Abstract
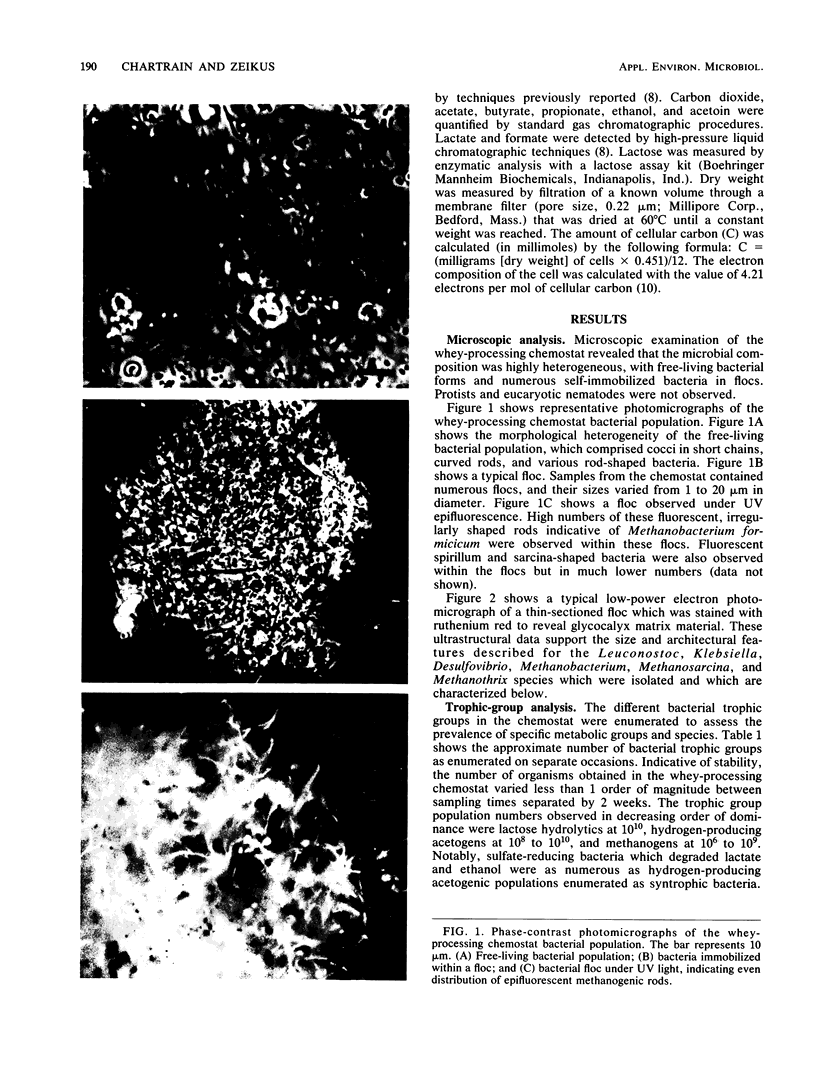
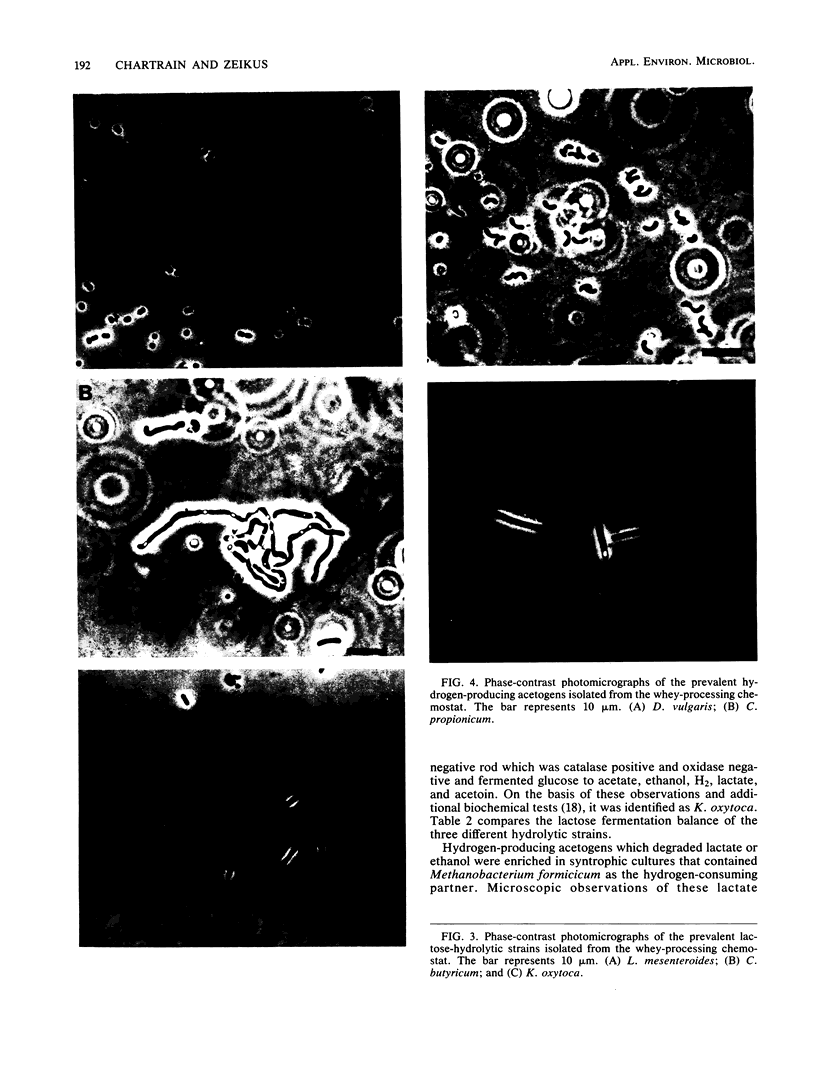
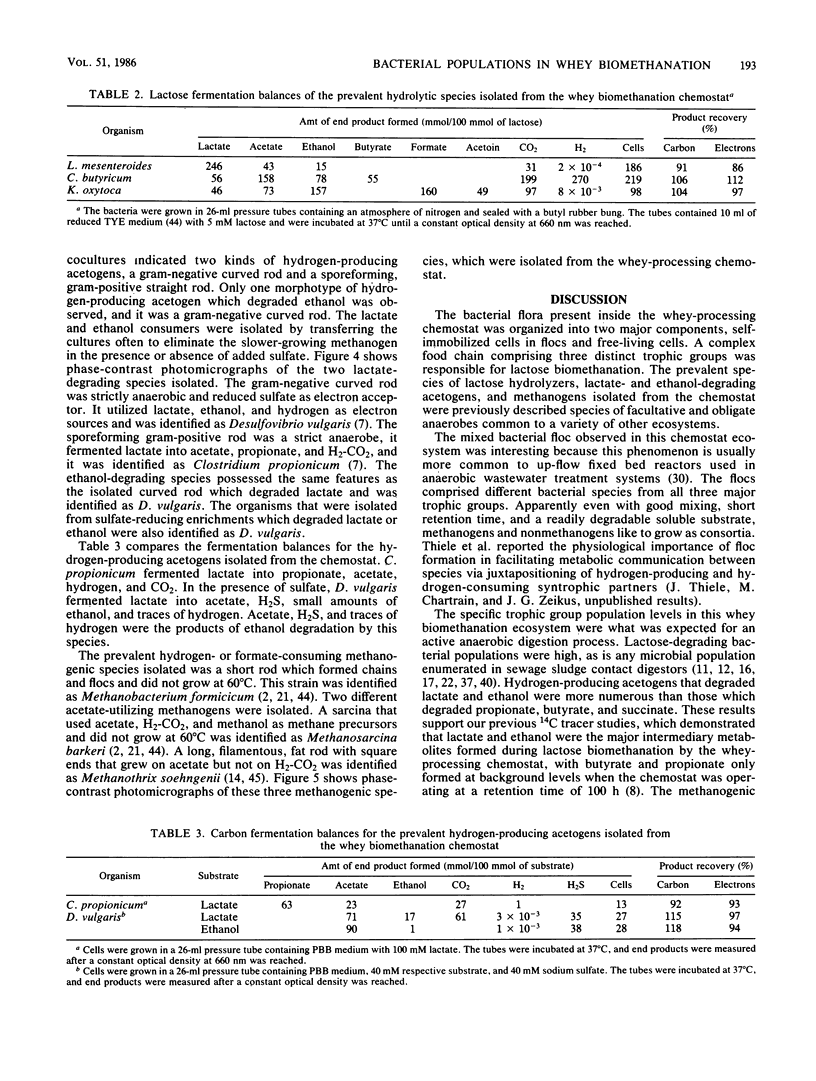
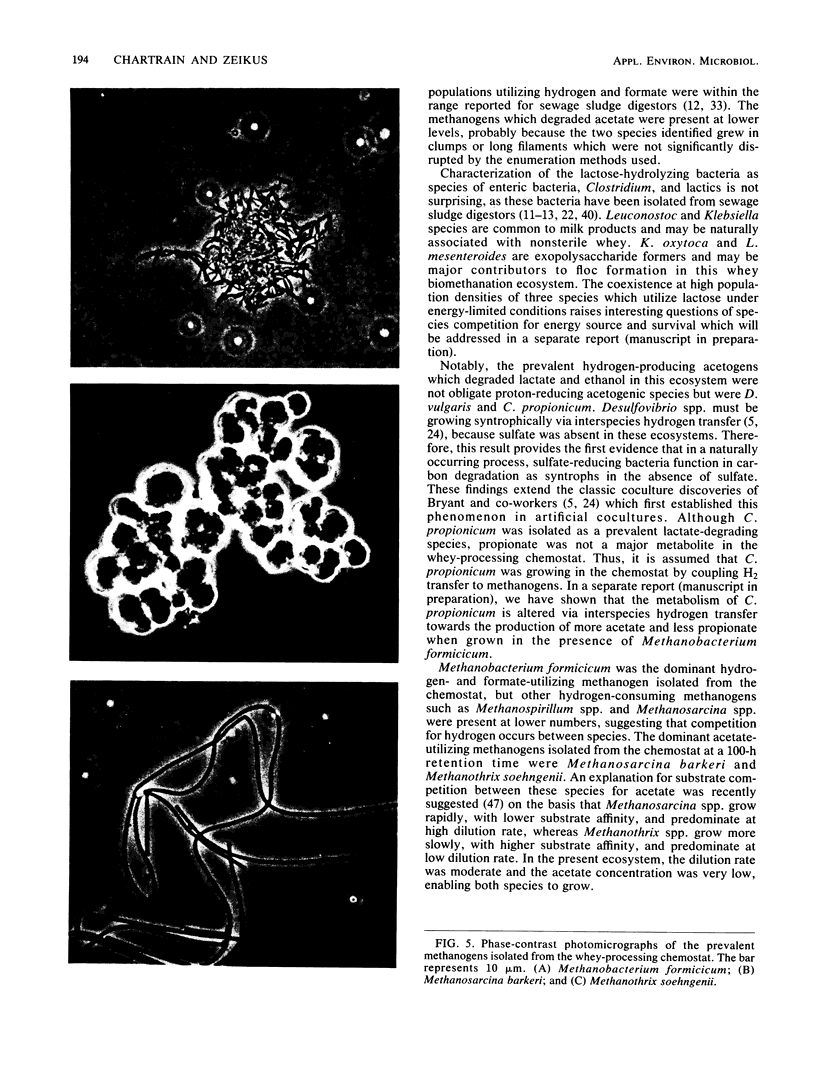
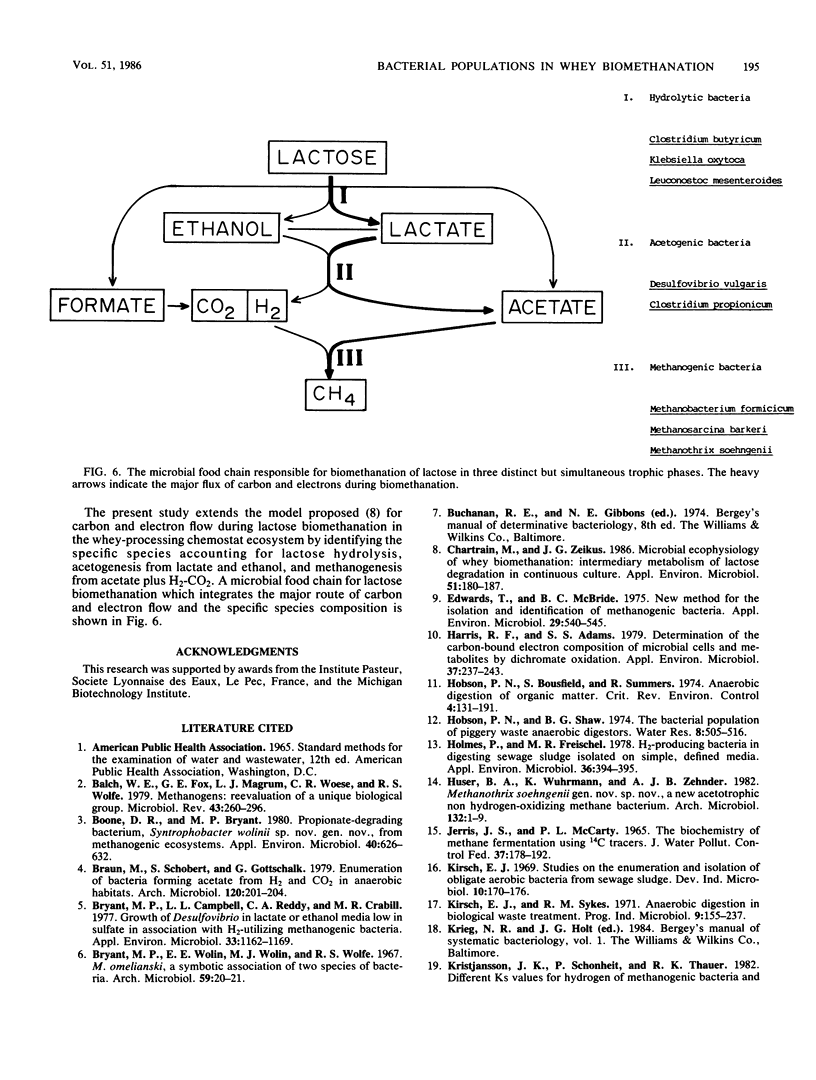
The organization and species composition of bacterial trophic groups associated with lactose biomethanation were investigated in a whey-processing chemostat by enumeration, isolation, and general characterization studies. The bacteria were spatially organized as free-living forms and as self-immobilized forms appearing in flocs. Three dominant bacterial trophic group populations were present (in most probable number per milliliter) whose species numbers varied with the substrate consumed: hydrolytic, 1010; acetogenic, 107 to 1010; and methanogenic, 106 to 109. The three prevalent species utilizing lactose were identified as Leuconostoc mesenteroides, Klebsiella oxytoca, and Clostridium butyricum. Clostridium propionicum and Desulfovibrio vulgaris were the dominant lactate-consuming, hydrogen-producing acetogenic bacteria, while D. vulgaris was the only significant ethanol-degrading species. Methanosarcina barkeri and Methanothrix soehngenii were identified as the dominant acetate-utilizing methanogens, and Methanobacterium formicicum was the prevalent hydrogen-utilizing methanogen. A microbial food chain is proposed for lactose biomethanation that comprises multiple species in three different groups, with the major hydrogen-producing acetogen being a sulfate-reducing species, D. vulgaris, which functioned in the absence of significant levels of environmental sulfate.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Balch W. E., Fox G. E., Magrum L. J., Woese C. R., Wolfe R. S. Methanogens: reevaluation of a unique biological group. Microbiol Rev. 1979 Jun;43(2):260–296. doi: 10.1128/mr.43.2.260-296.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone D. R., Bryant M. P. Propionate-Degrading Bacterium, Syntrophobacter wolinii sp. nov. gen. nov., from Methanogenic Ecosystems. Appl Environ Microbiol. 1980 Sep;40(3):626–632. doi: 10.1128/aem.40.3.626-632.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun M., Schoberth S., Gottschalk G. Enumeration of bacteria forming acetate from H2 and CO2 in anaerobic habitats. Arch Microbiol. 1979 Mar 12;120(3):201–204. doi: 10.1007/BF00423066. [DOI] [PubMed] [Google Scholar]
- Bryant M. P., Campbell L. L., Reddy C. A., Crabill M. R. Growth of desulfovibrio in lactate or ethanol media low in sulfate in association with H2-utilizing methanogenic bacteria. Appl Environ Microbiol. 1977 May;33(5):1162–1169. doi: 10.1128/aem.33.5.1162-1169.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant M. P., Wolin E. A., Wolin M. J., Wolfe R. S. Methanobacillus omelianskii, a symbiotic association of two species of bacteria. Arch Mikrobiol. 1967;59(1):20–31. doi: 10.1007/BF00406313. [DOI] [PubMed] [Google Scholar]
- Chartrain M., Zeikus J. G. Microbial ecophysiology of whey biomethanation: intermediary metabolism of lactose degradation in continuous culture. Appl Environ Microbiol. 1986 Jan;51(1):180–187. doi: 10.1128/aem.51.1.180-187.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards T., McBride B. C. New method for the isolation and identification of methanogenic bacteria. Appl Microbiol. 1975 Apr;29(4):540–545. doi: 10.1128/am.29.4.540-545.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris R. F., Adams S. S. Determination of the carbon-bound electron composition of microbial cells and metabolites by dichromate oxidation. Appl Environ Microbiol. 1979 Feb;37(2):237–243. doi: 10.1128/aem.37.2.237-243.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes P., Freischel M. R. H2-producing bacteria in digesting sewage sludge isolated on simmple, defined media. Appl Environ Microbiol. 1978 Aug;36(2):394–395. doi: 10.1128/aem.36.2.394-395.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsch E. J., Sykes R. M. Anaerobic digestion in biological waste treatment. Prog Ind Microbiol. 1971;9:155–237. [PubMed] [Google Scholar]
- Mah R. A., Sussman C. Microbiology of anaerobic sludge fermentation. I. Enumeration of the nonmethanogenic anaerobic bacteria. Appl Microbiol. 1968 Feb;16(2):358–361. doi: 10.1128/am.16.2.358-361.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerney M. J., Bryant M. P. Anaerobic Degradation of Lactate by Syntrophic Associations of Methanosarcina barkeri and Desulfovibrio Species and Effect of H(2) on Acetate Degradation. Appl Environ Microbiol. 1981 Feb;41(2):346–354. doi: 10.1128/aem.41.2.346-354.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerney M. J., Bryant M. P., Hespell R. B., Costerton J. W. Syntrophomonas wolfei gen. nov. sp. nov., an Anaerobic, Syntrophic, Fatty Acid-Oxidizing Bacterium. Appl Environ Microbiol. 1981 Apr;41(4):1029–1039. doi: 10.1128/aem.41.4.1029-1039.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohwaki K., Hungate R. E. Hydrogen utilization by clostridia in sewage sludge. Appl Environ Microbiol. 1977 Jun;33(6):1270–1274. doi: 10.1128/aem.33.6.1270-1274.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- REYNOLDS E. S. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J Cell Biol. 1963 Apr;17:208–212. doi: 10.1083/jcb.17.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson R. W., Akin D. E., Nordstedt R. A., Thomas M. V., Aldrich H. C. Light and electron microscopic examinations of methane-producing biofilms from anaerobic fixed-bed reactors. Appl Environ Microbiol. 1984 Jul;48(1):127–136. doi: 10.1128/aem.48.1.127-136.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith P. H., Mah R. A. Kinetics of acetate metabolism during sludge digestion. Appl Microbiol. 1966 May;14(3):368–371. doi: 10.1128/am.14.3.368-371.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurr A. R. A low-viscosity epoxy resin embedding medium for electron microscopy. J Ultrastruct Res. 1969 Jan;26(1):31–43. doi: 10.1016/s0022-5320(69)90033-1. [DOI] [PubMed] [Google Scholar]
- Winfrey M. R., Zeikus J. G. Effect of sulfate on carbon and electron flow during microbial methanogenesis in freshwater sediments. Appl Environ Microbiol. 1977 Feb;33(2):275–281. doi: 10.1128/aem.33.2.275-281.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehnder A. J., Huser B. A., Brock T. D., Wuhrmann K. Characterization of an acetate-decarboxylating, non-hydrogen-oxidizing methane bacterium. Arch Microbiol. 1980 Jan;124(1):1–11. doi: 10.1007/BF00407022. [DOI] [PubMed] [Google Scholar]
- Zeikus J. G. The biology of methanogenic bacteria. Bacteriol Rev. 1977 Jun;41(2):514–541. doi: 10.1128/br.41.2.514-541.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinder S. H., Cardwell S. C., Anguish T., Lee M., Koch M. Methanogenesis in a Thermophilic (58 degrees C) Anaerobic Digestor: Methanothrix sp. as an Important Aceticlastic Methanogen. Appl Environ Microbiol. 1984 Apr;47(4):796–807. doi: 10.1128/aem.47.4.796-807.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]