Abstract
High hydrostatic pressures (1–2 kbar), combined with low, nondenaturing concentrations of guanidine hydrochloride (GdmHCl) foster disaggregation and refolding of denatured and aggregated human growth hormone and lysozyme, and β-lactamase inclusion bodies. One hundred percent recovery of properly folded protein can be obtained by applying pressures of 2 kbar to suspensions containing aggregates of recombinant human growth hormone (up to 8.7 mg/ml) and 0.75 M GdmHCl. Covalently crosslinked, insoluble aggregates of lysozyme could be refolded to native, functional protein at a 70% yield, independent of protein concentration up to 2 mg/ml. Inclusion bodies containing β-lactamase could be refolded at high yields of active protein, even without added GdmHCl.
In spite of recent progress in theoretical and computational approaches (1) to understanding protein folding and refolding, efforts to manipulate folding in vitro often are plagued by competing off-pathway aggregation processes. Protein aggregation is the subject of intense investigation in disciplines including human medicine, fundamental protein chemistry, and biotechnology. For example, aggregation can have severe consequences in human diseases (e.g., Alzheimer’s disease and Parkinson’s disease) (2) and in the manufacturing, shipping, storage, and delivery of protein therapeutics (3).
In particular, exploitation of the unique medical benefits of recombinant protein therapeutics often is hindered by the formation of non-native protein aggregates from native protein molecules. If even a minor fraction (e.g., 1%) of a parenterally delivered protein is aggregated, adverse reactions, including anaphylactic shock, can be induced (4–6). Aggregation can occur during refolding, purification, concentration, vial filling, freeze-thawing, lyophilization/rehydration, and delivery to patients. Particularly dramatic manifestations of the competition between proper folding pathways and off-pathway formation of non-native aggregates occur during attempts to obtain native recombinant proteins from precipitates formed during processing or inclusion bodies, both of which are essentially completely aggregated protein with substantial non-native structure (7–10).
Currently, refolding proteins from non-native aggregates and inclusion bodies requires proteins to be disaggregated and then refolded into their native conformation. Most commonly, aggregates are solubilized in a strong chaotrope, such as 8 M guanidine hydrochloride (GdmHCl) (11–14), which results in nearly complete unfolding of the protein molecules. Once soluble and unfolded, the proteins are first diluted with additional GdmHCl solution and then refolded by removing the chaotrope by dialysis or additional dilution. The refolding step, however, is difficult and depends strongly on renaturing conditions (14, 15). For example, redox conditions, pH, rates of dialysis, and protein concentration all must be empirically optimized for each protein (16). Furthermore, because the process of protein folding is first order in protein concentration, and the overall aggregation process is at least second order, aggregation is favored over refolding at higher protein concentrations. Hence, achieving acceptable yield (e.g., >10%) of refolded protein often requires protein to be refolded at very low concentrations (10–100 μg/ml) (16, 17, 19). As a result, once a therapeutic protein is refolded, it must be concentrated (typically 100- to 1,000-fold) to final dosage concentration. Losses of native protein also can occur during this concentration step. In addition, yield of properly folded protein upon renaturation is often low regardless of refolding conditions. Finally, the large volumes of waste chaotrope solution generated are expensive to dispose of properly.
The purpose of this study is to investigate the use of high pressure as an alternative to high concentrations of strong chaotropes for protein disaggregation and refolding. Pressures between 1 and 3 kbar will reversibly dissociate oligomeric proteins into subunits (20–23). Pressures above 4 kbar begin to denature the secondary structure of proteins (24–26). Pressure has been shown to reduce aggregation rates during refolding from fully soluble, denatured protein (27). Also, aggregation of P22 tailspike protein was reduced from 41% to 18% by pressure treatment of a 100 μg/ml solution (28). However, pressure has not been used as a tool to obtain native protein from insoluble aggregates at relatively high protein concentrations, with high yields, nor from covalently crosslinked aggregates or inclusion bodies. We hypothesize that if the pressure effects on aggregates of non-native protein molecules are similar to those on native multimeric proteins, then there must exist a “pressure window” where pressure is high enough to solubilize aggregates, but still allow refolding to the native conformation. The model systems chosen to test this hypothesis were agitation-induced insoluble aggregates of recombinant human growth hormone (rhGH), chaotrope-induced aggregates of hen egg white lysozyme that were covalently crosslinked through non-native disulfides, and inclusion bodies containing β-lactamase produced in Escherichia coli.
Materials and Methods
Growth Hormone Aggregates.
rhGH was supplied as a generous donation from Genentech. rhGH was aggregated by rotating 10 ml of rhGH solution in a 50-ml Falcon tube on a cell suspender for 24 hr at 8 rpm. Samples were prepared in 10 mM sodium citrate buffer (1 mM EDTA/0.1% sodium azide, pH 6.0, 24°C) at a concentration of 2 mg/ml rhGH. Before pressurization, samples containing aggregated rhGH were diluted to the desired protein concentrations with appropriate GdmHCl solutions in buffer to yield desired refolding conditions.
Concentrations of soluble rhGH were determined by UV spectroscopy with extinction coefficients of 18,890 (cm mol/liter)−1 at 278 nm (29) on a Hewlett–Packard 8452A diode array spectrophotometer. Insoluble aggregates were removed by centrifugation (13,000 g, room temperature) for 15 min. Contributions caused by soluble aggregates were subtracted from the signal as reported (30).
Lysozyme Aggregates.
Lysozyme (Sigma) was aggregated by first unfolding and reducing 40 mg/ml of lysozyme in 8 M GdmCl and 40 mM DTT. Samples were shock-diluted between 10- and 40-fold with 50 mM Tris buffer (1 mM EDTA, pH 8.0, 24°C) to induce aggregation. Tris buffer containing DTT, oxidized glutathione (GSSG), and GdmHCl then was added to yield a refolding buffer of 50 mM Tris (pH 8.0, 24°C), 0.8 M GdmCl, 5 mM GSSG, and 2 mM DTT.
Concentrations of both denatured and renatured lysozyme were determined with UV spectroscopy. Samples were centrifuged (13,000 g, room temperature) for 15 min to eliminate insoluble aggregates. Extinction coefficients for denatured and native lysozyme are 2.37 and 2.63 (cm mg/ml)−1, respectively (31). Contributions caused by soluble aggregates were subtracted from the signal as reported (30).
Lysozyme catalytic activity was measured by a method similar to the one described by Jolles (32). A total of 0.25 mg/ml of Micrococcus lysodeikticus cells were suspended in 67 mM potassium phosphate buffer, pH 6.2. All samples were diluted in Tris buffer (pH 8.0, 1 mM EDTA, 24°C) to a concentration between 0.05 and 0.25 mg/ml. Ten microliters of the diluted samples then was mixed into 990 μl of cell suspension to initiate the reaction. Absorbance of the sample was measured at 450 nm for 90 sec at 24°C. Slopes of an absorbance vs. time plot were calculated on points between 30 and 90 sec. Lysozyme concentrations were calculated based on the specific activity of fully native lysozyme.
β-Lactamase Inclusion Bodies.
E. coli strain RB791 cells transfected with pGB1 were grown as described (33). β-lactamase synthesis was induced by the addition of 0.1 mM isopropyl β-d-thiogalactoside at a cell culture optical density between 0.35 and 0.4 (600 nm). Cell cultures were grown overnight.
Upon completion of cell growth, cells were centrifuged (6,000 g, 4°C) and resuspended in washing buffer (50 mM Tris⋅HCl, pH 8.0, 24°C, 0.2% lysozyme). After 30 min incubation, cells were lysed with a homogenizer (Vertishear 2000, Vertis, Gardiner, NY) and centrifuged as above. The pellet was washed and centrifuged again. The pellet then was resuspended in washing buffer and incubated at room temperature for 20 min. Then, 2% deoxycholate was added and mixed at room temperature for an additional 20 min before final centrifugation to yield purified inclusion bodies.
Enzymatic activity of β-lactamase was determined spectroscopically by using penicillin G as a substrate (34).
Sample Pressurization.
Pressure was generated by using high-pressure nitrogen (400 bar) connected to a 10-fold hydraulic intensifier equipment (High Pressure Equipment, Erie, PA). Samples were prepared in heat-sealed bulbs of SAMCO transfer pipettes and placed into a 2-liter cloverleaf reactor rated to 2,000 bar and filled with water. Samples were slowly pressurized (over 10 min) to final desired pressure to minimize pressurization-induced heating; the depressurization rate was approximately 10 bar per min. Unless otherwise stated, all pressure experiments were performed at 24°C.
Fourier Transform IR Spectroscopy (FTIR).
FTIR was used to determine the secondary structure of rhGH aggregates in solution. All spectra were collected on a Nicolet Magna model 550 spectrometer equipped with a dTGS (deuterized triglycine sulfate) detector. A 256-scan interferogram was acquired in single-beam mode with a 4-cm−1 resolution. Aggregate slurries were placed in an adjustable path-length IR cell set at 8 μm. The same cell was used to collect a buffer blank (the buffer used for the blank was identical to the buffer in the aggregate slurry). Buffer and water vapor contributions were subtracted from the spectrum by using the Nicolet software. Second derivative analysis was used for peak narrowing and resolution. A seven-point smoothing was used to remove white noise, and baseline correction was performed over the amide I region. Finally, spectra were normalized by their total area over the amide I region (35).
Size Exclusion Chromatography.
HPLC analysis of soluble protein fractions was performed on a Beckman Gold HPLC system, equipped with a TosoHaas (Montgomeryville, PA) 2000 SW size exclusion column, and a 0.2-μm prefilter. A mobile phase of 10 mM sodium citrate buffer, pH 6.0 (rhGH samples) or 1.2 M KCl (β-lactamase samples) at 0.8 ml/min was used, and 20-μl samples were injected with an autosampler. Protein elution was monitored by absorbance at 214 nm. To analyze the amount of β-lactamase remaining in the insoluble fraction after pressurization, the insoluble pellets first were dissolved in 4 M GdmHCl, 4 mM DTT, and then injected on the column.
Total Protein Assay.
Protein concentrations reported were measured with the BCA total protein assay distributed by Pierce. Standard concentrations of BSA were used to calibrate the method.
Results
Disaggregation and Refolding of rhGH.
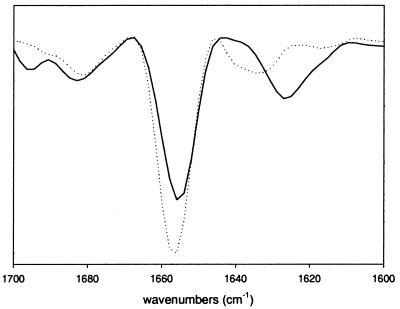
rhGH is a 22-kDa protein formed into a four α-helical bundle (13). Its surface contains two relatively large regions with a high percentage of hydrophobic residues (36, 37). Even gentle agitation rhGH solutions results in aggregation and precipitation (38). After 24 hr of agitation, 95% of the protein forms insoluble aggregates. To confirm that the structure of the precipitates is non-native, the secondary structure of rhGH aggregates was determined by IR spectroscopy. The second derivative spectra, in the conformationally sensitive amide I region, of native and aggregated growth hormone are given in Fig. 1. The spectrum of native rhGH is dominated by a strong band at 1,654 cm−1, which is the result of the high proportion of α-helix (13). The spectrum for the aggregated protein shows a substantial absorbance at 1,654 cm−1, demonstrating partial retention of α-helix and prominent non-native bands at 1,627 cm−1 and 1,695 cm−1. The latter bands are caused by intermolecular β-sheet structure, a common structural motif in non-native protein aggregates, precipitates, fibrils, and inclusion bodies (39).
Figure 1.
Second derivative Fourier transform IR spectroscopy spectra of native rhGH (dashed line) and aggregated growth hormone (solid line).
The effect of pressure on disaggregation and refolding of rhGH was examined at protein concentrations between 0.87 and 8.7 mg/ml. Various concentrations of GdmHCl were tested on 0.87 mg/ml samples to determine whether chaotropes augment the effects of pressure. The range of GdmHCl tested (0–1 M) was chosen so as to remain well below the concentrations where, at atmospheric pressure, rhGH begins to unfold or form molten globules (29). At atmospheric pressure, unfolding is detected first at 3.5 M GdmHCl. Thus, assuming that pressure (up to 2 kbar) does not greatly increase the effectiveness of GdmHCl as a denaturant, native rhGH is thermodynamically favored in the presence of the GdmHCl concentrations tested here.
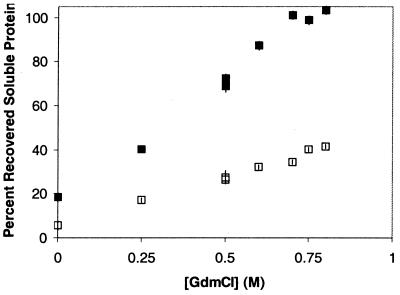
At atmospheric pressure and in the absence of GdmHCl, no protein refolding was detected (i.e., 95% of the protein remains aggregated) after 24 hr. Even in the presence of up to 0.8 M GdmHCl, control samples maintained at atmospheric pressure had at most 40% recovery of soluble protein (Fig. 2). In the absence of GdmHCl, pressurization to 2 kbar for 24 hr and return to atmospheric pressure increased soluble protein to 18% (Fig. 2). As the concentration of GdmHCl increased to 0.7 M, the amount of soluble protein recovered after 24-hr incubation at 2 kbar increased to a maximum of 100%. The soluble protein was fully native based on examination of far- and near-UV CD spectra (data not shown), documenting, respectively, that native secondary and tertiary structures were recovered. In addition, size-exclusion chromatography documented that the soluble protein was monomeric.
Figure 2.
Percent recovered soluble rhGH as a function of GdmHCl concentration. Outlined points (□) represent atmospheric samples, and solid points (■) represent samples pressurized to 2 kbar for 24 hr. In this and the following figures, error bars represent ± 1 SD, with data points taken at least in triplicate. Points with no apparent error bars have error bars smaller than the size of the data marker.
Remarkably, high pressure refolding of rhGH appears to be independent of protein concentration. Once optimal refolding conditions were determined, we increased protein concentrations up to 8.7 mg/ml, orders of magnitude higher than conditions typically used for refolding studies. Samples pressurized for 24 hr at 2 kbar, 1 M GdmHCl also achieved 100% recovery of rhGH from aggregates (data not shown).
These results document that neither pressure nor GdmHCl alone (<1 M) is sufficient to allow 100% recovery of native, soluble rhGH. Furthermore, the effect of the combination of factors is synergistic, for reasons that are not clear at this point. However, the current results show that, with pressure treatment, native rhGH can be recovered from aggregates by using GdmHCl concentrations an order of magnitude lower, and at protein concentrations several orders of magnitude higher, than those typically used for refolding.
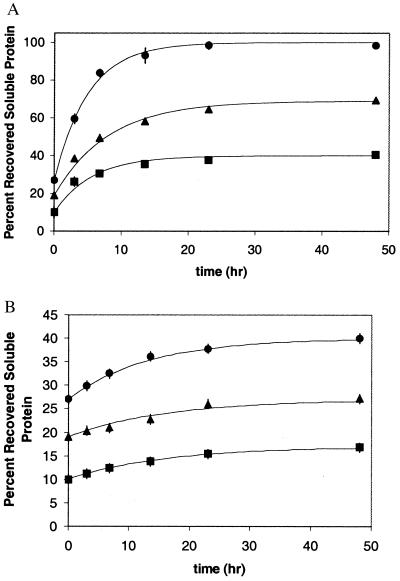
To determine whether the observed refolding yields represent equilibrium values, we investigated the effects of duration of pressure exposure on the degree of refolding measured after return to atmospheric pressure (Fig. 3). At 0.87 mg/ml rhGH and a given concentration of GdmHCl, refolding yields reach a plateau at approximately 24 hr in samples exposed to pressure (Fig. 3A) as well as those held at atmospheric pressure (Fig. 3B). Thus, the values previously shown in Fig. 2 represent results taken after the refolding had reached a maximum for the respective conditions. The existence of the plateaus implies that a steady-state condition is achieved within 24 hr for each GdmHCl concentration. At the system pressure and GdmHCl concentrations tested, the native state presumably is favored. If equilibrium were achieved, then there should be essentially the same level of recovery of native protein at all GdmHCl concentrations. There are, thus, undefined kinetic barriers to refolding, such as incomplete disaggregation, in samples with less than 0.75 M GdmHCl.
Figure 3.
(A) Percent recovered soluble rhGH as a function of time at 2 kbar. ■ represent samples refolded in 0.25 M GdmHCl, ▴ represent samples refolded in 0.5 M GdmHCl, and ● represent samples refolded in 0.75 M GdmHCl. The lines are for visual effect only. (B) Percent recovered soluble rhGH as a function of time under atmospheric pressure. ■ represent samples refolded in 0.25 M GdmHCl, ▴ represent samples refolded in 0.5 M GdmHCl, and ● represent samples refolded in 0.75 M GdmHCl. The lines are for visual effect only.
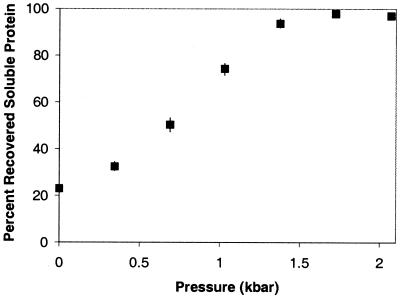
We also examined the effects of varying pressure on rhGH refolding in 0.75 M GdmHCl solutions. At 0.87 mg/ml, after 24 hr of incubation at pressure and return to atmospheric pressure, recovery of soluble protein as a function of pressure increases until 100% recovery is achieved at approximately 1.75 kbar (Fig. 4).
Figure 4.
Percent recovered soluble rhGH as a function of pressure. Samples were refolded in 0.75 M GdmHCl and pressurized for 24 hr.
Disaggregation and Refolding of Covalently Modified Aggregates.
Proteins containing cysteine can form covalent aggregates with intermolecular disulfide bonds and, potentially, non-native intramolecular disulfide bonds (i.e., “disulfide scrambling”). Clearly, proper refolding of these covalent aggregates cannot be achieved by treatment with chaotrope alone and can be much more complicated than refolding from noncovalent aggregates. Proper refolding of these proteins requires complete reduction of the protein to perform the solubilization step. Then, the correct disulfide bonds must be reformed via air oxidation, an oxido-shuffling system, or exposure to mixed disulfides (16). Most frequently, a mixture of low molecular weight thiols, such as glutathione (oxidized and reduced), is used to reshuffle disulfide bonds during refolding (31). Although these conditions have been shown to be useful in attempts to refold lysozyme (whose native conformation contains four disulfides) from initially soluble, unfolded molecules, studies on lysozyme inclusion bodies have been less successful (40).
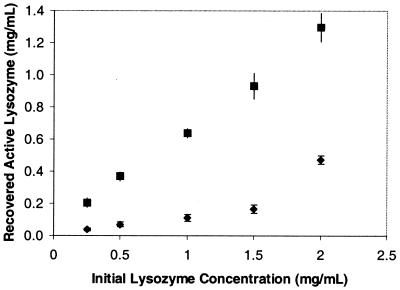
Precipitates of denatured, covalently crosslinked lysozyme were prepared by completely unfolding and reducing the protein, followed by shock dilution in buffer and reoxidation. IR spectroscopy revealed that the precipitates had a high proportion of non-native intermolecular β-sheet structure (data not shown). Aggregates were pressurized to 2 kbar for 48 hr and tested for soluble protein and catalytic activity after depressurization. Previous studies of pressure-induced unfolding of native lysozyme (reviewed by Jonas and Jonas in ref. 41) indicate that the midpoint of the pressure-induced unfolding transition is above 5 kbar, even at 68.5°C, which is near the thermal denaturation temperature. Recovery of catalytic activity is a sensitive indicator of proper refolding and requires reforming of the four proper disulfides in the native conformation (42). Recovered activity after pressurization increased linearly with lysozyme concentration. Remarkably, at all concentrations tested up to 2 mg/ml, yields of catalytically active protein were approximately 70% (Fig. 5), with 100% recovery of soluble protein. No attempt was made to optimize ratios of oxidized glutathione to DTT for maximum refolding. Conceivably, the 30% unrecoverable protein activity is the result of suboptimal redox conditions. Less than 30% recovery of activity was noted with samples held at atmospheric pressure.
Figure 5.
Recovered active lysozyme vs. initial lysozyme concentration. Refolding buffer of 50 mM Tris (pH 8.0), 0.8 M GdmHCl, 1 mM EDTA, 5 mM oxidized glutathione, and 2 mM DTT was used. ⧫ represent atmospheric samples, and ■ represent samples pressurized to 2 kbar for 48 hr.
Refolding of Protein from Inclusion Bodies.
During the overexpression of recombinant protein in E. coli, dense, insoluble particles of aggregated protein often are formed. These structures, referred to as inclusion bodies, contain a high level of non-native intermolecular β-sheet structure and pose a serious obstacle to efficient production of recombinant proteins. To test the applicability of high pressures to obtaining native proteins from inclusion bodies, we studied a cytoplasmic inclusion body system in E. coli (15) that contains β-lactamase as a major protein component. IR spectroscopy of harvested and washed (15) β-lactamase inclusion bodies documented that this system contains a large fraction of non-native intermolecular β-sheet structure (data not shown). As shown in Table 1, application of 2-kbar pressure for 48 hr results in significant levels of recovered catalytic activity, even in the absence of GdmHCl. In contrast, inclusion bodies held at atmospheric pressure show no catalytic activity below 1 M GdmHCl, consistent with earlier results (15). In addition, at atmospheric pressure, GdmHCl at concentrations below 1 M has little effect on protein solubility.
Table 1.
Recovery of β-lactamase from cytoplasmic inclusion bodies produced in E. coli RB791 (pGB1) as a function of [GdmHCl]
| [GdmHCl] (M) | Activity, units/mg soluble protein (SD) | Total soluble protein, mg/ml* | Percent β-lactamase recovered in soluble fraction† |
|---|---|---|---|
| 0 | 2,200 (400) | 0.10 | 85 |
| 0.3 | 2,000 (330) | 0.12 | 85 |
| 0.6 | 1,200 (200) | 0.20 | 85 |
| 0.9 | 700 (200) | 0.22 | 85 |
| 1.2 | 400 (100) | 0.43 | 84 |
Purified inclusion bodies (32) were held at 37°C, 2 kbar for 48 hr before analysis.
Total soluble protein by total protein assay relative to BSA standard.
† Recovery of soluble β-lactamase based on size exclusion chromatography. Percent recovery is determined as ×100 (height of β-lactamase peak in the soluble fraction, divided by the sum of the peak heights of β-lactamase in the soluble fraction and in the remaining insoluble pellet). All peak heights were corrected appropriately for dilution.
Interestingly, application of pressure in the absence of GdmHCl served not only to recover β-lactamase activity, but also to purify the protein. At higher GdmHCl concentrations, the total amount of soluble protein increases, although the amount of soluble β-lactamase and its recovered activity remain roughly constant.
Discussion
We conclude that pressure provides a powerful tool for obtaining native protein molecules from insoluble aggregates, inclusion bodies, and even covalently crosslinked aggregates. This process allows proteins to be refolded from such aggregates at concentration orders of magnitude greater than reported previously and at yields approaching 100%. In fact, recovery of both rhGH and lysozyme exhibited independence of protein concentration in the ranges studied. This finding is consistent with previous reports that the pressure-induced dissociation of erythrocruorin is concentration independent (43), as is the dissociation at 3.5 kbar of myoglobin aggregates initially formed by unfolding at 12 kbar (44). Previous refolding studies at atmospheric conditions report strong negative dependence of recovery of native protein on protein concentration, even when concentrations were very low (e.g., usually 1–50 μg/ml) (14, 33). For example, a study on lysozyme recovery from inclusion bodies required protein concentrations of less than 40 μg/ml (40), while another study on refolding from soluble, denatured lysozyme reported that recovery of active dropped from ca. 95% at 50 μg/ml to ca. 25% at 1 mg/ml (31). Refolding from a denatured (but not aggregated) protein at concentrations above 1 mg/ml has been achieved (45, 46). Only one case has been reported for inclusion body processing at high protein concentrations (47). Methionyl bovine somatotropin inclusion bodies were dissolved and refolded in 4.5 M urea at protein concentrations in the 5–15 mg/ml range with yields of 80% (47).
High pressure has been found to promote formation of molten globule intermediates (48–53). Although it is customary to think of molten globules as highly aggregation-competent conformations, high pressure appears to inhibit this tendency. Work by Smeller et al. (44) has shown that although high pressures (12 kbar) denature myoglobin, aggregates of the unfolded protein do not form until pressure is released. They suggest that intermediate pressures (ca. 3 kbar) populate a folding intermediate that is aggregation prone under atmospheric conditions, but prevented from aggregating by pressure. This suggestion is consistent with the volume change from native state to molten globule intermediate of apomyoglobin of −70 ml/mol (18). Presumably, in the present case, the conformational state(s) of rhGH, lysozyme, and β-lactamase that are populated at pressures near 2 kbar all have partial molar volumes that are less than the aggregated state.
Acknowledgments
We acknowledge the generous gifts of rhGH from Genentech and E. coli strain RB791 cells transfected with pGB1 from Prof. G. Georgiou (University of Texas, Austin). This work was supported by the National Science Foundation (BES 9505301 and BES 9816975).
Abbreviations
- GdmHCl
guanidine hydrochloride
- rhGH
recombinant human growth hormone
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Alm E, Baker D. Curr Opin Struct Biol. 1999;9:189–196. doi: 10.1016/S0959-440X(99)80027-X. [DOI] [PubMed] [Google Scholar]
- 2.Lansbury P T., Jr Proc Natl Acad Sci USA. 1999;96:3342–3344. doi: 10.1073/pnas.96.7.3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carpenter J F, Pikal M J, Chang B S, Randolph T W. Pharm Res. 1997;14:969–975. doi: 10.1023/a:1012180707283. [DOI] [PubMed] [Google Scholar]
- 4.Moore W V, Leppert P. J Clin Endocrinol Metab. 1980;51:691–697. doi: 10.1210/jcem-51-4-691. [DOI] [PubMed] [Google Scholar]
- 5.Ratner R E, Phillips T M, Steiner M. Diabetes. 1990;39:728–733. doi: 10.2337/diab.39.6.728. [DOI] [PubMed] [Google Scholar]
- 6.Thornton C A, Ballow M. Arch Neurol. 1993;50:135–136. doi: 10.1001/archneur.1993.00540020013009. [DOI] [PubMed] [Google Scholar]
- 7.Bowden G, Angel P M, Georgiou G. Bio/Technology. 1991;9:725–730. doi: 10.1038/nbt0891-725. [DOI] [PubMed] [Google Scholar]
- 8.Mitraki A, King J. Bio/Technology. 1989;7:690–697. [Google Scholar]
- 9.Oberg K, Chrunyk B A, Wetzel R, Fink A L. Biochemistry. 1994;33:2628–2634. doi: 10.1021/bi00175a035. [DOI] [PubMed] [Google Scholar]
- 10.Przybycien T, Dunn J P, Valax P, Georgiou G. Protein Eng. 1994;7:131–136. doi: 10.1093/protein/7.1.131. [DOI] [PubMed] [Google Scholar]
- 11.Mitraki A, Betton J, Desmadril M, Yon J. Eur J Biochem. 1987;163:29–34. doi: 10.1111/j.1432-1033.1987.tb10732.x. [DOI] [PubMed] [Google Scholar]
- 12.Vandenbroeck K, Martens E, D’Andrea S, Billiau A. Eur J Biochem. 1993;215:481–486. doi: 10.1111/j.1432-1033.1993.tb18057.x. [DOI] [PubMed] [Google Scholar]
- 13.DeLoskey R, Van Dyk D, Van Aken T, Campbell-Burk S. Arch Biochem Biophys. 1994;311:72–78. doi: 10.1006/abbi.1994.1210. [DOI] [PubMed] [Google Scholar]
- 14.Rudolph R, Lilie H. FASEB J. 1996;10:49–56. [PubMed] [Google Scholar]
- 15.Valax P, Georgiou G. Biotechnol Progr. 1993;9:539–547. doi: 10.1021/bp00023a014. [DOI] [PubMed] [Google Scholar]
- 16.Rudolph R. In: Modern Methods in Protein and Nucleic Acid Research. Tschesche H, editor. New York: de Gruyter; 1990. pp. 149–172. [Google Scholar]
- 17.Goldberg M, Rudolph R, Jaenicke R. Biochemistry. 1991;30:2790–2797. doi: 10.1021/bi00225a008. [DOI] [PubMed] [Google Scholar]
- 18.Vidugiris G J A, Royer C A. Biophys J. 1998;75:463–470. doi: 10.1016/S0006-3495(98)77534-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maachupalli-Reddy J, Kelley B, DeBernardez-Clark E. Biotechnol Prog. 1997;13:144–150. doi: 10.1021/bp970008l. [DOI] [PubMed] [Google Scholar]
- 20.Paladini A, Weber G. Biochemistry. 1981;20:2587–2593. doi: 10.1021/bi00512a034. [DOI] [PubMed] [Google Scholar]
- 21.Muller K, Ludemann H, Jaenicke R. Biophys Chem. 1982;16:1–7. doi: 10.1016/0301-4622(82)85001-1. [DOI] [PubMed] [Google Scholar]
- 22.Weber G. In: High Pressure Chemistry and Biochemistry. van Eldik R, Jonas J, editors. Norwell, MA: Reidel Publishing; 1987. pp. 401–420. [Google Scholar]
- 23.Ruan K, Weber G. Biochemistry. 1988;27:3295–3301. doi: 10.1021/bi00409a026. [DOI] [PubMed] [Google Scholar]
- 24.Lange R, Bee N, Frank J, Balny C. Prog Biotechnol. 1996;13:135–140. [Google Scholar]
- 25.Tang G, Ruan K. Prog Biotechnol. 1996;13:163–166. [Google Scholar]
- 26.Yamaguchi T, Yamada H, Akasaka K. Prog Biotechnol. 1996;13:141–146. [Google Scholar]
- 27.Gorovits B M, Horowitz P M. Biochemistry. 1998;37:6132–6135. doi: 10.1021/bi9730137. [DOI] [PubMed] [Google Scholar]
- 28.Foguel D, Robinson C R, Caetano de Sousa P, Jr, Silva J L, Robinson A S. Biotech Bioeng. 1999;63:552–558. doi: 10.1002/(sici)1097-0290(19990605)63:5<552::aid-bit5>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 29.Bam N B, Cleland J L, Randolph T W. Biotechnol Prog. 1996;12:801–806. doi: 10.1021/bp960068b. [DOI] [PubMed] [Google Scholar]
- 30.Leach S J, Scheraga H A. J Am Chem Soc. 1960;82:4790–4792. [Google Scholar]
- 31.De Bernardez Clark E, Hevehan D, Szela S, Maachupalli-Reddy J. Biotechnol Progr. 1998;14:47–54. doi: 10.1021/bp970123w. [DOI] [PubMed] [Google Scholar]
- 32.Jolles P. Methods Enzymol. 1962;5:137–140. [Google Scholar]
- 33.Valax P, Georgiou G. In: Biocatalyst Design for Stability & Specificity. Himmel M, Georgiou G, editors. Washington, DC: Am. Chem. Soc.; 1993. pp. 126–139. [Google Scholar]
- 34.Waley S G. Biochem J. 1974;139:789–790. doi: 10.1042/bj1390789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dong A, Prestrelski S J, Allison S D, Carpenter J F. J Pharm Sci. 1995;84:415–424. doi: 10.1002/jps.2600840407. [DOI] [PubMed] [Google Scholar]
- 36.de Vos A M, Ultcsh M, Kossiakoff A A. Science. 1992;255:306–312. doi: 10.1126/science.1549776. [DOI] [PubMed] [Google Scholar]
- 37.Ultsch M H, Somers W, Kossiakoff A A, de Vos A M. J Mol Biol. 1994;236:286–299. doi: 10.1006/jmbi.1994.1135. [DOI] [PubMed] [Google Scholar]
- 38.Bam N B, Cleland J L, Manning M C, Carpenter J F, Kelley R F, Randolph T W. J Pharm Sci. 1998;87:1554–1559. doi: 10.1021/js980175v. [DOI] [PubMed] [Google Scholar]
- 39.Fink A L. Folding Des. 1998;3:R9–R23. doi: 10.1016/S1359-0278(98)00002-9. [DOI] [PubMed] [Google Scholar]
- 40.Fischer B, Perry B, Sumner I, Goodenough P. Protein Eng. 1992;5:593–596. doi: 10.1093/protein/5.6.593. [DOI] [PubMed] [Google Scholar]
- 41.Jonas J, Jonas A. Annu Rev Biophys Biomol Struct. 1994;23:287–318. doi: 10.1146/annurev.bb.23.060194.001443. [DOI] [PubMed] [Google Scholar]
- 42.Jolles P. In: Lysozymes: Model Enzymes in Biochemistry and Biology. Jolles P, editor. Boston: Birkhauser; 1996. pp. 143–161. [Google Scholar]
- 43.Silva J L, Villasboas M, Bonafe C F S, Meirelles N C. J Biol Chem. 1989;264:15863–15868. [PubMed] [Google Scholar]
- 44.Smeller L, Rubens P, Heremans K. Biochemistry. 1999;38:3816–3820. doi: 10.1021/bi981693n. [DOI] [PubMed] [Google Scholar]
- 45.Hevehan D L, De Bernardez Clark E. Biotechnol Bioeng. 1997;54:221–230. doi: 10.1002/(SICI)1097-0290(19970505)54:3<221::AID-BIT3>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 46.Maeda Y, Ueda T, Imoto T. Protein Eng. 1996;9:95–100. doi: 10.1093/protein/9.1.95. [DOI] [PubMed] [Google Scholar]
- 47.Storrs S B, Przybycien T M. ACS Symp Ser. 1991;470:197–205. [Google Scholar]
- 48.Clery C, Renault F, Masson P. FEBS Lett. 1995;370:212–214. doi: 10.1016/0014-5793(95)00787-a. [DOI] [PubMed] [Google Scholar]
- 49.Zhang J, Peng X, Jonas A, Jonas J. Biochemistry. 1995;34:8631–8641. doi: 10.1021/bi00027a012. [DOI] [PubMed] [Google Scholar]
- 50.Vidugiris C A J. Biochemistry. 1995;34:4909–4912. doi: 10.1021/bi00015a001. [DOI] [PubMed] [Google Scholar]
- 51.Peng X D, Jonas J, Silva J L. Proc Natl Acad Sci USA. 1993;90:1776–1780. doi: 10.1073/pnas.90.5.1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Prevelige P E, King J, Silva J. Biophys J. 1994;66:1631–1641. doi: 10.1016/S0006-3495(94)80955-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tamura Y, Gekko K. Biochemistry. 1995;34:1878–1884. doi: 10.1021/bi00006a008. [DOI] [PubMed] [Google Scholar]