Abstract
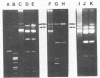
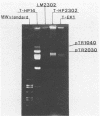
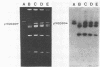
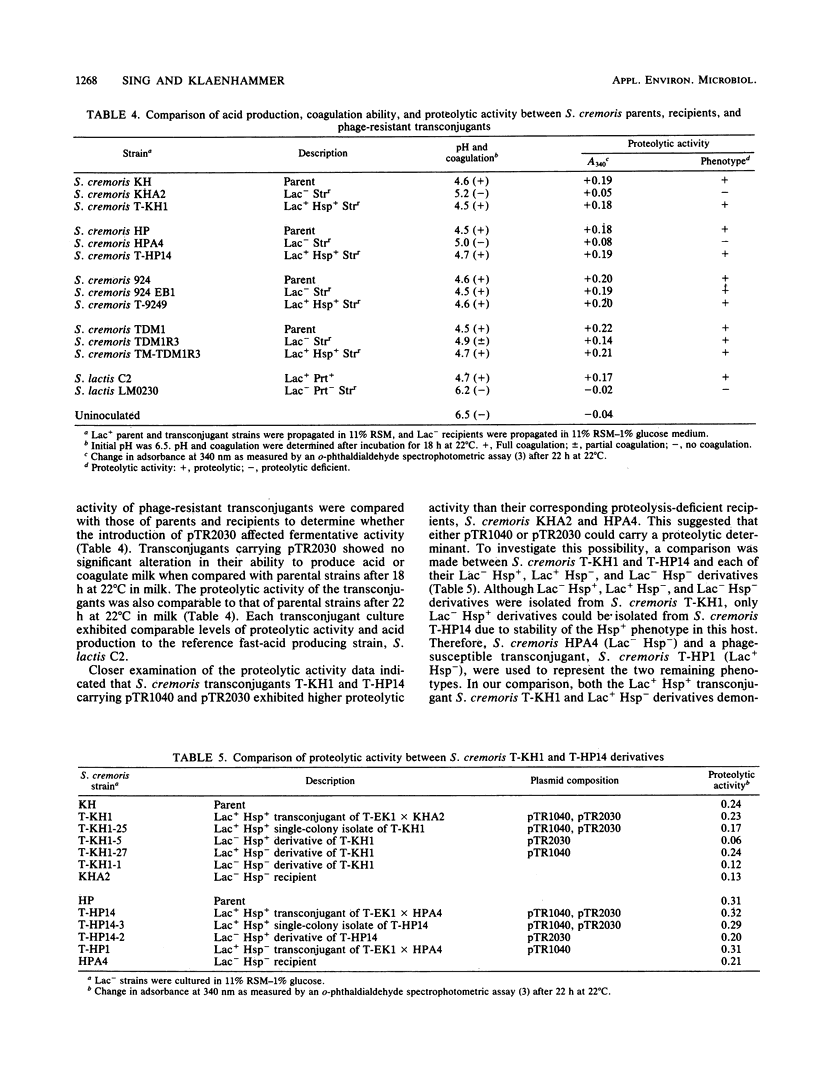
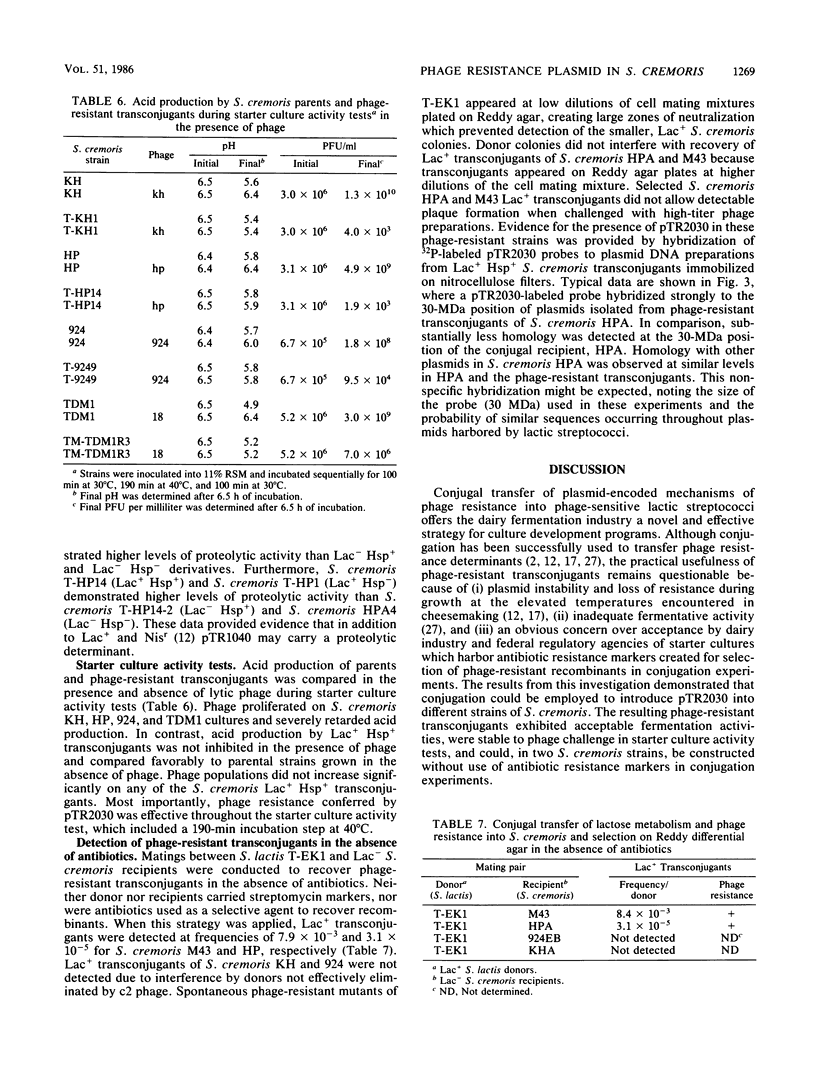
Agar surface conjugal matings were used to introduce heat-sensitive phage resistance (Hsp+) determinants carried on the conjugal plasmid pTR2030 into Streptococcus cremoris KH, HP, 924, and TDM1. Lactose-fermenting (Lac+) transconjugants were selected from matings of Lac− variants of S. cremoris KH, HP, 924, and TDM1 with Streptococcus lactis ME2 or a high-frequency donor, S. lactis T-EK1 (pTR1040, Lac+; pTR2030, Hsp+). For all of the S. cremoris strains examined, select Lac+ transconjugants were completely resistant to plaquing by their homologous lytic phages. In all cases the plaquing efficiencies were less than 10−9. Acquisition of a 30-megadalton plasmid (pTR2030) in the S. cremoris phage-resistant transconjugants was demonstrated by direct plasmid analysis, by hybridization with 32P-labeled probes, or by conjugal transfer of pTR2030 out of the phage-resistant transconjugants into a plasmid-cured recipient, S. lactis LM2302. Acid production, coagulation ability, and proteolytic activity of phage-resistant transconjugants in milk were comparable to those of their phage-sensitive parents. Further, S. cremoris phage-resistant transconjugants were not attacked by phage in starter culture activity tests, which included a 40°C incubation period. The results demonstrated that phage resistance determinants on pTR2030 could be conjugally transferred to a variety of S. cremoris strains and confer resistance to phage under conditions encountered during cheese manufacture. Phage-resistant transconjugants of S. cremoris M43 and HP were also constructed without the use of antiblotic markers to select conjugal recipients from mating mixtures.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson D. G., McKay L. L. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl Environ Microbiol. 1983 Sep;46(3):549–552. doi: 10.1128/aem.46.3.549-552.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopin A., Chopin M. C., Moillo-Batt A., Langella P. Two plasmid-determined restriction and modification systems in Streptococcus lactis. Plasmid. 1984 May;11(3):260–263. doi: 10.1016/0147-619x(84)90033-7. [DOI] [PubMed] [Google Scholar]
- Exterkate F. A., de Veer G. J. Partial Isolation and Degradation of Caseins by Cell Wall Proteinase(s) of Streptococcus cremoris HP. Appl Environ Microbiol. 1985 Feb;49(2):328–332. doi: 10.1128/aem.49.2.328-332.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis A. W. Differentiation of lactic streptococcal phages into phage species by DNA-DNA homology. Appl Environ Microbiol. 1984 Feb;47(2):343–349. doi: 10.1128/aem.47.2.343-349.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis A. W., Klaenhammer T. R. Bacteriophage Resistance Conferred on Lactic Streptococci by the Conjugative Plasmid pTR2030: Effects on Small Isometric-, Large Isometric-, and Prolate-Headed Phages. Appl Environ Microbiol. 1986 Jun;51(6):1272–1277. doi: 10.1128/aem.51.6.1272-1277.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaenhammer T. R., McKay L. L., Baldwin K. A. Improved lysis of group N streptococci for isolation and rapid characterization of plasmid deoxyribonucleic acid. Appl Environ Microbiol. 1978 Mar;35(3):592–600. doi: 10.1128/aem.35.3.592-600.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaenhammer T. R., Sanozky R. B. Conjugal transfer from Streptococcus lactis ME2 of plasmids encoding phage resistance, nisin resistance and lactose-fermenting ability: evidence for a high-frequency conjugative plasmid responsible for abortive infection of virulent bacteriophage. J Gen Microbiol. 1985 Jun;131(6):1531–1541. doi: 10.1099/00221287-131-6-1531. [DOI] [PubMed] [Google Scholar]
- Kondo J. K., McKay L. L. Plasmid transformation of Streptococcus lactis protoplasts: optimization and use in molecular cloning. Appl Environ Microbiol. 1984 Aug;48(2):252–259. doi: 10.1128/aem.48.2.252-259.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen L. D., McKay L. L. Isolation and characterization of plasmid DNA in Streptococcus cremoris. Appl Environ Microbiol. 1978 Dec;36(6):944–952. doi: 10.1128/aem.36.6.944-952.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macrina F. L., Kopecko D. J., Jones K. R., Ayers D. J., McCowen S. M. A multiple plasmid-containing Escherichia coli strain: convenient source of size reference plasmid molecules. Plasmid. 1978 Jun;1(3):417–420. doi: 10.1016/0147-619x(78)90056-2. [DOI] [PubMed] [Google Scholar]
- McKay L. L., Baldwin K. A. Conjugative 40-megadalton plasmid in Streptococcus lactis subsp. diacetylactis DRC3 is associated with resistance to nisin and bacteriophage. Appl Environ Microbiol. 1984 Jan;47(1):68–74. doi: 10.1128/aem.47.1.68-74.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay L. L., Baldwin K. A., Walsh P. M. Conjugal transfer of genetic information in group N streptococci. Appl Environ Microbiol. 1980 Jul;40(1):84–89. doi: 10.1128/aem.40.1.84-91.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay L. L., Baldwin K. A., Zottola E. A. Loss of lactose metabolism in lactic streptococci. Appl Microbiol. 1972 Jun;23(6):1090–1096. doi: 10.1128/am.23.6.1090-1096.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay L. L., Cords B. R., Baldwin K. A. Transduction of lactose metabolism in Streptococcus lactis C2. J Bacteriol. 1973 Sep;115(3):810–815. doi: 10.1128/jb.115.3.810-815.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy M. S., Vedamuthu E. R., Washam C. J., Reinbold G. W. Differential agar medium for separating Streptococcus lactis and Streptococcus cremoris. Appl Microbiol. 1969 Nov;18(5):755–759. doi: 10.1128/am.18.5.755-759.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders M. E., Klaenhammer T. R. Characterization of Phage-Sensitive Mutants from a Phage-Insensitive Strain of Streptococcus lactis: Evidence for a Plasmid Determinant that Prevents Phage Adsorption. Appl Environ Microbiol. 1983 Nov;46(5):1125–1133. doi: 10.1128/aem.46.5.1125-1133.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders M. E., Klaenhammer T. R. Evidence for Plasmid Linkage of Restriction and Modification in Streptococcus cremoris KH. Appl Environ Microbiol. 1981 Dec;42(6):944–950. doi: 10.1128/aem.42.6.944-950.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders M. E., Klaenhammer T. R. Phage Resistance in a Phage-Insensitive Strain of Streptococcus lactis: Temperature-Dependent Phage Development and Host-Controlled Phage Replication. Appl Environ Microbiol. 1984 May;47(5):979–985. doi: 10.1128/aem.47.5.979-985.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders M. E., Klaenhammer T. R. Restriction and modification in group N streptococci: effect of heat on development of modified lytic bacteriophage. Appl Environ Microbiol. 1980 Sep;40(3):500–506. doi: 10.1128/aem.40.3.500-506.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Steenson L. R., Klaenhammer T. R. Streptococcus cremoris M12R transconjugants carrying the conjugal plasmid pTR2030 are insensitive to attack by lytic bacteriophages. Appl Environ Microbiol. 1985 Oct;50(4):851–858. doi: 10.1128/aem.50.4.851-858.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzaghi B. E., Sandine W. E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975 Jun;29(6):807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh P. M., McKay L. L. Recombinant plasmid associated cell aggregation and high-frequency conjugation of Streptococcus lactis ML3. J Bacteriol. 1981 Jun;146(3):937–944. doi: 10.1128/jb.146.3.937-944.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]