-313x.1996.10060955.x. [DOI] [PubMed] [Google Scholar]
- 21.Aukerman M J, Schmidt R J, Burr B, Burr F A. Genes Dev. 1991;5:310–20. doi: 10.1101/gad.5.2.310. [DOI] [PubMed] [Google Scholar]
- 22.Cone K C, Burr F A, Burr B. Proc Natl Acad Sci USA. 1986;83:9631–9635. doi: 10.1073/pnas.83.24.9631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mena M, Ambrose B, Meeley R B, Briggs S P, Yanofsky M F, Schmidt R J. Science. 1996;274:1537–1540. doi: 10.1126/science.274.5292.1537. [DOI] [PubMed] [Google Scholar]
- 24.Christensen A H, Quail P H. Plant Mol Biol. 1989;12:619–632. doi: 10.1007/BF00044153. [DOI] [PubMed] [Google Scholar]
- 25.Pysh L D, Aukerman M J, Schmidt R J. Plant Cell. 1993;5:227–236. doi: 10.1105/tpc.5.2.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gyuris J, Golemis E, Chertkov H, Brent R. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- 27.Patel L, Abate C, Curran T. Nature (London) 1990;347:572–575. doi: 10.1038/347572a0. [DOI] [PubMed] [Google Scholar]
- 28.Unger E, Parsons R L, Schmidt R J, Bowen B, Roth B A. Plant Cell. 1993;5:831–841. doi: 10.1105/tpc.5.8.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hammond-Kosack M C U, Holdsworth M J, Bevan M W. EMBO J. 1993;12:545–554. doi: 10.1002/j.1460-2075.1993.tb05686.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Müller M, Knudsen S. Plant J. 1993;4:343–355. doi: 10.1046/j.1365-313x.1993.04020343.x. [DOI] [PubMed] [Google Scholar]
- 31.Albani D, Hammond-Kosack M C U, Smith C, Conlan S, Colot V, Holdsworth M, Bevan M W. Plant Cell. 1997;9:171–184. doi: 10.1105/tpc.9.2.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bensen R J, Johal G S, Crane V C, Tossberg J T, Schnable P S, Meeley R B, Briggs S P. Plant Cell. 1995;7:75–84. doi: 10.1105/tpc.7.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]


