Abstract
Protein–protein interacting surfaces are usually large and intricate, making the rational design of small mimetics of these interfaces a daunting problem. On the basis of a structural similarity between the CDR2-like loop of CD4 and the β-hairpin region of a short scorpion toxin, scyllatoxin, we transferred the side chains of nine residues of CD4, central in the binding to HIV-1 envelope glycoprotein (gp120), to a structurally homologous region of the scorpion toxin scaffold. In competition experiments, the resulting 27-amino acid miniprotein inhibited binding of CD4 to gp120 with a 40 μM IC50. Structural analysis by NMR showed that both the backbone of the chimeric β-hairpin and the introduced side chains adopted conformations similar to those of the parent CD4. Systematic single mutations suggested that most CD4 residues from the CDR2-like loop were reproduced in the miniprotein, including the critical Phe-43. The structural and functional analysis performed suggested five additional mutations that, once incorporated in the miniprotein, increased its affinity for gp120 by 100-fold to an IC50 of 0.1–1.0 μM, depending on viral strains. The resulting mini-CD4 inhibited infection of CD4+ cells by different virus isolates. Thus, core regions of large protein–protein interfaces can be reproduced in miniprotein scaffolds, offering possibilities for the development of inhibitors of protein–protein interactions that may represent useful tools in biology and in drug discovery.
The interaction of the gp120 envelope glycoprotein of HIV-1 with CD4 represents the initial step of virus entry into its target cells (1, 2) and triggers gp120 conformational changes that increase binding affinity for CCR5 (3, 4), the chemokine receptor used as coreceptor by most HIV-1 primary isolates (5, 6). X-ray structure analysis (7) of gp120 in complex with CD4 and the Fab of a neutralizing monoclonal antibody has revealed that CD4 binds to a large (800-Å2) depression on gp120 by using a 742-Å2 surface involving 22 residues in the span of amino acids 25–64 of the CD4 D1 domain and centered on the CDR2-like 36–47 loop (Fig. 1A). Phe-43, the side chain of which protrudes from the 36–47 loop and plugs the entrance of a deep cavity in gp120, Arg-59 (at the end of strand D), which makes multiple contacts with gp120, and strand C“, which interacts with strand β15 of gp120 in a β-sheet alignment, are critical elements in the CD4 interface (7). The involvement of these CD4 structural elements in gp120 binding is in agreement with previous mutational analyses that identified most residues of CDR2-like loop as important recognition elements (8–12).
Figure 1.
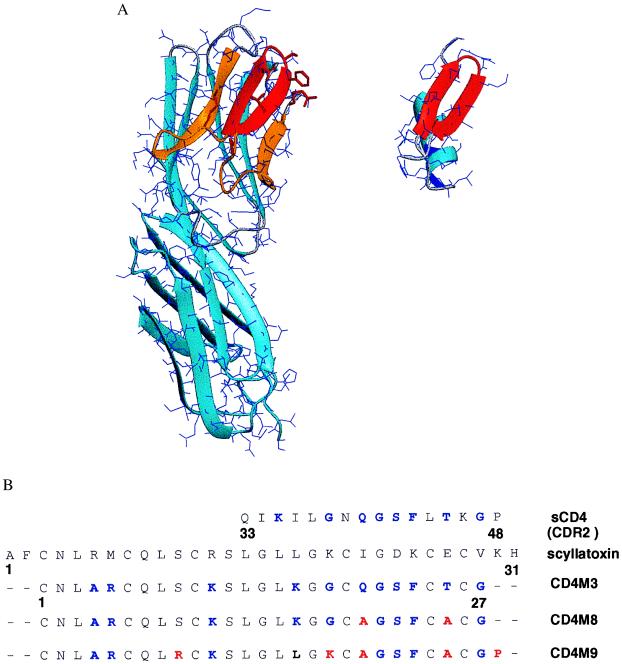
Structural comparison of CD4 and the scorpion scyllatoxin. (A) Three-dimensional structure of CD4 D1–D2 domains (Left; PDB code 1cdh; ref. 35) and of scyllatoxin (Right; PDB code 1scy; ref. 22). Backbone traces are in light blue ribbons (27). The 25–64 region of CD4, binding to gp120 (7), is in orange, and the 36–47 CDR2-like loop, corresponding to the C’C“ β-hairpin, is in red; the 18–29 β-hairpin of scyllatoxin, structurally similar to the CDR2-like loop, is also in red. CD4 side chains of the CDR2-like loop and R59, at the end of strand D, transferred to scyllatoxin, appear as red sticks. (B) Sequence alignment of the CDR2-like loop of CD4, scyllatoxin, CD4M3, double-mutant CD4M8, and quintuple-mutant CD4M9. Transferred amino acid residues of CD4 and amino acid changes of the engineered scaffolds are in blue; the additional changes that increase affinity for gp120 and antiviral activity are in red.
The large size and complexity of the CD4 surface interacting with gp120 (7) explains the difficulties encountered in deriving gp120 ligands based on CD4 structure that could effectively block virus entry. Peptides derived from the envelope glycoprotein gp41 (13) have been shown to trap gp41 in a fusion-noncompetent conformation after gp120 has interacted with cellular receptors (14), thereby preventing infection. Synthesis of constrained peptides based on CDR-like loops (15–17) or selection from linear and cyclic peptide libraries (18), however, failed to identify an effective inhibitor of CD4-gp120 interaction that was able to prevent virus attachment to cells and infection.
As a means to reproduce, on a small molecular system, the native-like conformation and binding function of the CDR2-like CD4 domain—including Arg-59—we applied a strategy already used in other systems (19, 20) and transferred the critical functional elements of the CD4-binding site to a structurally compatible small scaffold. In a preliminary study (21), we showed that transfer of the solvent-exposed residues of the CDR2-like loop to the structurally similar loop of the scorpion charybdotoxin scaffold produced a low-affinity inhibitor of CD4-gp120 interaction. For this study, we selected a different scorpion toxin scaffold, scyllatoxin (22), which could more favorably reproduce the CDR2-like loop of CD4. We show that transfer of the residues of the CD4 36–47 β-hairpin, together with Arg-59, to equivalent regions of this stable and permissive miniprotein scaffold allows for the reproduction of a substantial gp120-binding activity in a miniprotein system. Structural and functional characterization of the first miniprotein suggested sequence modifications that, once incorporated, substantially improved its apparent affinity for gp120. This improved chimeric miniprotein prevented HIV-1 attachment to cells and, consequently, infection. We suggest that the strategy used for this study may have a general application in protein–protein interaction systems and allows for the production of chimeric miniproteins as unique tools in biology and in drug discovery.
Materials and Methods
Peptide Synthesis.
CD4M3, its derivatives, and other peptides were synthesized on an Applied Biosystems Synthesizer (model 433; Perkin–Elmer) by the solid-phase method by using fluorenylmethyloxycarbonyl-protected amino acids and 2-(1-H-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate coupling (21). Ala and Val mutants were simultaneously synthesized on an Advanced ChemTech 357 Multisynthesizer. Disulfide bonds were formed with the peptides dissolved at 0.1 mg/ml in 50 mM phosphate buffer (pH 7.8) in the presence of 5 mM/0.5 mM oxidized/reduced glutathione. Synthetic peptides were purified by reverse-phase HPLC, and their identity was verified by amino acid analysis and electrospray mass spectrometry.
Structure Determination.
Structures were determined from two-dimensional homonuclear NMR experiments [nuclear Overhauser effect (NOE) spectroscopy; total correlation spectroscopy; and double quantum filtered correlated spectroscopy] at 600 MHz (Bruker AMX600) collected on a 4 mM protein sample (pH 3.5 at 25°C). The standard strategy described by Wüthrich (23) was used for assignment. The data sets were processed with gifa software (24) utilities. NOE intensities used for the structure calculations were obtained from the NOE spectrum recorded with a 150-ms mixing time on the fully protonated sample and checked for spin diffusion on spectra recorded at shorter mixing times (50 and 100 ms). NH-CαH and CαH-CβH coupling constants used to define angular restraints were measured in the double quantum filtered correlated spectra. To compensate for the overestimation of the 3JNH-Hα caused by the broad line width of the antiphase multiplets, we used the method previously described by Ludvingsen et al. (25).
The 239 collected NOEs were partitioned into five categories of intensities that were converted into distances ranging from a common lower limit of 1.8 Å (the sum of the van der Waals radii) to upper limits of 2.4 Å, 2.8 Å, 3.6 Å, 4.4 Å, and 4.8 Å; 40 three-dimensional structures were generated from these distance restraints as well as 46 additional angular restraints (φ and χ1), and the usual distance restraints were used to enforce the three disulfide bridges, i.e., ranges of 2.0–2.1 Å for d(Sγ,Sγ); 3.0–3.1 Å for d(Cβ,Sγ); and 3.5–4.5 Å for d(Cβ,Cβ) by using the standard force-field parameters of x-plor 3.1 (26). Of the 40 structures generated, 20 had no violation of NOE or angular restraints exceeding 0.2 Å and 5°, respectively, and were thus selected for analysis.
Structure Analysis and Modeling.
All structures were displayed, analyzed, and compared on a Silicon Graphics 4D/25 station by using the sybyl package (Tripos Associates, St. Louis). Molecular graphics were produced with molmol (27).
Competitive Binding Assays.
Inhibition of rgp120-CD4 interaction was measured in a competitive ELISA as described (21). Briefly, 250 ng per well of soluble recombinant CD4 (sCD4 D1–D4 domains; a gift of R. W. Sweet, SmithKline Beecham, King of Prussia, PA) was coated overnight at 4°C in 96-well plates (Maxisorb, Nunc); 80 ng per well of rgp120LAI (Intracel, Issaquah, WA) was then added, followed by addition of different concentrations of soluble competitors, anti-gp120 NEA mAb (DuPont/NEN), a goat anti-mouse peroxidase-conjugated antibody (Jackson ImmunoResearch), and the 3,3′,5,5′-tetramethylbenzidine substrate (Sigma) for revelation. Inhibition of binding was calculated from the OD at 450 nm by using the formula: percentage of inhibition = 100 × (ODgp120 − ODgp120+comp)/ODgp120. Results are means of triplicate experiments. Similar results were obtained with a different ELISA with plates coated with antibody D7324 (Aalto Bio Reagents, Dublin) to adsorb rgp120, followed by addition of sCD4 and the competitors, anti-CD4 mAb L120 (a gift of the Centralized Facility for AIDS Reagents, National Institute for Biological Standards and Control, Herts, U.K.) and a goat anti-mouse peroxidase-conjugated antibody for revelation (28). The other rgp120s tested were from strains IIIB (Intracel); JR-FL, BaL (a gift of R. W. Sweet); and MN, W61D (a gift of the Centralized Facility for AIDS Reagents, National Institute for Biological Standards and Control, Herts, U.K.)
HIV-1 Infection of HeLa Cells and Peripheral Blood Lymphocytes (PBL).
HeLa/CD4/CCR5/LacZ cells (4,500 cells per well; a gift of P. Charneau, Institut Pasteur, Paris, France; ref. 29) were cultured for 24 h in DMEM/10% (vol/vol) FCS (Roche Molecular Biochemicals)/500 μg/ml G418 (geneticin; GIBCO/BRL)/1 μg/ml puromycin (Sigma). Washed cells were infected with HIV in the presence or absence of different concentrations of CD4M9 or sCD4 (gifts of D. Klatzmann, Pitié-Salpêtrière Hospital). After 24 h, cells were washed and cultured for another 48 h before measuring β-galactosidase activity by a chemiluminescent assay according to the manufacturer’s instructions (Roche Molecular Biochemicals). The HIV-1 strains, used at 500 tissue culture ID50, were HIV-1LAI (Diagnostics Pasteur, Marne la Coquette, France), HIV-1BaL (a gift of B. Asjo, Gade Institute, Bergen University, Bergen, Norway), and HIV-1Ada (a gift of the National Institutes of Health AIDS Reagent Program, Bethesda, MD).
PBL from healthy HIV-negative donors (Pitié-Salpêtrière blood bank) were infected with HIV as described (30), after being stimulated with phytohemagglutinin (Difco) for 3 days in RPMI medium 1640/10% (vol/vol) FCS (R10)/50 μg/ml penicillin/streptomycin/2 mM glutamine (GIBCO/BRL). Different concentrations of CD4M9 or control sCD4 were incubated for 30 min with 100 tissue culture ID50 of HIV-1LAI or HIV-1BaL, which were then added to 5 × 105 PBL in IL-2-supplemented (10 units/ml; Roche Molecular Biochemicals) R10 for 3 h at 37°C. All compounds were maintained at initial concentrations during the whole culture period. Virus production was assessed by measuring p24 in culture supernatants by ELISA according to the manufacturer’s instructions (Coulter).
Results
A β-hairpin motif, structurally equivalent to the CD4 CDR2-like loop and central in gp120 binding, is present in short scorpion toxins (21). These small proteins, presenting an evolutionary conserved α/β-structural motif stabilized by three internal disulfide bridges (31, 32), seem particularly attractive as host structures to present exogenous sequences in a well defined conformation because of the proteins’ permissiveness in sequence mutations and stability, even after multiple sequence mutations (19, 20, 33). In a previous study, we transferred the solvent-exposed residues of the CDR2-like loop of CD4 on the 25–37 β-hairpin of the scorpion charybdotoxin triple-stranded β-sheet (21). The toxin scaffold was truncated of the N-terminal β-strand, which did not have a structural equivalent in CD4 and could therefore interfere with gp120 binding. This construction inhibited CD4-gp120 interaction in the 10−4–10−5 M range (21). Scyllatoxin (ref. 22; Fig. 1A) is a 31-residue scorpion toxin, presenting a double-stranded β-sheet 18–29 that can superimpose its backbone atoms on those of the CDR2-like loop 36–47 of CD4 with an rms deviation of only 1.10 Å. As compared with charybdotoxin, this shorter scaffold has no N-terminal β-strand and presents a shorter loop joining the helix to the first β-strand; both differences allow full access of the toxin β-hairpin to a large-molecule probe and make it a better host structure for the CD4 C′C" β-hairpin, which in the complex with gp120 is in a β-sheet alignment with a β-strand of gp120 (7). Furthermore, the two β-hairpins present a similar spatial orientation in their solvent-exposed side chains, and the scyllatoxin Cys-21, Cys-26, and Cys-28 side chains, which are engaged in disulfide bonds, are oriented similarly to the buried CD4 side chains Asn-39, Leu-44, and Lys-46, respectively. Based on these structural considerations, we modeled a chimeric miniprotein that preserved the structurally important Cys residues of the scaffold but included the functional Gly-38, Gln-40 to Phe-43, Thr-45, and Gly-47 of CD4 in structurally equivalent regions of the scyllatoxin β-hairpin (Fig. 1B). To increase the structural mimicry with CD4 further, an Arg and a Lys were included at positions 7 and 18, topologically equivalent to the functional Arg-59 and Lys-35 of CD4, respectively. To destroy the original K+ channel-binding function of scyllatoxin (33) and to allow specific labeling in future work, Arg-6 and Arg-13 were mutated into Ala and Lys, respectively. Finally, two residues, each at the N and C terminus, were deleted. The resulting miniprotein, dubbed CD4M3, contained 27 residues, of which 9 were topologically positioned as in CD4, and only 16 of the 31 amino acids (52%) of the scyllatoxin sequence remained (Fig. 1B). The new miniprotein was chemically synthesized. Despite the numerous mutations, its folding efficiency and CD spectrum (not shown) were similar to those of scyllatoxin.
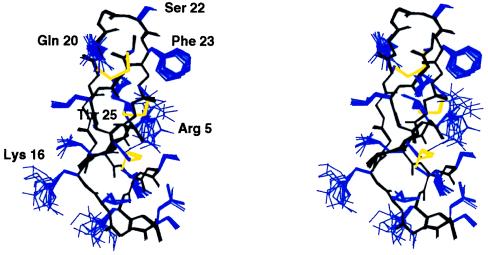
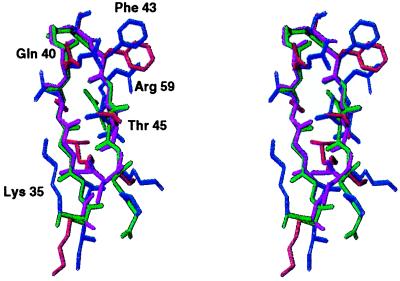
CD4M3 complete structure was solved by 1H NMR spectroscopy and molecular modeling. Complete 1H resonance assignments were obtained by standard two-dimensional NMR methods. A total of 239 NOE-derived distance restraints were obtained from two-dimensional NOE spectra and 46 additional angular restraints (φ and χ1) obtained from J coupling constants analysis; 40 three-dimensional structures were generated from these distance and angular restraints in molecular dynamics calculations, and 20 structures were selected for analysis. They satisfied all experimental restraints and had good energy (Etotal = −105.5 kcal, Eimp = 3.16 kcal, Ebond = 3.523 kcal, EVdW = −28.256 kcal, ENOE = 9.331 kcal, Eangle = 9.035 kcal, and Eelect = −103.1 kcal, for the best structure). The 20 structures ensemble is shown in Fig. 2. Global rms deviations as low as 0.2 Å were calculated for all the backbone atoms (0.8 Å for all the heavy atoms). The high definition of the solution structure of CD4M3, in conjunction with the high number of NOEs per residue, indicated an excellent sampling of the conformational space during the simulated annealing procedure. The final 20 structures possessed the expected scyllatoxin α/β-fold, with a helix in the region 2–13, an antiparallel β-sheet in the region 16–26, and an rms deviation of only 0.21 Å between the average structure and the corresponding region of scyllatoxin. The structure of the site transferred from CD4 was also well defined and superimposed on the native CD4 site with striking precision (Fig. 3); the rms deviation between the backbone atoms of the 17–26 sequence of the CD4M3 average structure and the 37–46 sequence of CD4 (34) was only 0.61 Å [0.66 Å with CD4 structure (ref. 7) bound to gp120]. The orientations of the side chains of Gln-20, Ser-22, Phe-23, and Thr-25 were similar to those of the corresponding side chains in CD4. In particular, the Phe-23 side chain, which is well defined in the 20 structures because of many long-range contacts, protrudes into the solvent in a conformation that is unusual for a hydrophobic moiety but is reminiscent of that of Phe-43 of CD4, which, in the crystal structure of the CD4-gp120 complex, plugs the entrance of the gp120 “Phe-43 cavity” (7). However, the position of the Arg-5 and Lys-16 side chains and of the C-terminal Gly-27 diverged from that of the corresponding Arg-59, Lys-35, and Gly-47 of CD4 by 8.9, 7.7, and 1.5 Å on their Cδ, Cɛ, and Cα, respectively, suggesting that these regions should be modified further to improve the structural reproduction of the CD4 active site.
Figure 2.
Stereo-view of NMR structure ensemble of CD4M3; 20 models are shown superimposed by using the backbone atoms (in black); disulfide bridges are yellow, and other side chains are blue. Critical side chains are labeled.
Figure 3.
Comparison of the 35–48 β-hairpin structure of CD4 with the corresponding 16–27 β-hairpin of CD4M3. Note the good superposition of CD4 backbone structure (green) with that of CD4M3 (magenta) and the similar conformation of the CD4 side chains Gln-40, Phe-43, and Thr-45 (blue) with the corresponding side-chains of CD4M3 (red).
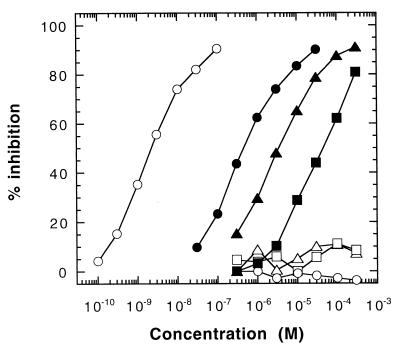
In competitive ELISA, CD4M3 inhibited the interaction of sCD4 and recombinant rgp120LAI with a 40 μM IC50 (Fig. 4; Table 1). CD4M3 also bound to rgp120s from different X4 (IIIB, MN), R5 (BaL, JR-FL), and R5X4 (W91D) HIV-1 strains, with affinities in the 20–350 μM range (Table 1). The native toxin, control linear 37–53 CD4 peptides, or cyclic 37–46 CD4 peptides did not bind rgp120, confirming the presence of a CD4-like site in CD4M3. To probe the functional role of the introduced residues more precisely, we individually substituted the solvent-exposed side chains of the β-hairpin region by an Ala residue and the Gly residues by a bulkier Val. Mutations at Gly-21 and Phe-23 (equivalent to CD4 Gly-41 and Phe-43) produced the most dramatic effect and abolished binding, whereas mutation of Ser-22 decreased the apparent affinity by 2.5-fold; substitution of Lys-16, Gly-18, and Gly-27 had no noticeable effect on the miniprotein-binding properties (Table 2). In contrast, two substitutions, Gln20Ala and Thr25Ala, increased the apparent binding affinity by about 5-fold. Interestingly, the corresponding two mutations in sCD4 also increased its binding affinity for rgp120 (9, 10). Altogether, these results suggest that the miniprotein may bind to rgp120 in a manner similar to that of the CDR2 region of sCD4 and that four positions, Gln-20, Gly-21, Phe-23, and Thr-25, are critical to reproduce a substantial and specific gp120-binding activity in the miniprotein.
Figure 4.
Effect of CD4M9 on gp120-CD4 interaction. Inhibition of gp120LAI-binding to coated sCD4 in the ELISA. sCD4 (○), CD4M3 (■), double-mutant CD4M8 (▴), and quintuple mutant CD4M9 (●); control peptides are scyllatoxin (▵), linear 37–53 (□), and cyclic 37–46 CD4 peptides (○).
Table 1.
Summary of binding data of sCD4, CD4M3, and its mutants to recombinant gp120s
| Competitor | IC50
|
|||||
|---|---|---|---|---|---|---|
| gp120LAI | gp120IIIB | gp120MN | gp120BaL | gp120JR-FL | gp120W91D | |
| sCD4, nM | 4.0 | 2.0 | 4.5 | 5.0 | 5.0 | 7.0 |
| CD4M3, μM | 40 | 100 | 150 | 350 | 200 | 20 |
| CD4M8, μM | 2.3 | — | — | — | — | — |
| CD4M9, μM | 0.4 | 1.0 | 0.4 | 1.0 | 1.0 | 0.1 |
IC50 for each molecule was obtained in competetive ELISA. Data are means of triplicate assays; SDs were within 15% of the mean.
Table 2.
gp120 binding activity of rCD4, CD4M3, and its mutants
| Competitor | IC50†, M |
|---|---|
| sCD4 | 1.4 ± 0.1 × 10−9 |
| CD4M3 | 4.0 ± 0.5 × 10−5 |
| Lys16Ala | 2.0 ± 0.1 × 10−5 |
| Gly18Val | 7.5 ± 0.5 × 10−5 |
| Gln20Ala | 7.5 ± 0.6 × 10−6 |
| Gly21Val | >1 × 10−3 |
| Ser22Ala | 1.0 ± 0.5 × 10−4 |
| Phe23Ala | >1 × 10−3 |
| Thr25Ala | 8.5 ± 0.9 × 10−6 |
| Gly27Val | 3.5 ± 0.2 × 10−5 |
Mutants of mini-CD4 are indicated by the residue mutation.
† Concentration (±SD) of competitor required for 50% inhibition of gp120 binding to coated rCD4 in competitive ELISA.
The above functional and structural analyses suggested two possible sets of changes to increase binding affinity for gp120. First, the two Ala mutations that led to increased apparent affinity for gp120LAI (Table 2) were included in a double Ala-20, Ala-25 mutant, dubbed CD4M8 (Fig. 1B). Second, to mimic better the native structure of Lys-35, Arg-59, and the C terminus of strand C“ in the miniprotein, Lys-16 was shifted to position 18; an Arg was incorporated at position 9; and a Pro was added to the C terminus; this mutant was dubbed CD4M9 (Fig. 1B). Both synthesized mutants folded correctly, with CD spectra that were highly similar to that of CD4M3 (data not shown). As determined in the ELISA, CD4M8 had a 16-fold increase of gp120-binding affinity (Fig. 4; Table 1), whereas CD4M9 showed a 4.0 × 10−7 M IC50 for gp120LAI, which corresponds to a 100-fold affinity increase relative to the original CD4M3 (Fig. 4; Table 1). Similar improvement in binding affinity was also noted for rgp120 from one R5X4 (W61D), two X4 (IIIB, MN), and two R5 (BaL, JR-FL) HIV-1 strains (Table 1).
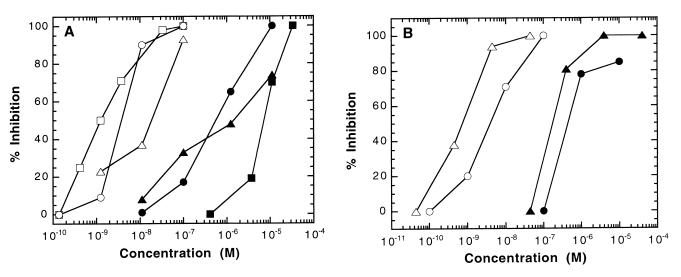
CD4M9 was tested for its ability to prevent infection of HeLa cells stably expressing CD4 and the CCR5 and CXCR4 coreceptors of HIV-1. Laboratory-adapted X4 (LAI) and R5 (BaL, Ada) strains were used. Independently of coreceptor usage, all viruses were inhibited effectively by CD4M9 in the 0.4–5.0 μM range (Fig. 5A), confirming the large spectrum antiviral activity of this construct. This engineered improved miniprotein also prevented infection of PBL by HIV-1LAI and HIV-1BaL (Fig. 5B) in the same concentration range as that inhibiting infection of HeLa cells. This result indicates that the miniprotein is also active in regards to infection of primary cells. Of note, the CD4M3 derivative was at least 100-fold less effective at inhibiting HIV-1 infection, whereas the Gly21Val and Phe23Ala mutants, which did not bind rgp120 in the ELISA, were inactive (data not shown). Interestingly, protection from infection was effective only when the miniprotein was added before but not after the virus, suggesting that the miniprotein prevented virus attachment to cells.
Figure 5.
Effect of CD4M9 on HIV-1 infection. (A) Inhibition of HeLa/CD4/CCR5/LacZ cell infection by HIV-1. Closed symbols, CD4M9; open symbols, control sCD4. X4 (LAI, ●, ○) and R5 (BaL, ▴, ▵; Ada, ■, □) HIV-1 strains were tested. Data from one of three experiments are presented as percentage of inhibition of infection. (B) Inhibition of PBL infection by HIV-1. Closed symbols, CD4M9; open symbols, sCD4. The strains were LAI (●, ○) and BaL (▴, ▵). Data from one of three experiments are presented as percentage of inhibition determined on day 10 after infection.
Discussion
The present work describes a rational process for reproducing a significant portion of gp120-binding affinity of CD4 in a miniprotein system. The strategy that we used included three steps: (i) transfer of the side chains of CD4 gp120-binding core to the structurally equivalent region of a stable and tolerant scaffold; (ii) NMR and mutational analysis of the miniprotein active site to assess its structural and functional mimicry with the CD4 gp120-binding core; (iii) incorporation of sequence changes suggested by the structural and functional analyses, which indeed improved the affinity of the engineered miniprotein. This strategy, which includes an optimization process after structure-function analysis, represents an obvious improvement on that of active site transfer to a stable, previously used scaffold (19, 20, 33). Most likely, this strategy can be applied to other protein–protein interfaces. No overall homology was needed between the structure of the large protein (an all β-structure) and the host scaffold (an α/β-motif)—the only guide being the structural similarity between the β-hairpins of both proteins. Such similarity is surely not unique to the system studied. More generally, the functional site of most large proteins may have a structural equivalent in a small protein, which could therefore be used as a host scaffold. The advantages of the scaffold chosen here are its conformational stability, folding efficiency, and tolerance of sequence mutations (33), which allowed multiple sequence changes, resulting in effective improvement of the initial design.
Protein-binding domains of the hormone atrial natriuretic peptide (36) and of protein A (37) have been minimized to about half their original size with retention of full activity by combining a structure-based design and phage display methods. In this study, we downsized the D1 gp120-binding domain of CD4 to about one-fourth of its original size by a structure-based approach, implying the transfer of the “core” of a protein–protein interface to a “miniprotein system,” and an optimization process based on a structure-function analysis. Further improvement to reduce the existing gap (≈100-fold) in affinity with native CD4 can probably be achieved on the basis of new additional crystallographic and NMR data on the interaction of the miniprotein with gp120. Use of the library approach on this stable and sequence-permissive scaffold may represent a further opportunity for functional improvement, as attested by successes where this approach has been applied (36–38).
However, because of its reasonably high gp120-binding affinity, its well defined three-dimensional structure, and its specific antiviral activity, the presently optimized miniprotein may represent a unique tool to study the complex process of HIV-1 entry into CD4+ cells. For example, binding of sCD4 to gp120 has been shown to induce envelope conformational changes that increase its affinity for CCR5 (3, 4), which is believed to be essential for efficient virus entry. A small molecule binding to the CD4-binding cleft of gp120 may be unable to induce such conformational changes, and it may block the envelope in a conformation that is not competent for fusion. Alternatively, the mini-CD4 can induce gp120 conformational changes similar to those induced by sCD4. In this case, such a small molecule may be useful in the context of vaccine strategies, in studies aiming at gp120 epitopes that are exposed only on interaction with CD4 (39–41).
The small size and the ease with which such miniproteins can be synthesized chemically and manipulated to include unnatural amino acids, fluorescent probes, or other useful probes may represent important and practical advantages. Such engineered miniproteins may become practical tools to probe biological processes and to test new therapeutic strategies.
Acknowledgments
We thank the Agence Nationale de Recherches sur le SIDA and the Ensemble Contre le SIDA for financial support.
Abbreviations
- NOE
nuclear Overhauser effect
- PBL
peripheral blood lymphocytes
- sCD4
soluble recombinant CD4
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1D5Q).
References
- 1.Dalgleish A G, Beverley A C, Clapham P R, Crawford D H, Greaves M F, Weiss R A. Nature (London) 1984;312:763–767. doi: 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- 2.Klatzmann D, Champagne E, Chamaret S, Gruest J, Guetard D, Hercend T, Gluckman J C, Montagnier L. Nature (London) 1984;312:767–768. doi: 10.1038/312767a0. [DOI] [PubMed] [Google Scholar]
- 3.Wu L, Gerard N P, Wyatt R, Choe H, Parolin C, Ruffing A, Borsetti A, Cardoso A A, Desjardin E, Newman W, et al. Nature (London) 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 4.Trkola A, Dragic T, Arthos J, Binley J M, Olson W C, Allaway G P, Cheng-Mayer C, Robinson J, Maddon P J, Moore J P. Nature (London) 1996;384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 5.Berger E A. AIDS. 1997;11:S3–S16. [PubMed] [Google Scholar]
- 6.Littman D R. Cell. 1998;93:677–680. doi: 10.1016/s0092-8674(00)81429-4. [DOI] [PubMed] [Google Scholar]
- 7.Kwong P D, Wyatt R, Robinson J, Sweet R W, Sodroski J, Hendrickson W A. Nature (London) 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arthos J, Deen K C, Chaikin M A, Fornwald J A, Sathe G, Sattentau Q J, Clapham P R, Weiss R A, McDougal J S, Pietropaolo C, et al. Cell. 1989;57:469–481. doi: 10.1016/0092-8674(89)90922-7. [DOI] [PubMed] [Google Scholar]
- 9.Ashkenazi A, Presta L G, Marsters S A, Camerato T R, Rosenthal K A, Fendley B M, Capon D J. Proc Natl Acad Sci USA. 1990;87:7150–7154. doi: 10.1073/pnas.87.18.7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moebius U, Clayton L K, Abraham S, Harrison S C, Reinherz E L. J Exp Med. 1992;176:507–517. doi: 10.1084/jem.176.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sweet R W, Truneh A, Hendrickson W A. Curr Opin Biotechnol. 1991;2:622–633. doi: 10.1016/0958-1669(91)90089-n. [DOI] [PubMed] [Google Scholar]
- 12.Ryu S E, Trueh A, Sweet R W, Hendrickson W A. Structure. 1994;2:59–74. doi: 10.1016/S0969-2126(00)00008-3. [DOI] [PubMed] [Google Scholar]
- 13.Wild C T, Shugars D C, Greenwell T K, McDanal C B, Matthews T J. Proc Natl Acad Sci USA. 1994;90:9770–9774. doi: 10.1073/pnas.91.21.9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Furuta R A, Wild C T, Weng Y, Weiss C D. Nat Struct Biol. 1998;5:276–279. doi: 10.1038/nsb0498-276. [DOI] [PubMed] [Google Scholar]
- 15.Lifson J D, Hwang K M, Nara P L, Fraser B, Padgett M, Dunlop N M, Eiden L E. Science. 1988;241:712–716. doi: 10.1126/science.2969619. [DOI] [PubMed] [Google Scholar]
- 16.Chen S, Chrusciel A, Nakanishi H, Raktabutr A, Johnson M E, Sato A, Weiner D, Hoxie J, Saragovi H U, Greene M I, et al. Proc Natl Acad Sci USA. 1992;89:5872–5876. doi: 10.1073/pnas.89.13.5872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang X, Gaubin M, Briant L, Srikantan V, Ramachandran M, Saragoni U, Weiner D, Devaux C, Autiero M, Platier-Tonneau D, et al. Nat Biotechnol. 1997;15:150–154. doi: 10.1038/nbt0297-150. [DOI] [PubMed] [Google Scholar]
- 18.Ferrer M, Harrison S C. J Virol. 1999;73:5795–5802. doi: 10.1128/jvi.73.7.5795-5802.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vita C, Roumestand C, Toma F, Ménez A. Proc Natl Acad Sci USA. 1995;92:6404–6408. doi: 10.1073/pnas.92.14.6404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Drakopoulou E, Zinn-Justin S, Guenneugues M, Gilquin B, Ménez A, Vita C. J Biol Chem. 1996;271:11979–11987. doi: 10.1074/jbc.271.20.11979. [DOI] [PubMed] [Google Scholar]
- 21.Drakopoulou E, Vizzavona J, Vita C. Lett Pept Sci. 1998;5:241–245. [Google Scholar]
- 22.Martins J C, Zhang W, Tartar A, Lazdunski M, Borremans F A M. FEBS Lett. 1990;260:249–253. doi: 10.1016/0014-5793(90)80115-y. [DOI] [PubMed] [Google Scholar]
- 23.Wüthrich K. NMR Proteins and Nucleic Acids. New York: Wiley Interscience; 1986. [Google Scholar]
- 24.Pons J L, Malliavin T E, Delsuc M A. J Biomol NMR. 1996;8:445–452. doi: 10.1007/BF00228146. [DOI] [PubMed] [Google Scholar]
- 25.Ludvingsen S, Andersen K V, Poulsen F M. J Mol Biol. 1991;217:731–736. doi: 10.1016/0022-2836(91)90529-f. [DOI] [PubMed] [Google Scholar]
- 26.Brunger A T. x-plor. New Haven, CT: Yale Univ.; 1992. , Version 3.1. [Google Scholar]
- 27.Koradi R, Billeter M, Wütrich K. J Mol Graphics. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 28.Moore J P. AIDS. 1990;4:297–303. doi: 10.1097/00002030-199004000-00003. [DOI] [PubMed] [Google Scholar]
- 29.Charneau P, Mirambeau G, Roux P, Paulous S, Buc H, Clavel F. J Mol Biol. 1994;241:651–662. doi: 10.1006/jmbi.1994.1542. [DOI] [PubMed] [Google Scholar]
- 30.Benjouad A, Seddiki N, Ylisastigui L, Gluckman J C. AIDS Res Hum Retroviruses. 1997;13:219–226. doi: 10.1089/aid.1997.13.219. [DOI] [PubMed] [Google Scholar]
- 31.Bontems F, Roumestand C, Gilquin B, Ménez A, Toma F. Science. 1991;254:1521–1523. doi: 10.1126/science.1720574. [DOI] [PubMed] [Google Scholar]
- 32.Ménez A, Bontems F, Roumestand C, Gilquin B, Toma F. Proc R Soc Edinb [Biol] 1992;99:83–103. [Google Scholar]
- 33.Vita C, Vizzavona J, Drakopoulou E, Zinn-justin S, Gilquin B, Ménez A. Biopolymers. 1998;47:93–100. doi: 10.1002/(SICI)1097-0282(1998)47:1<93::AID-BIP10>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 34.Auguste P, Hugues M, Mourre C, Moinier D, Tartar A, Lazdunski M. Biochemistry. 1992;31:648–654. doi: 10.1021/bi00118a003. [DOI] [PubMed] [Google Scholar]
- 35.Ryu S E, Kwong P D, Truneh A, Porter T G, Arthos J, Rosenberg M, Dai X, Xuong N, Axel R, Sweet R W, et al. Nature (London) 1990;348:419–426. doi: 10.1038/348419a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li B, Tom J Y K, Oare D, Yen R, Fairbrother W J, Wells J A, Cunningham B C. Science. 1995;270:1657–1660. doi: 10.1126/science.270.5242.1657. [DOI] [PubMed] [Google Scholar]
- 37.Braisted A C, Wells J A. Proc Natl Acad Sci USA. 1996;93:5688–5692. doi: 10.1073/pnas.93.12.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nygren P Å, Uhlén M. Curr Opin Struct Biol. 1997;7:463–469. doi: 10.1016/s0959-440x(97)80108-x. [DOI] [PubMed] [Google Scholar]
- 39.Wyatt R, Sodroski J. Science. 1998;280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 40.Sattentau Q J. Structure. 1998;6:945–949. doi: 10.1016/s0969-2126(98)00096-3. [DOI] [PubMed] [Google Scholar]
- 41.LaCasse R A, Follis K E, Trahey M, Scarborough J D, Littman D R, Nunberg J H. Science. 1999;283:357–362. doi: 10.1126/science.283.5400.357. [DOI] [PubMed] [Google Scholar]