Abstract
The bacterial iron response regulator (Irr) protein mediates iron-dependent regulation of heme biosynthesis. Pulse–chase and immunoprecipitation experiments showed that Irr degraded in response to 6 μM iron with a half-life of ≈30 min and that this regulated stability was the principal determinant of control by iron. Irr contains a heme regulatory motif (HRM) near its amino terminus. A role for heme in regulation was implicated by the retention of Irr in heme synthesis mutants in the presence of iron. Addition of heme to low iron (0.3 μM) cultures was sufficient for the disappearance of Irr in cells of the wild-type and heme mutant strains. Spectral and binding analyses of purified recombinant Irr showed that the protein bound heme with high affinity and caused a blue shift in the absorption spectrum of heme to a shorter wavelength. A Cys29 → Ala substitution within the HRM of Irr (IrrC29A) abrogated both high affinity binding to heme and the spectral blue shift. In vivo turnover experiments showed that, unlike wild-type Irr, IrrC29A was stable in the presence of iron. We conclude that iron-dependent degradation of Irr involves direct binding of heme to the protein at the HRM. The findings implicate a regulatory role for heme in protein degradation and provide direct evidence for a functional HRM in a prokaryote.
Heme carries out a wide range of biological functions as the prosthetic group of proteins involved in oxygen metabolism, electron transfer reactions, and nitric oxide synthesis. Heme is also an effector molecule that can modulate transcription (1), translation (2), and protein targeting (3). Furthermore, heme is the active moiety of proteins that sense and respond to diatomic gases (4–10).
The biosynthesis of heme (protoheme) involves seven sequential enzymatic steps from the universal heme precursor δ-aminolevulinic acid (ALA). Ferrochelatase catalyzes the insertion of ferrous iron into protoporphyrin IX to form protoheme in the final step of the pathway. Iron may be a limiting nutrient, and protoporphyrin and other porphyrins are toxic to cells (11–13). Thus, coordination of the heme biosynthetic pathway with iron availability presumably prevents the deleterious accumulation of porphyrins under iron limitation. Translation of mRNA encoding the erythroid heme synthesis enzyme ALA synthase is inhibited by the iron regulatory proteins under iron limitation (14, 15). In the bacterium Bradyrhizobium japonicum, the Irr protein mediates iron-dependent regulation of the heme biosynthetic pathway (16). Loss of function of the irr gene is sufficient to uncouple the pathway from iron-dependent control as discerned by protoporphyrin accumulation under iron limitation. Irr mediates iron-dependent control of the hemB gene encoding ALA dehydratase, resulting in repression under low iron conditions (16, 17). Furthermore, a separate role for Irr as a positive regulator ferric citrate transport was inferred from analysis of an irr mutant (16).
The Irr protein is structurally homologous to bacterial Fur (16, 18), a transcriptional regulator that represses genes involved in iron metabolism in the presence of iron. Very recently, several other Fur-like proteins have been identified that are not functional Fur homologs but instead are active in the presence of zinc or manganese and have roles in maintenance of zinc homeostasis (19, 20) or response to oxidative stress (21), respectively. Additional fur-like genes have been identified from genome sequencing and from screens for genes involved in pathogenesis (22, 23). There now appears to be a family of Fur proteins that are functionally diverse but are all involved in metal-dependent regulation. Irr may be the most divergent of these proteins described thus far in that it is active in the absence of metal rather than its presence and contains only a single cysteine residue rather than multiple cysteines found in the other proteins. Moreover, cellular Irr levels are strongly affected by the iron status whereas those of Fur are essentially constitutive.
Iron regulates irr gene expression at both a transcriptional and posttranscriptional step (16). The transcriptional control is mediated by Fur, but this regulation is weak and, thus, substantial irr mRNA remains even under repressing conditions (ref. 16; I.H. and M.R.O., unpublished observations). Irr protein, however, disappears rapidly to negligible levels upon addition of iron to the cell medium (16), and this response is also observed in a fur strain. Thus, B. japonicum can sense and respond to iron in a Fur-independent manner. In the present study, we show that the stability of Irr depends on the iron status and that heme binding to the protein is necessary for its turnover. This mechanism represents a regulatory role for heme in protein degradation.
Materials and Methods
Strains and Media.
B. japonicum I110 was the parent strain used in the present work. Mutant strains MLG1 (24) and I110ek4 (25) are defective in hemA and hemH, respectively. Strain ΔhemAH is a double mutant defective in the hemA and hemH genes. Strain IrrC29A contains an irr allele that encodes an Irr protein with a Cys29 → Ala mutation. B. japonicum strains were routinely grown at 29°C in GSY medium (26) with the appropriate antibiotics at 50 μg/ml. All four mutant strains were grown in media containing kanamycin. In addition, strain ΔhemAH was grown with spectinomycin and streptomycin, and culture medium was supplemented with tetracycline and streptomycin for growth of strain IrrC29A. Furthermore, medium was supplemented with 15 μM hemin for strains I110ek4 and ΔhemAH to fulfill their heme auxotrophy. Escherichia coli strains DH5α or XL1-Blue were used for propagation of plasmids in LB medium containing 100 μg/ml ampicillin.
Site-Directed Mutagenesis of the irr Gene.
A mutant irr gene encoding a protein with a Cys29 → Ala mutation was constructed by PCR using a primer containing the nucleotide changes. An oligonucleotide with the sequence 5′-CGTGCCACGGGGCGCCGGTCAAAGCCG-3′ was used as the reverse primer along with the pUC19 forward primer to amplify DNA in pUCΔSES7, which contains the irr gene within pUC19. The PCR product was denatured and used as a primer in a second round of PCR with the same template along with the M13 reverse primer. This product was cloned into pBluescript SKII (Stratagene) to construct pSKMES8. The nucleotide sequence of the modified irr gene (irrC29A) was confirmed by sequencing. The mutation changed the single cysteine codon of the gene (TGC) to an alanine codon (GCC) used highly in B. japonicum.
Construction of Mutant Strains.
Strain ΔhemAH was constructed by site-directed mutagenesis of hemA in the hemH strain I110ek4. To do this, a 0.6-kilobase XhoI fragment within the coding region of hemA was removed and replaced with a Ω cassette encoding resistance to spectinomycin and streptomycin. The ends were blunted before ligation. The disrupted hemA gene and flanking DNA were introduced into strain I110ek4 by conjugation followed by homologous recombination as described (27). The construction was confirmed by Southern blot analysis. Strain IrrC29A was constructed by ligating the irrC29A gene into pBR322 and integrating the plasmid into the mutant irr locus by single recombination as described previously (28).
Overexpression and Purification of Recombinant Irr Proteins.
The IMPACT I system (New England BioLabs) was used for synthesis of Irr and IrrC29A in E. coli and initial purification. The coding regions of irr or irrC29A were modified by PCR using the primers 5′-CTGCATATGAGCGAGAATACCGC-3′ and 5′-TAGGCTCTTCCGCAGCGCGCTTCTTGCGCAGG-3′ to introduce NdeI and SapI sites at the 5′ and 3′ ends, respectively, and then were ligated into the NdeI/SapI sites of pCYB1. Overexpression of the irr-chitin binding domain gene fusion in E. coli strain BL21(DE3)(pLysS) was carried out by growth of cells to OD550 of 0.5, followed by induction at 15°C overnight in the presence of 1 mM IPTG. Cells were disrupted by passage through a French pressure cell and were clarified by centrifugation at 20,000 × g for 30 min. The lysate was passed through a chitin affinity column, followed by self-cleavage of the fusion between the Irr and CBP domains at the intein linker with 30 mM DTT at 4°C overnight to release Irr. The eluted Irr protein was further purified by FPLC using a MonoQ column (Amersham Pharmacia).
Irr Turnover Experiments.
The turnover of Irr in cells was examined by pulse–chase experiments followed by immunoprecipitation with anti-Irr antibodies. Cells were grown to OD540 of 0.5 in defined media or in modified GSY. The defined media is as follows (per liter of media): 670 mg of yeast nitrogen base, 243 mg of amino acid dropout powder (29), 0.2 mg of riboflavin, 0.08 mg of p-aminobenzoic acid, 0.5 mg of nicotinic acid, 0.12 of mg biotin, 0.8 mg of thiamin⋅HCl, 0.8 mg of calcium pantothenate, 0.48 mg of inositol, 300 mg of KH2PO4, 300 mg of Na2HPO4, 100 mg of MgSO4⋅7H2O, 50 mg of CaCl2⋅2H2O, 10 mg of H3BO3, 1 mg of ZnSO4⋅2H2O, 0.5 mg of CuSO4⋅5H2O, 0.5 mg of MnCl2⋅4H2O, 0.5 mg of Na2MoO4⋅2H20, and 4 g of glycerol (pH 6.8). Cultures (50 ml) then were centrifuged, were resuspended in 50 ml of defined media lacking cysteine and methionine, and were incubated with shaking at 28°C. After 1 h, cells were radiolabeled by addition of 20 μCi/ml Trans35S-label l-(35S)methionine and l-(35S)cysteine (ICN) and were allowed to incubate for 2.5 h. Cells then were washed and resuspended in 50 ml of growth medium including unlabeled methionine and cysteine. At time zero of the chase, either 6 μM ferric chloride or an equivalent volume of metal free buffer was added to cells and then incubated with shaking in a 28°C water bath. Aliquots (5 ml) were removed at various times and were washed once in PBS buffer (10 mM Na2HPO4/1.8 mM KH2PO4/2.7 mM KCl/140 mM NaCl, pH 7.3). Cell pellets were resuspended in 10 cell volumes of media containing 50 mM glucose, 10 mM EDTA, 25 mM Tris (pH 8), 4 mg/ml lysozyme, and 1 mM PMSF, followed by incubation at room temperature for 10 min. The lysed cells were combined with 1 ml RIPA buffer [50 mM Tris⋅HCl, pH 8/1 mg/ml SDS/5 mg/ml deoxycholate/150 mM NaCl/1% (vol/vol) NP40] containing 1 mM PMSF and were incubated on ice for 30 min with occasional mixing. The suspension was centrifuged, and the supernatant was used for preclearing and immunoprecipitation by using a modification of a protocol described previously (30). Lysate (1 ml) was precleared with preimmune serum, then was incubated with 50 μl of primary antibody (1:2000) for 1 h followed by incubation with 100 μl protein A-Sepharose for another hour. A control for nonspecific immunoprecipitation was carried out in the absence of primary antibody. The primary antibodies used for immunoprecipitations were affinity-purified by using Irr bound to CNBr-activated Sepharose-4B. The immunoprecipitate was washed four times with RIPA buffer, then was placed into 25 μl of SDS sample buffer, and 15 μl of it was analyzed by SDS/PAGE followed by autoradiography using x-ray film or a Bio-Rad Phosphorimager. The Phosphoimager was used for quantitation of radiolabeled protein.
Heme Binding Experiments.
The effects of Irr or IrrC29A on the spectral properties of heme were determined by recording the absorption spectrum of 4 μM heme in the presence or absence of 8 μM protein monomer. To quantify heme binding, protein was incubated with various concentrations of heme in PBS buffer and then was passed through a Sephadex G25 column to separate the bound from unbound heme. Heme bound to protein was determined by using the pyridine hemochromagen assay of dithionite-reduced heme as described (31).
Detection of Irr by Immunoblot Analysis.
Irr levels were measured in cells grown under varying conditions of iron, heme, or protoporphyrin. The medium used for culturing cells under iron limitation was a modified GSY medium in which 0.5 g/liter yeast extract (Difco) was used instead of 1 g/liter, and no exogenous iron source was added. The actual iron concentration of the media was 0.3 μM as determined with a Perkin–Elmer model 1100B atomic absorption spectrometer. Wild-type and mutant strains grew well in this medium. The medium was supplemented with 6 μM FeCl3 for high iron media and with 15 μM hemin or protoporphyrin where indicated. Aliquots of cells grown to mid-log phase were added to SDS sample buffer and were analyzed by immunoblots using anti-Irr antibodies as described (16).
Measurement of Irr and Overall Protein Synthesis in Tetracycline-Treated Cells.
To determine the effects of tetracycline on protein synthesis, cells were grown to mid-log phase in modified GSY medium, and then 500 μCi of 35S-labeled methionine and cysteine were added to 25-ml cultures. Fifteen minutes after addition of radiolabel, 200 μg/ml tetracycline or an equivalent volume of buffer lacking the antibiotic was added to the cultures and was allowed to incubate at 28°C with shaking. Aliquots were removed at various times, and protein was precipitated with 10% (vol/vol) trichloroacetic acid. Incorporated radiolabel was counted with a Wallac (Gaithersburg, MD) 1409 liquid scintillation counter. To determine the effects of tetracycline on iron-dependent Irr levels, cells were grown in low iron media to mid-log phase, followed by addition of 200 μg/ml tetracycline or an equivalent volume of buffer lacking the antibiotic, and were allowed to incubate at 28°C with shaking. Fifteen minutes after tetracycline addition, 6 μM FeCl3 or an equivalent volume of metal-free buffer was added to the cultures. Aliquots were removed at various times and were analyzed by immunoblots using anti-Irr antibodies as described (16).
Results
Iron Regulates the Stability of Irr.
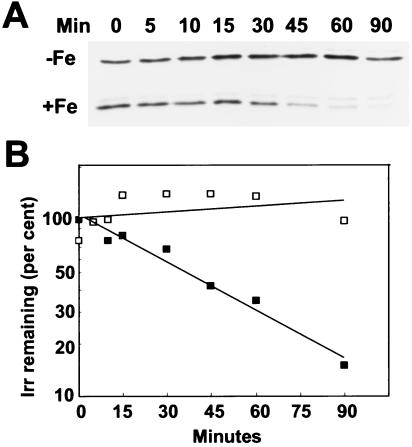
We previously found that addition of iron to cells grown in iron-limited media resulted in the rapid disappearance of Irr as discerned by immunoblot analysis (16). In principle, those observations could be caused by iron-dependent degradation of Irr or by iron-dependent synthesis of a protein with a constitutively rapid turnover rate. To examine iron-dependent degradation, cells were pulse-labeled with (35S)methionine and (35S)cysteine in low iron media, and the fate of labeled Irr was followed during the chase by immunoprecipitation at various times after addition of 6 μM ferric chloride (Fig. 1). In the absence of added iron, Irr was stable throughout the 90-min time course of the experiment. However, addition of iron resulted in turnover of Irr, with little labeled protein remaining after 90 min. The faint band above Irr was also present in control experiments in which primary antibody was omitted during the immunoprecipitation (data not shown). Detection of smaller degradation products required longer exposures of autoradiograms, suggesting that complete degradation of Irr was rapid. The decay of Irr followed first order kinetics, with a half-life of ≈30 min (Fig. 1B). When the experiment was carried by using modified GSY media rather than the defined media shown in Fig. 1, the results were qualitatively similar, but the half-life of the protein was ≈20 min (data not shown). The findings show that Irr is a conditionally unstable protein that depends on the iron status.
Figure 1.
Iron-dependent turnover of Irr in vivo. Immunoprecipitation of Irr from 35S-radiopulsed cells was carried out at various times of the chase after treatment of cells with 6 μM FeCl3 (+Fe) or an equivalent volume of iron-free buffer (−Fe) at time zero. (A) Autoradiograms of immunoprecipitated Irr at various times during the chase. (B) Quantification of the autoradiogram shown in A of iron-treated (closed squares) or untreated (open squares) cells. The data are typical of three independent experiments. The quantity of labeled protein at time zero was set at 100%, and the ordinate is a log scale.
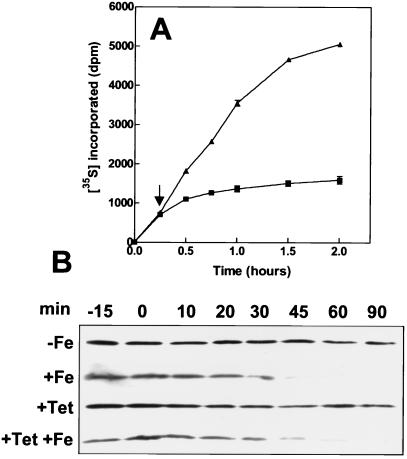
To determine whether iron levels also control synthesis of Irr, the effect of the metal on Irr was determined in the presence of the translation inhibitor tetracycline. We first established whether tetracycline effectively inhibited protein synthesis overall in B. japonicum as determined by incorporation of (35S)methionine and (35S)cysteine into total cellular protein in the presence of 200 μg/ml of the antibiotic (Fig. 2A). Tetracycline strongly inhibited protein synthesis. Furthermore, labeled protein levels did not decrease in the presence of tetracycline during the time course, indicating that, like other bacteria (32), B. japonicum proteins are stable in general.
Figure 2.
Effect of tetracycline on overall protein synthesis and on iron-dependent levels of Irr. (A) 35S-labeled methionine and cysteine were added to cultures at time zero, followed by addition of 200 μg/ml tetracycline (squares) or an equivalent volume of buffer (triangles) after 15 min (arrow). Cellular protein was precipitated at various times and was counted. Each point is the average of three trials ± the standard deviation. (B) Cells were treated with tetracycline (+Tet) or an equivalent volume of buffer (−Tet) 15 min before the addition of 6 μM FeCl3 (+Fe) or an equivalent volume of buffer (−Fe) at time zero. Aliquots were taken at various times, and Irr was detected by immunoblotting using anti-Irr antibodies. Thirty micrograms of protein was loaded per lane.
We then determined the effects of iron on Irr levels by immunoblots in cells preincubated with tetracycline for 15 min (Fig. 2B). In the absence of tetracycline, the Irr level in cells decreased with time upon exposure to iron but was stable in the absence of iron, as was reported previously (16). Preincubation of cells with tetracycline had little effect on the response of Irr to iron, suggesting that the observed dependence of Irr levels on iron is not at the level of synthesis. If the observed disappearance of Irr in the presence of iron were caused primarily by decreased synthesis of a constitutively unstable protein, then we would expect Irr to disappear under iron-limited conditions in the presence of tetracycline. These findings, as well as the turnover experiments (Fig. 1), argue in favor of iron-dependent regulation of Irr primarily at the level of protein stability.
Evidence for Heme Involvement in Iron-Dependent Turnover of Irr in Vivo.
Inspection of the Irr amino acid sequence identified a region near the N terminus with similarity to heme regulatory motifs (HRM) found in several eukaryotic proteins. HRMs are involved in heme-dependent transcriptional activity (33), organelle targeting (3), and other regulatory functions (34). Putative HRM sequences have been identified in bacterial proteins as well (33) but have not been characterized. The putative motif in Irr contains the sequence Gly28-Cys-Pro-Trp-His-Asp. The Cys-Pro residues are invariant in HRMs, and there is a tendency for a hydrophobic amino acid in the fourth position. Iron regulates Irr turnover and is a component of heme; thus, the putative HRM within Irr led us to ask whether heme participates in iron-dependent turnover.
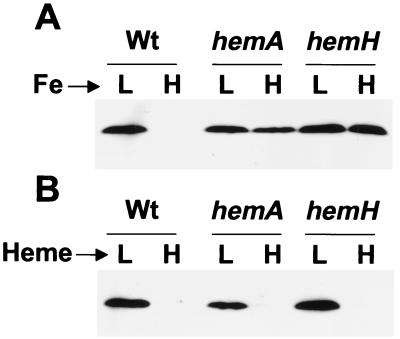
The effects of iron on Irr levels were measured in mutants defective in heme biosynthesis enzymes. ALA synthase, encoded by hemA, and ferrochelatase, encoded by hemH, catalyze the first and final steps of the heme biosynthetic pathway, respectively. Cells of hemA strain MLG1 (24) and hemH strain I110ek4 (25) were grown in media containing high or low iron concentrations, and Irr levels were detected by immunoblots using anti-Irr antibodies (Fig. 3A). Whereas Irr was present in parent strain I110 only under iron limitation, the protein persisted in the hemA or hemH mutant even in the presence of iron (added as ferric chloride), indicating a role for heme in response to iron in wild-type cells. The lack of iron responsiveness cannot be attributable to the status of a heme precursor because both the hemA and hemH strains gave similar results. Furthermore, the heme mutants grow, respire, and take up iron as well as wild-type cells (25, 26), and thus the persistence of Irr in those cells cannot be an indirect consequence of an energy deficiency or an iron uptake defect.
Figure 3.
Effects of iron and heme on Irr in cells of the wild-type and of heme biosynthesis mutants. (A) Cells of strains I110 (Wt), MLG1 (hemA), or I110ek4 (hemH) were grown in modified GSY medium containing either low (L) or high (H) iron media. Heme (0.05 μM) was present in the media to satisfy the heme requirement of hemH strain I110ek4. Irr in cells was analyzed by immunoblots using anti-Irr antibodies. Thirty micrograms of protein was loaded per lane. (B) Cells were grown in 0.05 μM (L) or 15 μM (H) heme, and Irr was analyzed as described for A. No iron was added to the media.
B. japonicum cells take up exogenous heme (25), and thus we tested whether addition of heme would affect cellular Irr levels. Cells were grown in low iron media supplemented with either 0.05 μM heme (low heme) or 15 μM heme (high heme), and Irr was detected by immunoblot analysis (Fig. 3B). Irr was observed in cells of the wild-type and the heme synthesis mutants grown in low heme media, but the protein was nearly undetectable when any of the strains were supplemented with 15 μM heme (Fig. 3B). The data in Fig. 3 indicate that heme is necessary for Irr turnover and suggest that the direct form of iron to which Irr responds may be heme.
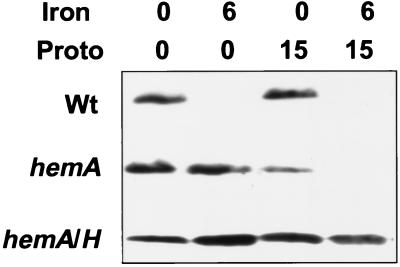
Iron supplied as ferric chloride is readily taken up by wild-type or heme mutant cells of B. japonicum (25), and therefore it is unlikely that heme is simply serving as an iron source for the heme synthesis mutant strains. To investigate this further, we examined the effects of iron (ferric chloride) on Irr levels in heme mutants in media supplemented with protoporphyrin IX, the immediate heme precursor (Fig. 4). Whereas Irr accumulated to high levels in the hemA strain in media containing 6 μM FeCl3, addition of 15 μM protoporphyrin and iron to the growth media resulted in the disappearance of Irr (Fig. 4). Ferrochelatase converts protoporphyrin and iron into heme, and thus it appears that the two compounds were taken up by the cells and converted to heme. Protoporphyrin alone caused some decrease in Irr levels in the hemA strain, which may be the result of some synthesis of heme from the low quantity of iron in the media (0.3 μM). Non-enzymatic conversion to heme in media does not occur to any detectable extent (data not shown). If the effects of iron and protoporphyrin are attributable to cellular conversion to heme, then a mutation in the hemH gene encoding ferrochelatase should counter the effects of iron plus protoporphyrin on Irr disappearance. A hemA hemH double mutant did not respond to iron or iron plus protoporphyrin with respect to Irr disappearance (Fig. 4), indicating that ferrochelatase is indeed necessary for the observed response in the hemA strain. We conclude that iron must be enzymatically incorporated into heme to exert its affect on Irr.
Figure 4.
Effects of iron, protoporphyrin, and a combination of both on Irr levels in cells of the wild-type and of heme biosynthesis mutants. Cells strain I110 (Wt), MLG1 (hemA), or ΔhemAH (hemA hemH; labeled hemA/H in the figure) were grown in modified media containing 6 μM ferric chloride, 15 μM protoporphyrin, both, or neither. Irr in cells was analyzed by immunoblots using anti-Irr antibodies. Thirty micrograms of protein was loaded per lane.
Evidence for Heme Binding to Irr at the Heme Regulatory Motif.
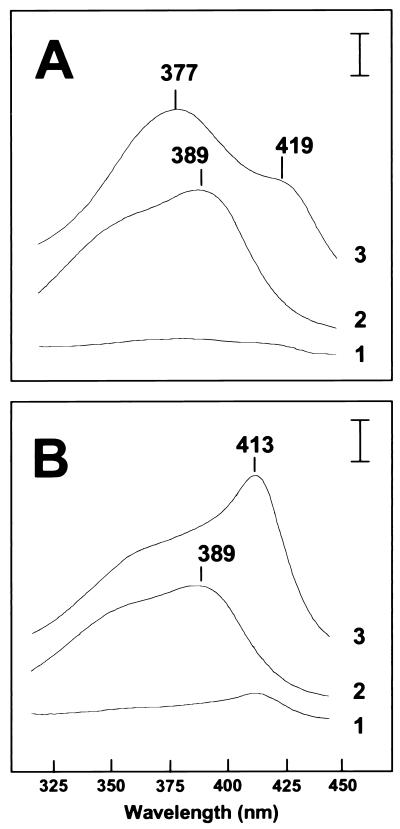
HRMs bind to heme through the conserved cysteine, and this binding is distinct from that found in globins or cytochromes (33). A synthetic decapeptide containing an HRM is sufficient to bind heme and, unlike cytochromes or globins, causes a shift in the absorbance of heme to shorter wavelengths (blue shift) (33). The effects of Irr on the absorption spectrum of heme (hemin) were examined by using recombinant purified wild-type Irr and a mutant protein with a Cys29→Ala change within the putative HRM (IrrC29A) (Fig. 5). The absorption peak of aqueous free heme at 389 nm was blue-shifted to 377 nm in the presence of Irr (Fig. 5A), which is consistent with heme binding to an HRM (33). The blue shift was not observed with the mutant protein IrrC29A (Fig. 5B). These observations indicate that Irr binds heme, resulting in a shift in the absorption spectrum to a shorter wavelength, and that this binding involves Cys29 within the putative HRM.
Figure 5.
Effect of Irr or IrrC29A on the absorption spectrum of heme. (A) Wild-type Irr. Absorption spectra were taken between 320 and 450 nm for 8 μM wild-type Irr alone (scan 1), 4 μM heme alone (scan 2), and Irr plus heme (scan 3). (B) Mutant IrrC29A. Absorption spectra were taken between 320 and 450 nm for 8 μM IrrC29A alone (scan 1), 4 μM heme alone (scan 2), and IrrC29A plus heme (scan 3). From scan 2, some heme may be bound to the purified recombinant IrrC29A protein. The wavelengths of the relevant features of the spectra are marked. The vertical bar represents a ΔA of 0.03.
Although IrrC29A did not cause a blue shift of the heme spectrum, it did result in a shift to longer wavelengths (red shift) with a peak at 413 nm (Fig. 5B). A red shift is typical of conventional heme proteins with a methionine/histidine or bis-histidine coordination at the fifth and sixth positions of the heme iron, and the 413-nm peak is almost identical to the spectrum of heme coordinated with imidizole, which mimics bis-histidine coordination (33). Furthermore, the heme spectrum with wild-type Irr showed a shoulder at 419 as well as the 377 peak (Fig. 5A). Interestingly, Zhang and Guarente (33) observed a similar spectrum of heme in the presence of both an HRM decapeptide and imidizole. The data suggest that Irr has a second heme binding site that does not involve the HRM cysteine. No attempts were made to identify residues involved in the second binding site.
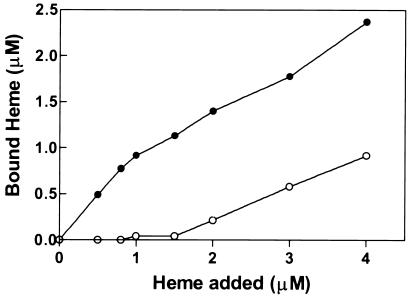
Binding of heme (hemin) to purified recombinant Irr proteins was determined as a function of heme concentration. For wild-type Irr, the binding did not saturate but appeared to be biphasic (Fig. 6). Unlike the wild-type protein, IrrC29A bound heme poorly below 2 μM, but binding was observed at higher concentrations (Fig. 6). Thus, Irr has a heme binding site that is absent in IrrC29A, thereby strongly implicating the HRM in high affinity binding. We did not attempt to establish whether the low affinity binding was nonspecific, as may occur with an amphipathic molecule such as heme, or whether it has physiological relevance. However, addition of 0.5 M NaCl drastically diminished the red shift in the heme spectrum without affecting the peak at 377 nm (data not shown); thus, we suggest that the low affinity site is responsible for the red shift in the heme spectrum (Fig. 5). The data in Figs. 5 and 6 support the conclusion that Irr is a heme-binding protein with a high affinity binding site at the HRM.
Figure 6.
Binding of Irr or Irr C29A to heme. Shown is bound heme to wild-type Irr (closed circles) or IrrC29A (open circles) as a function of the heme concentration. Irr-heme complex was separated from free heme and quantified as described in Materials and Methods.
Evidence for a Role for the Heme Regulatory Motif in Iron-Dependent Turnover of Irr in Vivo.
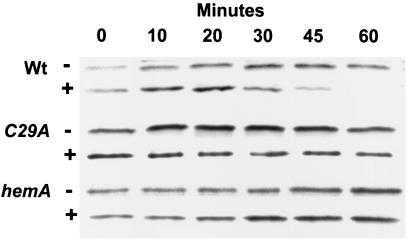
To determine whether the heme regulatory motif is involved in iron-dependent turnover of Irr, a B. japonicum mutant was constructed that contains irrC29A in the genome as the only expressed irr allele. The stability of IrrC29A in cells was assessed by pulse–chase experiments whereby the cells were 35S-labeled under low iron conditions in which Irr accumulates, followed by a chase with unlabeled amino acids in the presence of 6 μM ferric chloride and immunoprecipitation of labeled Irr at various times throughout the chase. IrrC29A persisted in cells throughout the 90-min time course whereas the wild-type protein degraded during that time (Fig. 7). Immunoblots showed that IrrC29A eventually disappeared 4 h after addition of iron, suggesting a slow response that does not involve the HRM (data not shown). The disappearance was accompanied by accumulation of ALA dehydratase, a protein under the negative control of Irr; thus, IrrC29A is an active protein.
Figure 7.
Irr turnover in vivo in irrC29A and hemA mutant strains. Immunoprecipitation of Irr from 35S-radiopulsed cells was carried out at various times of the chase after treatment of cells with 6 μM FeCl3 (+) or an equivalent volume of iron-free buffer (−) at time zero. The experiments were carried out in strains I110 (Wt), MLG1 (hemA), or IrrC29A (C29A). Shown are autoradiograms of immunoprecipitated, radiolabeled Irr at various times of the chase.
For comparison, we observed that wild-type Irr did not turnover in 90 min in hemA strain MLG1 in the presence of iron (Fig. 7). Thus, mutations that should prevent formation of a heme-Irr complex, either because the heme-binding site of Irr was abolished (irrC29A) or because the cell is heme-deficient (hemA), resulted in the loss of iron-dependent turnover of the protein. We conclude that heme binding to the HRM is required for iron-dependent degradation of Irr.
Discussion
In the current study, we demonstrate that Irr is a conditionally stable protein that depends on the iron status. Irr accumulates under iron limitation but degrades when cells are exposed to the metal. There are only a few known examples of conditionally stable regulatory proteins in bacteria (reviewed in ref. 35), and to our knowledge none are controlled in a metal-dependent manner. Furthermore, the mechanism of control of Irr by iron is fundamentally different from the bacterial iron sensor/regulators Fur and DtxR, which are essentially constitutive proteins but are active only when bound to iron (36). Thus, the strategies by which bacteria respond to iron may be more diverse than has been appreciated.
We propose that the destabilization of Irr in response to iron involves the binding of heme to a heme regulatory motif within Irr, demonstrating a functional HRM in a prokaryote. Exogenous heme was sufficient to cause Irr disappearance in cells of the wild-type and heme mutants, suggesting that heme is the form of iron to which Irr directly responds. Heme is iron-protoporphyrin, and therefore the simplest view is that the quantity of heme available to bind Irr is related to iron availability. A direct relationship between iron and heme would be expected when iron is the limiting substrate in heme formation, which is the condition under which Irr is present and functional (16). Although heme plays numerous regulatory roles in eukaryotes and prokaryotes, it is uncertain whether heme is actually free in a cell (reviewed in ref. 37), and therefore it will be important to understand the status of heme available to Irr and to other proteins that are regulated by heme.
The current study identifies a role for heme as an effector molecule that binds directly to a protein to promote its destabilization in a regulated manner. Very recently, Wang et al. showed that stability of Salmonella typhimurium glutamyl-tRNA reductase depends on cellular heme levels, but direct binding to the protein was not demonstrated (38). Nevertheless, further understanding of that system and of the one described here may reveal common mechanisms. Binding of heme to HRMs likely alters the conformation or folding of Saccharomyces cerevisiae HAP1 (1) and erythroid ALA synthase presequence (3). Thus, it is feasible that the conformation of Irr changes upon heme binding such that a portion of the protein becomes accessible to proteolysis. Another possibility is that heme catalyzes the formation of reactive oxygen species locally, as has been described for metal-catalyzed oxidation of other proteins, leading to protein damage and subsequent degradation. Indeed, the ability of heme to generate superoxide has been documented (39). Iron-dependent degradation of mammalian iron regulatory protein 2 (IRP2) entails iron binding through cysteine residues and metal-catalyzed oxidation of proximal susceptible residues (40). We note that one of the four cysteines in the degradation domain of human IRP2 is in the context Cys201-Pro-Phe-His (41, 42), which is similar to Cys29-Pro-Trp-His found in the Irr HRM. Although Iwai et al. (40) have shown convincingly that free iron is sufficient to degrade pure IRP2 in the presence of air and reducing agent, it may be fruitful to ascertain its sensitivity to heme as well.
Acknowledgments
This work was supported by National Science Foundation Grant MCB-9722974 to M.R.O.
Abbreviations
- ALA
δ-aminolevulinic acid
- HRM
heme regulatory protein
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Pfeifer K, Kim S, Kogan S, Guarente L. Cell. 1989;56:291–301. doi: 10.1016/0092-8674(89)90903-3. [DOI] [PubMed] [Google Scholar]
- 2.Mendez R, Moreno A, de Haro C. J Biol Chem. 1992;267:11500–11507. [PubMed] [Google Scholar]
- 3.Lathrop J T, Timko M P. Science. 1993;259:522–525. doi: 10.1126/science.8424176. [DOI] [PubMed] [Google Scholar]
- 4.Gilles-Gonzalez M A, Ditta G S, Helinski D R. Nature (London) 1991;350:170–172. doi: 10.1038/350170a0. [DOI] [PubMed] [Google Scholar]
- 5.Lowenstein C J, Snyder S H. Cell. 1992;70:705–707. doi: 10.1016/0092-8674(92)90301-r. [DOI] [PubMed] [Google Scholar]
- 6.Verma A, Hirsch D J, Glatt C E, Ronnett G V, Snyder S H. Science. 1993;259:381–384. doi: 10.1126/science.7678352. [DOI] [PubMed] [Google Scholar]
- 7.Bunn H F, Poyton R O. Physiol Rev. 1996;76:839–885. doi: 10.1152/physrev.1996.76.3.839. [DOI] [PubMed] [Google Scholar]
- 8.Crawford M J, Goldberg D E. J Biol Chem. 1998;273:34028–34032. doi: 10.1074/jbc.273.51.34028. [DOI] [PubMed] [Google Scholar]
- 9.Kwast K E, Burke P V, Staahl B T, Poyton R O. Proc Natl Acad Sci USA. 1999;96:5446–5451. doi: 10.1073/pnas.96.10.5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang L E, Willmore W G, Gu J, Goldberg M A, Bunn H F. J Biol Chem. 1999;274:9038–9044. doi: 10.1074/jbc.274.13.9038. [DOI] [PubMed] [Google Scholar]
- 11.Kappas A, Sassa S, Galbraith R A, Nordman Y. In: The Metabolic and Molecular Bases of Inherited Disease. Scriver C R, Beaudet A L, Sly W S, Valle D, Stanbury J B, Wyngaarden J B, Frederickson D S, editors. New York: McGraw–Hill; 1995. pp. 2103–2159. [Google Scholar]
- 12.Mock H-P, Grimm B. Plant Physiol. 1997;113:1101–1112. doi: 10.1104/pp.113.4.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakahigashi K, Nishimura K, Miyamoto K, Inokuchi H. Proc Natl Acad Sci USA. 1991;88:10520–10524. doi: 10.1073/pnas.88.23.10520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hentze M W, Kuhn L C. Proc Natl Acad Sci USA. 1996;93:8175–8182. doi: 10.1073/pnas.93.16.8175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rouault T A, Klausner R D. Trends Biochem Sci. 1996;21:174–177. [PubMed] [Google Scholar]
- 16.Hamza I, Chauhan S, Hassett R, O’Brian M R. J Biol Chem. 1998;273:21669–21674. doi: 10.1074/jbc.273.34.21669. [DOI] [PubMed] [Google Scholar]
- 17.Chauhan S, Titus D E, O’Brian M R. J Bacteriol. 1997;179:5516–5520. doi: 10.1128/jb.179.17.5516-5520.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamza I, Hassett R, O’Brian M R. J Bacteriol. 1999;181:5843–5846. doi: 10.1128/jb.181.18.5843-5846.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaballa A, Helmann J D. J Bacteriol. 1998;180:5815–5821. doi: 10.1128/jb.180.22.5815-5821.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patzer S I, Hantke K. Mol Microbiol. 1998;28:1199–1210. doi: 10.1046/j.1365-2958.1998.00883.x. [DOI] [PubMed] [Google Scholar]
- 21.Bsat N, Herbig A, Casillas-Martinez L, Setlow P, Helmann J D. Mol Microbiol. 1998;29:189–198. doi: 10.1046/j.1365-2958.1998.00921.x. [DOI] [PubMed] [Google Scholar]
- 22.Camilli A, Mekalanos J J. Mol Microbiol. 1995;18:671–683. doi: 10.1111/j.1365-2958.1995.mmi_18040671.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang J, Mushegian A, Lory S, Jin S. Proc Natl Acad Sci USA. 1996;93:10434–10439. doi: 10.1073/pnas.93.19.10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guerinot M L, Chelm B K. Proc Natl Acad Sci USA. 1986;83:1837–1841. doi: 10.1073/pnas.83.6.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frustaci J M, O’Brian M R. J Bacteriol. 1992;174:4223–4229. doi: 10.1128/jb.174.13.4223-4229.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frustaci J M, Sangwan I, O’Brian M R. J Bacteriol. 1991;173:1145–1150. doi: 10.1128/jb.173.3.1145-1150.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chauhan S, O’Brian M R. J Bacteriol. 1993;175:7222–7227. doi: 10.1128/jb.175.22.7222-7227.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chauhan S, O’Brian M R. J Biol Chem. 1995;270:19823–19827. doi: 10.1074/jbc.270.34.19823. [DOI] [PubMed] [Google Scholar]
- 29.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley Interscience; 1994. [Google Scholar]
- 30.Harlow E, Lane D. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 31.Smith K M. Porphyrins and Metalloporphyrins. Amsterdam: Elsevier Science; 1975. [Google Scholar]
- 32.Gottesman S, Maurizi M R. Microbiol Rev. 1992;56:592–621. doi: 10.1128/mr.56.4.592-621.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang L, Guarente L. EMBO J. 1995;14:313–320. doi: 10.1002/j.1460-2075.1995.tb07005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCoubrey W K, Huang T J, Maines M D. J Biol Chem. 1997;272:12568–12574. doi: 10.1074/jbc.272.19.12568. [DOI] [PubMed] [Google Scholar]
- 35.Gottesman S. Annu Rev Genet. 1996;30:465–506. doi: 10.1146/annurev.genet.30.1.465. [DOI] [PubMed] [Google Scholar]
- 36.Helmann J D. Metal Ions in Gene Regulation. New York: Chapman & Hall; 1997. pp. 45–76. [Google Scholar]
- 37.Smith A. In: Biosynthesis of Heme and Chlorophylls. Dailey H A, editor. New York: McGraw–Hill; 1990. pp. 435–490. [Google Scholar]
- 38.Wang L, Elliott M, Elliott T. J Bacteriol. 1999;181:1211–1219. doi: 10.1128/jb.181.4.1211-1219.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aft R L, Mueller G C. J Biol Chem. 1984;259:301–305. [PubMed] [Google Scholar]
- 40.Iwai K, Drake S K, Wehr N B, Weissman A M, La Vaute T, Minato N, Klausner R D, Levine R L, Rouault T A. Proc Natl Acad Sci USA. 1998;95:4924–4928. doi: 10.1073/pnas.95.9.4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Samaniego F, Chin J, Iwai K, Rouault T A, Klausner R D. J Biol Chem. 1994;269:30904–30910. [PubMed] [Google Scholar]
- 42.Guo B, Brown F M, Phillips J D, Yu Y, Leibold E A. J Biol Chem. 1995;270:16529–16535. doi: 10.1074/jbc.270.28.16529. [DOI] [PubMed] [Google Scholar]