Abstract
Geranyl diphosphate synthase, which catalyzes the condensation of dimethylallyl diphosphate and isopentenyl diphosphate to geranyl diphosphate, the key precursor of monoterpene biosynthesis, was purified from isolated oil glands of spearmint. Peptide fragments generated from the pure proteins of 28 and 37 kDa revealed amino acid sequences that matched two cDNA clones obtained by random screening of a peppermint-oil gland cDNA library. The deduced sequences of both proteins showed some similarity to existing prenyltransferases, and both contained a plastid-targeting sequence. Expression of each cDNA individually yielded no detectable prenyltransferase activity; however, coexpression of the two together produced functional geranyl diphosphate synthase. Antibodies raised against each protein were used to demonstrate that both subunits were required to produce catalytically active native and recombinant enzymes, thus confirming that geranyl diphosphate synthase is a heterodimer.
Keywords: monoterpene biosynthesis, isoprenoid biosynthesis
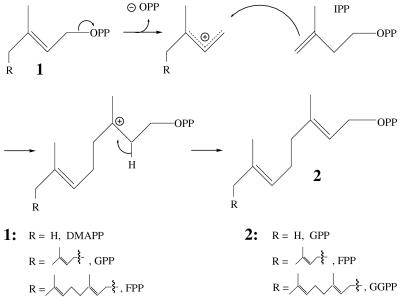
The prenyl diphosphate synthases (prenyltransferases) catalyze the condensation of isopentenyl diphosphate (IPP) with an allylic (prenyl) diphosphate to produce the next higher C5 homolog (Fig. 1) (1, 2). The chain-length specificity of these enzymes ranges broadly from geranyl diphosphate (GPP) synthase, which catalyzes a single transfer of dimethylallyl diphosphate (DMAPP) to IPP to generate the C10 product, to rubber transferase, which catalyzes multiple elongations with IPP to produce large polymers (3, 4). The family of “short-chain prenyltransferases” consists of GPP synthase, farnesyl diphosphate (FPP; C15) synthase, and geranylgeranyl diphosphate (GGPP, C20) synthase (2, 3) (Fig. 1). Genes encoding FPP synthase and GGPP synthase have been isolated from a range of organisms in which they play a central role in both primary and secondary isoprenoid metabolism (2, 4). These two homodimeric enzymes (FPP synthase and GGPP synthase) have been the object of extensive structure–function studies directed to understanding the electrophilic reaction mechanism and the determinants of chain-length specificity (3–6).
Figure 1.
Mechanism of the prenyltransferase reaction illustrating the condensations catalyzed by GPP synthase, FPP synthase, and GGPP synthase.
Thus far, GPP synthase is known only at the enzyme level, having been isolated from several plant sources (7–10) where it appears to participate primarily in the plastidial biosynthesis of monoterpenes (11, 12) by supplying the essential precursor of this family of natural products (13, 14). The reported properties of GPP synthase vary somewhat; however, in mechanism and many reaction parameters, the GPP synthase resembles both FPP synthase and GGPP synthase (2, 3, 13). In spite of these apparent similarities, a homology-based cloning approach, founded on consensus sequences of FPP synthases and GGPP synthases (2, 4), failed to yield the GPP synthase gene target.
In this paper, we describe the purification of GPP synthase from isolated oil glands of spearmint (15) and the use of amino acid sequence information to identify two distinct cDNA clones obtained by random sequencing of a peppermint-oil gland library. Expression studies, combined with the use of antibodies directed to each gene product, demonstrated that GPP synthase is a functional heterodimer, unlike other homodimeric short-chain prenyltransferases (2, 4, 12).
Materials and Methods
Plant Materials, Substrates, and Reagents.
Peppermint (Mentha × piperita L., cv. Black Mitcham) and spearmint (Mentha spicata L.) (16) were used for the preparation of glandular trichome (oil gland) secretory cells as the source of the enzyme (17) and of the mRNA used for cDNA library construction (18). [4-14C]IPP (54 Ci/mol) was purchased from DuPont/NEN. DMAPP was synthesized as previously described (19), as were GPP (20) and FPP (21). Terpenol standards were from our own collection.
Prenyltransferase Assay.
To 10 μl of enzyme suspension was added 70 μl of Mopso buffer (25 mM, pH 7.0) containing 10% glycerol, 10 mM MgCl2, and 1 mM DTT. DMAPP (10 μM) and [4-14C]IPP (7 μM) were added (100 μl total volume) to initiate the reaction, and the contents were overlaid with 1 ml pentane and incubated for 1 h at 31°C. After incubation, 10 μl of 3 N HCl was added and, after acid hydrolysis was complete (20 min at 31°C), the contents were mixed to partition into the pentane layer products derived from the acid labile allylic diphosphates. The pentane was removed, the remaining aqueous phase extracted with diethyl ether (2 ml), and the radioactive products contained in the combined extract were measured by liquid scintillation counting. For the assay based on enzymatic, rather than acid, hydrolysis, the products and remaining substrates of the reaction mixture were hydrolyzed with 1 unit each of wheat germ alkaline phosphatase and potato apyrase (both from Sigma), added in 1 ml of 200 mM Tris buffer (pH 9.5), followed by incubation for 8 h at 30°C. The organic extract was then prepared for analysis as before. To prepare sufficient material for product identification, the assay was scaled up by a factor of five. After the hydrolysis step, the combined organic extract was dried over anhydrous Na2SO4, diluted with internal standards (see Fig. 2) and concentrated for radio–GLC analysis by published protocols (22).
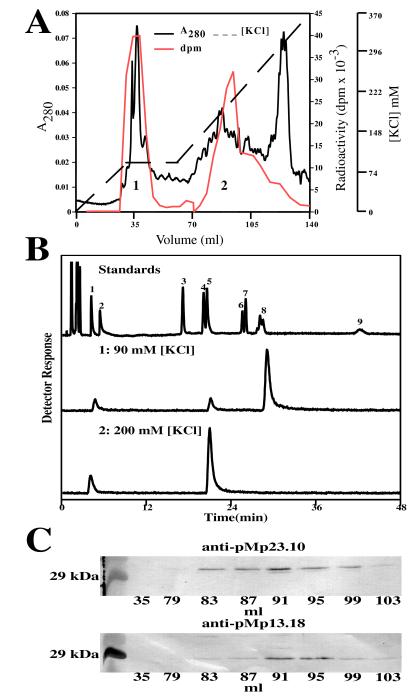
Figure 2.
Purification of GPP synthase from spearmint-oil gland secretory cells, product identification, and immunoblotting. (A) Anion-exchange chromatography of the prenyltransferase activity showing separation of FPP synthase (eluting at 90 mM KCl, region 1) from GPP synthase (eluting at 200 mM KCl, region 2). Absorbance at A280, the KCl gradient, and the prenyltransferase assay values (in dpm) are indicated. (B) Radio-GC separation of the labeled dephosphorylated reaction products of FPP synthase (region 1) and GPP synthase (region 2). The upper tracing is the detector response to the authentic standards separated on the polydimethylsiloxane column: isopentenol (peak 1), dimethylallyl alcohol (peak 2), linalool (peak 3), nerol (peak 4), geraniol (peak 5), cis-nerolidol (peak 6), trans-nerolidol (peak 7), mixture of cis- and trans-farnesol (peak 8), and geranylgeraniol (peak 9). (C) Immunodetection of the large and small subunits of the native GPP synthase in the corresponding fractions separated by anion-exchange chromatography (A) using the antibodies raised against the recombinant proteins expressed from pMp13.18 and pMp23.10. The marker (Left) is carbonic anhydrase (29 kDa).
GPP Synthase Purification from Mint Glandular Trichomes.
Glandular trichome secretory cell clusters were isolated from 40-g batches of spearmint leaves, and the enzyme extract was prepared by sonication and filtration as previously described (17). The filtrate was then centrifuged at 12,000 × g (30 min), then at 195,000 × g (90 min), and the supernatant was utilized as the enzyme source.
The supernatant was exhaustively dialyzed in Mes buffer (25 mM, pH 6.2) containing 10% glycerol, 1 mM DTT, and 10 mM MgCl2, and then subjected batchwise to dye-ligand interaction chromatography by using DyeMatrex Red A Gel (Amicon) equilibrated with dialysis buffer. After 1 h of gentle mixing, the gel was gravity drained and washed with dialysis buffer. GPP synthase was then eluted with Hepes buffer (25 mM, pH 7.2) containing 10% glycerol, 5 mM potassium phosphate, 1 mM DTT, and 1 mM EDTA.
The combined eluate from above was then loaded onto an HR 10/10 FPLC column containing Source 15Q anion-exchange medium (Pharmacia Biotech) equilibrated in Hepes buffer (25 mM, pH 7.5) containing 10% glycerol, 10 mM MgCl2 and 1 mM DTT. GPP synthase, determined by assay and monitored by SDS/PAGE (23), eluted at ≈200 mM KCl after a discontinuous salt gradient (see Fig. 2). Gel permeation chromatography of this material was conducted on a calibrated Superdex 75 6/60 column (Pharmacia FPLC).
Protein Microsequencing.
The partially purified GPP synthase, pooled from several purification runs (≈1 mg), was thermally denatured, dialyzed against distilled water, lyophilized, and separated by SDS/PAGE (23) on a 12.5% acrylamide gel. The two protein bands at ≈28 kDa and ≈37 kDa, which had been shown to track GPP synthase activity, were visualized with Coomassie blue stain, excised, and individually proteolyzed “in-gel” with trypsin (24). The resulting peptides were purified by reversed-phase HPLC and subjected to N-terminal sequence analysis at the Washington State University Laboratory for Biotechnology and Bioanalysis.
GPP Synthase Cloning and Expression.
Comparison of the amino acid microsequence data from above to translated sequences obtained by random sequencing of a peppermint oil gland cDNA library (18) allowed matching to two distinct “prenyltransferase-like” clones. These full-length clones, corresponding to the 28-kDa protein (designated pMp13.18) and to the 37-kDa protein (designated pMp23.10), were initially obtained as expressed sequence tag fragments, from which full-length versions were acquired by screening of the library with 5′-probes (25). The full-length ORF of pMp13.18 (initially as a pBluescript phagemid) was directionally cloned into pSBET (26) for expression, via the addition of an NdeI site at the starting methionine by site-directed mutagenesis (QuickChange, Stratagene) and by the use of a convenient BamHI site (8 bp downstream of the stop codon). The vector and derivative of pMp13.18 were doubly digested with BamHI and NdeI, and the resulting DNA fragments were purified and ligated, then transformed into Escherichia coli XL1-blue cells. The resulting plasmid, designated pSBET13.18, was purified and verified by sequencing. The full-length ORF of pMp23.10 was similarly modified for cloning into pSBET to yield pSBET23.10. Sequencing revealed that no errors had been introduced during mutagenesis.
For coexpression studies, the ORF of pMp23.10 was similarly subcloned into pET32a (Novagen) to yield pET23.10. For expression of clone pET23.10 alone, this plasmid was cotransformed with pSBETa (26) to take advantage of the ArgU gene of the latter, which encodes a tRNA specifying rare codon usage for arginine. The above plasmids, as well as control pSBET and control pET plasmids (without insert), were transformed into E. coli BLR(DE3) for expression. For coexpression, E. coli BLR(DE3) was doubly transformed with pSBET13.18 and pET23.10 to provide pSBET13.18-pET23.10/BLR. Each transformant was grown to A600 = 0.5 in 1 l of Luria–Bertani medium (with 1% glucose) with kanamycin selection (for pSBET) or carbenecillin selection (for pET), or with dual antibiotic selection (for coexpression by using both plasmids). The transformed bacteria were then induced with 1 mM isopropyl-d-thiogalactoside and allowed to express for 24 h at 15°C.
Purification of Recombinant Proteins and Antibody Preparation.
After expression, the bacteria were harvested by centrifugation, washed with Tris buffer (pH 7.0) containing 50 mM KCl, resuspended in 25-ml sonication buffer (25 mM Hepes, pH 7.2/10 mM MgCl2/10% glycerol/1 mM DTT/1 mM EDTA/1 mM benzamidine) and disrupted by brief sonication (VirSonic, 25% power/two 30-s bursts at 4°C). The sonicate was centrifuged at 12,000 × g (30 min), then at 195,000 × g (30 min), and the supernatant was loaded onto an HR 5/5 FPLC column containing Source 15Q anion-exchange chromatography medium (Pharmacia Biotech) that had been equilibrated with Hepes buffer (25 mM, pH 7.5) containing glycerol, MgCl2, DTT, and benzamidine as before. A KCl gradient (0–600 mM) was applied to separate the various recombinant proteins from endogenous E. coli FPP synthase; fractions were assayed for prenyltransferase activity by using [4-14C]IPP and each allylic cosubstrate (DMAPP, GPP, and FPP) and were monitored for protein content by SDS/PAGE (23). Gel permeation chromatography of the recombinant proteins was performed on a calibrated Sephacryl-S100 column (Pharmacia FPLC).
The combination of chromatography and SDS/PAGE was sufficient to resolve the recombinant proteins expressed from pSBET13.18 and pET23.10; these proteins were easily recognized in extracts of transformed cells because they were absent in preparations from the empty vector controls. Each purified protein (1.5 mg, in gel) was used to generate polyclonal antibodies in rabbits (Alpha Diagnostic, San Antonio, TX). For immunoblotting, protein samples were prepared by standard protocol (27) and separated on a 4–15% polyacrylamide gradient gel (Bio-Rad). After electrotransfer to supported nitrocellulose membranes (Bio-Rad), the antigens were detected by using the rabbit antibodies with goat anti-rabbit IgG conjugated to alkaline phosphatase as secondary antibody (Boehringer Mannheim).
Results
Unlike the ubiquitous prenyltransferases, FPP synthase and GGPP synthase (2), GPP synthase is largely restricted to plant species that produce abundant quantities of monoterpenes (7–12). The epidermal oil glands are the exclusive site of monoterpene biosynthesis in mint (Mentha) species (17). Thus, the secretory cells of these structures represent a highly enriched source of the enzymes of monoterpene biosynthesis, including GPP synthase (15), and of the messages that encode these specialized catalysts (18). Simultaneously with the purification of GPP synthase from this source, a random sequencing effort using a peppermint-oil gland cDNA library was undertaken, as was an attempt at homology-based cloning of GPP synthase. The latter approach was founded on the assumption that GPP synthase would resemble the mechanistically similar FPP and GGPP synthases (2, 4). Various PCR primers based on consensus regions of FPP synthases and GGPP synthases failed to amplify GPP synthase gene fragments from the oil gland library, thus indicating that a protein-based cloning strategy was required (28).
Purification of GPP Synthase from Mint.
Assessment of the level of GPP synthase activity in oil gland preparations from peppermint and spearmint (16, 17) indicated the latter of these closely related species (29) to be the quantitatively superior starting material. After preliminary purification by dye-ligand interaction chromatography, the GPP synthase and FPP synthase were separated by anion-exchange chromatography (Fig. 2A). Assay of region 1 (eluting at 90 mM KCl) coupled to radio-GC analysis of the dephosphorylated reaction products (Fig. 2B, region 1), confirmed this fraction to contain principally FPP synthase; the production of small amounts of GPP (as evidenced by the detection of geraniol on radio-GC analysis) is an expected consequence of the prenyltransferase assay (with IPP and DMAPP) in which this intermediate of the FPP synthase reaction is observed.
Region 2 (eluting at 200 mM KCl) (Fig. 2A) was shown, by similar assay and radio-GC analysis of the dephosphorylated products (Fig. 2B, region 2), to be comprised largely of GPP synthase; this fraction did not produce prenols longer than C10 as determined by assay with each of the allylic cosubstrates (DMAPP, GPP, and FPP). Further attempts to purify the GPP synthase by hydrophobic interaction or hydroxyapatite chromatography were thwarted by the instability of the enzyme. Nevertheless, the size of the native enzyme was determined to be 68 ± 6 kDa by gel permeation chromatography.
Microsequencing of Putative GPP Synthase Proteins.
Because further efforts to purify the GPP synthase from mint oil glands were unsuccessful, active fractions separated by anion-exchange chromatography were evaluated for protein content by SDS/PAGE. Because of the enriched starting material and the partial purification obtained, a simple banding pattern of only 10 proteins was observed, and only two of these, at 28 kDa and 37 kDa, tracked closely the GPP synthase activity (data not shown). Because the native enzyme was shown to possess a molecular weight of roughly 68,000, dimeric architecture was implied. Therefore, the 28-kDa and 37-kDa proteins were isolated by SDS/PAGE for trypsinization (because both proteins were N-blocked). Separation of the resulting peptides by HPLC and Edman degradative sequencing yielded two sequences from the 28-kDa protein and four from the 37-kDa protein (Table 1).
Table 1.
Peptide sequences derived by trypsinization of putative GPP synthase
| Protein | Peptide | Deduced sequence correspondence
|
|
|---|---|---|---|
| pMp13.18 | pMp23.10 | ||
| 28 kDa | VIIEIS | 184–189 | — |
| FGLYQGTL | 253–260 | — | |
| 37 kDa | LIGVE | — | 333–337 |
| YIAYR | — | 371–375 | |
| EAVETLLHF | — | 349–357 | |
| TAALLTGSVVLGAIL | — | 263–277 | |
A dash indicates that this sequence element was not found.
Molecular Cloning of GPP Synthase.
Comparison of the peptide sequences of the purified proteins to the deduced amino acid sequences of two full-length prenyltransferase-like clones, which had been acquired by random sequencing of the peppermint-oil gland cDNA library, allowed matching of the 28-kDa protein to clone pMp13.18 and of the 37-kDa protein to clone pMp23.10. Clone pMp13.18 (1,131 nt) encodes an ORF of 939 nt, corresponding to a protein of 313 amino acids (Fig. 3) with a calculated molecular weight of 33,465 and pI of 6.18. The first 48 deduced amino acid residues show the expected characteristics of an N-terminal plastid-targeting sequence [i.e., the sequence is rich in serine residues and small hydrophobic amino acids and is low in acidic residues (30)]. The presence of such a targeting sequence is consistent with the plastidial origin of monoterpene biosynthesis in plant cells (13, 14) and with biochemical localization studies (11). By excluding the transit peptide of the preprotein, the amino acid sequence corresponds to a deduced mature (processed) protein of molecular weight 28,485, in full agreement with a size of 28 ± 1 kDa determined for this presumptive subunit of the native enzyme by SDS/PAGE. Clone pMp13.18 exhibits 33% amino acid identity to its presently closest apparent relative, a putative GGPP synthase-like protein from A. thaliana (Fig. 3); however, the peppermint clone is missing the conserved DD(X)2–4D motifs, which comprise essential substrate binding elements of all short-chain prenyltransferases thus far defined (2–6).
Figure 3.
Alignment of the deduced amino acid sequences of pMp13.18 and pMp23.10 to each other and to their nearest homologues (A. thaliana GGPP synthase-like protein, AL035540, for pMp13.18, and Catharanthus roseus GGPP synthase, X92893, for pMp23.10). The DDXD motif of pMp13.18 is underlined in green. The red underlines indicate the DD(X)2–4D motifs of the other sequences. Identical residues (three or more) are shown in black; similar residues are shaded.
Clone pMp23.10 (1,341 nt) encodes an ORF of 1,131 nt, corresponding to a preprotein of 377 amino acids with a calculated molecular weight of 40,800 and pI of 6.93. The first 40 deduced amino acid residues show the expected characteristics of an N-terminal plastidial targeting sequence (30). By excluding the putative transit peptide in this case, the sequence corresponds to a deduced mature (processed) protein of molecular weight 36,400, in full agreement with a size of 37 ± 1 kDa determined for this presumptive subunit of the native enzyme by SDS/PAGE. Clone pMp23.10 shares a surprising level of amino acid identity with GGPP synthases of plant origin (62–75%) and contains the DD(X)2–4D motifs anticipated for a prenyltransferase (3, 4) (Fig. 3). Clones pMp13.18 and pMp23.10 exhibit only 26% amino acid identity to each other.
Heterologous Expression of GPP Synthase.
To evaluate the possible function of the proteins expressed from pMp13.18 and pMp23.10, each cDNA insert was subcloned into an expression vector that would allow high-level bacterial (E. coli) production, as well as the ability to coexpress both clones in a single bacterial cell. Clone 13.18 was expressed from pSBET in E. coli BLR(DE3), and efficient translation was verified by SDS/PAGE of the soluble protein fraction; however, much of the expressed protein remained in inclusion bodies (data not shown). Assay of the crude soluble fraction, with DMAPP and IPP as cosubstrates, evidenced little prenyltransferase activity above that of endogenous E. coli FPP synthase activity. Purification by anion-exchange chromatography demonstrated the presence of essentially only FPP synthase derived from the bacterial host (i.e., eluting at 85 mM of the salt gradient; region 1 of Fig. 4A), as determined by radio-GC of the hydrolyzed product (Fig. 4B, region 1). This chromatographic separation procedure, along with SDS/PAGE and comparison to empty vector controls, allowed purification of the expressed protein as antigen for polyclonal antibody preparation, and also permitted direct demonstration that this partially purified protein possessed negligible prenyltransferase activity when assayed with [14C]IPP and the three allylic cosubstrates (DMAPP, GPP, FPP). Truncation of the cDNA to remove the transit peptide (by installation of a starting methionine, via an NdeI site, immediately upstream of Gln49) and expression and partial purification of this “pseudomature” form of the enzyme yielded the same result, namely no detectable prenyltransferase activity.
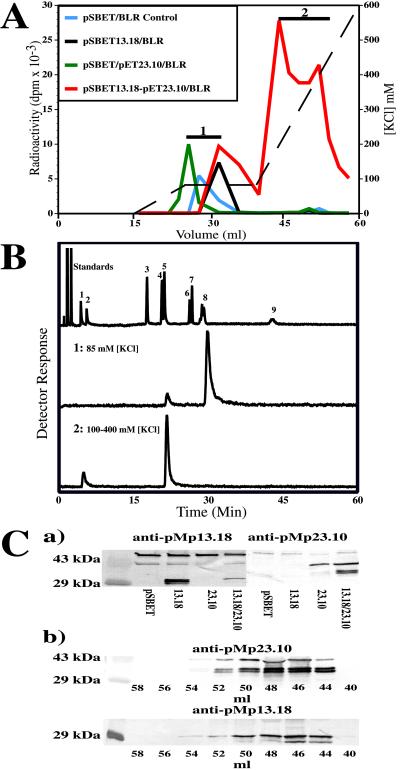
Figure 4.
Purification of recombinant GPP synthase expressed in E. coli, product identification and immunoblotting. (A) Anion-exchange chromatography of prenyltransferase activity (in dpm) expressed from pSBET13.18 (alone), pET23.10 (alone) and the combination of pSBET13.18-pET23.10, showing the separation of host-derived FPP synthase (eluting at 85 mM KCl, region 1) from GPP synthase (eluting at ≈200 mM KCl, region 2) obtained by coexpression. The KCl gradient is indicated by the dashed line. (B) Radio-GC separation of the labeled dephosphorylated reaction products of FPP synthase (1) and GPP synthase (2). The standards shown are the same as those described in Fig. 2B. (C) Immunodetection of the small and large subunits of GPP synthase (crude extracts) expressed alone and in combination (a), and of the functional heterodimer in the corresponding fractions (b) separated by anion-exchange chromatography (A), using the antibodies raised against the individual recombinant proteins expressed from pMp13.18 and pMp23.10. Empty vector controls (pSBET) and molecular mass markers, carbonic anhydrase (29 kDa) and ovalbumin (43 kDa), are included. Note that anti-pMp13.18 and anti-pMp23.10 detect other proteins (≈43 kDa) from the E. coli host (that are also detected by preimmune serum), and that, on coexpression of pMp13.18 and pMp23.10, truncated versions of both preproteins are produced.
Clone 23.10 was similarly evaluated by expression from pET3a in E. coli BLR(DE3), in this case with the assistance of cotransformed pSBETa to improve translation efficiency of rare arginine codons (26). Translation of soluble protein was verified as before by SDS/PAGE (significant inclusion bodies were also formed), and, as before, assay of the crude soluble protein fraction yielded no measurable prenyltransferase activity above that resulting from endogenous FPP synthase of the host. Similar purification of this material by anion exchange chromatography again demonstrated the presence of only E. coli FPP synthase activity (Fig. 4A, region 1), allowed the preparation of antigen (after SDS/PAGE) suitable for generating polyclonal antibodies, and also permitted the direct demonstration that the expressed protein, freed of competing enzymes, possessed no detectable prenyltransferase activity when assayed with all combinations of cosubstrates. It is notable that, although the sequence of pMp23.10 rather closely resembles those of GGPP synthases of plant origin (up to 75% identity), the expressed protein was devoid of this activity. Truncation of the cDNA to delete the targeting sequence and partial purification of this expressed “pseudomature” form, again, as with the 13.18 clone, did not yield a functional prenyltransferase.
Interestingly, when both clones were coexpressed (as pSBET13.18-pET23.10/BLR), assay for prenyltransferase in the resulting soluble fraction of the bacterial extract gave levels of activity greatly in excess of host-derived FPP synthase. After verification of expression of both recombinant protein species by immunodetection, separation of the preparation by anion-exchange chromatography revealed a new activity, distinct from E. coli FPP synthase, that bound to the matrix more tightly than did either the 13.18- or 23.10-derived proteins when expressed alone (i.e., eluting at 200 mM KCl; region 2 of Fig. 4A). The biosynthetic products of the assay with [14C]IPP and DMAPP were dephosphorylated and analyzed by radio-GC (Fig. 4B, region 2) to reveal essentially only geraniol, thereby indicating the presence of GPP synthase. More detailed analysis of this protein fraction demonstrated the presence of only GPP synthase, i.e., this preparation was inactive with GPP and FPP as cosubstrates and thus was devoid of FPP synthase and GGPP synthase activity. Gel-permeation chromatography indicated this recombinant GPP synthase to elute at a volume corresponding to a size of 85 ± 8 kDa (the approximate sum of the two expressed preproteins), suggesting that the enzyme produced by coexpression of cDNA inserts 13.18 and 23.10 was a functional dimer. A rough estimate of kcat, obtained by assay coupled to SDS/PAGE and densitometry, gave a value of 0.5 sec−1, which is close to the value estimated for the native enzyme by similar means (0.8 sec−1). These results strongly indicated that an authentic recombinant GPP synthase had been obtained via the interaction of the two proteins resulting from coexpression of pSBET13.18 and pET23.10.
Demonstration that GPP Synthase Is a Heterodimer.
With the availability of specific polyclonal antibodies directed to each of the pMp13.18- and pMp23.10-derived proteins (Fig. 4Ca), it became possible to directly examine the subunit architecture of both native and recombinant GPP synthases by immunoblotting. Evaluation of the elution behavior of the native enzyme on anion-exchange chromatography (Fig. 2A, region 2) indicated the presence of both proteins in comparable amounts in the active fractions at an abundance proportional to the amount of GPP synthase activity (Fig. 2C). This evidence, in conjunction with the size of the native enzyme (68 ± 6 kDa), indicated the native GPP synthase to be a heterodimer comprised of the 28-kDa and the 37-kDa subunits. Evaluation of the elution behavior of the functional recombinant enzyme on anion-exchange chromatography (Fig. 4A, region 2), by identical immunoblotting protocols, revealed the essentially identical result, in this case for the preproteins (Fig. 4Cb). Thus, both expressed proteins were present in comparable amounts in the active fractions at an abundance proportional to the amount of GPP synthase activity. This evidence, in conjunction with the size of the recombinant enzyme (85 ± 8 kDa), indicated the recombinant GPP synthase to be a heterodimer comprised of the two preprotein subunits of 33.5 kDa and 41 kDa. Truncated species of both preprotein subunits are observed in the coexpressed functional heterodimer (Fig. 4Cb) that are not produced when the individual preproteins are expressed alone (Fig. 4Ca). Such proteolysis by the host has been observed previously with related plastid-directed enzymes (31). Apparent differences in proteolytic susceptibility in the present case may be related to protein-folding differences between the functional heterodimer compared with that of either subunit when expressed alone.
Discussion
It is notable that the GPP synthase subunits more closely resemble GGPP synthases than FPP synthases. Thus, the small subunit exhibits ≈25% identity to homodimeric GGPP synthase preproteins but only ≈17% identity to homodimeric FPP synthases. For the large subunit, the resemblance is more striking; 62–75% identity to GGPP synthase preproteins but only ≈25% identity to FPP synthases. These observations suggest the evolutionary origin of both subunits of GPP synthase from GGPP synthase, which is also plastidial, not from FPP synthase, which is a cytosolic enzyme (2).
The heterodimeric subunit architecture of GPP synthase was unanticipated, because the other short-chain prenyltransferases are homodimers (2, 4). However, precedent for heterodimeric prenyltransferases is established by hexaprenyl diphosphate synthase (32) and heptaprenyl diphosphate synthase (33) of microbial origin. These “medium chain-length” prenyltransferases produce C30 and C35 products, respectively, and consist of two distinct subunits, the large one exhibiting the sequence motifs found in other prenyltransferases, and the small one bearing little homology to extant prenyltransferases; no detailed functional mechanism for the interaction between these disparate subunits has been proposed (2, 3). Although GPP synthase resembles the medium chain-length prenyltransferases in general architecture, the similarity between the respective small and large subunits is limited (<26% and <36% identity, respectively). In this context, the structure–function relationships of the GPP synthase subunits are of interest, especially because this enzyme represents the “minimal” prenyltransferase in catalyzing but a single condensation reaction between the two smallest natural cosubstrates (IPP and DMAPP) to strictly control product chain length. One possibility is that the small subunit modifies a GGPP synthase-like protein to direct a single reaction cycle with the formation of the C10 chain, although it should be emphasized that the large subunit, GGPP synthase-like, protein is, by itself, apparently catalytically inactive. The alternative is that the small subunit participates directly in binding and catalysis, in spite of the fact that it is only the large subunit that bears the signature DD(X)2–4D motifs (Fig. 3) that are seemingly necessary for interaction with the diphosphate ester cosubstrates (4, 5). The small subunit, however, does possess a DDXD sequence (Fig. 3), which might serve as a surrogate element in substrate binding interactions. Experiments to explore the structural and functional properties of this novel prenyltransferase are in progress.
Acknowledgments
We thank D. Tholl for technical assistance. This investigation was supported by grants from the U.S. Department of Energy, the Mint Industry Research Council, and Pioneer Hi-Bred International.
Abbreviations
- GPP
geranyl diphosphate
- FPP
farnesyl diphosphate, GGPP, geranylgeranyl diphosphate
- IPP
isopentenyl diphosphate
- DMAPP
dimethylallyl diphosphate
Footnotes
References
- 1.Poulter C D, Rilling H C. In: Biosynthesis of Isoprenoid Compounds. Porter J W, Spurgeon S L, editors. Vol. 1. New York: Wiley; 1981. pp. 161–224. [Google Scholar]
- 2.Ogura K, Koyama T. Chem Rev. 1998;98:1263–1276. doi: 10.1021/cr9600464. [DOI] [PubMed] [Google Scholar]
- 3.Koyama T, Ogura K. In: Comprehensive Natural Products Chemistry. Cane D E, editor. Vol. 2. Oxford: Pergammon/Elsevier Science; 1999. pp. 69–96. [Google Scholar]
- 4.Wang, K. & Ohnuma, S.-I. (1999) Trends Biochem. Sci., in press. [DOI] [PubMed]
- 5.Tarshis L C, Yan M, Poulter C D, Sacchettini J C. Biochemistry. 1994;33:10871–10877. doi: 10.1021/bi00202a004. [DOI] [PubMed] [Google Scholar]
- 6.Tarshis L C, Proteau P J, Kellogg B A, Sacchettini J C, Poulter C D. Proc Natl Acad Sci USA. 1996;93:15018–15023. doi: 10.1073/pnas.93.26.15018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Croteau R, Purkett P T. Arch Biochem Biophys. 1989;271:524–535. doi: 10.1016/0003-9861(89)90304-4. [DOI] [PubMed] [Google Scholar]
- 8.Heide L, Berger U. Arch Biochem Biophys. 1989;273:331–338. doi: 10.1016/0003-9861(89)90491-8. [DOI] [PubMed] [Google Scholar]
- 9.Suga T, Endo T. Phytochemistry. 1991;30:1757–1761. [Google Scholar]
- 10.Clastre M, Bantignies B, Feron G, Soler E, Ambid C. Plant Physiol. 1993;102:205–211. doi: 10.1104/pp.102.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soler E, Feron G, Clastre M, Dargent R, Gleizes M, Ambid C. Planta. 1992;187:171–175. doi: 10.1007/BF00201934. [DOI] [PubMed] [Google Scholar]
- 12.Sommer S, Severin K, Camara B, Heide L. Phytochemistry. 1995;38:623–627. [Google Scholar]
- 13.Wise M L, Croteau R. In: Comprehensive Natural Products Chemistry. Cane D E, editor. Vol. 2. Oxford: Elsevier Science; 1999. pp. 97–153. [Google Scholar]
- 14.Turner G, Gershenzon J, Nielson E E, Froehlich J E, Croteau R. Plant Physiol. 1999;120:879–886. doi: 10.1104/pp.120.3.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McConkey, M., Gershenzon, J. & Croteau, R. (2000) Plant Physiol., in press. [DOI] [PMC free article] [PubMed]
- 16.Alonso W R, Rajaonarivony J I M, Gershenzon J, Croteau R. J Biol Chem. 1992;267:7582–7587. [PubMed] [Google Scholar]
- 17.Gershenzon J, McCaskill D, Rajaonarivony J I M, Mihaliak C, Karp F, Croteau R. Anal Biochem. 1992;200:130–138. doi: 10.1016/0003-2697(92)90288-i. [DOI] [PubMed] [Google Scholar]
- 18.Lupien S, Karp F, Wildung M, Croteau R. Arch Biochem Biophys. 1999;368:181–192. doi: 10.1006/abbi.1999.1298. [DOI] [PubMed] [Google Scholar]
- 19.Davisson V J, Woodside A B, Poulter C D. Methods Enzymol. 1985;110:130–144. doi: 10.1016/s0076-6879(85)10068-6. [DOI] [PubMed] [Google Scholar]
- 20.Croteau R, Alonso W R, Koepp A E, Johnson M A. Arch Biochem Biophys. 1994;309:184–192. doi: 10.1006/abbi.1994.1101. [DOI] [PubMed] [Google Scholar]
- 21.Dixit V M, Laskovics F M, Noall W I, Poulter C D. J Org Chem. 1981;46:1964–1969. [Google Scholar]
- 22.Croteau R, Satterwhite D M. J Chromatogr. 1990;500:349–354. doi: 10.1016/s0021-9673(00)96076-x. [DOI] [PubMed] [Google Scholar]
- 23.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 24.Coligan J E. In: Current Protocols in Protein Science. Coligan J E, Dunn B M, Ploegh H L, Speicher D W, Wingfield P T, editors. Vol. 1. New York: Wiley; 1996. pp. 11.3.1–11.3.13. [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 26.Schenk P M, Baumann S, Mattes R, Steinbiss H. BioTechniques. 1995;19:196–200. [PubMed] [Google Scholar]
- 27.Garfin D E, Bers G. In: Protein Blotting: Methodology, Research and Diagnostic Applications. Baldo B A, Tovey E R, editors. Basel: Karger; 1989. pp. 5–42. [Google Scholar]
- 28.Croteau, R., Wildung, M. R., Burke, C. & Gershenzon, J. (1999) U.S. Patent 5,876,964.
- 29.Croteau R, Gershenzon J. Recent Adv Phytochem. 1994;28:193–229. [Google Scholar]
- 30.von Heijne G, Steppuhn J, Herrmann R G. Eur J Biochem. 1989;180:535–545. doi: 10.1111/j.1432-1033.1989.tb14679.x. [DOI] [PubMed] [Google Scholar]
- 31.Colby S M, Alonso W R, Katahira E J, McGarvey D J, Croteau R. J Biol Chem. 1993;268:23016–23024. [PubMed] [Google Scholar]
- 32.Shimizu N, Koyama T, Ogura K. J Bacteriol. 1998;180:1578–1581. doi: 10.1128/jb.180.6.1578-1581.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koike-Takeshita A, Koyama T, Ogura K. J Biol Chem. 1997;272:12380–12383. doi: 10.1074/jbc.272.19.12380. [DOI] [PubMed] [Google Scholar]