Abstract
Small molecules that bind their biological receptors with high affinity and selectivity can be isolated from randomized pools of combinatorial libraries. RNA-protein interactions are important in many cellular functions, including transcription, RNA splicing, and translation. One example of such interactions is the mechanism of trans-activation of HIV-1 gene expression that requires the interaction of Tat protein with the trans-activation responsive region (TAR) RNA, a 59-base stem-loop structure located at the 5′ end of all nascent HIV-1 transcripts. Here we demonstrate the isolation of small TAR RNA-binding molecules from an encoded combinatorial library. We have made an encoded combinatorial tripeptide library of 24,389 possible members from d-and l-alpha amino acids on TentaGel resin. Using on-bead screening we have identified a small family of mostly heterochiral tripeptides capable of structure-specific binding to the bulge loop of TAR RNA. In vitro binding studies reveal stereospecific discrimination when the best tripeptide ligand is compared with diastereomeric peptide sequences. In addition, the most strongly binding tripeptide was shown to suppress transcriptional activation by Tat protein in human cells with an IC50 of ≈50 nM. Our results indicate that tripeptide RNA ligands are cell permeable, nontoxic to cells, and capable of inhibiting expression of specific genes by interfering with RNA-protein interactions.
Protein-nucleic acid interactions are involved in many cellular functions, including transcription, RNA splicing, and translation. Readily accessible synthetic molecules that can bind with high affinity to specific sequences of single- or double-stranded nucleic acids have the potential to interfere with these interactions in a controllable way, making them attractive tools for molecular biology and medicine. Successful approaches used thus far include duplex-forming (antisense) (1) and triplex-forming (antigene) oligonucleotides (2–4), peptide nucleic acids (5), and pyrrole-imidazole polyamide oligomers (6, 7). Each class of compounds uses a readout system based on simple rules for recognizing the primary or secondary structure of a linear nucleic acid sequence. Another approach uses carbohydrate-based ligands, calicheamicin oligosaccharides, which interfere with the sequence-specific binding of transcription factors to DNA and inhibit transcription in vivo (8, 9). Although antisense oligonucleotides and peptide nucleic acids use the familiar Watson–Crick base-pairing rules, two others, the triplex-forming oligonucleotides and the pyrrole-imidazole polyamides, take advantage of straightforward rules to read the major and minor grooves, respectively, of the double helix itself.
In addition to its primary structure, RNA has the ability to fold into complex tertiary structures consisting of such local motifs as loops, bulges, pseudoknots, and turns (10, 11). It is not surprising that, when they occur in RNAs that interact with proteins, these local structures are found to play important roles in protein-RNA interactions (12). This diversity of local and tertiary structure, however, makes it impossible to design synthetic agents with general, simple-to-use recognition rules analogous to those for the formation of double- and triple-helical nucleic acids. Because RNA-RNA and protein-RNA interactions can be important in viral and microbial disease progression, it would be advantageous to have a general method for rapidly identifying synthetic compounds for targeting specific RNA structures. A particular protein-binding RNA structure can be considered as a molecular receptor not only for the protein with which it interacts but also for synthetic compounds, which may prove to be antagonists of the protein-RNA interaction.
One of our laboratories recently has been involved in the design and combinatorial testing of synthetic receptors for homochiral and heterochiral peptides (13). This method for finding receptors for specific peptides has led us to approach the problem of RNA recognition, and in particular the recognition of protein-binding RNAs, by treating a particular RNA structure as a receptor for an unknown small molecule ligand. Peptides are well suited to this task not only because they are made from the same building blocks as the natural protein ligand but also because they can be coupled in high yield on a solid-phase resin, which might then be used directly in screening. In lieu of a reliable set of nucleic acid recognition rules, then, the combinatorial synthesis of many diverse potential antagonists might be used to find new lead compounds for disruption of a particular RNA-protein interaction.
One example of such an interaction is the mechanism of trans-activation of HIV-1 gene expression. Transcriptional up-regulation requires binding of the Tat protein to the trans-activation response region (TAR) RNA (see Fig. 1), a 59-base stem-loop structure located at the 5′ end of all nascent HIV-1 transcripts (14). Inhibition of Tat-TAR interactions is thus a potential approach for the development of anti-HIV therapeutics (15–17). In the work presented here, we have made an encoded (18) combinatorial tripeptide library of 24,389 possible members from d-and l-alpha amino acids on TentaGel resin (19). Using on-bead screening we have identified a small family of mostly heterochiral tripeptides capable of structure-specific binding to the bulge loop of TAR RNA. In vitro binding studies reveal stereospecific discrimination when the best tripeptide ligand is compared with diastereomeric peptide sequences. In addition, the most strongly binding tripeptide was shown to repress Tat-induced trans activation in vivo with an IC50 of approximately 50 nM.
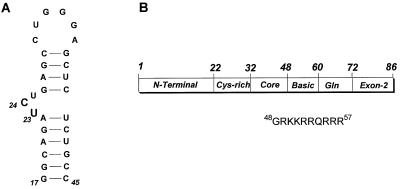
Figure 1.
(A) Sequence and secondary structure of TAR RNA used in this study. TAR RNA spans the minimal sequences that are required for Tat responsiveness in vivo (37) and for in vitro binding of Tat-derived peptides (38). Wild-type TAR contains two non-wild-type base pairs to increase transcription by T7 RNA polymerase. Nucleotides that interact with the RNA-binding ligand 1 are highlighted. (B) Regions of the HIV-1 Tat protein and the sequence of the Tat (residues 48–57) peptide that binds TAR RNA with high affinity.
Materials and Methods
Encoded Combinatorial Library.
We prepared a tripeptide library on TentaGel (TentaGel S-NH2 from Rapp Polymere, Tübingen, Germany; ref. 19) by encoded split-synthesis (18). The library had the general structure Ac-AA3-AA2-AA1-NH(CH2)2-O-TentaGel, and contained the following amino acids: Gly, d-Ala, l-Ala, d-Val, l-Val, d-Leu, l-Leu, d-Pro, l-Pro, d-Phe, l-Phe, d-Gln, l-Gln, d-Asn, l-Asn, d-Lys, l-Lys, d-His, l-His, d-Arg, l-Arg, d-Glu, l-Glu, d-Asp, l-Asp, d-Ser, l-Ser, d-Thr, and l-Thr. The library did not contain Cys, Trp, Tyr, Met, or Ile. The library was encoded by using a set of 15 photocleavable tags (five tags per AAn). After the synthesis of the library, side chains were deprotected with trifluoroacetic acid. One copy of the library contained 24,389 possible tripeptide sequences.
RNA Synthesis.
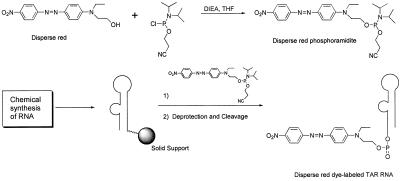
RNAs were synthesized by chemical and enzymatic methods. Modified TAR RNA was synthesized on an Applied Biosystems model 392 DNA/RNA synthesizer by using 2-cyanoethyl phosphoramidite chemistry. All the monomers of (2-cyanoethyl)phosphoramidites were obtained from Glen Research (Sterling, VA). Disperse red phosphoramidite (Scheme S1) (0.15 M solution in CH3CN) was used to incorporate a dye label at the 5′ end of TAR RNA. Synthesis of the disperse red phosphoramidite (Scheme S1) was accomplished according to standard phosphoramidite chemistry methods (20–22). RNA (1 μmol) containing disperse red was deprotected by treatment with NH3-saturated methanol (2 ml) at 25°C for 17 h. Product was filtered and dried in Speedvac (Savant). To deprotect 2′-OH silyl groups, the red pellet was dissolved in 50% triethylamine trihydrofluoride in DMSO (0.5 ml) and left at room temperature for 16 h. Deprotected RNA was precipitated by the addition of 2 ml of isopropyl alcohol. After deprotection, RNA was purified and characterized as described (21, 23, 24).
Scheme 1.
Wild-type and mutant TAR RNAs were prepared by in vitro transcription (25, 26). All DNAs were synthesized on an Applied Biosystems ABI 392 DNA/RNA synthesizer. The template strands encode the sequences for wild-type and mutant TAR RNAs. The top strand is a short piece of DNA complementary to the 3′ end of all template DNAs having the sequence 5′-TAATACGACTCACTATAG-3′. The template strand of DNA was annealed to an equimolar amount of top-strand DNA, and transcriptions were carried out in transcription buffer (40 mM Tris⋅HCI, pH 8.1/1 mM spermidine/0.01% Triton X-100/5 mM DTT) and 4.0 mM NTPs at 37°C for 2–4 h. For reactions (20 μl) containing 8.0 pmol template DNA, 40–60 units of T7 polymerase (Promega) was used. Transcription reactions were stopped by adding an equal volume of sample loading buffer. RNA was purified on 20% acrylamide 8 M urea denaturing gels and stored in diethyl pyrocarbonate water at −20°C.
Enzymatically transcribed RNAs were 5′ dephosphorylated by incubation with calf intestinal alkaline phophatase (Promega) for 1 h at 37°C in 50 mM Tris⋅Cl, pH 9.0/1 mM MgCl2/0.1 mM ZnCl2/1 mM spermidine. The RNAs were purified by multiple extractions with Tris-saturated phenol and one extraction with 24:1 chloroform/isoamyl alcohol followed by ethanol precipitation. Chemically synthesized RNA contains only free 3′-OH groups and does not require dephosphorylation procedures. The RNAs were 5′ end-labeled with 0.5 μM [γ-32P]ATP (6,000 Ci/mmol) (ICN) per 100 pmol RNA by incubating with 16 units of T4 polynucleotide kinase (New England Biolabs) in 70 mM Tris⋅HCl (pH 7.5), 10 mM MgCl2, 5 mM DTT (26, 27). The RNAs were labeled at the 3′ end by ligation to cytidine 3′,5′-[5′-32P]bisphosphate ([32P]pCp) by using T4 RNA ligase. Reaction mixtures (50 μl) contained 250 pmol RNA, 65 μCi [32P]pCp (3,000 Ci/mmol, New England Nuclear), and 40 units of T4 RNA ligase (New England Biolabs) in a buffer containing 50 mM Tris⋅HCl (pH 8.0), 3 mM DTT, 10 mM MgCl2, 25 mM NaCl, 50 mM ATP, 25 μg/ml BSA, and 10% DMSO (vol/vol). After incubation at 4°C overnight, the labeled RNAs were purified by phenol-chloroform extraction and ethanol precipitation. 3′- and 5′-End-labeled RNAs were gel-purified on a denaturing gel, visualized by autoradiography, eluted out of the gels, and desalted on a reverse-phase cartridge.
The sequence of RNAs was determined by base hydrolysis and nuclease digestion. Alkaline hydrolysis of RNAs was carried out in hydrolysis buffer for 8–12 min at 85°C. RNAs were incubated with 0.1 units of RNase from Bacillus cereus (Amersham Pharmacia) per pmol RNA for 4 min at 55°C in 16 mM sodium citrate (pH 5.0), 0.8 mM EDTA, and 0.5 mg/ml yeast tRNA (GIBCO/BRL). This enzyme yields U- and C-specific cleavage of RNA. Sequencing products were resolved on 20% denaturing gels and visualized by PhosphorImager analysis.
Screening the Library with TAR RNA.
The library beads (three copies of library) were placed in an Eppendroff tube and washed with water (500 μl × 5) and TK buffer (50 mM Tris⋅HCl, pH 7.4/20 mM KCl/0.1% Triton X-100, 500 μl × 4). Three copies of the library contain more than 95% of the members of the library. TK buffer is a commonly used buffer for the formation of Tat-TAR complexes in vitro (28). After filtration, the resin was suspended in TK buffer (400 μl) and BSA (0.1 mg/ml) at room temperature for 1 h to reduce nonspecific binding. The aqueous phase was removed, and the resin was washed with TK buffer (500 μl × 3). After filtration, bulge mutant (containing a single nucleotide bulge) TAR RNA (2.5 μM) was added to the resin in TK buffer (600 μl), and the suspension was stirred at 4°C for 1 day. The buffer was drained, and TK buffer (600 μl), disperse red dye-labeled-TAR RNA (250 nM), and bulge mutant TAR RNA (2.5 μM) were added to the resin. After 2 days stirring at 4°C the solution was drained, and the remaining beads were washed with water (500 μl × 3). Two red beads were selected under a microscope and placed in capillary tubes individually. The library was further incubated with red dye-labeled TAR and mutant TAR RNA for 3 days under the conditions described above. During a second screening, eight beads became red, which were picked and placed in capillary tubes.
Decoding the Structures of RNA-Binding Ligands.
An encoded bead was placed in a 25-μl micro capillary tube with dimethylformamide (DMF) (2 μl), and the bead was washed with DMF (5 μl × 4). After draining the solvent, the bead was resuspended in DMF (2 μl), and the micro capillary was centrifuged for 4 min and then sealed by flame. The capillary containing a bead was irradiated by UV (350 nm) for 4 h, and the capillary tube was centrifuged for 5 min. The capillary was opened, and the cleaved tag alcohols were silylated with N,O-bis(trimethylsilyl)acetamide in a micro syringe. The trimethylsilyl derivatives (1 μl) were analyzed by electron capture GC. A Hewlett–Packard 6890 series Gas Chromatography system, equipped with a micro electron capture detector (μ-ECD), and a Hewlett–Packard Chemstation operating system, were used for all decoding analyses. The GC was operated in splitless inlet mode, using helium as carrier gas. A 35-m × 0.2-mm i.d. × 0.33-μm film thickness Hewlett–Packard Ultra 1 column was used with a temperature program of 1 min isothermal at 200°C followed by heating at 10°C/min to 320°C. The μ-ECD make-up gas was nitrogen.
Solid-Phase RNA-Peptide Binding Assays.
RNA-binding ligands (shown in Table 1) were synthesized on TentaGel S-NH2 (4.6 μmol). All fluorenylmethoxycarbonyl-amino acids were purchased from Bachem. 1-Hydroxybenzotriazole and diisopropylcarbodiimide were obtained from Aldrich. Piperidine and trifluoroacetic acid were purchased from the Applied Biosystems Division, Perkin–Elmer. All the ligands were synthesized manually according to standard solid-phase peptide synthesis protocols. Coupling efficiencies of residues at each step were monitored by Kaiser test. Deprotection of trimer peptides was carried out in 10% water in trifluoroacetic acid (1 ml) for 16 h at room temperature. After filtration of solvents, the resin was washed with water (1 ml × 4), dimethylformamide (1 ml × 4), dichloromethane (1 ml × 4), and dried under reduced pressure. The ligand attached to the resin (10 beads) was placed in an Eppendroff tube and washed with TK buffer (200 μl × 3). The beads were suspended in TK buffer (300 μl) and incubated with TAR RNA (1.95 μM) overnight at 4°C. The suspension was centrifuged, and the supernatant containing unbound RNA was transferred to a cuvette for OD260 measurements. The UV absorbance was measured by a Shimadzu UV-1601 spectrometer. Equilibrium concentrations of RNA were determined from these measurements. Given that the initial concentrations of ligand and RNA are known, and assuming simple bimolecular receptor/substrate binding, dissociation constant (KD) can be calculated from straightforward equations.
Table 1.
RNA-binding ligands
| ID# | Structures | Frequency | Color | KD, nM | KREL |
|---|---|---|---|---|---|
| 1 | NH2-(l)Lys-(d)Lys-(l)Asn-OH | 2 | Red | 420 ± 44 | 1.73 |
| 2 | NH2-(l)Lys-(d)Lys-(d)Asn-OH | 1 | Pink | 4,173 ± 208 | 0.17 |
| 3 | NH2-(l)Lys-(l)Lys-(l)Asn-OH | 2 | Pink | 3,224 ± 183 | 0.23 |
| 4 | NH2-(l)Arg-(d)Lys-(l)Ala-OH | 1 | Pink | 2,640 ± 219 | 0.28 |
| 5 | NH2-(l)Arg-(d)Lys-(d)Val-OH | 1 | Pink | 10,434 ± 594 | 0.07 |
| 6 | NH2-(l)Arg-(d)Lys-(l)Arg-OH | 1 | Pink | 878 ± 80 | 0.83 |
| 7 | NH2-(d)Thr-(d)Lys-(l)Asn-OH | 1 | Pink | 564 ± 80 | 1.29 |
| 8 | NH2-(d)Thr-(d)Lys-(l)Phe-OH | 1 | Pink | 2,087 ± 244 | 0.35 |
KD values were determined from four independent experiments. KREL = KD of a basic Tat peptide (727 ± 74 nM)/KD of inhibitor. The bead library peptides all were N-acetylated. Peptides used for in vitro and in vivo experiments were not N-acetylated and contained free amino termini.
Inhibition of Tat Trans-Activation in Cellular Assays.
We used HL3T1 cells, a HeLa cell line derivative containing an integrated HIV-1 long terminal repeat promoter and chloramphenicol acetyltransferase (CAT) reporter gene. Cells were grown in 2 ml of DMEM supplemented with 10% FCS in 60-mm dishes at 37°C in 5% CO2 in a tissue culture incubator. Cells were refreshed by 2 ml of DMEM before transfection. Transfection was started by dropwise addition of 250 μl 2 × Hepes-buffered saline and then kept at room temperature for 10 min. Approximately 10 μg of plasmids (pSV2Tat and pAL) and increasing amounts of ligand 1 were transfected in the presence of CaCl2 (final concentration 125 mM), and the cells were incubated for 4 h at 37°C in a tissue culture incubator. The medium then was discarded, and the cells were subjected to glycerol (1.5 ml of 15%) shock for 45 sec. Finally, the cells were washed twice with PBS (5 ml) and were grown in 3 ml of DMEM. The cells were harvested after 48 h posttransfection with changing with fresh DMEM at 24 h and lysed in reporter lysis buffer (Promega). Aliquots were used for CAT and luciferase assays. Both activities were normalized to protein concentration determined by using a modified Bradford assay (Bio-Rad).
Results and Discussion
On-Bead Selection of Structure-Specific TAR-Binding Ligands.
Previous studies using combinatorial chemistry to identify new ligands to block the TAT-TAR interaction have relied on a variety of complex methods that are labor intensive or require expensive robotics equipment (29). For the most part, these methods originated in the study of individual protein-nucleic acid interaction experiments. Moreover, in some cases time-consuming deconvolution strategies also are needed to identify the individual compounds responsible for the properties found in a mixture of compounds tested together (15, 16). We decided to try methods that we previously have used with success on small organic receptors. This entailed covalently attaching the dye disperse red to the TAR RNA (Scheme S1) and incubating it in a suspension of library beads made from the split synthesis method. Diffusion of low molecular weight receptors into a bead of TentaGel resin is known to be rapid, while one might expect that a macromolecule such as a protein or large nucleic acid might be excluded from the bead interior where the bulk of the peptide is displayed. Nevertheless, we have found that the dye-TAR conjugate was able to enter the beads and bind in a structure-dependent manner. The dye’s red color was clearly distributed evenly throughout the translucent bead. Broken beads were not selected. Peptides specific for portions of TAR other than the bulge region were blocked by using a relatively large concentration of an unlabeled TAR analog lacking the natural 3-nt bulge. A small amount of detergent and the use of a low RNA concentration (250 nM) also minimizes nonspecific binding. Another advantage to our method is the use of chemically encoded beads (18). Once a dye-stained bead is selected, the identification of the peptide sequence is rapid and straightforward. Although many binding experiments are conducted simultaneously, the compounds remain discrete, each in its own assay vessel, the bead produced by the split-synthesis method.
Ligand Sequence Analysis.
Upon incubating the dye-TAR conjugate with the library we initially found that only two beads took on a deep red color. We picked these two beads and allowed the incubation to continue 3 days at 4°C. The first two deepest red beads were found to be the same sequence, ligand 1, (l)Lys-(d)Lys-(l)Asn. At the end of the second incubation we picked eight more beads that had turned pink-red during this second incubation period. It is important to note that there were only eight beads that turned pink-red during the second screening. The sequences of these beads are listed in Table 1.
To verify that our assay reflected RNA-trimer interaction and to determine the affinity of these trimer ligands for TAR RNA, we resynthesized trimer peptides 1–8 (Table 1) and measured their dissociation constants with wild-type TAR RNA. The results in Table 1 confirm that the on-bead assay mimics RNA binding because ligand 1 has the highest affinity for TAR RNA (KD = 420 ± 44 nM). To compare the RNA-binding affinities of eight ligands to natural Tat peptide, we synthesized a Tat-derived peptide (Gly-48 to Arg-57) containing the RNA-binding region of Tat protein (Fig. 1). Dissociation constants of the Tat peptide-RNA complexes were determined under the same conditions used for trimer ligand-TAR RNA complexes. These experiments showed that the Tat peptide (residues 48–57) binds TAR RNA with a KD of 727 ± 74 nM. A relative dissociation constant (KREL) can be determined by measuring the ratios of the Tat peptide to trimer ligand dissociation constants (KD) for TAR RNA. These results are shown in Table 1. Ligand 1 binds TAR RNA with affinities higher than the wild-type Tat peptide. These results indicate that selection frequency reflects ligand activity and, if a large enough library sample is used, could be used as an indicator of ligand affinity for RNA.
Although these studies provide useful information about the relative binding affinities of the TAR RNA ligands, it is important to note that these assays were performed on immobilized ligands and cannot represent physiological interactions. To determine activities of these inhibitors of Tat-TAR interactions under physiological conditions, we have developed a system based on fluorescence resonance energy transfer (FRET) containing TAR RNA and Tat peptides uniquely labeled with donor and acceptor dye molecules (J. Zhang, N.T., S.H., and T.M.R., unpublished results). We monitored inhibition of FRET between a fluorescein-labeled TAR RNA and a rhodamine-labeled Tat peptide (amino acids 38–72) in the presence of increasing concentration of ligand 1. The Ki values were calculated by fitting data to quadratic equation (30), which showed that ligand 1 inhibited the RNA-protein complex formation with a Ki of 34 ± 2.4 nM (data not shown).
Several control experiments further support these observations and demonstrate the specificity of ligand-TAR interactions. First, incubating the library with free dye did not stain any beads, indicating that there was no interaction between the trimer peptides and the dye molecule (data not shown). Second, the equilibrium interaction between dye-labeled TAR RNA and a tripeptide tethered to beads was examined by incubating a suspension of beads containing ligand 1 in TK buffer (400 μl) with dye-labeled TAR RNA (1 μM) at 4oC for 5 h. As shown in Fig. 2C, beads were stained red upon TAR RNA binding. The interaction between red dye-labeled TAR RNA and ligand 1 is reversible and can be abolished by the addition of unlabeled TAR RNA or ligand 1 as a competitor. Finally, screening the library with dye-labeled TAR RNA was carried out in the presence of excess unlabeled bulge-mutant TAR RNA, therefore inhibiting nonspecific TAR RNA-peptide interactions.
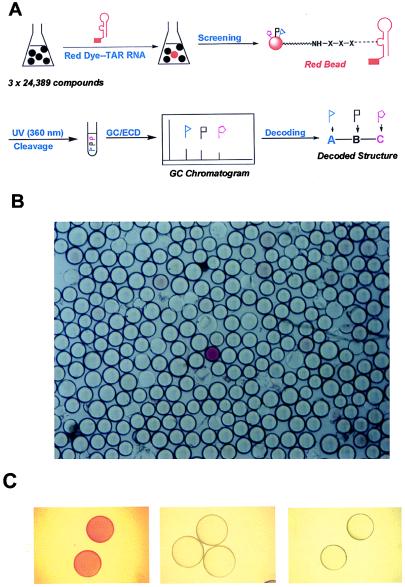
Figure 2.
(A) Schematic presentation of screening and decoding of the combinatorial library. TAR RNA was labeled with a red dye, disperse red, at its 5′ end by chemical synthesis and incubated with the trimer library as described in the text. (B) A portion of the beads in the library after incubating with red dye-labeled TAR RNA. The dark bead in the center was identified as ligand 1. (C) The equilibrium interaction between dye-labeled TAR RNA and a tripeptide tethered to beads. A suspension of beads containing ligand 1 in TK buffer (400 μl) was incubated with dye-labeled TAR RNA (1 μM) at 4oC for 5 h. Beads were stained red upon TAR RNA binding (Left). Red beads became colorless when excess of unlabeled TAR RNA (Middle), or ligand 1 (Right) was added, indicating the displacement of red-TAR RNA from the beads.
As shown in Table 1, ligands 1 [(l)Lys-(d)Lys-(l)Asn] and 7 [(d)Thr-(d)Lys-(l)Asn] were the tightest binding tripeptides found, suggesting a consensus sequence of X-(d)Lys-(l)Asn. Furthermore, two diastereomers of ligand 1, peptides 2 and 3, were found in the assay, peptide 3 being the only homochiral sequence. RNA-peptide binding measurements revealed that the dissociation constants for these two diastereomeric sequences were approximately seven times higher than for the strongest sequence, 1. This sharp loss of binding energy among diastereomers indicates that the binding interaction is highly stereospecific and not merely the result of a nonspecific lysine-phosphate backbone attraction. This finding is not surprising because the TAR RNA, and hence the binding site, is necessarily chiral. Another interesting feature of our results is that all but one of the TAR-binding sequences found were heterochiral and would have been missed by other techniques, such as phage display, which only use the proteinogenic amino acids. Use of d- and l-amino acids together yields a richer stereochemical variety of ligands, in addition to the diversity imparted by using the alpha-amino acids.
NMR Spectroscopy.
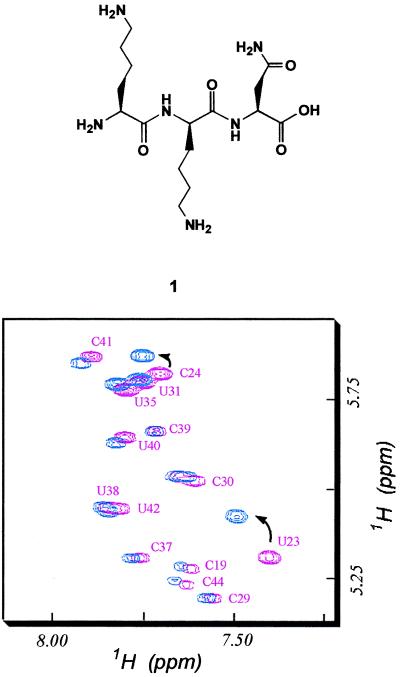
To determine the interaction sites of our trimer ligand 1 on TAR RNA, we performed NMR experiments. NMR spectra of free TAR and TAR complex with ligand 1 were recorded. Because of the spectral overlap, it was impossible to follow all but a few well-isolated resonances by conventional one-dimensional (1D) experiments. Therefore, we carried out two-dimensional (2D) nuclear Overhauser enhancement spectroscopy (NOESY) and total correlated spectroscopy (TOCSY) experiments. All spectra were recorded on a Bruker AMX-500 NMR spectrometer operating at 500 MHz for 1H equipped with triple resonance probe and H-broadband inverse detection probes. The 1D and 2D 1H spectra were recorded at ≈1–2 mM RNA and ligand concentrations in 5 mM phosphate buffer (pH 5.5), with up to 100 mM NaCl. All other conditions were the same as described by Aboul-ela et al. (31). We obtained a set of complete TAR RNA assignments. Increasing amounts of ligand 1 were added to TAR RNA, and the spectral changes were monitored by 2D TOCSY experiments. The TOCSY spectrum contains a region where only pyrimidine H5-H6 resonances are found, and this region has a well-dispersed 2D spectrum. Resonances in the free RNA and RNA-ligand complexes were assigned by NOESY experiments. Results of our TOCSY experiments are shown in Fig. 3. Resonances in the bulge region, U23 and C24, were shifted. All other resonances were not affected significantly by the addition of the ligand. To address the question whether spectral changes at U23 and C24 were caused by specific ligand 1 binding or were the result of perturbation by a nonspecific exogeneous ligand, we performed NMR experiments in the presence of a basic tripeptide containing l-Lys amino acids. Our results showed that the Lys peptide did not cause shift of resonances in the bulge region including U23 and C24 (data not shown), indicating that ligand 1 specifically interacts with TAR RNA at the bulge region. Interestingly, ligand 1-TAR RNA interactions are different from TAR RNA-peptide or TAR-Arg complexes reported in previous NMR studies (31, 32) because there were no detectable interactions with G26 and A27 regions as observed in previous studies. Another TAR-binding ligand, CGP64222, which contains Arg side chains in its sequence, also causes conformational change in TAR and creates an RNA structure that is similar to TAR-peptide structure (15). These results indicate that ligand 1 is the first ligand that binds specifically to the bulge of TAR RNA in a manner different from previously reported TAR ligands and Tat peptides. These findings suggest an intriguing possibility that small molecules that interact with TAR RNA and induce a conformational change in TAR, resulting in a structure different from that of Tat-TAR complex, could be used to lock RNA in a nonfunctional structure.
Figure 3.
Titration of wild-type TAR RNA by increasing amounts of ligand 1, (l)Lys-(d)Lys-(l)Asn (Upper). (Lower) Superposition of TOCSY spectra at increasing concentration of ligand 1 (free RNA, pink and 1:1 ligand to RNA ratio, blue) shows that only resonances at bulge U23 and C24 are affected by ligand 1 binding.
Inhibition of Tat Trans-Activation in Vivo.
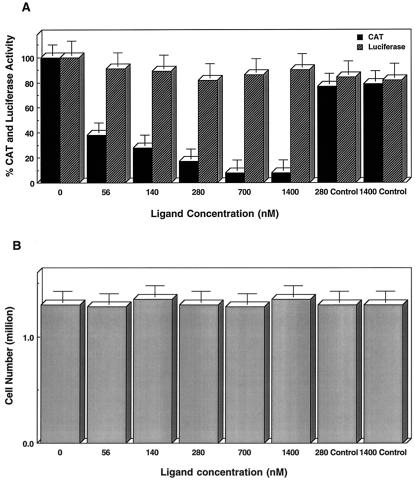
To test whether this small molecule-RNA interaction could be used to control HIV-1 gene expression in vivo, we used HL3T1 cells, a HeLa cell line derivative containing an integrated HIV-1 long terminal repeat promoter and CAT reporter gene (33). We added different amounts of ligand 1 during transfection of pSV2-Tat (34) and pAL (35) plasmids into HL3T1 cells. Plasmids pSV2Tat and pAL express the first exon of Tat protein and luciferase enzyme, respectively. Luciferase reporter gene provides an internal control. Transfection of HeLa cells with pSV2Tat enhanced transcription as determined by CAT activity. As shown in Fig. 4, increasing amounts of ligand 1 resulted in a decrease of CAT activity whereas luciferase activity was not affected. In the presence of 700 nM concentrations of ligand 1, more than 90% of Tat trans-activation was inhibited. To rule out the possibility that the observed inhibition of trans-activation could be caused by some nonspecific toxicity of ligand 1 or reduction of the pSV2Tat plasmid uptake, transcription of luciferase gene was monitored (Fig. 4). Transcription of luciferase gene was not affected by ligand 1. Cell viability assays showed that ligand 1 treatment was not toxic to the cells (Fig. 4B). Further control experiments showed that weaker TAR RNA-binding ligands such as l-argininamide (Fig. 4) and a scrambled Tat peptide containing d-amino acids had no inhibitory effect on Tat trans-activation (36). Thus, we conclude that ligand 1 binds TAR RNA and inhibits Tat-TAR interactions in vivo.
Figure 4.
Inhibition of Tat trans-activation by ligand 1 in vivo. CAT activity expressed from the integrated HIV-1 long terminal repeat of HL3T1 cells with increasing amounts of ligand 1 is shown. Monitoring luciferase activity was a control experiment to quantify the transfection efficiency and nonspecific inhibition of gene expression by the addition of ligand 1. Transfection and enzymatic activity (CAT and luciferase) assays were performed as described (34, 35). CAT and luciferase activities were measured from five experiments and normalized to 100%. Error bars represent the estimated SD. Control lanes contain l-argininamide as an RNA ligand.
We have described the application of encoded combinatorial library and on-bead screening methods to identify ligands that interact specifically with target RNA and can be used to design small molecules for controlling gene expression in vivo. The results also establish an example of the application of small molecules as artificial regulators of cellular processes involving RNA-protein interactions in vivo.
Acknowledgments
We thank Prof. R. M. J. Liskamp at Utrecht University for his advice on RNA-peptide binding experiments. This work was supported in part by National Institutes of Health Grants AI 41404, AI 43198, and TW 00702 (to T.M.R.). T.M.R. is a recipient of a Research Career Development Award from the National Institutes of Health. K.R. is a recipient of a National Research Service Award from the National Institutes of Health.
Abbreviations
- TAR
trans-activation responsive region
- CAT
chloramphenicol acetyltransferase
- TOCSY
total correlated spectroscopy
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Miller P S. Progr Nucleic Acid Res Mol Biol. 1996;52:261–291. doi: 10.1016/s0079-6603(08)60969-1. [DOI] [PubMed] [Google Scholar]
- 2.Beal P A, Dervan P B. Science. 1991;251:1360–1363. doi: 10.1126/science.2003222. [DOI] [PubMed] [Google Scholar]
- 3.Maher L J D, Wold B, Dervan P B. Antisense Res Dev. 1991;1:277–281. [PubMed] [Google Scholar]
- 4.Helene C, Thuong N T, Harel-Bellan A. Ann NY Acad Sci. 1992;660:27–36. doi: 10.1111/j.1749-6632.1992.tb21054.x. [DOI] [PubMed] [Google Scholar]
- 5.Nielsen P E. Curr Opin Biotechnol. 1999;10:71–75. doi: 10.1016/s0958-1669(99)80013-5. [DOI] [PubMed] [Google Scholar]
- 6.Gottesfeld J M, Neely L, Trauger J W, Baird E E, Dervan P B. Nature (London) 1997;387:202–205. doi: 10.1038/387202a0. [DOI] [PubMed] [Google Scholar]
- 7.White S, Szewczyk J W, Turner J M, Baird E E, Dervan P B. Nature (London) 1998;391:468–471. doi: 10.1038/35106. [DOI] [PubMed] [Google Scholar]
- 8.Ho S N, Boyer S H, Schreiber S L, Danishefsky S J, Crabtree G R. Proc Natl Acad Sci USA. 1994;91:9203–9207. doi: 10.1073/pnas.91.20.9203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu C, Smith B M, Ajito K, Komatsu H, Gomez-Paloma L, Li T, Theodorakis E A, Nicolaou K C, Vogt P K. Proc Natl Acad Sci USA. 1996;93:940–944. doi: 10.1073/pnas.93.2.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chastain M, Tinoco I., Jr Progr Nucleic Acid Res Mol Biol. 1991;41:131–177. doi: 10.1016/S0079-6603(08)60008-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chow C S, Bogdan F M. Chem Rev. 1997;97:1489–1514. doi: 10.1021/cr960415w. [DOI] [PubMed] [Google Scholar]
- 12.Weeks K M, Crothers D M. Science. 1993;261:1574–1577. doi: 10.1126/science.7690496. [DOI] [PubMed] [Google Scholar]
- 13.Still W C. Acc Chem Res. 1996;29:155–163. [Google Scholar]
- 14.Jones K A, Peterlin B M. Annu Rev Biochem. 1994;63:717–743. doi: 10.1146/annurev.bi.63.070194.003441. [DOI] [PubMed] [Google Scholar]
- 15.Hamy F, Felder E, Heizmann G, Lazdins J, Aboulela F, Varani G, Karn J, Klimkait T. Proc Natl Acad Sci USA. 1997;94:3548–3553. doi: 10.1073/pnas.94.8.3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamy F, Brondani V, Florsheimer A, Stark W, Blommers M J J, Klimkait T. Biochemistry. 1998;37:5086–5095. doi: 10.1021/bi972947s. [DOI] [PubMed] [Google Scholar]
- 17.Mei H-Y, Cui M, Heldsinger A, Lemrow S, Loo J, Sannes-Lowery K, Sharmeen L, Czarnik A. Biochemistry. 1998;37:14204–14212. doi: 10.1021/bi981308u. [DOI] [PubMed] [Google Scholar]
- 18.Ohlmeyer M H J, Swanson R N, Dillard L W, Reader J C, Asouline G, Kobayashi R, Wigler M, Still W C. Proc Natl Acad Sci USA. 1993;90:10922–10926. doi: 10.1073/pnas.90.23.10922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bayer E. Angew Chem. 1991;30:113–129. [Google Scholar]
- 20.Scaringe S A, Francklyn C, Usman N. Nucleic Acids Res. 1990;18:5433–5441. doi: 10.1093/nar/18.18.5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shah K, Wu H, Rana T M. Bioconjugate Chem. 1994;5:508–512. doi: 10.1021/bc00030a005. [DOI] [PubMed] [Google Scholar]
- 22.Misiura K, Durrant I, Evans M R, Gait M J. Nucleic Acids Res. 1990;18:4345–4354. doi: 10.1093/nar/18.15.4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shah K, Neenhold H, Wang Z, Rana T M. Bioconjugate Chem. 1996;7:283–289. doi: 10.1021/bc960023w. [DOI] [PubMed] [Google Scholar]
- 24.Ping Y-H, Liu Y, Wang X, Neenhold H R, Rana T M. RNA. 1997;3:850–860. [PMC free article] [PubMed] [Google Scholar]
- 25.Milligan J F, Groebe D R, Witherell G W, Uhlenbeck O C. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Z, Rana T M. Biochemistry. 1996;35:6491–6499. doi: 10.1021/bi960037p. [DOI] [PubMed] [Google Scholar]
- 27.Neenhold H R, Rana T M. Biochemistry. 1995;34:6303–6309. doi: 10.1021/bi00019a007. [DOI] [PubMed] [Google Scholar]
- 28.Churcher M J, Lamont C, Hamy F, Dingwall C, Green S M, Lowe A D, Butler P J C, Gait M J, Karn J. J Mol Biol. 1993;230:90–110. doi: 10.1006/jmbi.1993.1128. [DOI] [PubMed] [Google Scholar]
- 29.Mei H, Mack D, Galan A, Halim N, Heldsinger A, Loo J, Moreland D, Sannes-Lowery K, Sharmeen L, Truong H, Czarnik A. Bioorg Med Chem. 1997;5:1173–1184. doi: 10.1016/s0968-0896(97)00064-3. [DOI] [PubMed] [Google Scholar]
- 30.Muller B, Restle T, Reinstein J, Goody R S. Biochemistry. 1991;30:3709–3715. doi: 10.1021/bi00229a017. [DOI] [PubMed] [Google Scholar]
- 31.Aboul-ela F, Karn J, Varani G. J Mol Biol. 1995;253:313–332. doi: 10.1006/jmbi.1995.0555. [DOI] [PubMed] [Google Scholar]
- 32.Puglisi J D, Tan R, Calnan B J, Frankel A D, Williamson J R. Science. 1992;257:76–80. doi: 10.1126/science.1621097. [DOI] [PubMed] [Google Scholar]
- 33.Felber B K, Pavlakis G N. Science. 1988;239:184–187. doi: 10.1126/science.3422113. [DOI] [PubMed] [Google Scholar]
- 34.Frankel A D, Pabo C O. Cell. 1988;55:1189–1194. doi: 10.1016/0092-8674(88)90263-2. [DOI] [PubMed] [Google Scholar]
- 35.Nordeen S K. BioTechniques. 1988;6:454–457. [PubMed] [Google Scholar]
- 36.Huq I, Ping Y-H, Tamilarasu N, Rana T M. Biochemistry. 1999;38:5172–5177. doi: 10.1021/bi982638h. [DOI] [PubMed] [Google Scholar]
- 37.Jakobovits A, Smith D H, Jakobovits E B, Capon D J. Mol Cell Biol. 1988;8:2555–2561. doi: 10.1128/mcb.8.6.2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cordingley M G, La Femina R L, Callahan P L, Condra J H, Sardana V V, Graham D J, Nguyen T M, Le Grow K, Gotlib L, Schlabach A J, Colonno R J. Proc Natl Acad Sci USA. 1990;87:8985–8989. doi: 10.1073/pnas.87.22.8985. [DOI] [PMC free article] [PubMed] [Google Scholar]