Abstract
HIV integrase, the enzyme that inserts the viral DNA into the host chromosome, has no mammalian counterpart, making it an attractive target for antiviral drug design. As one of the three enzymes produced by HIV, it can be expected that inhibitors of this enzyme will complement the therapeutic use of HIV protease and reverse transcriptase inhibitors. We have determined the structure of a complex of the HIV-1 integrase core domain with a novel inhibitor, 5ClTEP, 1-(5-chloroindol-3-yl)-3-hydroxy-3-(2H-tetrazol-5-yl)-propenone, to 2.1-Å resolution. The inhibitor binds centrally in the active site of the integrase and makes a number of close contacts with the protein. Only minor changes in the protein accompany inhibitor binding. This inhibitor complex will provide a platform for structure-based design of an additional class of inhibitors for antiviral therapy.
Integration of viral DNA into the host chromosome is a necessary process in the HIV replication cycle (1). The key steps of DNA integration are carried out by the viral integrase protein, which, along with protease and reverse transcriptase, is one of three enzymes encoded by HIV. Combination antiviral therapy with protease and reverse transcriptase inhibitors has demonstrated the potential therapeutic efficacy of antiviral therapy for treatment for AIDS (2). However, the ability of HIV to rapidly evolve drug resistance, together with toxicity problems, requires the development of additional classes of antiviral drugs. Integrase is an attractive target for antivirals because it is essential for HIV replication and, unlike protease and reverse transcriptase, there are no known counterparts in the host cell. Furthermore, integrase uses a single active site to accommodate two different configurations of DNA substrates, which may constrain the ability of HIV to develop drug resistance to integrase inhibitors. However, unlike protease and reverse transcriptase, for which several classes of inhibitors have been developed and cocrystal structures have been determined, progress with the development of integrase inhibitors has been slow. A major obstacle has been the absence of good lead compounds that can serve as the starting point for structure-based inhibitor development. Although numerous compounds have been reported to inhibit integrase activity in vitro, most of these compounds exhibit little specificity for integrase and are not useful as lead compounds (3).
HIV-1 integrase is a 32-kDa enzyme that carries out DNA integration in a two-step reaction (1). In the first step, called 3′ processing, two nucleotides are removed from each 3′ end of the viral DNA made by reverse transcription. In the next step, called DNA strand transfer, a pair of transesterification reactions integrates the ends of the viral DNA into the host genome. Integrase is comprised of three structurally and functionally distinct domains, and all three domains are required for each step of the integration reaction (4). The isolated domains form homodimers in solution, and the three-dimensional structures of all three separate dimers have been determined (5–10). Although little is known concerning the organization of these domains in the active complex with DNA substrates, integrase is likely to function as at least a tetramer (5). Extensive mutagenesis studies mapped the catalytic site to the core domain (residues 50–212), which contains the catalytic residues Asp-64, Asp-116, and Glu-152 (11, 12). The structure of this domain of HIV-1 integrase has been determined previously in several crystal forms (5–7). Refs. 6 and 7 describe structures with bound magnesium and predict that these structures would provide a more suitable platform for inhibitor binding than the earlier structure of the apoenzyme. Here we report the structure of an integrase inhibitor in complex with the active site of the core domain of HIV integrase. This integrase/inhibitor complex provides a platform for the design of an additional class of HIV inhibitors.
Materials and Methods
The soluble HIV-1 core domain mutant, F185K, was engineered to contain an additional site-specific mutation, W131E, to further increase the solubility. The double mutant was expressed in Escherichia coli and purified as described (6). Crystallization conditions were the same as in ref. 6 for crystal form III, but with 1% glycerol in the precipitant solution, which appeared to facilitate nucleation. Crystals were soaked in a 5 mM solution of the inhibitor in the mother liquor. For data collection at cryotemperatures, glycerol was added gradually in small amounts until its concentration reached 20% (vol/vol). Data were collected at 95 K on a Raxis IIC image plate detector mounted on a Rigaku (Tokyo) RU200 rotating anode source operated at 50 kV 100 mA with double-mirror focused CuKα radiation. All diffraction data were integrated and scaled with the hkl suite (13). A 94.2% complete data set contained 26,564 unique reflections to 2.1-Å resolution with Rsym 5.1%. The high-resolution shell was 81% complete with Rsym 27.4% and I/σ ratio 2.86. To position the inhibitor, (Fcomp − Fnative) maps were calculated with native protein phases, using the software package cns (14). Location of the chlorine atom of the inhibitor at the A site was confirmed by the observation of a 3-σ high peak in the anomalous difference map. The final model contains only the inhibitor molecule at the A binding site and was refined to an R factor of 20.8% (Rfree 25.7%) at 2.1-Å resolution.
Results
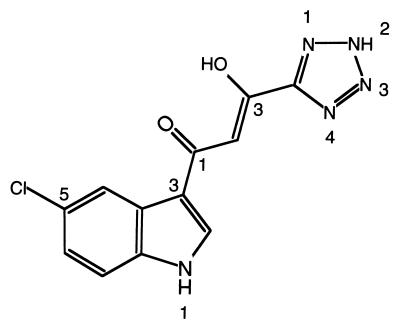
The integrase inhibitor 5ClTEP, 1-(5-chloroindol-3-yl)-3-hydroxy-3-(2H-tetrazol-5-yl)-propenone (Fig. 1), is one of a group of Shionogi integrase inhibitors. Its 50% inhibition concentrations (IC50) by our multiple well plate integration assay (MIA) and multiple well plate preincubation assay (MPA) were 2.3 ± 0.1 μM and 2.1 ± 0.1 μM, respectively. These assays using 96-well plates, which basically resemble the assays reported by Hazuda et al. (15), monitor 3′ processing together with DNA strand transfer. MPA differs from MIA in that, in the former, blunt end substrate DNA is preincubated with integrase before the addition of inhibitor and target DNA. We find many inhibitors that are active in the MIA are inactive in the MPA (details to be published elsewhere). In this respect, like assays with preintegration complexes (16), MPA is a more stringent assay for identification of integrase inhibitors. Because the structure of 5ClTEP is quite different from previously reported integrase inhibitors, together with its similar activity in MIA and MPA, led us to probe the mechanism of inhibition. Additionally, 5ClTEP inhibits the disintegration reaction catalyzed by the isolated core domain (data not shown), indicating a possible interaction with the active site.
Figure 1.
Chemical structure of the inhibitor 5ClTEP.
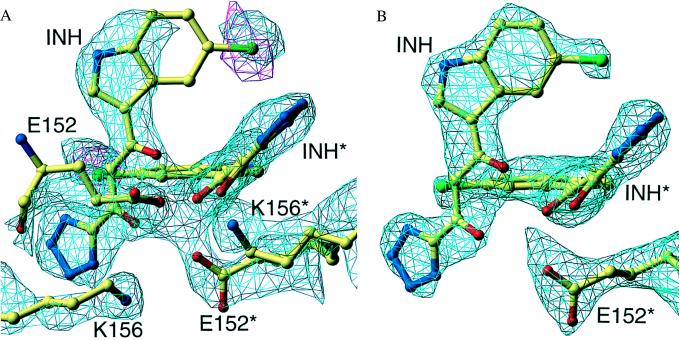
The crystals of HIV-1 integrase catalytic domain soaked with 5ClTEP contain three molecules of the integrase core domain within the crystallographic asymmetric unit, designated A, B, and C. The active site of A is close to the active site of a fourth subunit, A′, which is related to A by a crystallographic 2-fold axis. Molecule B and A together form the core domain dimer, with molecule C as the third member of the asymmetric unit (6). The crystals were soaked in the inhibitor, and the position of the inhibitor was determined by calculating (Fcomp − Fnative) maps with native protein phases (14). The quality of the inhibitor electron density was different for the three crystallographically independent subunits in the asymmetric unit. The strongest density was associated with subunit A, where the active site was in the vicinity of the crystallographic 2-fold axis. Much weaker density was observed at the other two subunits, B and C, indicating that there was preferential binding of the inhibitor at the A site. Samples of the difference electron density maps in the vicinity of the inhibitors are shown for molecule A in Fig. 2 A and B. Difference maps, with (Fcomp − Fcalc) coefficients exhibited fragments of density at sites B and C that coincided with parts of the inhibitor model, positioned relative to the protein as in the A site, suggesting that binding occurs with similar orientations in all three potential binding sites within the asymmetric unit. However, the density at sites B and C was quite low and led to unacceptably high B values during refinement so that these coordinates subsequently were omitted from the refinement.
Figure 2.
Electron density in the active site region of subunit A. (A) The cyan contours show the difference map between the experimental data sets of the inhibitor-soaked and the unsoaked crystals at the 2-σ level. The red contours represent the anomalous density for the chlorine atom at the 2-σ level. The final model is superimposed on this density. Note the high density for the side chains of E152 and K156, attributed to the big decrease in the B values for these residues in the soaked crystals. (B) The same active site region of subunit A, with density from a 2Fo-Fc map of the inhibitor-soaked crystal, contoured at the 1-σ level.
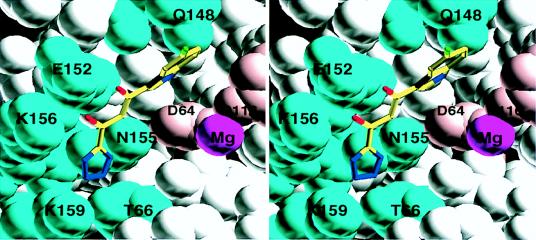
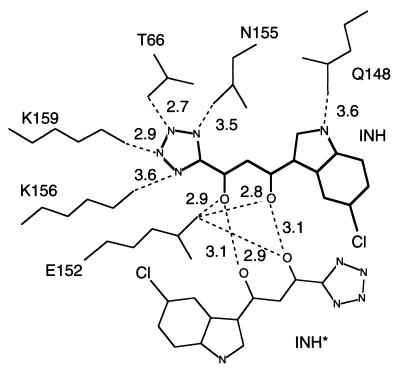
The electron density map clearly reveals the inhibitor bound in the middle of the active site of subunit A of the enzyme, lying between the three catalytic acidic residues, Asp-64, Asp-116, and Glu-152. A stereo view of the inhibitor bound at the active site of the enzyme is shown in Fig. 3, and a schematic of the contacting residues is presented in Fig. 4. They involve several residues that are known to be important for catalysis or DNA binding. Gln-148 forms a hydrogen bond to the nitrogen of the indole ring; Glu-152 is within hydrogen-bonding distance of the enol hydroxyl. All four nitrogen atoms of the tetrazole ring are hydrogen-bonded to Asn-155, Thr-66, Lys-159, and Lys-156. Although no direct contacts with the two catalytically essential aspartates are observed, one of the four water molecules coordinating the magnesium ion is close enough to the plane of the indole ring to be regarded as in van der Waals contact.
Figure 3.
Stereo image of the inhibitor/protein contacts. Contacting protein side chains are shown in cyan, with magnesium in purple and the catalytic residues Asp-64 and Asp-116 in pink.
Figure 4.
A schematic drawing of the inhibitors at the two active sites related by a crystallographic 2-fold axis. The contacting distances corresponding to potential hydrogen bonds are shown. Note that for this subunit the inhibitor makes two hydrogen bonds with its symmetry-related neighbor. Also, one of the oxygens of Glu-152 is within contact distance of both inhibitors.
No substantial conformational changes in the protein were observed in the complex, except possibly for a change in the orientation of the side chain of Glu-152. However, the temperature factors of the side chains of Glu-152 and Lys-156 are noticeably lower in the complex structure than in the free protein structure, changing from 79 to 54 A2 for Glu-152 and from 71 to 52 A2 for Lys-156, indicating an increased degree of order upon binding of the inhibitor.
At the crystallographic 2-fold axis two active sites are positioned close to each other. Each inhibitor molecule makes most of its protein contacts with one subunit. The only contacts between an inhibitor molecule and the symmetry related subunit are a van der Waals interaction of the chlorine at the C5 position of the indole with the main chain of Thr-66* and a possible hydrogen bond of the enol group of the inhibitor to Glu-152*. However, the inhibitor bound at one site makes two symmetrically positioned hydrogen bonds to the inhibitor bound in the second site via their keto and enol groups, forming an eight-membered ring structure (Fig. 4). These additional contacts, which are not observed in the other two crystallographically independent sites, could provide an explanation for the enhanced binding observed at this 2-fold axis site as evidenced by the better defined density. Gln-148, which is within hydrogen-bonding distance of the N1 of the indole, adopts a different conformation in the other two subunits, consistent with the failure to observe significant inhibitor binding at these sites.
Discussion
It is striking that the inhibitor is located centrally in the active site between the two aspartates, D64 and D116, and the glutamate E152, all three of which are required for catalysis. It is also notable that the inhibitor binding does not displace the bound magnesium ion, which remains complexed to the two aspartates. It is tempting to speculate that the interactions between the inhibitor and integrase at least partially mimic the normal interactions with viral DNA substrate during the 3′ processing reaction. Site-directed mutagenesis and photo-crosslinking experiments have identified several residues near the active site, including Lys-156, Lys-159, Gln-148, and Tyr-143, that are critical for binding viral DNA substrate (17, 18). Many of these residues, including Lys-156, Lys-159, and Gln-148, also are involved in binding the inhibitor.
The inhibitor binding seems to mimic the DNA substrate/integrase interaction. The distance between the indole and tetrazole ring systems on the inhibitor is approximately 8 Å. This is a distance that could easily be spanned by two nucleotides, with the sugar phosphate backbone containing the scissile bond passing between the active site carboxylates in close proximity to the bound magnesium ion. In such a model the two bases adjacent to the scissile phosphate potentially would overlap the pockets in which the indole and tetrazole rings of the inhibitor are buried. The enhanced in vitro activity of viral DNA substrates with mutations near the DNA terminus that disrupt base-pairing supports the idea that integrase recognizes the viral DNA ends in a conformation in which the bases are extensively unstacked near the active site (19). Because many of the side chains that contact the inhibitor are likely to play a role in the recognition of substrate DNA, there is a good possibility that resistant mutants will have to pay a significant cost in decreased catalytic activity.
A structure of the avian sarcoma virus integrase core domain in complex with a HIV integrase inhibitor, Y-3, has been reported (20). The authors state that Y-3 binds in close proximity to the active site, but the location of Y-3 is quite distant from the site to which 5ClTEP binds and is located on the other side of the flexible loop from the catalytic residues.
Here we report an example of a structure of the HIV-1 integrase core domain complexed with an inhibitor. The inhibitor binds in the middle of the multifunctional active site of integrase, possibly making it more difficult to evolve inhibitor-resistant mutants without compromising integrase activity. This enzyme/inhibitor complex provides a platform for the design of an additional class of inhibitors to complement those directed at the HIV protease and reverse transcriptase.
Acknowledgments
We acknowledge the expert assistance of Dr. Sangkee Rhee with the preparation of the figures.
Abbreviations
- 5ClTEP
1-(5-chloroindol-3-yl)-3-hydroxy-3-(2H-tetrazol-5-yl)-propenone
- MPA
multiple well plate preincubation assay
Footnotes
Data deposition: The coordinate reported in this paper has been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1QS4).
References
- 1.Brown P O. In: Retroviruses. Coffin J M, Hughes S H, Varmus H E, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. pp. 161–203. [Google Scholar]
- 2.Vandamme A M, Van Vaerenbergh K, De Clercq E. Antiviral Chem Chemother. 1998;9:187–203. doi: 10.1177/095632029800900301. [DOI] [PubMed] [Google Scholar]
- 3.Pommier Y, Pilon A A, Bajaj K, Mazumder A, Neamati N. Antiviral Chem Chemother. 1997;8:463–485. [Google Scholar]
- 4.Engelman A, Bushman F D, Craigie R. EMBO J. 1993;12:3269–3275. doi: 10.1002/j.1460-2075.1993.tb05996.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dyda F, Hickman A B, Jenkins T M, Engelman A, Craigie R, Davies D R. Science. 1994;266:1981–1986. doi: 10.1126/science.7801124. [DOI] [PubMed] [Google Scholar]
- 6.Goldgur Y, Dyda F, Hickman A B, Jenkins T M, Craigie R, Davies D R. Proc Natl Acad Sci USA. 1998;95:9150–9154. doi: 10.1073/pnas.95.16.9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maignan S, Guilloteau J-P, Zhou-Liu Q, Clement-Mella C, Mikol V. J Mol Biol. 1998;282:359–368. doi: 10.1006/jmbi.1998.2002. [DOI] [PubMed] [Google Scholar]
- 8.Lodi P J, Ernst J A, Kuszewski J, Hickman A B, Engelman A, Craigie R, Clore G M, Gronenborn A M. Biochemistry. 1995;34:9826–9833. doi: 10.1021/bi00031a002. [DOI] [PubMed] [Google Scholar]
- 9.Eijkelenboom A P, Lutzke R A, Boelens R, Plasterk R H, Kaptein R, Hard K. Nat Struct Biol. 1995;2:807–810. doi: 10.1038/nsb0995-807. [DOI] [PubMed] [Google Scholar]
- 10.Cai M L, Zheng R, Caffrey M, Craigie R, Clore G M, Gronenborn A M. Nat Struct Biol. 1997;4:839–840. doi: 10.1038/nsb0797-567. [DOI] [PubMed] [Google Scholar]
- 11.Engelman A, Craigie R. J Virol. 1992;66:6361–6369. doi: 10.1128/jvi.66.11.6361-6369.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kulkosky J, Jones K S, Katz R A, Mack J P, Skalka A M. Mol Cell Biol. 1992;12:2331–2338. doi: 10.1128/mcb.12.5.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 14.Brunger A T, Adams P D, Clore G M, DeLano W L, Gros P, Grosse-Kunstleve R W, Jiang J-S, Kuszewski J, Nilges M, Pannu N S, et al. Acta Crystrallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 15.Hazuda D, Blau C U, Felock P, Hastings J, Pramanik B, Wolfe A, Bushman F, Farnet C, Goetz M, Williams M, et al. Antiviral Chem Chemother. 1999;10:63–70. doi: 10.1177/095632029901000202. [DOI] [PubMed] [Google Scholar]
- 16.Farnet C M, Wang B B, Lipford J R, Bushman F D. Proc Natl Acad Sci USA. 1996;93:9742–9747. doi: 10.1073/pnas.93.18.9742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jenkins T M, Esposito D, Engelman A, Craigie R. EMBO J. 1997;16:6849–6859. doi: 10.1093/emboj/16.22.6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Esposito D, Craigie R. EMBO J. 1998;17:5832–5843. doi: 10.1093/emboj/17.19.5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scottoline B P, Chow S, Ellison V, Brown P O. Genes Dev. 1997;11:371–382. doi: 10.1101/gad.11.3.371. [DOI] [PubMed] [Google Scholar]
- 20.Lubkowski J, Yang F, Alexandratos J, Wlodawer A, Zhao H, Burke T R, Neamati N, Pommier Y, Merkel G, Skalka A M. Proc Natl Acad Sci USA. 1998;95:4831–4836. doi: 10.1073/pnas.95.9.4831. [DOI] [PMC free article] [PubMed] [Google Scholar]