Abstract
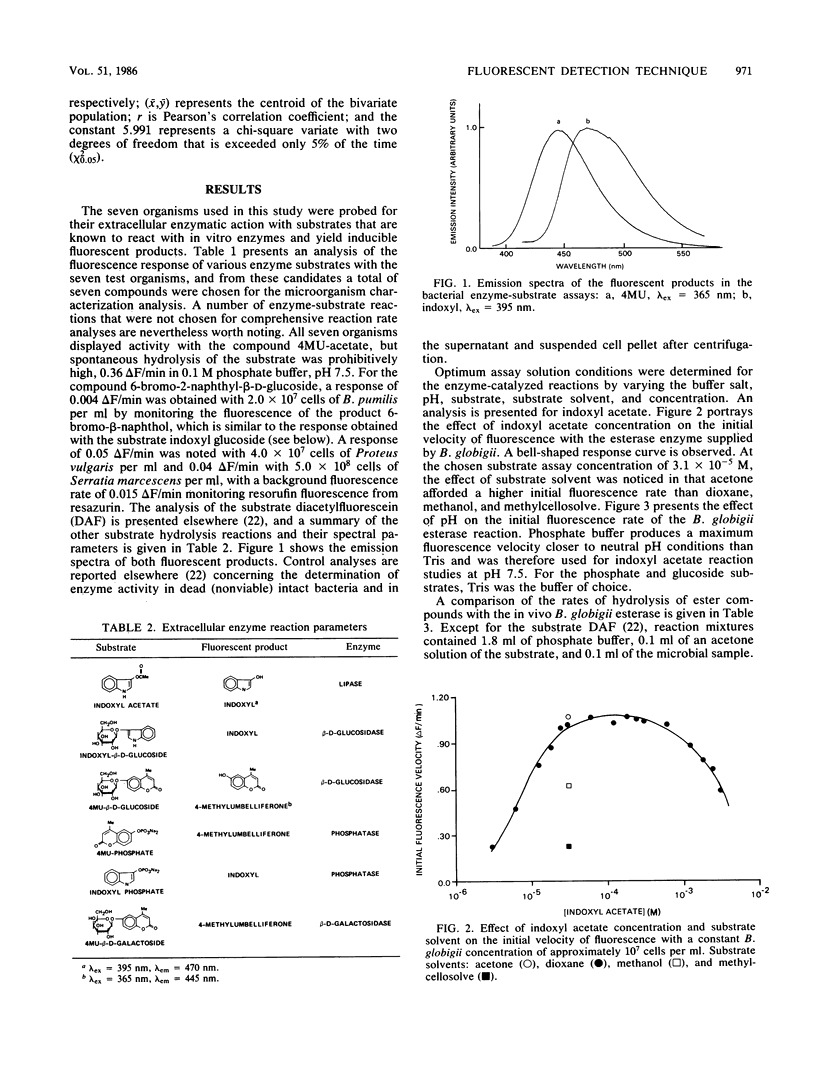
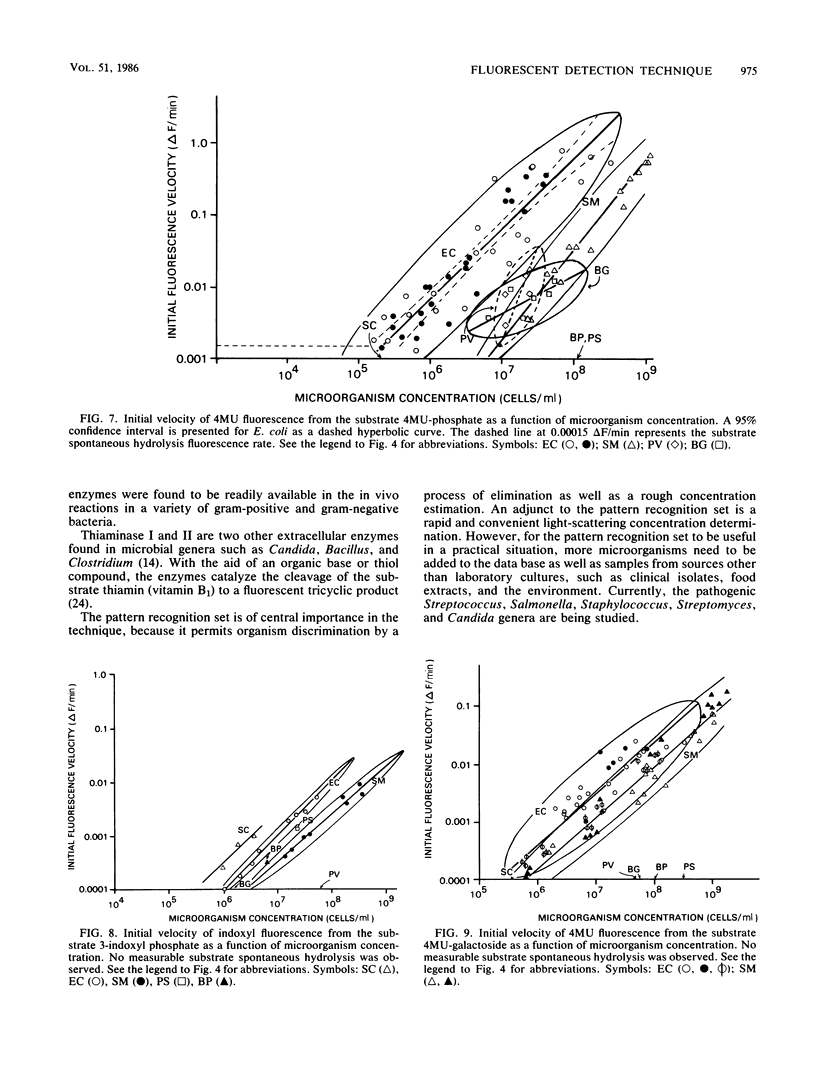
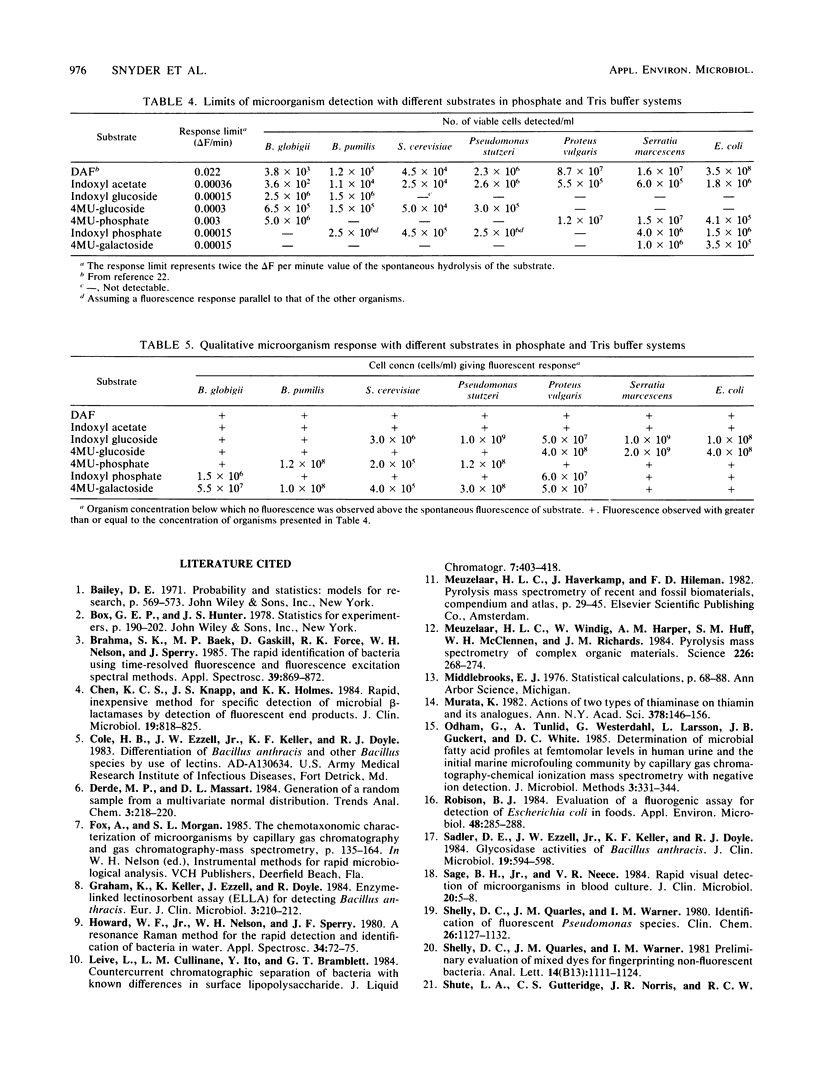
A spectrometric technique is presented that combines most of the important criteria necessary for efficient detection and identification of microorganisms. These criteria include simplicity of experimental design, various degrees of sensitivity and selectivity, convenience, and total reaction times of less than 15 min. The study takes advantage of the inherent extracellular enzymes present in living as opposed to dead, non-enzyme-producing organisms. Sequentially these are harnessed in in vivo reactions with a substrate containing a select organic functional group that is known to be cleaved or hydrolyzed by a certain enzyme. The substrate is tailored so that one of the products can be induced to fluoresce, and by using a conventional spectrofluorimeter the rate at which the fluorescence appears can be recorded. By subjecting the same bacterial sample to a number of different enzyme substrates, a pattern of fluorescence response rates emerges from a 7 by 7 microorganism-substrate matrix. Detection limits ranged from 3.6 X 10(2) to 3.5 X 10(8) cells per ml for the Bacillus globigii-indoxyl acetate and Escherichia coli-diacetylfluorescein pairs, respectively. The specificity and versatility of the method for bacterial determination is demonstrated in probing different bacterial enzymes through their spectrally active metabolic products.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chen K. C., Knapp J. S., Holmes K. K. Rapid, inexpensive method for specific detection of microbial beta-lactamases by detection of fluorescent end products. J Clin Microbiol. 1984 Jun;19(6):818–825. doi: 10.1128/jcm.19.6.818-825.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham K., Keller K., Ezzell J., Doyle R. Enzyme-linked lectinosorbent assay (ELLA) for detecting Bacillus anthracis. Eur J Clin Microbiol. 1984 Jun;3(3):210–212. doi: 10.1007/BF02014881. [DOI] [PubMed] [Google Scholar]
- Meuzelaar H. L., Windig W., Harper A. M., Huff S. M., McClennen W. H., Richards J. M. Pyrolysis mass spectrometry of complex organic materials. Science. 1984 Oct 19;226(4672):268–274. doi: 10.1126/science.6484572. [DOI] [PubMed] [Google Scholar]
- Murata K. Actions of two types of thiaminase on thiamin and its analogues. Ann N Y Acad Sci. 1982;378:146–156. doi: 10.1111/j.1749-6632.1982.tb31193.x. [DOI] [PubMed] [Google Scholar]
- Robison B. J. Evaluation of a fluorogenic assay for detection of Escherichia coli in foods. Appl Environ Microbiol. 1984 Aug;48(2):285–288. doi: 10.1128/aem.48.2.285-288.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler D. F., Ezzell J. W., Jr, Keller K. F., Doyle R. J. Glycosidase activities of Bacillus anthracis. J Clin Microbiol. 1984 May;19(5):594–598. doi: 10.1128/jcm.19.5.594-598.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage B. H., Jr, Neece V. R. Rapid visual detection of microorganisms in blood culture. J Clin Microbiol. 1984 Jul;20(1):5–8. doi: 10.1128/jcm.20.1.5-8.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelly D. C., Quarles J. M., Warner I. M. Identification of fluorescent Pseudomonas species. Clin Chem. 1980 Jul;26(8):1127–1132. [PubMed] [Google Scholar]
- Shute L. A., Gutteridge C. S., Norris J. R., Berkeley R. C. Curie-point pyrolysis mass spectrometry applied to characterization and identification of selected Bacillus species. J Gen Microbiol. 1984 Feb;130(2):343–355. doi: 10.1099/00221287-130-2-343. [DOI] [PubMed] [Google Scholar]



