Abstract
Protein kinases play central roles in the regulation of eukaryotic and prokaryotic cell growth, division, and differentiation. The Caulobacter crescentus divL gene encodes a novel bacterial tyrosine kinase essential for cell viability and division. Although the DivL protein is homologous to the ubiquitous bacterial histidine protein kinases (HPKs), it differs from previously studied members of this protein kinase family in that it contains a tyrosine residue (Tyr-550) in the conserved H-box instead of a histidine residue, which is the expected site of autophosphorylation. DivL is autophosphorylated on Tyr-550 in vitro, and this tyrosine residue is essential for cell viability and regulation of the cell division cycle. Purified DivL also catalyzes phosphorylation of CtrA and activates transcription in vitro of the cell cycle-regulated fliF promoter. Suppressor mutations in ctrA bypass the conditional cell division phenotype of cold-sensitive divL mutants, providing genetic evidence that DivL function in cell cycle and developmental regulation is mediated, at least in part, by the global response regulator CtrA. DivL is the only reported HPK homologue whose function has been shown to require autophosphorylation on a tyrosine, and, thus, it represents a new class of kinases within this superfamily of protein kinases.
Protein phosphorylation is a common form of posttranslational modification in biological systems ranging from bacteria to mammals, and protein kinases are central components of the signal transduction pathways controlling cell physiology, growth, and development. These proteins have been classified into two large superfamilies. Serine, threonine, and tyrosine kinases play major roles in cellular regulation in eukaryotes (1, 2). Although members of this kinase family have also been described in prokaryotes (3, 4), their function is not well understood. The second kinase superfamily is made up of the histidine protein kinases (HPKs), which are almost ubiquitous in bacteria. All previously studied HPKs are autophosphorylated on a conserved histidine residue (5–8). Autophosphorylation is followed by phosphate transfer from the histidyl-phosphate residue of the kinase to an invariant aspartate residue of a cognate response regulator (9), which typically functions as a transcription factor. This family of His-Asp proteins also functions in multicomponent phosphorelay pathways in bacteria (10, 11), lower eukaryotes (12), mitochondria (13), and plants (14).
HPK-mediated signal transduction pathways are important in the control of cellular responses to changing environmental conditions. In the dimorphic bacterium Caulobacter crescentus they also play crucial roles in coordinating developmental events with cell cycle progression (15, 16). The Caulobacter stalked cell divides repeatedly to produce the stalked cell and a new swarmer cell, and during the course of the stalked cell cycle the predivisional cell assembles the polar structures required to build a new swarmer cell. This tightly ordered sequence of developmental events includes flagellum biosynthesis and initiation of flagellum rotation, when the cell gains motility. After cell division, development at the flagellated pole of the swarmer cell continues with the assembly of pili, loss of motility, and stalk formation after the flagellum is ejected (17, 18).
Several of the Caulobacter His-Asp signal transduction proteins are thought to regulate developmental events, apparently in response to cell cycle cues. These include the HPK PleC, which is required for gain of motility (19, 20), and a novel response regulator PleD, which is required for flagellar rotation, loss of motility, and stalk formation (21). Other signal transduction proteins are involved directly in cell cycle control. The HPK DivJ (22) and the single-domain response regulator DivK (20) have been identified by genetic analysis as members of a phosphorelay pathway regulating CtrA (23), a global response regulator required for multiple cell cycle events (24). We have now identified another protein kinase, DivL, and report here that this protein is essential for cell viability and also functions in a signal transduction network controlling CtrA activity. DivL represents a new class of proteins within the bacterial protein kinase superfamily in that it is autophosphorylated on a tyrosine residue instead of histidine.
Materials and Methods
Bacterial Strains, Plasmids, and Media.
Escherichia coli strain DH5α was used for propagating plasmids and grown on ML medium (25) supplemented with ampicillin (100 μg/ml) or tetracycline (10 μg/ml) as necessary. All Caulobacter strains were derived from strain CB15 (ATCC 19089) and grown in either peptone–yeast extract medium or M2 minimal glucose medium supplemented with antibiotics as required (25).
Cloning and Sequencing of divL Gene.
A genomic library in cosmid pLAFR1–7 containing random inserts of DNA from Caulobacter (26) was used to identify DNA fragments that complemented the temperature-sensitive divL346 mutation in the strain PC4403 at 37°C. The DNA fragment complementing the divL346 allele in a recA− (27) background was subcloned into pBluescript to generate plasmid pJW35 and sequenced.
Site-Directed Mutagenesis.
The BamHI/HindIII DNA fragment from plasmid pJW35 containing the divL gene was subcloned into pALTER-1 (Promega), creating plasmid pJW362. Single-stranded DNA made from pJW362 was used as a template for mutagenesis. Primer (no. 1), 5′-CGGCAATGTCTCCTTCGAACTGCGCACGCC-3′ and primer (no. 2), 5′-CGTCGGCAATGTCTCCCACGAGCTGCGCAC-3′ were used to generate DivLY550F and DivLY550H mutations, yielding plasmids pJW364 and pJW365, respectively.
Overexpression and Purification of DivL Protein.
To produce the DivL299 protein, a SstI/NcoI DNA fragment from pJW35 encoding the C-terminal 299 aa of DivL was subcloned into pRSET(A) to generate pJW375. To produce DivL286, the DNA fragment encoding C-terminal 286 aa of DivL was amplified by PCR by using the primer (no. 4) 5′-TCGGAGACGTCCAGCCGCACCTCG-3′ and the primer (no. 16) 5′-CGGACGGCGCTAGCCTGATCGCCT-3′. The PCR product was cloned into the NheI/NcoI sites of plasmid pRSET(A) to generate pJW9. Plasmids producing the C-terminal domains of DivL fused in-frame to the poly(His) were overexpressed in the E. coli strain BL21 as described (28). The inclusion bodies formed were solubilized in 6 M guanidine hydrochloride, purified on a nickel chelate column (29), and renatured as described (30). DivJ, DivK, and CtrA proteins were isolated and purified as described previously (20, 23).
Assays of Protein Phosphorylation.
For kinase autophosphorylation assays, 100 pmol of each protein was incubated at 37°C for 30 min in 20 μl of kinase buffer (50 mM Tris⋅HCl, pH 7.5/5 mM MnCl2/5 mM MgCl2) containing 2 pmol [γ-32P]ATP. For transphosphorylation assay of response regulators, the kinase reactions were continued after the addition of 1 μmol of each protein for 20 min. The reaction was stopped by the addition of 5 μl of SDS/PAGE sample buffer containing 50 mM EDTA, subjected to gel electrophoresis on 6% polyacrylamide gel, and visualized by autoradiography. Phospho-CtrA was prepared by incubating 2 μmol CtrA in the kinase buffer containing 20 pmol [γ-32P]ATP and 100 pmol DivL for 30 min at 37°C and purified by passing the reaction mixture over a 10-ml Sephadex G50 column.
Assay of Transcription Activity in Vitro.
The formation of open complexes at the σ73-dependent promoters from Caulobacter fliF gene was measured in single-cycle run-off transcription assays (23) by using reconstituted Caulobacter Eσ73 holoenzyme (31), and the products were analyzed as described previously (23).
Results
DivL Is a Novel Tyrosine Kinase Required for Cell Cycle Regulation.
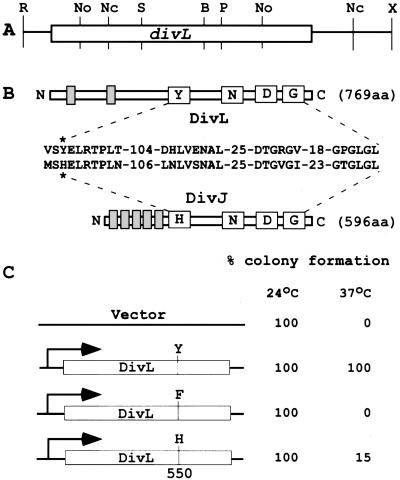
Mutations in divL were isolated as conditional alleles that suppress the motility defect in the Caulobacter HPK gene pleC (32). The temperature-sensitive divL346 strain PC4403 (33) is motile and divides normally at 24°C, but it displays a severe cell division defect at the nonpermissive temperature of 37°C, where it forms extremely long filaments (see Fig. 2 A and B). The cell division phenotype of divL346 was fully complemented in a recA− background by a 3.6-kb EcoRI-XhoI DNA fragment isolated from a Caulobacter cosmid clone (Fig. 1A). This subcloned DNA fragment contains a single, complete ORF encoding a predicted protein of 769 aa with two putative transmembrane sequences near the N terminus (Fig. 1B). The C-terminal region of DivL contains the highly conserved H-, N-, D-, and G-box sequences present in the bacterial HPK superfamily (5), including DivJ from Caulobacter (22) (Fig. 1B). Surprisingly, however, the so-called H-box sequence of DivL (referred to here as the Y-box) contains a tyrosine at position 550 (Tyr-550) instead of the expected histidine (Fig. 1B), which is the normal site of autophosphorylation (5, 9).
Figure 2.
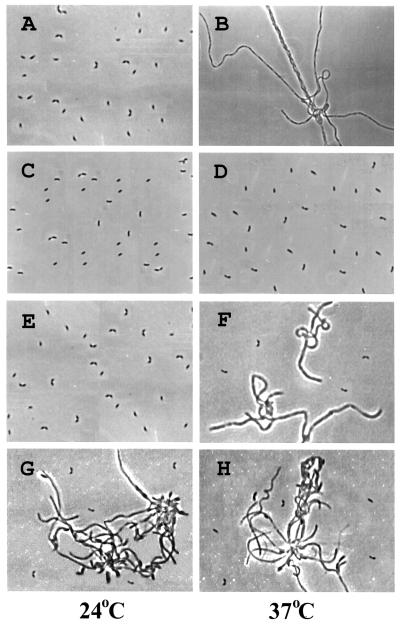
Requirement of Tyr-550 for cell division. Plasmid pRK2L10 (A and B) and its derivatives carrying the DNA encoding DivL (WT) (C and D), DivLY550F (E and F), and DivLY550H (G and H), respectively, were transformed into strain PC4403 at 24°C. Cells were grown in peptone–yeast extract medium overnight at 24°C, diluted 25-fold in duplicates, and incubated for ca. 12 hr at either 24°C (A, C, E, and G) or 37°C (B, D, F, and H). Samples were photographed under a phase-contrast microscope at ×630 magnification (23).
Figure 1.
DivL is a novel tyrosine kinase required for cell division and differentiation. (A) The 3,595-bp EcoRI/XhoI DNA fragment contains an ORF (open box) of 759 aa. R, EcoRI; No, NotI; Nc, NcoI; S, SstI; B, BamHI; P, PstI; and X, XhoI. (B) The conserved catalytic motifs, including the H- (Y- for DivL), N-, D-, and G-box, of the HPK superfamily (5), are for DivL and DivJ (22). An asterisk indicates the presumptive sites of autophosphorylation. (C) The replicating vector and hybrid plasmids encoding either wild-type [DivL (WT)] or mutant DivL proteins DivLY550F or DivLY550H (see text) were transferred by conjugation (25) to a recA-deficient strain carrying the temperature-sensitive divL346 allele. Cells were plated onto minimal medium at both 24°C and 37°C as indicated, and complementation was measured by colony-forming units. Numbers of colonies formed were normalized to 100% for DivL (WT).
We verified that the identification of Tyr-550 in DivL did not result from an artifact of cloning or DNA sequencing by examining independent clones of the 590-bp BamHI/NotI DNA fragment containing divL (Fig. 1A). Each of four clones generated directly from Caulobacter chromosomal DNA by PCR amplification encoded a sequence containing the Tyr-550 residue. Using the same primers, we also amplified 590-bp BamHI/NotI DNA fragments from Caulobacter strains CB2, CB4, CB13, and CV115 and from the related genera Asticcacaulis excentricus and A. biprosthecum, which display a different pattern of polar differentiation (34). The resulting DNA fragments displayed 95–100% sequence identity to the divL sequence from strain CB15 at the amino acid level, and the translated sequences contained a Tyr residue instead of a His residue within the Y-box at the predicted site of autophosphorylation (data not shown). Thus, the novel DivL tyrosine kinase is not singular to Caulobacter strains and may represent a member of a previously uncharacterized subfamily of prokaryotic protein kinases.
The Tyr-550 Residue Is Essential for DivL Function in Colony Formation, Cell Division, and Viability.
To examine the role of Tyr-550 in DivL function we determined the ability of proteins containing either phenylalanine (DivLY550F) or histidine (DivLY550H) at this position to complement a conditional divL mutation. Plasmids encoding either the wild-type DivLWT, DivLY550F, or DivLY550H protein were introduced into a temperature-sensitive divL346, rec− strain, and the resulting merodiploids were assayed for growth and viability by colony formation on plates at 24°C and 37°C. The temperature-sensitive defect of the divL346 strain at 37°C was fully complemented by the wild-type gene, not at all by the plasmid encoding DivLY550F, and only to a limited extent by the plasmid encoding DivLY550H (Fig. 1C).
We also determined the effect of the mutant proteins on cell division by light microscopy (Fig. 2). The wild-type DivL protein complemented the filamentous phenotype of divL346 at 37°C (compare Fig. 2 B and D), but the DivLY550F (Fig. 2F) and DivLY550H (Fig. 2H) proteins did not, although a few, single cells were apparent in these cultures. We noted that some portion of the cells carrying DivLY550H displayed a cell division defect at 24°C (Fig. 2G), suggesting that the mutant DivLY550H protein is expressed in these cells and partially dominant to the mutant protein in strain PC4403 under permissive conditions. Thus, Phe-550 cannot substitute for Tyr-550 in DivL function, and His and Tyr are not fully interchangeable as measured by colony formation and division (see Discussion). We conclude that the Tyr-550 residue in DivL is required for normal growth and cell division in Caulobacter.
The failure of the divL346 mutant to form colonies or divide normally at the nonpermissive temperature suggested that the gene is essential. To examine this possibility we constructed a disruption of the gene. A null mutation of a plasmid-borne copy of divL was generated by replacing a 1.8-kb HincII-SmaI fragment, which includes most of the 2.3-kb coding sequence and the catalytic domain of DivL, with the gentamycin-resistance gene cassette aacC1 (35). This recombinant plasmid was integrated into the chromosome of the wild-type Caulobacter genome by a single cross-over, and a gene disruption was constructed by bacteriophage φCr30-mediated transduction, as described previously in an analysis of divK (20). We obtained divL∷aacC1 (GmR) replacement strains only when a copy of the wild-type divL+ gene was present either on the chromosome or on the plasmid pJZ98. We further confirmed that divL is essential by showing that φCr30 lysates prepared on the divL∷aacC1/pJZ98 strain yielded GmR transductants on the CB15/pJZ98 strain with several copies of the divL+ gene, but not on a wild-type strain carrying only the vector plasmid (data not shown).
DivL Protein Is Autophosphorylated on Tyr-550.
To examine the biochemical activity of DivL we purified the catalytic domain of the protein containing the C-terminal 299 aa (DivL299). This DivL299 polypeptide was autophosphorylated by [γ-32P]ATP (Fig. 3B, lane 1), but not by [α-32P]ATP (data not shown). The 32P label on the DivL protein (Fig. 3B, lane 1) was removed with nonspecific alkaline phosphatase, to which the phosphate was transferred (Fig. 3B, lane 2). These findings demonstrate that DivL is labeled as a result of phosphorylation and not simply the noncovalent binding of ATP or covalent adenylation (37, 38).
Figure 3.
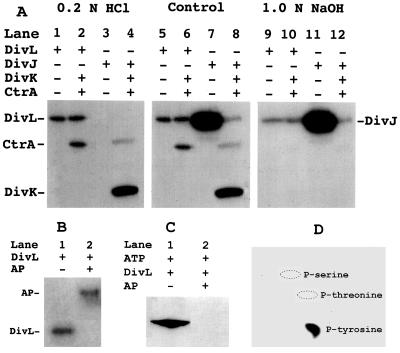
DivL is autophosphorylated on Tyr-550. (A) Stability of phosphoproteins to acid and base treatment. Phosphorylation of DivL, DivJ (21), DivK (21), and CtrA was carried out as described previously (23). The presence or absence of each protein is indicated by “+” or “−,” respectively. After phosphorylation and electrophoresis, three identical gels were treated with 0.2 M HCl (lanes 1–4), 10% methanol (lanes 5–8), or 1.0 M NaOH (lanes 9–12), respectively, as described (10). Phosphoproteins were visualized by autoradiography. (B) Phospho-DivL was treated without (lane 1) or with (lane 2) 0.5 unit of calf thymus alkaline phosphatase (AP) for 30 min at 37°C. (C) Purified DivL was incubated with ATP and treated without (lane 1) or with (lane 2) 1 unit of calf thymus alkaline phosphatase (AP). Samples were separated by SDS/PAGE and detected by Western blot analysis by using an antiphosphotyrosine mAb (HR20) as described by the manufacture (Transduction Laboratories, Lexington, KY). (D) Identification of phosphotyrosine in DivL by two-dimension electrophoresis. 32P-labeled DivL was hydrolyzed in 6 M HCl at 110°C for 2 hr. The hydrolysate then was analyzed on a thin-layer cellulose plate by electrophoresis at pH 1.9 in the first dimension and pH 3.5 in the second dimension, as described (36).
Several lines of evidence indicate that DivL is autophosphorylated on Tyr-550. (i) The 32P label on DivL was acid-resistant (Fig. 3A, lanes 1 and 2) and partially base-resistant (lanes 9 and 10), which is expected for a relatively stable phosphotyrosine (5). In contrast, the 32P label on DivJ (20) was acid-labile (lanes 3 and 4) and base-resistant (lanes 11 and 12). The 32P label on response regulators CtrA and DivK was base-labile (lanes 10 and 12) and acid-resistant, as expected for an aspartyl-phosphate (lanes 2 and 4). (ii) Purified DivL in the presence of ATP was recognized on Western blots by a mAb against phosphotyrosine (Fig. 3C, lane 1), but not when the DivL protein had been treated with alkaline phosphatase (Fig. 3C, lane 2). (iii) The only phosphoamino acid detected by two-dimensional electrophoresis after acid hydrolysis of phosphorylated DivL was phosphotyrosine (Fig. 3D). (iv) A shorter form of DivL containing the C-terminal 246 aa of the protein (DivL246), in which the Tyr-550 is the only tyrosine residue, was autophosphorylated to the same extent as the DivL299 polypeptide examined above (data not shown). We conclude from these results (Fig. 3) and the analysis of mutant DivL protein considered below (see Fig. 5A) that DivL is autophosphorylated specifically on Tyr-550.
Figure 5.
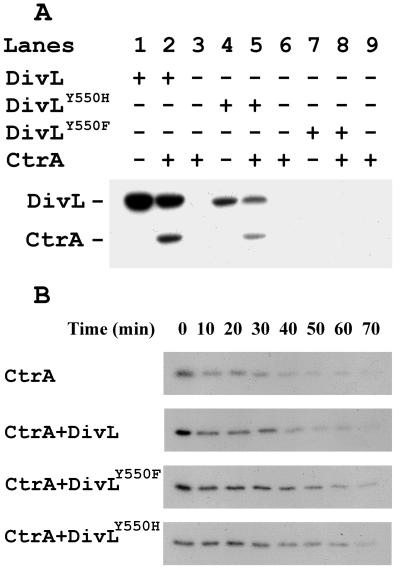
Activities of purified DivL (WT), DivLY550H, and DivLY550H proteins. (A) Kinase activity assays. Experiments were carried out with C-terminal 299 aa of DivL (WT) (lanes 1 and 2), DivLY550H (lanes 4 and 5), or DivLY550F (lanes 7 and 8). The conditions for autophosphorylation of DivL and for phosphorylation of CtrA by DivL are the same as in Figs. 3 and 4. (B) Phosphatase activity assay. The CtrA protein was 32P-labeled by the DivL (WT) protein and purified as described in Materials and Methods. The dephosphorylation of 32P-CtrA was measured in the presence or absence of the DivL protein (200 pmol) as indicated. The reaction mixtures were incubated at 25°C, and samples (15 μl) were withdrawn at the time intervals indicated.
CtrA Is a Downstream Target of the DivL Kinase both in Vitro and in Vivo.
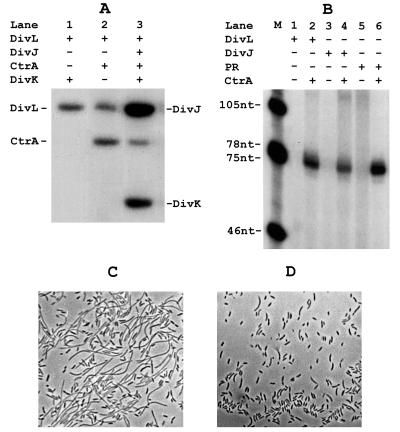
Strains with conditional mutations in divL, like those with mutant alleles of one of the essential response regulator genes divK (20) or ctrA (24), are blocked early in cell division and form long, unpinched filamentous cells (Fig. 2B). This similarity in phenotype suggested that the DivL kinase could function to phosphorylate CtrA either directly or indirectly through DivK in the previously described phosphorelay pathway DivJ → DivK → X → CtrA (23). To determine whether DivK or CtrA is a downstream target of the DivL kinase, we first assayed the ability of DivL to phosphorylate the two response regulators. As shown above (Fig. 3A, lane 5), DivL is autophosphorylated in the presence of [γ-32P]ATP, and it catalyzes the phosphorylation of CtrA when it was also present (Fig. 4A, lane 2). However, DivL did not catalyze the phosphorylation of DivK under the same conditions (Fig. 4A, lane 1). Consistent with this specificity, when both CtrA and DivK were present, DivL phosphorylated CtrA exclusively (Fig. 3A, lane 6), whereas DivJ predominantly phosphorylated DivK in the presence of the two response regulators (Fig. 3A, lane 8; ref. 23). Thus, DivL is unlikely to regulate cell division through the DivK-dependent phosphorelay by the phosphorylation of DivK (see Discussion).
Figure 4.
Cell cycle regulation by DivL is mediated by CtrA. (A) Phosphorylation of DivK and CtrA by DivJ286 and DivL299. The presence or absence of each protein was indicated as “+” or “−,” respectively. Results of autophosphorylation and phosphotransfer were visualized by autoradiography. (B) Activation of CtrA by DivL kinase. In vitro run-off transcription from the class II flagellar gene fliF promoter was carried out in the presence of reconstituted Caulobacter RNA polymerase containing the sigma factor σ73 (final concentration at 1 μM) as described previously (23). Final concentration of proteins were 10 μM for CtrA and 1 μM for DivL (lanes 1 and 2) and DivJ (lanes 3 and 4). The low-molecular-weight phosphoryl group donor phosphoramidate (PR) was added to the reactions (lanes 5 and 6) at the final concentration of 1 mg/ml. (C) Phase-contrast photomicrograph of cold-sensitive strain PC3407 (divL342; ref 32) at 24°C. (D) Suppression of divL342 by the ctrA allele sokA301 in strain PC3303 (divL342 sokA301 zzz471∷Tn5) at 24°C.
We also examined the possibility that DivL catalyzes phosphotransfer from DivK to CtrA in the proposed phosphorelay pathway (23). When DivL was added to a reaction containing DivJ, DivK, and CtrA (see Fig. 3A, lane 8), most of the transferred phosphate accumulated in DivK (Fig. 4A, lane 3). Moreover, we were unable to detect transfer of radiolabeled phosphate to CtrA from purified phospho-DivK either in the presence or absence of the DivL protein (data not shown). We conclude that DivL does not act as the unidentified phosphotransfer protein X.
As a genetic approach to determining whether CtrA is a downstream target of DivL, we examined whether the sokA301 (suppressor of divK) allele, which was isolated originally as a divK suppressor mapping to ctrA (23), would also bypass the cell division defect of a divL mutant. The filamentous phenotype of the cold-sensitive divL342 mutation (Fig. 4C; ref. 32) was suppressed effectively by sokA301 (Fig. 4D). This result provides direct genetic evidence that cell cycle regulation by the tyrosine kinase DivL is mediated, at least in part, by CtrA.
Phosphorylation of CtrA by DivL Activates Transcription of a Developmental Gene.
Transcription from the class II fliF flagellar gene promoter by reconstituted RNA polymerase containing σ73 (Eσ73) requires activation by phospho-CtrA (23). We have used this in vitro transcription assay to determine whether phosphorylation of CtrA by the DivL kinase also activates this response regulator. Purified DivL in the presence of CtrA protein and ATP activated transcription from the fliF promoter by Eσ73 and produced the expected 76-nt run-off transcript (Fig. 4B, lane 2; ref. 23). The DivJ kinase (lane 4) or the low-molecular-weight phosphoryl group donor phosphoramidate (lane 6) also stimulated production of the 76-nt transcript. Recognition of the fliF promoter by Eσ73 was completely dependent on CtrA, regardless of whether DivL (Fig. 4B, lane 1), DivJ (lane 3), or phosphoramidate (lane 5) was present. Thus, the DivL tyrosine kinase is equivalent to the histidine kinase DivJ in its ability to activate CtrA in vitro.
Activity of the DivLY550H and DivLY550F Proteins.
Because mutations at residue Tyr-550 of DivL failed to complement the severe defects of divL alleles in cell division in vivo (Figs. 1C and 2), we examined the activity of the mutant DivLY550H and DivLY550F proteins in vitro. DivLY550H was autophosphorylated weakly at a level ca. 6-fold lower than that of wild-type DivL protein, as judged by PhosphorImager analysis (compare Fig. 5A, lanes 1 and 4). The DivLY550H protein was also ca. 6-fold less active in the phosphorylation of CtrA (lanes 5) than the counterpart DivLWT protein (lane 2). In similar experiments with the mutant DivLY550F protein, no autophosphorylation (lanes 7 and 8) or phosphorylation of CtrA could be detected (lane 8). As expected, purified CtrA alone was not autophosphorylated under our assay conditions (lanes 3, 6, and 9).
We also considered the possibility that a phospho-CtrA phosphatase activity of DivL could account for the regulation of cell division by this HPK. Over a 70-min time course, the DivLWT protein had little or no effect on the stability of phosphorylated CtrA (Fig. 5B), which displayed a half-life of ca. 30 min. Essentially the same result was obtained for the DivLY550H and DivLY550F proteins (Fig. 5B). We draw two conclusions from these results. First, the failure of the two mutant proteins to complement fully cell division defects of divL alleles (compare Figs. 1C and 2 F and H) is not explained by an enhanced phospho-CtrA phosphatase activity, and, second, if DivL functions directly to regulate CtrA activity, it acts primarily as a kinase (see Discussion).
Discussion
The experiments described in this report lead to two important findings. The first is the identification of DivL as an essential protein kinase required for the regulation of cell division and developmental events during Caulobacter differentiation. Both genetic and biochemical results indicate that DivL functions in cell cycle and developmental regulation by controlling phosphorylation of the global response regulator CtrA. The second finding is that DivL represents a new class of the HPK superfamily that contains a tyrosine residue in the conserved H-box motif instead of histidine, which is the normal site of autophosphorylation. Our results show that Tyr-550 is the site of DivL autophosphorylation and that this residue is required for full DivL function in vivo (Figs. 1C and 2) and in vitro (Fig. 5A).
Activation of protein kinases by autophosphorylation on tyrosine is a ubiquitous and essential mechanism for regulation of signal transduction pathways in eukaryotic cells. Examples include the activation of membrane sensor kinases by interaction with growth factors (39, 40) and tyrosine autophosphorylation of the products of various protooncogenes (41). In the instances in which tyrosine phosphorylation of purified protein has been reported in bacteria (4), the proteins display sequence similarity to the eukaryotic serine/threonine/tyrosine kinase family (42–44).
Phosphorylation of sensor protein kinases in prokaryotic cells typically occurs on histidine residues in response to a variety of environmental conditions. The phosphate is transferred to the aspartyl residue of a cognate response regulator (5, 45, 46). In Caulobacter, signal transduction by these His-Asp proteins plays a central role in the regulation of cell division and differentiation, presumably in response to internal cues that track cell cycle progression (15, 16). However, DivL is the only reported member of the histidine kinase superfamily whose function has been shown to require autophosphorylation on a tyrosine residue. The identification of DivL homologues with tyrosine at the presumptive site of phosphorylation in several strains of Caulobacter and related genera suggests that DivL represents a new protein kinase subfamily (Fig. 1).
The mechanistic significance of the Tyr-550 residue in DivL is unclear, but the histidine and tyrosine residues at the site of autophosphorylation are not functionally equivalent. The DivLY550H mutant protein did not fully complement the division phenotype of a conditional divL allele (Fig. 2H) and showed only residual activity in the autophosphorylation assay (Fig. 5A). In a related study, an H243Y substitution at the active site of the E. coli EnvZ HPK has been shown to abolish kinase activity (47). Phosphotyrosine is known to be a high-energy phosphate bond (6), and phosphotransfer would seem to be the most probable mechanism of CtrA phosphorylation, but other mechanisms are possible. One example is illustrated by the Bacillus subtilis SpoIIAB protein. This antisigma factor is similar in sequence to members of the HPK family, but it does not contain the conserved H-box sequence and is not autophosphorylated. Instead, SpoIIAB is thought to act as a serine protein kinase in the phosphorylation of SpoIIAA (48, 49). This serine kinase differs from those described in the Gram-negative bacterium Myxococcus xanthus that are members of the eukaryotic Ser/Thr kinase family (50).
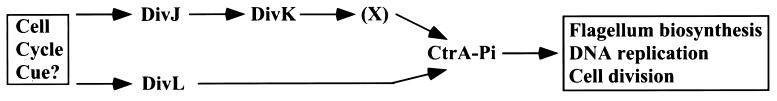
Several essential His-Asp signal transduction proteins now have been identified in Caulobacter and shown to play key roles in cell cycle and developmental regulation. One of these is CtrA, a global response regulator that coordinates multiple cell cycle-dependent events, including initiation of DNA replication (51), cell division (52), and flagellum biosynthesis (23, 24). Another two-component protein is DivK, an essential, single-domain response regulator that is also required for cell cycle control, as well as the regulation of cell motility (20). A genetic analysis of divK mutations and the results of phosphotransfer assays led to the conclusion that these proteins are members of a multicomponent phosphorelay pathway, DivJ → DivK → X → CtrA, in which X is an unidentified phosphotransferase protein (23).
Our biochemical studies (Figs. 4A and 5) and the ability of a ctrA allele (sokA301) to suppress the cell division defects of conditional divL mutants (Fig. 4D) indicate that the tyrosine kinase DivL regulates CtrA independently of DivJ-DivK phosphorelay. It thus seems likely that DivL functions to control CtrA activity either directly or indirectly via a second pathway (DivL → CtrA) and, as proposed in Fig. 6, two signal transduction pathways form part of an essential signaling network coordinating Caulobacter cell cycle regulation.
Figure 6.
Model for an essential signal transduction network regulating cell cycle events in Caulobacter. The cell cycle-regulated activity of CtrA is modulated by a signal transduction network, including the DivJ-dependent phosphorelay pathway described previously (23) and directly or indirectly by the DivL kinase described in this paper. As demonstrated for transcription of class II flagellar genes in vitro (23), we assume that CtrA is activated by phosphorylation.
While this manuscript was in preparation, the report of another CtrA kinase, CckA, appeared (53). It is not clear how this HPK functions in concert with the signal transduction pathways summarized in Fig. 6. CtrA is, however, a global transcription factor like the B. subtilis Spo0A protein, which responds to several protein kinases and other signal transduction components to control sporulation (54, 55). Given the complex sequence of cell cycle and developmental events that must be coordinated in Caulobacter, it is not surprising that multiple signal transduction pathways are necessary for the regulation of CtrA activity.
Acknowledgments
We thank Jean Poindexter for the Asticcacaulis spp. strains used in these studies and J. Stock and T. Silhavy for valuable suggestions and comments. This work was supported by Public Health Service Grants GM22299 and GM58794 from the National Institutes of Health to A.N.
Abbreviation
- HPK
histidine protein kinase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF083422).
References
- 1.Hanks K H, Quinn A M, Hunter T. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- 2.Hunter T. Cell. 1995;80:225–236. doi: 10.1016/0092-8674(95)90405-0. [DOI] [PubMed] [Google Scholar]
- 3.Cozzone A J. J Cell Biochem. 1993;5:7–13. doi: 10.1002/jcb.240510103. [DOI] [PubMed] [Google Scholar]
- 4.Zhang C-C. Mol Microbiol. 1996;20:9–15. doi: 10.1111/j.1365-2958.1996.tb02483.x. [DOI] [PubMed] [Google Scholar]
- 5.Stock J B, Ninfa A J, Stock A M. Microbiol Rev. 1989;53:450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stock J B, Stock A M, Mottonen J M. Nature (London) 1990;344:395–400. doi: 10.1038/344395a0. [DOI] [PubMed] [Google Scholar]
- 7.Alex L A, Simon M I. Trends Genet. 1994;10:133–138. doi: 10.1016/0168-9525(94)90215-1. [DOI] [PubMed] [Google Scholar]
- 8.Grebe, T. & Stock, J. (1999) Adv. Microb. Physiol.41, in press. [DOI] [PubMed]
- 9.Swanson R V, Alex L A, Simon M I. Trends Biochem Sci. 1994;19:485–490. doi: 10.1016/0968-0004(94)90135-x. [DOI] [PubMed] [Google Scholar]
- 10.Burbulys D, Trach K A, Hoch J A. Cell. 1991;64:545–552. doi: 10.1016/0092-8674(91)90238-t. [DOI] [PubMed] [Google Scholar]
- 11.Appleby J L, Parkinson J S, Bourret R B. Cell. 1996;86:845–848. doi: 10.1016/s0092-8674(00)80158-0. [DOI] [PubMed] [Google Scholar]
- 12.Wurgler-Murphy S M, Saito H. Trends Biochem Sci. 1997;22:172–176. doi: 10.1016/s0968-0004(97)01036-0. [DOI] [PubMed] [Google Scholar]
- 13.Popov K M, Hawes J W, Harris R A. Adv Second Messenger Phosphoprotein Res. 1997;31:105–111. [PubMed] [Google Scholar]
- 14.Miyata S, Urao T, Yamaguchi-Shinozaki K, Shinozaki K. FEBS Lett. 1998;437:11–14. doi: 10.1016/s0014-5793(98)01188-0. [DOI] [PubMed] [Google Scholar]
- 15.Ohta N, Newton A. Trends Microbiol. 1996;4:326–332. doi: 10.1016/0966-842x(96)10050-0. [DOI] [PubMed] [Google Scholar]
- 16.Ohta N, Grebe T W, Newton A. In: Prokaryotic Development. Brun Y, Shimkets L, editors. Washington, DC: Am. Soc. Microbiol.; 1999. , in press. [Google Scholar]
- 17.Newton A, Ohta N. Annu Rev Microbiol. 1990;44:689–719. doi: 10.1146/annurev.mi.44.100190.003353. [DOI] [PubMed] [Google Scholar]
- 18.Brun Y V, Marczynski G, Shapiro L. Annu Rev Biochem. 1994;63:419–450. doi: 10.1146/annurev.bi.63.070194.002223. [DOI] [PubMed] [Google Scholar]
- 19.Wang S P, Sharma P L, Schoenlein P V, Ely B. Proc Natl Acad Sci. 1993;90:630–634. doi: 10.1073/pnas.90.2.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hecht G B, Lane T, Ohta N, Sommer J N, Newton A. EMBO J. 1995;14:3915–3924. doi: 10.1002/j.1460-2075.1995.tb00063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hecht G B, Newton A. J Bacteriol. 1995;177:6223–6229. doi: 10.1128/jb.177.21.6223-6229.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohta N, Lane T, Ninfa E G, Sommer J M, Newton A. Proc Natl Acad Sci. 1992;89:10297–10301. doi: 10.1073/pnas.89.21.10297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu J, Ohta N, Newton A. Proc Natl Acad Sci. 1998;95:1443–1448. doi: 10.1073/pnas.95.4.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quon K C, Marczynski G T, Shapiro L. Cell. 1996;84:83–93. doi: 10.1016/s0092-8674(00)80995-2. [DOI] [PubMed] [Google Scholar]
- 25.Ohta N, Swanson E, Ely B, Newton A. J Bacteriol. 1984;158:897–904. doi: 10.1128/jb.158.3.897-904.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sommer J M. Ph.D. thesis. Princeton: Princeton Univ.; 1988. [Google Scholar]
- 27.Bender R A. Mol Gen Genet. 1985;197:399–402. doi: 10.1007/BF00329935. [DOI] [PubMed] [Google Scholar]
- 28.Tabor S, Richardson C. Proc Natl Acad Sci USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hochuli E, Dobeli H, Schacher A. J Chromatogr. 1995;411:177–184. doi: 10.1016/s0021-9673(00)93969-4. [DOI] [PubMed] [Google Scholar]
- 30.McCleary W R, Zusman D R. J Bacteriol. 1990;172:6661–6668. doi: 10.1128/jb.172.12.6661-6668.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu J, Ohta N, Benson A K, Ninfa A J, Newton A. J Biol Chem. 1997;272:21558–21564. doi: 10.1074/jbc.272.34.21558. [DOI] [PubMed] [Google Scholar]
- 32.Sommer J M, Newton A. Genetics. 1991;129:623–630. doi: 10.1093/genetics/129.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hecht G B. Ph.D. thesis. Princeton: Princeton Univ.; 1995. [Google Scholar]
- 34.Poindexter J S. Bacteriol Rev. 1964;28:231–295. doi: 10.1128/br.28.3.231-295.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schweizer H D. BioTechniques. 1993;15:831–834. [PubMed] [Google Scholar]
- 36.Boyle W J, Geer P V D, Hunter T. Methods Enzymol. 1977;201:110–149. doi: 10.1016/0076-6879(91)01013-r. [DOI] [PubMed] [Google Scholar]
- 37.Cortay J C, Duclos B, Cozzone A J. J Mol Biol. 1986;187:305–308. doi: 10.1016/0022-2836(86)90236-6. [DOI] [PubMed] [Google Scholar]
- 38.Foster R, Thorner J, Martin G S. J Bacteriol. 1989;171:272–279. doi: 10.1128/jb.171.1.272-279.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fantl W J, Johnson D E, Williams L T. Annu Rev Biochem. 1993;62:453–481. doi: 10.1146/annurev.bi.62.070193.002321. [DOI] [PubMed] [Google Scholar]
- 40.Sun H, Tonks N D. Trends Biochem Sci. 1994;19:480–485. doi: 10.1016/0968-0004(94)90134-1. [DOI] [PubMed] [Google Scholar]
- 41.Cantley L C, Auger K R, Carpenter C, Duckworth B, Graziani A, Kapeller R, Soltoff S. Cell. 1991;64:281–302. doi: 10.1016/0092-8674(91)90639-g. [DOI] [PubMed] [Google Scholar]
- 42.Ostrovsky P C, Maloy S. Genes Dev. 1995;9:2034–2041. doi: 10.1101/gad.9.16.2034. [DOI] [PubMed] [Google Scholar]
- 43.Grangeasse C, doublet P, Vaganay E, Vincent C, Deleage G, Diclos B, Cozzone A J. Gene. 1997;204:259–265. doi: 10.1016/s0378-1119(97)00554-4. [DOI] [PubMed] [Google Scholar]
- 44.Freestone P, Trinei M, Clarke S C, Nystrom T, Norris V. J Mol Biol. 1998;279:1045–1051. doi: 10.1006/jmbi.1998.1836. [DOI] [PubMed] [Google Scholar]
- 45.Parkinson J S. Cell. 1993;73:857–871. doi: 10.1016/0092-8674(93)90267-t. [DOI] [PubMed] [Google Scholar]
- 46.Hoch J A, Silhavy T J. Two-Component Signal Transduction. Washington, DC: Am. Soc. Microbiol.; 1995. [Google Scholar]
- 47.Hsing W, Silhavy T J. J Bacteriol. 1997;179:3729–3735. doi: 10.1128/jb.179.11.3729-3735.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Duncan L, Losick R. Proc Natl Acad Sci USA. 1993;90:2325–2329. doi: 10.1073/pnas.90.6.2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Errington J. Microbiol Rev. 1993;57:1–33. doi: 10.1128/mr.57.1.1-33.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Udo H, Munoz-Dorado J, Inouye M, Inouye S. Genes Dev. 1995;9:972–983. doi: 10.1101/gad.9.8.972. [DOI] [PubMed] [Google Scholar]
- 51.Quon K C, Yang B, Domian I J, Shapiro L, Marczynski G T. Proc Natl Acad Sci USA. 1998;95:120–125. doi: 10.1073/pnas.95.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kelly A J, Sackett M J, Din N, Quardokus E, Brun Y V. Genes Dev. 1998;12:880–893. doi: 10.1101/gad.12.6.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jacobs C, Domian I J, Maddock J R, Shapiro L. Cell. 1999;97:111–120. doi: 10.1016/s0092-8674(00)80719-9. [DOI] [PubMed] [Google Scholar]
- 54.Hoch J A. In: Two-Component Signal Transduction. Hoch J A, Silhavy T J, editors. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 129–144. [Google Scholar]
- 55.Grossman A D. Annu Rev Genetics. 1995;29:477–508. doi: 10.1146/annurev.ge.29.120195.002401. [DOI] [PubMed] [Google Scholar]