Abstract
We report the crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus, a major human pathogen, to 2.8-Å resolution. This enzyme is a key target for developing specific antiviral therapy. The structure of the catalytic domain contains 531 residues folded in the characteristic fingers, palm, and thumb subdomains. The fingers subdomain contains a region, the “fingertips,” that shares the same fold with reverse transcriptases. Superposition to the available structures of the latter shows that residues from the palm and fingertips are structurally equivalent. In addition, it shows that the hepatitis C virus polymerase was crystallized in a closed fingers conformation, similar to HIV-1 reverse transcriptase in ternary complex with DNA and dTTP [Huang H., Chopra, R., Verdine, G. L. & Harrison, S. C. (1998) Science 282, 1669–1675]. This superposition reveals the majority of the amino acid residues of the hepatitis C virus enzyme that are likely to be implicated in binding to the replicating RNA molecule and to the incoming NTP. It also suggests a rearrangement of the thumb domain as well as a possible concerted movement of thumb and fingertips during translocation of the RNA template-primer in successive polymerization rounds.
Hepatitis C virus (HCV) is a major human pathogen that has infected an estimated 3% of the population worldwide. The virus is capable of establishing a persistent infection in the majority of cases, leading to chronic hepatitis that often develops into cirrhosis and, in many cases, into hepatocellular carcinoma. There is no vaccine available against HCV, and current therapies are effective only in a minority of cases. There is thus an urgent need to develop HCV-specific antiviral agents to counteract this important public health problem.
HCV is a positive-strand RNA virus in the Flaviviridae family. The viral genome contains a single ORF encoding a polyprotein of ≈3,000 amino acids (1) that is the precursor to all viral proteins. Genome replication proceeds in two steps: synthesis of complementary minus-strand RNA using the genome as template and the subsequent synthesis of genomic RNA using this minus-strand RNA template. The key enzyme involved in both of these steps is a virally encoded RNA-dependent RNA polymerase (RdRp). The HCV NS5B protein, located at the extreme C terminus of the polyprotein, contains motifs shared by all RdRps, such as the GDD motif (2). Indeed, RdRp activity has been demonstrated with recombinant NS5B (3).
The catalytic domain of nucleic acid polymerases is organized around a central cleft in an arrangement that is reminiscent of a right hand, with subdomains that have been termed “palm,” “fingers,” and “thumb,” as originally described in the structure of the Klenow fragment of Escherichia coli DNA polymerase 1 (4). Several three-dimensional structures of polynucleotide polymerases have been determined; they show that the four different classes of polymerases, i.e., DNA-dependent DNA and RNA polymerases and RNA-dependent DNA and RNA polymerases, share the same fold for the palm subdomain, which contains a cluster of catalytically important residues (see ref. 5 for a recent review). An exception is polymerase β, which has a different topology in the palm (6, 7). The similarity of DNA-dependent DNA polymerases of the polymerase 1 family with the other classes of polymerases had actually been suggested earlier by sequence alignments (8). The particular fold adopted by the palm subdomain is shared by many proteins that bind nucleotides and/or nucleic acids (9). It contains two absolutely conserved aspartic acid residues that coordinate two Mg2+ ions, which actually carry out the polymerization reaction. Fidelity in replication is achieved by allowing only a nucleotide triphosphate that is correctly base-paired to the matching nucleotide in the template to adopt the right geometry for the esterification reaction to take place (10).
In the case of RNA-dependent polymerases, the structures of two retroviral polymerases are known: reverse transcriptase (RT) from HIV 1 (for which structures are available in the unliganded form and in complex with several ligands; see ref. 11 for a recent review) and a fragment of RT from murine Moloney leukemia virus (12). The only RdRp for which we have structural data is unliganded poliovirus polymerase (9). This structure shows that the palm is folded as in all of the other classes of polymerases and that the fold of the thumb is different. Large parts of the fingers were disordered in the crystals, so that the complete fold of this subdomain is not known.
The catalytic domain of the HCV RdRp consists of the 531 amino-terminal residues of NS5B. As a key step to developing specific anti-HCV drugs that interfere with viral replication, we have crystallized this catalytic fragment. We describe in the following sections the crystal structure determined by the multiple anomalous diffraction method (13) to a resolution of 2.8 Å.
Materials and Methods
Protein Preparation and Crystallization.
Details of the purification, screening of variants, and crystallization are described elsewhere (L.T., S.B., M.M., R.L.V., I.I., A. Biroccio, A. Lahm, F.A.R., and R.D.F., unpublished work). In brief, a variant of NS5B deleted of its 55 C-terminal residues was produced in E. coli. This construct retained the same RNA polymerization activity in vitro as full length NS5B (L.T., S.B., M.M., R.L.V., I.I., A. Biroccio, A. Lahm, F.A.R., and R.D.F., unpublished work). A selenomethionine derivative, which was equally active, was used for the structure determination. The sample was concentrated to 14 mg/ml in a buffer containing 20 mM Hepes (pH 7.5), 20 mM DTT, and 1 mM EDTA. Crystals were grown by vapor diffusion at 4°C in hanging drops by mixing 1:1 volumes of the protein and a reservoir solution containing 5% (wt/vol) polyethylene glycol 8,000, 0.1 M sodium citrate (pH 6.5), and 5% (vol/vol) 2-propanol. A cryoprotectant solution containing 8% (wt/vol) polyethylene glycol 8,000, 15% (vol/vol) 2-methyl-2-5 pentanediol (MPD), and 2% (wt/vol) trehalose in the same buffer was used for cryocooling the crystals. After a 1-minute soak in this solution, crystals were directly plunged into liquid ethane. The crystals are orthorhombic, space group P212121, with unit cell parameters a = 67.1 Å, b = 96.9 Å, and c = 194.4 Å. The solvent content is 47%, and there are two molecules in the asymmetric unit.
Structure Determination.
Crystals were characterized at synchrotron beamlines DW32 of the “Laboratoire pour l’Utilisation du Rayonnement Electromagnétique” (Orsay, France) and X11 of the “Deutsches Elektronen-Synchrotron” (Hamburg, Germany). Multiwavelength anomalous diffraction data were collected at 105 K at the European Synchrotron Radiation Facility (Grenoble, France) beamline BM14 on a MarCCD detector and were processed with denzo and scalepack (14). The positions of the 24 selenium atoms in the asymmetric unit were automatically determined and were used to calculate multiwavelength anomalous diffraction phases with solve (15). The phases were improved by density modification including 2-fold noncrystallographic symmetry averaging, solvent flattening, and histogram matching with dm (16). The molecular envelopes were determined automatically by dm, and the initial noncrystallographic symmetry matrix was calculated from the positions of the heavy atoms. The resulting electron density map at 3-Å resolution was of excellent quality and was used to build an initial model with turbo frodo (17). Places where the electron density was weak corresponded to exposed loops 147–153 and 400–406. The five C-terminal residues (532–536) are disordered, and, thus, our model ends at residue 531. Refinement of the model was done with the cns package (18) against reflections between 19 and 2.8 Å with a maximum-likelihood target using the experimental phase probability distribution (19). Noncrystallographic symmetry restraints were applied throughout. The main refinement statistics are listed in Table 1, along with data collection statistics.
Table 1.
Summary of data collection and refinement statistics
| MAD data collection and phasing | ||||||||
|---|---|---|---|---|---|---|---|---|
| Wl (Å) | f′ | f" | Res, Å | Nref | Red | Compl. (%)* | Rsym*† | I/σ* |
| 0.9791 (p) | −7.9 | 5.6 | 2.8 | 31167 | 3.4 | 97.3 (82.6) | 8.4 (13.7) | 13.3 (5.7) |
| 0.9794 (i) | −8.8 | 3.7 | 2.8 | 31187 | 3.4 | 97.4 (82.8) | 7.3 (13.1) | 15.5 (6.1) |
| 0.8857 (r) | −2.4 | 3.9 | 2.8 | 31327 | 3.5 | 97.8 (85.9) | 7.3 (12.7) | 15.3 (6.2) |
| Mean figure of merit, 30–3 Å: 0.56; Z-score: 106‡ | ||||||||
| Refinement | |||
|---|---|---|---|
| R/Rfree† | rms bonds, Å | rms angles, degrees | No. of atoms, protein/solvent |
| 0.237/0.286 | 0.008 | 1.4 | 8,244/184 |
Wl, wavelength; p, absorption peak; i, inflection point, r, remote Wl; Res, resolution; Nref, number of unique reflections; Red, redundancy.
*Overall value (highest resolution shell).
† Rsym = Σh,i|Ih,i − 〈Ih〉|/Σh,iIh,i, where Ih,i is the intensity of the ith observation of the reflection of index h, R = Σ|Fo − Fc|/ΣFo for all reflections, and Rfree is R for 6% of the reflections omitted from the refinement (1,985 reflections) (33).
‡ Z-score: assessment of the quality of phasing according to the program solve (15).
Illustrations.
Fig. 1a was prepared with turbo frodo (17), Figs. 1b and 4b with ribbons (20), and Figs. 3 and 4a with grasp (21).
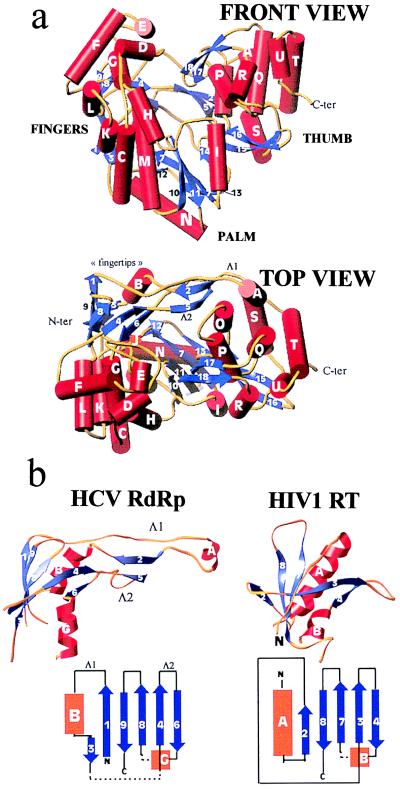
Figure 1.
(a) Front (Upper) and top (Lower) view of the HCV polymerase in a ribbons representation of the polypeptide chain. α-helices (labeled with capital letters) are shown as red cylinders, β-strands (labeled with numbers) as blue arrows, and connecting loops as yellow tubes. The front view has the palm subdomain at the base of the cleft, which is closed at either side by the fingers and thumb and at the back by loops Λ1 and Λ2. (b) The fingertips. Shown is the mixed barrel, composed of β-strands and an α-helix, that lies at the tip of the fingers and aligns structurally between RTs and RdRps. The left panel shows the ribbon diagram of this domain in the HCV polymerase along with a topology diagram. Two long loops (Λ1 and Λ2) emanate from the barrel to close the back of the enzyme. The right panel depicts the corresponding domain of HIV-1 RT in the same orientation. The topology diagram shows that there is a difference in connectivity in the left-most (N-terminal) strands. It is clear that Λ1 is absent but that Λ2 has its counterpart in RT. Broken lines indicate regions in which the polypeptide chain makes excursions into neighboring regions of the fingers to come back and complete the fold of the barrel.
Figure 4.
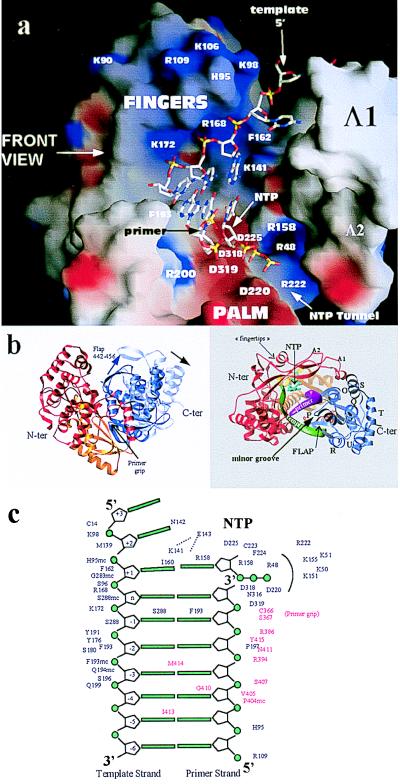
Interactions of the HCV RdRp with nucleic acid as inferred from the comparison with the structure of HIV-1 RT in a ternary complex with DNA and dTTP (26). (a) The template–primer duplex and dTTP molecule from the structure of the RT ternary complex superposed on a surface representation of the HCV polymerase colored according to electrostatic potential (as in Fig. 3 Upper). The view is a rotation of 90 degrees with respect to the front view of Figs. 1a and 3, looking from the thumb domain, which was removed for clarity. Some of the amino acids seen in the contacts are labeled. The transformation that superposes the palm and fingertips of the two enzymes has been applied to the nucleic acid coordinates for this figure. Five nucleotides of the template strand are shown, two of them base paired to the 3′ bases of the primer strand and a third to the incoming nucleotide, the triphosphate moiety of which points toward the tunnel mentioned in Fig. 3 and described in the text. (b) Proposed movement of the thumb on binding of RNA. The HCV polymerase is shown in a ribbon representation colored according to the different subdomains: fingers, red; palm, yellow; and thumb, blue. A rotation of ≈10 degrees (indicated by the black arrow in the left panel) about the connection between thumb and palm brings helix P in our structure into coincidence with helix H in HIV-1 RT, which tracks the minor groove of the A form DNA duplex. The resulting position of helix P in the complex is shown in the left panel, in a top view of the molecule where the template and primer backbone are displayed as green and magenta ribbons, respectively. The NTP molecule in the active site is colored cyan. The “flap” is a β-hairpin (strands 17 and 18), which has to move as well, as shown by the blue arrow in the left panel of the figure. (c) Diagram of the template–primer duplex in the catalytic site indicating the amino acid residues seen to contact the nucleic acid and nucleotide molecules after superposition to HIV-1 RT. These residues are very likely involved in binding RNA and nucleotide during polymerization. Residues behind an arc, around the triphosphate moiety of the dNTP molecule, are those lining the tunnel through which the NTP molecules are likely to access the active site of the enzyme. Residues indicated in red are those that come into contact after applying the rearrangement on the thumb shown in b.
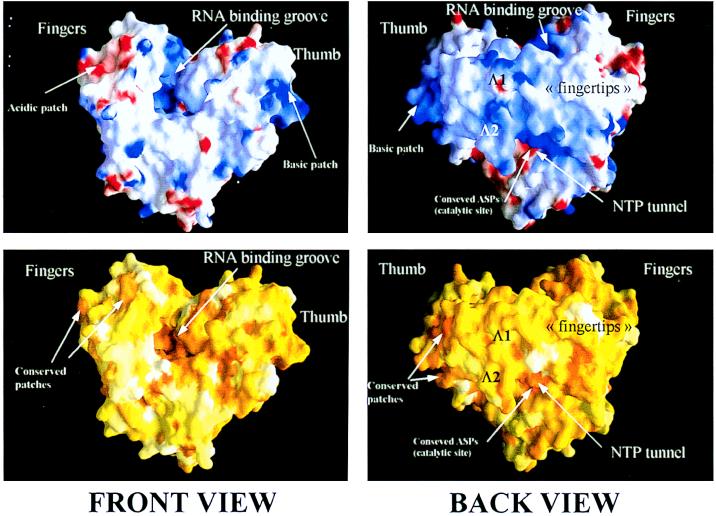
Figure 3.
The molecular surface of the HCV RdRp, front view (Left) and back view (Right). (Upper) Electrostatic potential calculated on the molecular surface. Positive and negative charges are indicated in blue and red, respectively. The positively charged groove along which RNA would bind during viral replication is apparent in the front view. The region of the thumb at the very C terminus of the catalytic domain of NS5B is also positively charged. In the back view, the lower ridge of the “bridge” formed by loops Λ1 and Λ2 is seen to have a strong positive charge. This area corresponds to the “tunnel” where NTP would access to the polymerizing complex. The red patch underneath the tunnel corresponds to the strictly conserved aspartic acids that are known to coordinate the catalytic Mg2+ ions. (Lower) The molecular surface is colored according to sequence conservation from an alignment that included one representative from each of the know HCV serotypes (1 through 6) and all three of the related GB viruses (A, B, and C) (32). The scoring ranges from 0 (white, no conservation) to 1 (dark orange, strict conservation). Note that both the RNA binding groove and the putative NTP tunnel are highly conserved. Note also the conserved ridges lining the acidic patch mentioned above and the conservation of the basic region at the back of the thumb.
Results and Discussion
Molecular Architecture.
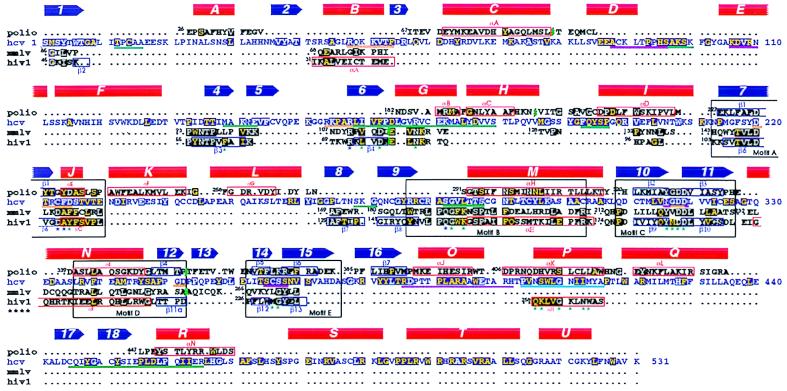
The HCV polymerase is a heart-shaped molecule (shown in Fig. 1a) with dimensions of 70 × 60 × 40 Å presenting a deep cleft in the middle with the palm at the base, as is the case for all other polymerases. The overall fold contains 21 α-helices and 18 β-strands. We use the standard secondary structure nomenclature used first for the Klenow fragment (4) and label β-strands with numbers and α-helices with capital letters proceeding from the N terminus. The fingers domain can be divided into a palm-proximal region, folded as a bundle of α-helices, and a distal region that is folded as a barrel, which we have named the “fingertips.” The latter is composed of a six-stranded, mixed directional β-sheet that is twisted to embrace helix B as an additional stave of the barrel, as shown in Fig. 1b. Two long loops emanate from the barrel, labeled Δ1 and Δ2. The longer loop, Δ1, extends away from the fingertips to reach the thumb domain from the back. A short α-helix at the tip of Δ1 (helix A) packs against the thumb domain, fitting in a groove between parallel α-helices O and Q (see below), thus closing the gap between the two domains (see Fig. 1a, Top view). In the poliovirus enzyme, where the top of the fingers subdomain was disordered, interpretable density was found for the corresponding N-terminal region of polypeptide chain (9), which remained ordered presumably through the interaction with the thumb. This region of the polypeptide chain, which was thought to be a separate domain unique to RdRp enzymes (22), is thus actually part of the fingers. The corresponding barrel of the fingertips is also found in retroviral polymerases, as shown in Fig. 1b, although the connectivity at the N-terminal end is different. The α-helix and four β-strands indeed align structurally (see Fig. 2). Loop Δ1 is missing in RT, but the loop corresponding to Δ2 (β3-β4 loop in HIV-1 RT) occupies the same region in space, even though it is shorter. Residues from these β-strands are implicated in contacts with the nucleic acid and incoming nucleotide, as will be discussed below. The fingers subdomain of the HCV polymerase (and very likely of many other RdRps) contains ≈200 more residues than the fingers of retroviral RTs, and as a consequence most of the proximal region is different, with the exception of the α-helices that extend into the palm subdomain. In contrast, the ordered regions of the fingers in the poliovirus RdRp match strikingly well their HCV counterparts, indicating that the whole fingers domain of RdRps from the Flaviviridae and Picornaviridae families very likely share the same fold. A structural alignment of NS5B with poliovirus polymerase and with the catalytic domain of RT from HIV-1 and Moloney leukemia virus is shown in Fig. 2.
Figure 2.
Structural alignment of RdRps and RTs. The HCV sequence is displayed in blue letters, with the poliovirus RdRp sequence above and the sequence of RT from Moloney leukemia virus and HIV-1 below. α-helices (red) and β-strands (blue) are indicated (with the corresponding label) as solid symbols above the sequences for HCV and as boxes surrounding the corresponding sequences for poliovirus and HIV-1. Conserved residues are indicated with a dark background (blue for HCV, black for the others). White letters denote strict conservation, which for HCV means conserved also in the related GB viruses (32). Yellow letters denote conservation to a lesser extent, which in the case of NS5B means conservation in all HCV strains and at least one of the GB viruses. In the case of poliovirus, we have not included residues that were disordered in the structure. The number of the N-terminal amino acid is shown next to each aligned segment. Green marks in the sequences indicate places in which there are insertions with respect to the HCV RdRp. The known polymerase “motifs,” A through E (28), are indicated by black boxes that comprise all four sequences. Blue and green stars underneath the HIV-1 RT sequences denote residues that contact the dNTP molecule and DNA, respectively. Underlines show regions of NS5B that would be implicated in such contacts in NS5B, as inferred from the superposition of the two structures. Orange, green, and magenta underlines mark regions that surround the NTP site, the template strand, and the primer strand, respectively. In the thumb, a cyan underline marks residues that would contact bases in the minor groove. In this domain, the underlined regions would contact the nucleic acid after a structural rearrangement as explained in the text.
The thumb is significantly bigger than the corresponding domain of the RNA dependent polymerases of known structure. It contains amino acids 370–531, at the carboxy-terminal end of the polypeptide chain, and folds as an independent domain. The fold is mainly α-helical, containing seven α-helices (labeled O through U; see Fig. 2) and two β-hairpins (β15–16 and β17–18). It is interesting to note that the T7 DNA polymerase has a β-hairpin in the thumb in a position that is very similar to β17–18, even though the overall fold of the respective domains is different. In the structure of the T7 polymerase ternary complex (30), this β-hairpin (labeled β6c-6d) drapes over the DNA primer strand.
The core of the NS5B thumb is formed by a repetition of a two helix motif that is similar to the “armadillo” (arm) repeats that have been found in a number of unrelated molecules that participate in protein–protein or protein–peptide recognition, like karyopherin α (23). Helices O and P, Q and R, and T and U can be superimposed to arm repeats 3–5 of karyopherin-α with an rms of 3.5 Å for 94 topologically equivalent α carbons and a dali (24) score of 3.5. Only the amino-terminal four α-helices in the NS5B thumb have their counterparts in the poliovirus enzyme, for which the corresponding domain is smaller. The connection between thumb and palm is similar to that found in poliovirus and in retroviral polymerases, going through a β-hairpin that has been called the “primer grip” (25), in our case β14–15.
As shown in Fig. 3, the central cleft is lined by basic amino acids and has the conserved aspartic acid residues (that are known to coordinate the catalytic Mg2+ ions) at the base. Conserved residues cluster in defined regions of the molecular surface: the RNA and NTP binding groove, the back of the thumb, the NTP tunnel (see below), and an acidic patch at the top-front of the fingers. The back surface of the thumb, with its “arm repeat”-like fold, could indeed be a site of interaction with other viral/cellular proteins because viral replication is likely to involve several partners.
Superposition to HIV-1 RT.
Superposition of the palm domain of NS5B with the corresponding region in HIV-1 RT brings the fingertips (shown in Fig. 1b) also into superposition in the case of RT in ternary complex (26) but not in the other, more open forms. The overall superposition results in an rms value of 1.4 Å for 101 topologically equivalent α carbons in the fingers and palm. This suggests that NS5B crystallized in the closed form, with the fingers in a conformation that resembles the ternary complex. It has been proposed that flexing of the fingers on binding of the nucleotide triphosphate is an important step that is necessary for translocation of the template to the next base because the structure adopted after the esterification reaction (that of the binary complex) is different, with the fingers more open (27). We cannot state with certainty, however, why the structure of this unliganded HCV polymerase is in the closed form. It may be because of the particular conditions under which the enzyme was crystallized.
The fact that our structure corresponds to the “closed fingers” conformation of the enzyme allows us to superpose it on the RT/dNTP/DNA structure and thus identify the residues in NS5B that come into proximity of the ligands. Fig. 4 shows that after such a superposition the nucleic acid fits nicely into the groove described in Fig. 3. Many strictly conserved residues of the HCV polymerase are seen in position to interact both with the nucleotide and the nucleic acid molecules. The superposition thus shows that in a replicating complex the NTP molecule would be wedged between (i) helix B, (ii) strand 6 and the preceding segment, where residue R158 is structurally equivalent to HIV-1-RT R72 (which in the structure of the ternary complex makes a number of interactions with the nucleotide), and (iii) strand 7 and helix J, which form the well known motif A of polymerases (28). The latter segment contains one of the strictly conserved aspartic acid residues, Asp 220 in NS5B, which coordinates the Mg2+ ions in the structures of ternary complexes. In agreement with earlier proposals inferred from the structure of the poliovirus polymerase (9), Asp 225 would be in position to hydrogen bond to the O2′ oxygen of the incoming NTP. This position is occupied by a tyrosine residue in RT and is responsible for discriminating against NTPs and selecting exclusively dNTPs (29). It remains to be seen what the determinants of the inverse selectivity are: i.e., NTPs instead of dNTPs. There is in addition a cluster of basic residues around the putative position of the triphosphate moiety of the NTP molecule; these are indicated in Fig. 4c and are R222, K155, K151, K51, and K50, all of them very conserved. These residues line a tunnel behind the molecule through which NTPs can access the binding site (visible in Fig. 3 underneath the “bridge” formed by Δ1 and Δ2, and also on Fig. 4a). The nucleotide would otherwise be quite buried in the complex, and it is difficult to imagine a way of access through the front of the molecule. In fact, during productive polymerization with concomitant elongation of the nascent chain, there must be an intense two-way traffic of pyrophosphate leaving from the previous round and NTP accessing the site for the new round through this tunnel.
As depicted in Fig. 4a, the superposition shows that the 5′ end of the template would enter the enzyme through a groove at the top of the fingers domain. The base that is paired to the incoming nucleotide would pack against the strictly conserved residue I160 in β strand 6 (which is structurally equivalent to HIV-1 RT residue L74 in β4 of the fingertips; see Figs. 1b and 2). The sugar-phosphate backbone of this nucleotide would contact the segment of polypeptide chain between amino acids 282 and 288, within the conserved motif B of polymerases (28). Continuing toward the 3′ end of the template strand, the sugar-phosphate backbone would pack against the C-terminal end of β-strand 6 and α-helices G and H. These two helices can be regarded as a long, curved helix with a break in the middle (at residue L175). The break allows the enzyme to adopt a shape in this region that is complementary to the curve made by the template strand. Before switching to contact the thumb, the template strand also would contact the region just before helix I (amino acids 193–197; see diagram in Fig. 4c and also see Fig. 2).
Proposed Rearrangement of the Thumb Domain on Binding of RNA.
A modest movement of the thumb domain on binding of substrate seems necessary because there would not be enough room for an RNA double-stranded molecule to fit in the structure as we see it. A rotation of ≈10 degrees about the connection to the palm (indicated in Fig. 4b) indeed allows superposition of helix P onto helix H of HIV-1-RT (which tracks the minor groove of the DNA molecule), superimposing the strictly conserved G410 on top of the strictly conserved G262 of RT. The movement is such that α-helix A on loop Δ1 is likely to follow and still pack the same way in this conformation because it is at the tip of a long loop that could adapt to this type of change. Residues from helix P in the thumb would thus track the minor groove of the newly synthesized RNA double helix. In this conformation, there is still a clash with β-hairpin 17–18, which has to move out of the way for the RNA molecule to fit. In our structure, the particular conformation of the hairpin is stabilized in part by a crystal contact. It is possible that this hairpin would act like a flap, with conserved P456 at the base of the β-hairpin as part of the hinge, and would drape over the RNA molecule in a similar way to the β-hairpin of the thumb of the T7 DNA-polymerase ternary structure (30). The modeling exercise of moving the flap is indicated in Fig. 4b. In this conformation, the sugar-phosphate backbone of the template and primer strands would be contacted by residues from α-helix R (and the flap) and by α-helix O, respectively.
Concluding Remarks.
This first structure of the complete catalytic domain of an RdRp supports the view that RNA-dependent polymerases of picornaviruses, flaviviruses, and retroviruses all evolved from the same ancestor, an issue that could not be resolved by sequence comparisons alone (31). Many other RNA-dependent polymerases also may turn out to belong to the same family. Superposition to the ternary complex of HIV-1-RT reveals most of the amino acids that are likely to be important for binding to the RNA template-primer duplex, which is believed to adopt a double-helical conformation similar to the A form DNA found in the ternary complex. This comparison also suggests certain conformational changes that might take place in NS5B on binding RNA. The connection between fingertips and thumb via loop Δ1 suggests the possibility of a concerted movement of the two domains for translocation of the nascent RNA molecule to the following base in the template during polymerization. This interaction also could contribute to keeping the fingers domain in the closed form in the absence of ligands, as the thumb itself is found in a closed conformation (it would have to open on binding RNA, a movement that could change, in turn, the conformation of the fingers). Loop Δ1 is thus a unique structural element of this enzyme that could be responsible for some of its special features.
Acknowledgments
We thank V. Stojanoff and J. Lescar of the European Synchrotron Radiation Facility for help during data collection, J. Janin for comments on the manuscript, and I. Petitpas for assistance. This work was funded in part through Human Frontier Science Program Grant RG-509 and Association pour la Recherche contre le Cancer Grant 9646 to F.A.R.
Abbreviations
- HCV
hepatitis C virus
- RdRp
RNA-dependent RNA polymerase
- RT
reverse transcriptase
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1csj).
References
- 1.Rice C M. In: Virology. Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, Straus S E, editors. Vol. 1. Philadelphia: Lippincott; 1996. pp. 931–959. [Google Scholar]
- 2.Miller R H, Purcell R H. Proc Natl Acad Sci USA. 1990;87:2057–2061. doi: 10.1073/pnas.87.6.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Behrens S E, Tomei L, De Francesco R. EMBO J. 1996;15:12–22. [PMC free article] [PubMed] [Google Scholar]
- 4.Ollis D L, Brick P, Hamlin R, Xuong N G, Steitz T A. Nature (London) 1985;313:762–766. doi: 10.1038/313762a0. [DOI] [PubMed] [Google Scholar]
- 5.Brautigam C A, Steitz T A. Curr Opin Struct Biol. 1998;8:54–63. doi: 10.1016/s0959-440x(98)80010-9. [DOI] [PubMed] [Google Scholar]
- 6.Pelletier H, Sawaya M R, Kumar A, Wilson S H, Kraut J. Science. 1994;264:1891–1903. [PubMed] [Google Scholar]
- 7.Sawaya M R, Pelletier H, Kumar A, Wilson S H, Kraut J. Science. 1994;264:1930–1935. doi: 10.1126/science.7516581. [DOI] [PubMed] [Google Scholar]
- 8.Delarue M, Poch O, Tordo N, Moras D, Argos P. Protein Eng. 1990;3:461–467. doi: 10.1093/protein/3.6.461. [DOI] [PubMed] [Google Scholar]
- 9.Hansen J L, Long A M, Schultz S C. Structure (London) 1997;5:1109–1122. doi: 10.1016/s0969-2126(97)00261-x. [DOI] [PubMed] [Google Scholar]
- 10.Steitz T A. Nature (London) 1998;391:231–232. doi: 10.1038/34542. [DOI] [PubMed] [Google Scholar]
- 11.Jäger, J. & D Pata, J. (1999) Curr. Opin. Struct. Biol.9, 21–28. [DOI] [PubMed]
- 12.Georgiadis M M, Jessen S M, Ogata C M, Telesnitsky A, Goff S P, Hendrickson W A. Structure (London) 1995;3:879–892. doi: 10.1016/S0969-2126(01)00223-4. [DOI] [PubMed] [Google Scholar]
- 13.Hendrickson W A. Science. 1991;254:51–58. doi: 10.1126/science.1925561. [DOI] [PubMed] [Google Scholar]
- 14.Otwinowski Z, Minor W. In: Macromolecular Crystallography: Part A. Carter C W, Sweet R M, editors. Vol. 276. London: Academic; 1996. pp. 307–326. [Google Scholar]
- 15.Terwilliger T C, Berendzen J. Acta Crystallogr D. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cowtan K, Main P. Acta Crystallogr D. 1998;54:487–493. doi: 10.1107/s0907444997011980. [DOI] [PubMed] [Google Scholar]
- 17.Roussel A, Cambillaud C. Silicon Graphics Geometry Patners Directory. Vol. 86. Mountain View, CA: Silicon Graphics; 1991. pp. 77–78. [Google Scholar]
- 18.Brünger A T, Adams P D, Clore G M, DeLano W L, Gros P, Grosse-Kunstleve R W, Jiang J S, Kuszewski J, Nilges M, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 19.Pannu N S, Murshudov G N, Dodson E J, Read R J. Acta Crystallogr D. 1998;54:1285–1294. doi: 10.1107/s0907444998004119. [DOI] [PubMed] [Google Scholar]
- 20.Carson M. J Mol Graphics. 1987;5:103–106. [Google Scholar]
- 21.Nichols A, Sharp K A, Honig B. Proteins Struct Funct Genet. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 22.O’Reilly E K, Kao C C. Virology. 1998;252:287–303. doi: 10.1006/viro.1998.9463. [DOI] [PubMed] [Google Scholar]
- 23.Conti E, Uy M, Leighton L, Blobel G, Kuriyan J. Cell. 1998;94:193–204. doi: 10.1016/s0092-8674(00)81419-1. [DOI] [PubMed] [Google Scholar]
- 24.Holm L, Sander C. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 25.Jacobo-Molina A, Ding J, Nanni R G, Clark A D, Jr, Lu X, Tantillo C, Williams R L, Kamer G, Ferris A L, Clark P, et al. Proc Natl Acad Sci USA. 1993;90:6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang H, Chopra R, Verdine G L, Harrison S C. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- 27.Doublie S, Sawaya M R, Ellenberger T. Structure Fold Des. 1999;7:R31–R35. doi: 10.1016/S0969-2126(99)80017-3. [DOI] [PubMed] [Google Scholar]
- 28.Koonin E V. J Gen Virol. 1991;72:2197–2206. doi: 10.1099/0022-1317-72-9-2197. [DOI] [PubMed] [Google Scholar]
- 29.Gao G, Orlova M, Georgiadis M M, Hendrickson W A, Goff S P. Proc Natl Acad Sci USA. 1997;94:407–411. doi: 10.1073/pnas.94.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Doublie S, Tabor S, Long A M, Richardson C C, Ellenberger T. Nature (London) 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 31.Zanotto P M, Gibbs M J, Gould E A, Holmes E C. J Virol. 1996;70:6083–6096. doi: 10.1128/jvi.70.9.6083-6096.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoshiba M, Okamoto H, Mishiro S. Lancet. 1995;346:1131–1132. doi: 10.1016/s0140-6736(95)91802-7. [DOI] [PubMed] [Google Scholar]
- 33.Brünger A T. Methods Enzymol. 1997;277:366–396. doi: 10.1016/s0076-6879(97)77021-6. [DOI] [PubMed] [Google Scholar]