Abstract
Background
NKX3.1 and PCAN1 are both prostate-specific genes related to prostate development and prostate cancer. So far, little is known about the regulatory mechanisms of the expression of these two genes. In the present study, we found that NKX3.1 upregulated PCAN1 gene transcription in LNCaP prostate cancer cells. To understand the regulatory mechanisms, our work focused on identifying the functional NKX3.1 binding sites upstream of the PCAN1 gene, which might be involved in the positive regulation of PCAN1 expression by NKX3.1.
Results
We cloned and characterized a 2.6 kb fragment upstream of the PCAN1 gene. Analysis of the 2.6 kb sequence with MatInspector 2.2 revealed five potential binding sites of NKX3.1 transcription factor. Luciferase reporter assays, electrophoretic mobility shift assays, chromatin immunoprecipitation and RNA interference were performed to study the effects of NKX3.1 on PCAN1 gene expression in prostate cancer cells. Our results showed that PCAN1 promoter activity and mRNA expression were increased by transfection with the NKX3.1 containing plasmid (pcDNA3.1-NKX3.1) and that PCAN1 mRNA expression was decreased by RNA interference targeting human NKX3.1 in LNCaP prostate cancer cells. The results of electrophoretic mobility shift assays and chromatin immunoprecipitation showed that NKX3.1 bound to NBS1 (-1848 to -1836) and NBS3 (-803 to -791) upstream of the PCAN1 gene. The luciferase reporter assays showed that NBS1 and NBS3 enhanced the promoter activity in pGL3-promoter vector with cotransfection of the NKX3.1 containing plasmid. Furthermore, the deletion of NBS1 or both NBS1 and NBS3 reduced PCAN1 promoter activity and abolished the positive regulation of PCAN1 expression by NKX3.1.
Conclusion
Our results suggested that two functional NKX3.1 binding sites located at -1848 to -1836 and -803 to -791 upstream of the PCAN1 gene were involved in the positive regulation of PCAN1 gene transcription by NKX3.1.
Background
Prostate cancer is the most frequently diagnosed neoplasia in men and one of the leading causes of cancer-related deaths in men over 60 [1]. Although early prostate specific antigen (PSA) detection and surgery have decreased the death rate, most of the patients still die of metastasis and recurrence of prostate cancer [2,3]. However, the mechanisms involved in the onset and progression of prostate cancer are poorly understood at the molecular level. It is important to understand the molecular biology of this cancer for its prevention, early diagnosis, and effective treatment.
PCAN1 (prostate cancer gene 1, also known as GDEP) is highly expressed in prostate epithelial tissue and frequently mutated in prostate tumors [4-6]. PCAN1 expression is undetectable in the highly undifferentiated DU145 and PC-3 prostate cancer cell lines and weakly detected in the more differentiated LNCaP cell line [5]. This gene is localized to chromosome 4q21, a region of the genome that experiences frequent loss of heterozygosity (LOH) in prostate cancer. It is mutated in 35% of the tumor samples [5]. Therefore,PCAN1 has been proposed to have tumor suppressing function in prostate cancer.
NKX3.1 is a prostate-specific homeobox gene that is thought to play an important role in the normal development of prostate and carcinogenesis. In mice Nkx3.1 is exclusively expressed in prostate epithelium [7,8]. Its targeted disruption leads to aberrations in prostate ductal morphogenesis and secretory protein production, and epithelial hyperplasia and dysplasia [9]. Notably Nkx3.1 mutant mice display the pathologic changes of prostatic intraepithelial neoplasia (PIN) [10] that is the presumed precursor to prostate cancer in humans, implying that loss of Nkx3.1 expression correlates with the initiation of prostate carcinogenesis. Human NKX3.1 has been mapped to human chromosome 8p21, a region with frequent loss of heterozygosity in human prostate cancer [11]. This gene has been proposed to have tumor suppressing function [12]. It also inhibits the growth of cultured prostate cancer cells [13-15] in vitro. The strong association of NKX3.1 with prostate development and prostate cancer makes this gene an attractive molecular target for further study.
NKX3.1 and PCAN1 are both prostate-specific genes strongly related to prostate development and prostate cancer. Studying the regulatory mechanisms of their expression is important for understanding their roles in prostate development and prostate cancer. In this study, we cloned and characterized a 2.6 kb fragment upstream of the PCAN1 gene. Analysis of the 2.6 kb sequence with MatInspector 2.2 revealed potential binding sites of some important transcription factors, including NKX3.1, P53, Sp1, cEBP and PPAR/RXR heterodimers. In our study, the eukaryotic expression plasmids containing NKX3.1, p53, Sp1, cEBPα and PPARγ were respectively used to study their effects on PCAN1 expression. We found that NKX3.1 upregulated PCAN1 promoter activity and mRNA expression. To explore the regulatory mechanisms of NKX3.1 on PCAN1 transcription, we focused on identifying the functional NKX3.1 binding sites (NBSs) upstream of the PCAN1 gene. We demonstrated that NKX3.1 upregulated PCAN1 gene transcription through direct binding with NBSs upstream of the PCAN1 gene. Our study provided a molecular mechanism for the regulation of PCAN1 gene expression.
Results
Effects of NKX3.1 on PCAN1 promoter activity and mRNA expression
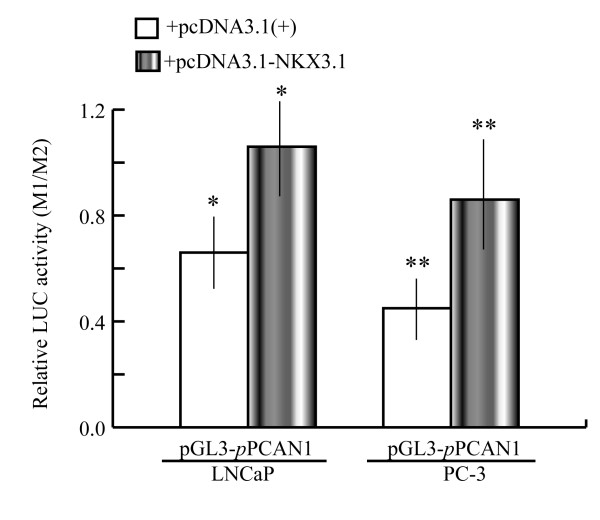
In our previous work, a 2.6 kb promoter fragment (+32 to -2598) of the PCAN1 gene amplified by PCR was inserted into pGL3-basic vector to form the PCAN1 promoter-luciferase reporter plasmid designated as pGL3-pPCAN1. Firefly luciferase expression driven by the 2.6 kb PCAN1 promoter was used to evaluate the promoter activity [16]. To detect the effects of NKX3.1 on PCAN1 gene expression, LNCaP and PC-3 cells were harvested 48 h after cotransfection with pGL3-pPCAN1 and pcDNA3.1-NKX3.1. The control cells were cotransfected with pGL3-pPCAN1 and pcDNA3.1 (+) plasmid. The PCAN1 promoter activity detected by dual-luciferase reporter assays was enhanced by 1.6-fold in LNCaP cells and by 1.8-fold in PC-3 cells with NKX3.1 cotransfection, compared with PCAN1 promoter activity in the control cells (Fig. 1).
Figure 1.
Effects of NKX3.1 on the PCAN1 promoter in LNCaP and PC-3 cells. LNCaP and PC-3 cells were cotransfected with pGL3-pPCAN1 and pcDNA3.1-NKX3.1 (grey bars). The control cells were cotransfected with pGL3-pPCAN1 and pcDNA3.1 (+) plasmid (blank bars). The cells were harvested 48 h after transfection and PCAN1 promoter activity was detected by dual-luciferase reporter assays. Results were expressed as relative luciferase activities (M1/M2). The data were represented as the mean of four individual values ± SD.*p < 0.05, **p < 0.05, grey bar vs blank bar.
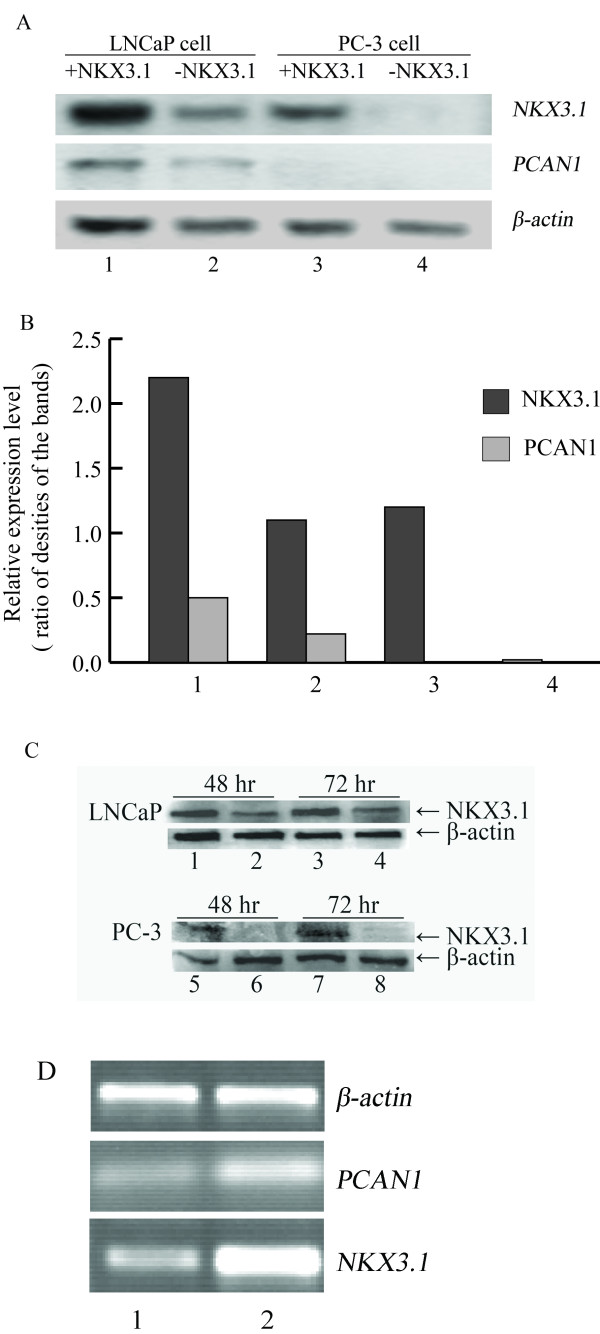
The PCAN1 mRNA expression level in LNCaP cells, as detected by RT-PCR, was increased significantly by NKX3.1 cotransfection, compared with that of the control cells transfected with pcDNA3.1 (+) plasmid (Fig. 2A and 2B). The PCAN1 mRNA expression level in PC-3 cells was undetectable by RT-PCR (Fig. 2A and 2B).
Figure 2.
Effects of NKX3.1 on PCAN1 mRNA expression in LNCaP and PC-3 cells. The effects of NKX3.1 on PCAN1 mRNA expression as detected by RT-PCR. A. PCAN1 mRNA expression following transfection of LNCaP cells with 1. pcDNA3.1-NKX3.1; 2. pcDNA3.1 (+); or following transfection of PC-3 cells with 3. pcDNA3.1-NKX3.1; or 4. pcDNA3.1 (+). B. Relative expression levels were presented as the ratio of densities of NKX3.1 or PCAN1 to β-actin bands. The results were expressed as mean ± SD (n = 3). C. The expression of NKX3.1 protein in LNCaP and PC-3 cells after transfection of pcDNA3.1-NKX3.1 (1, 3, 5, 7) or pcDNA3.1 (+) (2, 4, 6, 8).D. NKX3.1 and PCAN1 mRNA expression following stable transfection of LNCaP cells with 1. pRNAT-RNAi1 targeting human NKX3.1; 2. pRNAT-RiN as a control vector.
The expression of endogenous NKX3.1 in LNCaP cells was knocked down by RNAi, which made the PCAN1 mRNA expression decrease, as detected by RT-PCR (Fig. 2D).
Identification of specific NKX3.1 binding sites (NBSs) with nuclear extracts
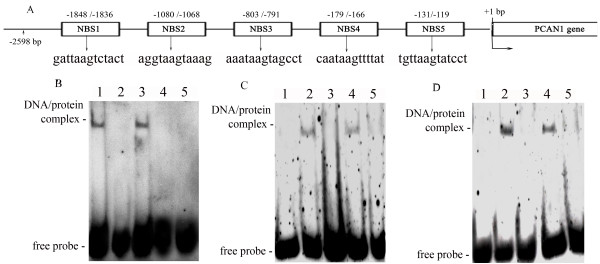
Analysis of the 2.6 kb promoter sequence with MatInspector 2.2 revealed five potential NKX3.1 transcription factor binding sites (sequences are shown in Fig. 3A). They were located at -1848 to -1836 (NBS1), -1080 to 1068 (NBS2), -803 to -791 (NBS3), -179 to 166 (NBS4) and -131 to -119 (NBS5), upstream of the PCAN1 gene. To investigate the binding activities of these five NBSs with NKX3.1 transcription factor, we carried out electrophoretic mobility shift assays (EMSA). It was performed with NKX3.1-transfected nuclear extracts from LNCaP cells and synthesized oligonucleotide probes containing these five NBS sequences. The results showed that DNA-protein binding complexes were identified for the NBS1 probe and the NBS3 probe (Fig 3B). The bindings of NBS1 and NBS3 with nuclear extracts proved to be specific, as they were blocked by a 250-fold excess of unlabeled NBS1 probe (Fig. 3C) or NBS3 probe (Fig. 3D) and by anti-NKX3.1 antibody (Fig. 3C and 3D), but not by unlabeled mutant NBS1 probe or mutant NBS3 probe (Fig. 3C and 3D).
Figure 3.
Binding assays of NKX3.1 binding sites with nuclear extracts. A. Locations and sequences of five potential NKX3.1 binding sites (NBSs) upstream of the PCAN1 gene. B. EMSA was performed to assay the binding activities of the five NBSs with the nuclear extracts from LNCaP cells. 1–5: labelled NBS1~NBS5 with LNCaP nuclear extracts respectively. A DNA-protein binding complex was formed when the labelled probe of NBS1 (lane 1) or NBS3 (lane 3) was reacted with LNCaP nuclear extracts. C and D. Competition binding and antibody blocking were used to detect the specific binding of the NBS1 (C) or NBS3 (D) with NKX3.1. Labelled NBS1 or NBS3 without nuclear extracts was used as the control (lane1); a specific DNA-protein complex was formed when labelled NBS1 or NBS3 was reacted with LNCaP nuclear extracts (lane 2). Competition binding was performed in the presence of a 250-fold molar excess of unlabelled NBS1 or NBS3 (lane 3), unlabelled mutant NBS1 or NBS3 (lane 4). The binding activity of probe NBS1 or NBS3 to nuclear extracts could be blocked by anti-NKX3.1 antibody (lane5).
NKX3.1 binds to NBS1 and NBS3 upstream of the PCAN1 gene in living cells
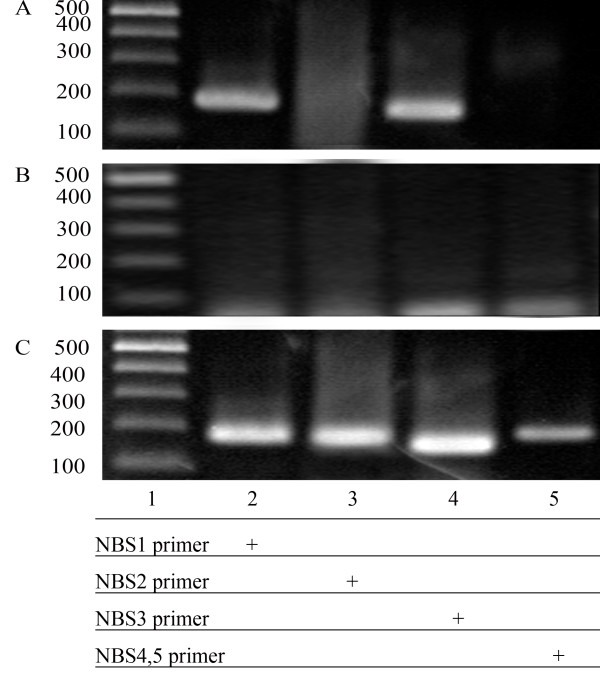
To determine whether NKX3.1 also binds to the NBSs in vivo, we performed chromatin immunoprecipitation (ChIP) assays, which are used to define interactions of proteins with specific DNA elements in living cells. ChIP was carried out in LNCaP cells transfected with pcDNA3.1-NKX3.1. After cross-linking with formaldehyde, cell lysates were immunoprecipitated with anti-NKX3.1 antibody or rabbit IgG (negative control). The DNA purified from this coprecipitation was analyzed by PCR with primers (sequences are shown in Table 1) spanning the NBSs in the PCAN1 promoter. As shown in Fig. 4, we observed a clear PCR product using NBS1 primer or NBS3 primer but no band was observed using NBS2 primer or NBS4,5 primer, and no PCR product was identified for all primers with rabbit IgG precipitation complexes, indicating that NKX3.1 bound to NBS1 and NBS3 upstream of the PCAN1 gene in living cells.
Table 1.
PCR primers used in chromatin immunoprecipitation
| Names | Sequences | Product sizes |
| NBS1 primers | F: GATTCTTTGACTGGTCTGGCACAC | |
| R: TTATCCATTGTGCCTGGAGCTGAG | 170 bp (spanning NBS1) | |
| BNS2 primers | F: TCCTACTAACGGCATGTAAGGAGG | |
| R: ATTGCCATGTCTGGACTGTGAGTG | 170 bp (spanning NBS2) | |
| NBS3 primers | F: AAGAATGAGCTGATCCTCCTATGC | |
| R: GGTTTAGTAATAGACTGGGCACCA | 150 bp (spanning NBS3) | |
| NBS4,5 primers | F: GTGTAGCAGGTAAATCAGTGTGAG | |
| R: TCAGCTGACGAGCAACTTCAATTC | 180 bp (spanning NBS4 and 5) |
NBS1 F and R span NBS1;
NBS2 F and R span NBS2; NBS3 F and R span NBS3; BNS4,5 F and R span NBS4 and NBS5
Figure 4.
NKX3.1 binds to the NBSs of the PCAN1 promoter in living cells. LNCaP cells transfected with pcDNA3.1-NKX3.1 were cross-linked by formaldehyde treatment and lysed. Cell lysates were subjected to immunoprecipitation with either an antibody to NKX3.1 (A) or rabbit IgG (B). Four primers (names and sequences are shown in Table 1) spanning five NBSs of PCAN1 promoter region were used for PCR of recovered DNA from the immunoprecipitation (lanes 2–5). Input DNA was used as positive control (C).
Interaction of NKX3.1 with NBS1 or NBS3 stimulates luciferase reporter expression driven by SV40 promoter
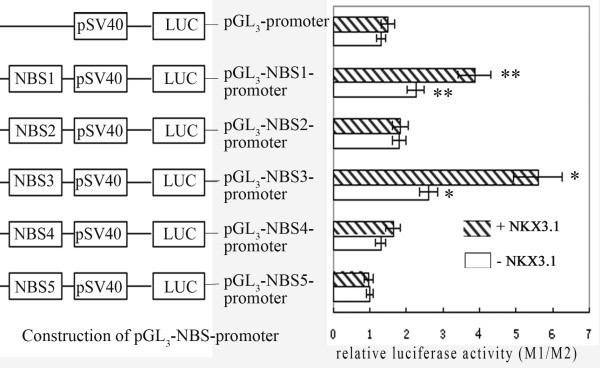
To investigate the interactions between NKX3.1 and these five potential NBSs in vivo, five pGL3-NBS-promoter luciferase plasmids were constructed (Fig. 5) and cotransfected with pcDNA3.1-NKX3.1 plasmid respectively into LNCaP cells, while the control cells were cotransfected with pcDNA3.1 (+) vector. The luciferase reporter assays showed that when cotransfected with NKX3.1 expression plasmid, NBS1 and NBS3 enhanced SV40 promoter activity by 1.7-fold and 2.1-fold, respectively, compared with that of the control cells that were cotransfected with pcDNA3.1 (+) vector. NBS2, NBS4, NBS5 showed no significant effects on SV40 promoter activity. These results suggested that NBS1 and NBS3 were the functional cis-elements in vivo for the upregulation by NKX3.1.
Figure 5.
Effects of NBSs on the SV40 heterogeneous promoter. The sequences of five NBSs were synthesized in vitro and inserted upstream of the SV40 promoter in pGL3-promoter vector generating the pGL3-NBS-promoter plasmids. The five recombinant pGL3-NBS-promoter plasmids or pGL3-promoter vector were then cotransfected with pcDNA3.1-NKX3.1 respectively, to study the effects of NBSs on the activity of the SV40 heterogeneous promoter. Cotransfection of pGL3-NBS-promoter with pcDNA3.1 (+) vector was used as the control. The promoter activities were determined by dual luciferase assays. Results were expressed as relative luciferase activity (M1/M2). The data were represented as the mean of four individual values ± SD. *p < 0.01,**p < 0.01, strip bar vs blank bar.
Deletion of NBS1 and NBS3 in the PCAN1 promoter abolishes the positive regulation by NKX3.1
The binding assays of five NBSs in vivo and in vitro suggested that NBS1 and NBS3 in the PCAN1 promoter were involved in the positive regulation of PCAN1 expression by NKX3.1. To further confirm this observation, the sequences of NBS1, or both NBS1 and NBS3 were deleted from pGL3-pPCAN1 to examine the effects of the deletions on PCAN1 promoter activity. The results in Fig. 6 showed that deletion of NBS1 (pGL3-NBS1id-pPCAN1) or both NBS1 and NBS3 (pGL3-NBS1,3idd-pPCAN1) reduced the promoter activity to 75% or 50% of the wild-type promoter (pGL3-pPCAN1). With cotransfection of the NKX3.1 expression plasmid, deletion of NBS1 partially abolished, and deletion of both NBS1 and NBS3 completely abolished NKX3.1 stimulation of the PCAN1 promoter. These findings suggested that NBS1 and NBS3 upstream of the PCAN1 gene were functional cis-elements mediating the positive regulation by NKX3.1 of PCAN1 gene transcription.
Figure 6.
Effects of deletion of NBS1 and NBS3 on PCAN1 promoter activity. pGL3-pPCAN1 is a luciferase reporter plasmid containing a 2.6 kb wild type PCAN1 promoter; pGL3-NBS-Id represented pGL3-NBS1id-pPCAN1 with deletion of NBS1 from the PCAN1 promoter; pGL3-NBS-Idd represented pGL3-NBS1,3idd-pPCAN1, in which both NBS1 and NBS3 were deleted from the PCAN1 promoter. They were cotransfected with pcDNA3.1-NKX3.1 (grey bar) or pcDNA3.1 (+) vector (blank bar) into LNCaP cells, and the promoter activity was analyzed by dual-luciferase reporter assays. Results were expressed as relative luciferase activities (M1/M2). The data were represented as the mean of four individual values ± SD. *p < 0.01, **p < 0.05, grey bar vs blank bar.
Discussion
NKX3.1 is a prostate-specific homeobox gene that is thought to play important roles in normal prostate development. NKX3.1 protein has been proposed to act as a specific tumor suppressor in prostate. Loss of NKX3.1 expression correlates with prostate carcinogenesis [15] and prostate tumor progression [17]. It was reported that NKX3.1 could collaborate with other transcription factors, such as serum response factor (SRF) [18], Sp-family protein [19] and prostate derived Ets factor (PDEF) [20], to regulate the expression of target genes. The potential for NKX3.1 to exert a differentiating and growth suppressing effect on prostatic epithelium was confirmed by targeted gene disruption of Nkx3.1 in mice [21]. Deletion of either one or both copies of Nkx3.1 resulted in prostatic epithelial hyperplasia and dysplasia that increased in severity with age. Magee et al. [21] has analyzed the expression profiles of prostate tissues from wild-type,Nkx3.1+/-, and Nkx3.1-/- mice and identified Nkx3.1 target genes. However, the genes directly regulated by human NKX3.1 have yet to be identified. In the present study, we found that NKX3.1 upregulated the expression of the PCAN1 gene in LNCaP prostate cancer cells and identified two functional NKX3.1 binding sites upstream of the PCAN1 gene.
PCAN1 is highly expressed in prostate epithelial tissue and was initially identified in a screen for prostate-specific genes. As an important gene in prostate cancer initiation or progression [22], it has been shown that PCAN1 is frequently mutated or deleted in prostate tumor samples [5] and differentially expressed in tumor versus normal prostate tissue, demonstrating a prostate tumor suppressor role [6]. So far, little is known about the regulatory mechanisms of PCAN1 gene expression as well as the relevant regulatory elements and factors. Cross et al. [4] has identified PCAN1 gene in prostate epithelial tissue and demonstrated its expression pattern in different cells. They did an initial characterization of 2.5 kb sequence upstream of the initiation sites with the MatInspector program, in which they found several important transcription factor binding sites, including NKX3.1 binding sites. In our study, we cloned a 2.6 kb PCAN1 promoter sequence and five NKX3.1 binding sites were found in this region with the MatInspector (core/matrix sim: 0.75/optimized). We further performed reporter assays, RNAi, EMSA and ChIP to demonstrate that NKX3.1 could directly bind to NKX3.1 binding sites in the PCAN1 promoter to enhance PCAN1 gene expression in prostate cancer cells. This finding provides a foundation for future studies of the regulatory mechanisms and roles of NKX3.1 on PCAN1 gene expression in prostate development and prostate cancer.
NKX3.1 is proposed to be a nuclear transcriptional factor. It preferentially binds the TAAGTA sequence [23] to regulate its target gene expression. It is reported that NKX3.1 acts as a transcription factor through recruitment of some corepressors [24,25] or coactivators [26,27] to repress or activate its target genes. Our study provided some evidences to show that NKX3.1 directly binds to NKX3.1 binding sites in the PCAN1 promoter to activate PCAN1 gene expression in prostate cancer cells. However, further studies will be required to explore the regulatory mechanisms and the cofactors involved in the regulation. NKX3.1 is a prostate-specific gene required for maintenance of the normal differentiated state of the prostate epithelium. It is proposed to have tumor suppressing function. However, it is not a classic tumor suppressor gene. Despite that loss of function of NKX3.1 predisposes to prostate cancer, it is not sufficient for tumorigenesis. Moreover, while one allele of NKX3.1 is lost by means of chromosomal deletion in prostate cancer, the other allele does not undergo mutational inactivation. These features are not consistent with activities of "classic" tumor suppressor genes. Instead, NKX3.1 appears to act more like a tumor modulator, serving as a regulator of differentiation, which in turn prevents cancer initiation. Our finding that NKX3.1 upregulates PCAN1 expression will give a clue for further exploring the relationships between NKX3.1 and PCAN1 and understanding the functional importance of NXK3.1 in regulating PCAN1 in prostate differentiation and carcinogenesis. Identification of the specific biological functions of NKX3.1 in prostate cancer may provide targets for the early diagnosis and prevention of prostate cancer.
NKX3.1 is an androgen regulated gene and its expression is upregulated by androgens. It is presumed that androgens upregulate PCAN1 expression through increasing NKX3.1 expression. In the experiments we have analyzed the effect of androgen (R1881) on endogenous PCAN1 and NKX3.1 expression in LNCaP cells. Our results showed that R1881 (10-8~10-10 M) increased NKX3.1 expression but have no significant effects on PCAN1 expression in RT-PCR (results are not shown in this paper) and PCAN1 promoter activity in luciferase reporter assays (results are shown in our paper in Cell Mol Biol Lett). So far, it is not very clear whether or not PCAN1 gene expression is regulated directly by androgens. It may be possible that PCAN1 is negatively regulated by androgens and NKX3.1 compensates this effect through positively regulating PCAN1. In addition, AR, NKX3.1 and PCAN1 play different roles in prostate development and cancer, and their interactions in prostate are very complicated and need further to be investigated.
Conclusion
In conclusion, we cloned and characterized the human PCAN1 promoter, and identified two functional NKX3.1 binding sites upstream of the PCAN1 gene, which were involved in the positive regulation by NKX3.1 of PCAN1 gene expression. Both NKX3.1 and PCAN1 are related to prostate development and prostate cancer. Our findings will contribute to the understanding of molecular regulatory mechanisms of PCAN1 gene expression in prostate development and cancer.
Methods
Construction of the NKX3.1 eukaryotic expression plasmid (pcDNA3.1-NKX3.1)
pCR2.1-NKX3.1, a T-clone of NKX3.1 cDNA containing the complete sequence of NKX3.1 (a gift from Dr. Charles Young, Mayo Clinics, USA), was digested with EcoR I (TakaRa, Shiga, Japan) to release the 971 bp fragment of NKX3.1 cDNA. The NKX3.1 cDNA sequence (971 bp) contains the CDS of NKX3.1 including a start codon, a stop codon and a partial 3' UTR. It was then inserted into pcDNA3.1 (+) vector (Invitrogen Life Technologies, San Diego, CA, USA), which had been digested with EcoR I and dephosphorylated with calf intestine alkaline phosphatase (TaKaRa), to generate the NKX3.1-cDNA eukaryotic expression plasmid pcDNA3.1-NKX3.1. The recombinant plasmid was digested with EcoR I to identify the size of the insert and digested with Not I (TaKaRa) to identify the correct insert orientation. The NKX3.1 cDNA was also confirmed by DNA sequencing.
Construction of the PCAN1 promoter-luciferase reporter plasmid (pGL3-pPCAN1)
The 2.6 kb promoter fragment (+32 to -2598) of the PCAN1 gene was amplified by PCR using human genomic DNA as the template. The primer pairs were PCANF 5'-CCCTAGCTAGCCATCTCTGCAGTCTCGAC-3' with a Nhe I site at the 5'-end and PCANR 5'-CCCAAGCTTCGCTCTGACTTCCTCTTC-3' with a Hind III site at the 5'-end. The PCR was conducted at 96°C for 2 min followed by 35 cycles at 98°C for 20 s, 68°C for 10 min. The amplified fragment was isolated and purified following agarose electrophoresis using a Gel Extract Kit (Omega Bil-Tek. Inc. Doraville, GA, USA), digested with Nhe I and Hind III (TaKaRa), and ligated into the equivalent sites of the pGL3-basic vector (Promega, Madison, WI, USA) to generate the pGL3-pPCAN1 construct. The resulting construct was confirmed by restriction enzyme digestion and DNA sequence analysis.
Construction of luciferase reporter plasmids of NKX3.1 binding sites (pGL3-NBS-promoter)
Analysis of the 2.6 kb promoter sequence using MatInspector 2.2 revealed five potential binding sites for the NKX3.1 transcription factor. To confirm the functional binding sites, we synthesized oligonucleotides corresponding to these five sequences (NBS1~NBS5) shown in Table 2. Each NBS sequence was synthesized with an overhanging Mlu I site (CGCGT) at the 5'-end of the sense strand and an overhanging Xho I site (TCGAG) at the 5'-end of the antisense strand. The double-stranded NBS was generated by annealing equal amounts of sense and antisense oligonucleotides at 95°C for 10 min, then cooling to room temperature. The double-stranded NBS was inserted upstream of the SV40 promoter in the pGL3-promoter (Promega) vector to generate the recombinant plasmid of NBS-SV40 promoter-luciferase reporter gene. All constructs were confirmed by DNA sequencing.
Table 2.
Probes used in electrophoretic mobility shift assays
| Names | Sequences (sense strand) |
| NBS1 | GATTAAGTCTACT |
| NBS2 | AGGTAAGTAAAG |
| NBS3 | AAATAAGTAGCCT |
| NBS4 | CAATAAGTTTTAT |
| NBS5 | TGTTAAGTATCCT |
| Mutant NBS1 | GATTCCTGCTACT |
| Mutant NBS3 | AAATCCTGAGCCT |
Construction of the NBS deletion plasmids of pGL3-pPCAN1
The construction of NBS1 or both NBS1 and NBS3 deletion plasmids was made by a two-step PCR procedure. In the first step, pGL3-pPCAN1 was used as the template for the construction of pGL3-NBS1id-pPCAN1, and two PCR fragments were generated with two primer pairs, NBS1id-up 5'-CCCTGACTTGGGGGGCTCC ACTGTAGTAC-3' and PCANF 5'-CCCTAGCTAGCCATCTCTGCAGTCTCGAC-3', NBS1id-down 5'-TGGAGCCCCCCAAGTCAGGGGGTTTAATC-3' and PCANR 5'-CCCAAGCTTCGCTCTGACTTCCTCTTC-3'. The PCR conditions were 94°C for 2 min, 98°C for 10 s, 68°C for 5 min, 32 cycles. The two PCR fragments were purified by agarose electrophoresis and used together as templates in the second step of PCR with primers PCANF and PCANR. The PCR conditions were 94°C for 2 min, 60°C for 30 s, 72°C for 5 min, followed by addition of PCANF and PCANR primers and PCR at 98°C for 10 s, 68°C for 6 min, 32 cycles. The PCR fragments and pGL3-basic were both digested with Hind III and Nhe I and ligated together to construct pGL3-NBS1id-pPCAN1. A similar approach was used to generate pGL3-NBS1, 3idd-pPCAN1, in which both NBS1 and NBS3 sequences were deleted. pGL3-NBS1id-pPCAN1 was used as the template and other two primer pairs, NBS3id-up 5'-GAATGCCTGGTATTTCTTATCAAGAGAAC-3' and PCANF, NBS3id-down 5'-ATAAGAAATACCAGGCATTCCCTTCCACCA-3' and PCANR, were used. The constructed deletion plasmids were validated by DNA sequencing.
Cell culture
The human prostate cancer cell lines LNCaP and PC-3 were obtained from the American Type Culture Collection (ATCC). They were grown at 37°C in 5% CO2 with RPMI 1640 media (Sigma, St. Louis, MO, USA) supplemented with 10% fetal bovine serum (Gibco, BRL Grand Island, NK, USA), ampicillin 100 U/ml and streptomycin 100 U/ml.
Transient transfection
For the promoter activity assay of pGL3-pPCAN1, LNCaP and PC-3 cells were transfected with lipofectamin™ 2000 (Invitrogen) in 24-well plates. Each well included 1.5 × 105 cells, 1.0 μg pGL3-pPCAN1, 0.04 μg internal control vector pRL-TK (Promega), 2 μl lipofectamin™ 2000 and 500 μl RPMI 1640 media without serum and antibiotics. The cells were analyzed using a dual-luciferase reporter assay system (Promega) 48 h after completion of the transfection procedure.
For cotransfection experiments of the NKX3.1 expression plasmid (pcDNA3.1-NKX3.1) with pGL3-construct (pGL3-pPCAN1, pGL3-NBS-promoter, pGL3-NBS1id-pPCAN1 or pGL3-NBS1,3idd-pPCAN1), LNCaP cells were transfected with lipofectamin™ 2000 in 24-well plates, and each well included 1.5 × 105 cells, 0.8 μg pGL3-construct, 0.4 μg pcDNA3.1-NKX3.1, 0.04 μg pRL-TK, 2 μl lipofectamin™ 2000 and 500 μl RPMI 1640 media without serum and antibiotics.
Dual-luciferase reporter assays
Forty-eight hours after the transfection, the activities of Firefly luciferase in pGL3-constructs and Renilla luciferase in pRL-TK were determined by the dual-luciferase reporter assays following the protocol of the manufacture (Promega). The cells were rinsed with phosphate-buffered saline, and lysed with 1 × passive lysis buffer. Twenty μl of cell lysate was transferred into the luminometer tube containing 100 μl luciferase assay reagent II. Firefly luciferase activity (M1) was measured first, and then Renilla luciferase activity (M2) was determined after the addition of 100 μl Stop & Glo reagent. M1/M2 was taken as the relative luciferase activity of the pGL3-constructs.
Reverse transcription-PCR
Total RNA was isolated from LNCaP and PC-3 cells using Trizol reagent (Invitrogen) 48 h after transfection with pcDNA3.1-NKX3.1, and expression of PCAN1 mRNA was determined by RT-PCR with M-MuL V reverse transcriptase (Promega) in the presence of random hexamer primers. PCR primers for PCAN1 were PCAN-F 5'-GCGATGTGCTGTGAAATCTA-3', PCAN-R 5'-CTTTCACATTCCCCGTGGT-3'; for NKX3,1 were NKX-F 5'-GTACCTGTCGGCCCCTGAACG-3', NKX-R 5'-GCTGTTATACACGGAGACCAGG-3'. A β-actin mRNA was amplified and used to normalize the quantity of the PCAN1 mRNA in RT-PCR. The primers were β-actinF 5'-GCTGTCAGAGTGGTTATGT-3', β-actinR 5'-ACATTGACGTACAGAG AGAG-3'. The PCR conditions were 94°C 3 min, 94°C 30 s, 56°C 30 s for PCAN1, 63°C 30 s for NKX3.1, 72°C 50s, 32 cycles for PCAN1, 26 cycles for NKX3.1, 72°C 6 min. The products were identified by 1.5% agarose gel electrophoresis.
Western blot analysis of NKX3.1 protein expression
Expression of NKX3.1 in prostate cancer cells was analyzed by Western blot analysis. Briefly, total protein was extracted from PC-3 cells or LNCaP cells using lysis buffer (containing 50 mM Tris-Cl, pH 8.0, 150 mM NaCl, 0.1%SDS, 1%NP-40, 100 μg/ml PMSF) after transfection with pcDNA3.1-NKX3.1 for 48 h to 72 h. The protein content of the samples was measured using the BCA protein assay kit (Shenergy Biocolor Bioscience & Technology Company, Shanghai, China). Thirty μg of each protein sample was used to detect the NKX3.1 protein expression by Western blot analysis. The primary antibody was rabbit anti-human NKX3.1 (RDI, Concord MA, USA) diluted 1: 2000; the second antibody was goat anti-rabbit IgG (Sigma) diluted 1: 2000. Relative protein levels were calculated in comparison to β-actin as standard. Immunoblots were detected using an ECL kit (Santa Cruz, CA, USA) and visualized after exposure to X-ray film.
Electrophoretic mobility shift assays (EMSA)
Nuclear extracts were prepared from LNCaP cells using a nuclear extraction kit (Active Motif, Carlsbad, CA, USA) following the manufacturer's instructions. Oligonucleotides corresponding to the five binding sites shown in Table 2 were synthesized as probes. Equal amounts of sense and antisense oligonucleotides of NBSs were mixed and annealed in a buffer (10 mM Tris-HCl, pH 8.0, 200 mM NaCl, 1 mM EDTA) by heating to 95°C for 5 min and cooling slowly to room temperature. The five double-stranded NBSs were labelled with digoxigenin (DIG) (Roche). Binding reactions were performed for 20 min at room temperature in a 20 μl mixture containing 0.2% (W/V) Tween-20, 1 mM EDTA, 1 mM dithiothreitol, 30 mM KCl, 20 mM HEPES (pH 7.6), 1 μg of poly (dI-dC), 0.1 μg of poly (L-Lys), 20 μg of nuclear extract and 0.8 ng of DIG labelled double-stranded NBS. For the competition experiment, unlabelled double-stranded NBS or mutant NBS (sequences are shown in Table 2) in 250-fold excess were added to the binding reaction mixture and incubated. For supershift assays, anti-NKX3.1 antibody was pre-incubated with the nuclear extracts at room temperature for 30 min in the binding buffer, followed by an additional incubation for 20 min at room temperature with the reaction mixtures. Bound and free oligonucleotide probes were resolved by electrophoresis on an 8% nondenaturing polyacrylamide gel in 0.25 × Tris-Boric acid (TBE) buffer. Electroblotting and chemiluminescence detection were performed based on the instructions of the manufacturer of the DIG gel shift kit (Roche, Penzberg, Germany).
Chromatin immunoprecipitation (ChIP)
In vivo binding of NKX3.1 to the NBSs in the upstream region of the PCAN1 gene was investigated using the ChIP assay kit (Upstate Biotechnology, Inc., Lake Placid, NY, USA). Confluent human LNCaP prostate cancer cells were transfected with pcDNA3.1-NKX3.1. Forty-eight hours after the transfection, cells were treated with formaldehyde (1% final concentration) to cross-link NKX3.1 to the DNA. Cells were washed with cold phosphate-buffered saline and lysed in SDS lysis buffer (1% SDS, 10 mM EDTA, and 50 mM Tris-HCl pH 8.1). The lysate was sonicated to shear DNA to a length between 200 and 1000 bp. The sonicated supernatant was diluted 10-fold with ChIP dilution buffer (0.01% SDS, 1% Triton X-100, 2 mM Tris-HCl pH 8.1), and 150 mM NaCl) and incubated with anti-NKX3.1 antibody (Santa Cruz) or rabbit IgG overnight at 4°C with rotation. To collect DNA/protein complexes, salmon sperm DNA/protein A-agarose slurry was added to the mixture and incubated for 1 h at 4°C with rotation, and the DNA/protein A-agarose was pelleted by centrifugation. After extensive washing of the pellet with a series of wash buffers, the pellet was dissolved with 250 μl of elution buffer and centrifuged to remove the agarose. The supernatant was treated with 20 μl of 5 M NaCl and heated to 65°C for 4 h to reverse the NKX3.1-DNA cross-link. After treatment with EDTA and proteinase K, the supernatant was extracted with phenol/chloroform and precipitated with ethanol to recover the DNA. For PCR using the chromatin-immunoprecipitated DNA, one-tenth of the DNA was PCR-amplified using four pairs of primers (primer names and sequences are shown in Table 1) that span the five NBSs in the upstream region of the PCAN1 gene respectively. Twenty-five cycles of PCR at 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s were performed. PCR products were analyzed in 1% agarose gels.
RNA interference (RNAi)
pRNAT-U6.1/Neo, containing human U6 promoter, was used to generate a series of RNAi expression vectors by inserting annealed oligonucleotides between BamH I and Hind III sites. The oligonucleotides RNAi1 (5'GATCCCCagttcagccatcagaagtaTT CAAGAGAtacttctgatggctgaactTTTTTGGAAA3' and 3'GGGtcaagtcggtagtcttcatAA GTTCTCTatgaagactaccgacttgaAAAAACCTTTTCGA5'), RNAi2 (5'GATCCCCtcca gaacagacgctataaTTCAAGAGAttatagcgtctgttctggaTTTTTGGAAA3' and 3'GGGagg tcttgtctgcgatattAAGTTCTCTaatatcgcagacaagacctAAAAACCTTTTCGA5'), RNAi3 (5'GATCCCCtataacagctatccttactTTCAAGAGAagtaaggatagctgttataTTTTTGGAAA3'and 3'GGGatattgtcgataggaatgaAAGTTCTCTtcattcctatcgacaatatAAAAACCTTTTC GA5') were used for the construction of pRNAT-RNAi (1~3) targeting human NKX3.1. The oligonucleotides RiN (5'GATCCCCttctccgaacgtgtcacgtTTCAAGAGA acgtgacacgttcggagaaTTTTTTGGAAA3' and 3' GGGaagaggcttgcacagtgcaAAGTTCT CTtgcactgtgcaagcctcttAAAAACCTTTTCGA5') were used for a control vector pRNAT-RiN producing a random sequence of RNAi. pRNAT-RNAi (1–3) and pRNAT-RiN were transfected into LNCaP cells, Forty-eight hours post-transfection, the cells were harvested to detect the silencing of NKX3.1 with RT-PCR and Western blot. The stable cell lines transfected with pRNAT-RNAi1 or pRNAT-RiN were selected with G418 (20 μg/ml) and individual clones were isolated.
Authors' contributions
WL assisted in the design of the study, carried out the cloning of expression vectors and reporter plasmid constructs, transfection studies, RT-PCR, EMSA and ChIP; helped draft the manuscript. PZ assisted in the design of the study, participated in the sequence alignment and use of computer database; assisted in the construction of recombinant vectors, EMSA, ChIP and RNAi; helped draft the manuscript.WC participated in maintenance of cell lines, performed luciferase reporter assays and assisted with the transfection experiments. CY carried out the isolation of RNA and purification of plasmids, assisted with the RT-PCR and ChIP. FC carried out Western blot and assisted with the purification of plasmids. FK assisted with the maintenance of cell lines and the transfection experiments. JZ participated in the design of the study, assisted with the revising of the manuscript. AJ participated in the design of the study and revision of the manuscript, helped draft the manuscript, assisted with the construction of recombinant vectors, EMSA and ChIP.
All authors have read and approved the final manuscript.
Acknowledgments
Acknowledgements
The authors are grateful to Dr. Charles Young for kindly providing pCR2.1-NKX3.1 plasmid used in this study. This work was supported by Natural Science Foundation of Shandong Province (No. Y2007C96) and the National Natural Science Foundation of China (No.30470952 and No.30670581).
Contributor Information
Wenwen Liu, Email: liuwenwen_87@126.com.
Pengju Zhang, Email: zhpj@sdu.edu.cn.
Weiwen Chen, Email: chenweiwen@sdu.edu.cn.
Chunxiao Yu, Email: Yu_chunxiao@tom.com.
Fuai Cui, Email: cuifuai@sdu.edu.cn.
Feng Kong, Email: kongfeng@sdu.edu.cn.
Jianye Zhang, Email: zhjy@sdu.edu.cn.
Anli Jiang, Email: jianganli@sdu.edu.cn.
References
- Jemal A, Siegel R, Ward E, Murray T, Xu J, Smigal C, Thun MJ. Cancer statistics, 2006. CA Cancer J Clin. 2006;56:106–130. doi: 10.3322/canjclin.56.2.106. [DOI] [PubMed] [Google Scholar]
- Saad F, Al Dejmah A, Perrotte P, McCormack M, Benard F, Valiquette L, Karakiewicz PI. Therapeutic approach to hormone-refractory prostate cancer. Can J Urol. 2006;13:52–56. [PubMed] [Google Scholar]
- Di Lorenzo G, De Placido S. Hormone refractory prostate cancer (HRPC): present and future approaches of therapy. Int J Immunopathol Pharmacol. 2006;19:11–34. [PubMed] [Google Scholar]
- Cross D, Reding DJ, Salzman SA, Zhang KQ, Catalona WJ, Burke J, Burmester JK. Expression and initial promoter characterization of PCAN1 in retinal tissue and prostate cell lines. Med Oncol. 2004;21:145–153. doi: 10.1385/MO:21:2:145. [DOI] [PubMed] [Google Scholar]
- Reding DJ, Zhang KQ, Salzman SA, Thomalla JV, Riepe RE, Suarez BK, Catalona WJ, Burmester JK. Identification of a gene frequently mutated in prostate tumors. Med Oncol. 2001;18:179–187. doi: 10.1385/MO:18:3:179. [DOI] [PubMed] [Google Scholar]
- Olsson P, Bera TK, Essand M, Kumar V, Duray P, Vincent J, Lee B, Pastan I. GDEP, a new gene differentially expressed in normal prostate and prostate cancer. Prostate. 2004;48:231–241. doi: 10.1002/pros.1102. [DOI] [PubMed] [Google Scholar]
- Sciavolino PJ, Abrams EW, Yang L, Austenberg LP, Shen MM, Abate-Shen C. Tissue-specific expression of murine Nkx 3.1 in the male urogenital system. Dev Dyn. 1997;209:127–138. doi: 10.1002/(SICI)1097-0177(199705)209:1<127::AID-AJA12>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Bieberich CJ, Fujita K, He WW, Jay G. Prostate-specific and androgen dependent expression of a novel homeobox gene. J Biol Chem. 1996;271:31779–31782. doi: 10.1074/jbc.271.50.31779. [DOI] [PubMed] [Google Scholar]
- Bhatia-Gaur R, Donjacour AA, Sciavolino PJ, Kim M, Desai N, Young P, Norton CR, Gridley T, Cardiff RD, Cunha GR, Abate-Shen C, Shen MM. Roles for Nkx3.1 in prostate cancer development and cancer. Genes Dev. 1999;13:966–977. doi: 10.1101/gad.13.8.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdulkadir SA, Magee JA, Peters TJ, Kaleem Z, Naughton CK, Humphrey PA, Milbrandt J. Conditional loss of Nkx3.1 in adult mice induces prostatic intraepithelial neoplasia. Mol Cell Biol. 2002;22:1495–1503. doi: 10.1128/MCB.22.5.1495-1503.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voeller HJ, Augustus M, Madike V, Bova GS, Carter KC, Gelmann EP. Coding region of NKX3.1, a prostate-specific homeobox gene on 8p21, is not mutated in human prostate cancers. Cancer Res. 1997;57:4455–4459. [PubMed] [Google Scholar]
- Dong JT. Chromosomal deletions and tumor suppressor genes in prostate cancer. Cancer Metastasis Rev. 2001;20:173–193. doi: 10.1023/A:1015575125780. [DOI] [PubMed] [Google Scholar]
- Jiang AL, Hu XY, Zhang PJ, He ML, Kong F, Liu ZF, Yuan HQ, Zhang JY. Up-regulation of NKX3.1 expression and inhibition of LNCaP cell proliferation induced by an inhibitory element decoy. Acta Biochim Biophys Sin (Shanghai) 2005;37:335–340. doi: 10.1111/j.1745-7270.2005.00047.x. [DOI] [PubMed] [Google Scholar]
- Jiang AL, Zhang PJ, Chen WW, Liu WW, Yu CX, Hu XY, Zhang XQ, Zhang JY. Effects of 9-cis retinoic acid on human homeobox gene NKX3.1 expression in prostate cancer cell line LNCaP. Asian J Androl. 2006;8:435–441. doi: 10.1111/j.1745-7262.2006.00171.x. [DOI] [PubMed] [Google Scholar]
- Kim MJ, Bhatia-Gaur R, Banach-Petrosky WA, Desai N, Wang Y, Hayward SW, Cunha GR, Cardiff RD, Shen MM, Abate-Shen C. Nkx3.1 mutant mice recapitulate early stages of prostate carcinogenesis. Cancer Res. 2002;62:2999–3004. [PubMed] [Google Scholar]
- Liu W, Chen W, Zhang P, Yu C, Kong F, Deng J, Zhang J, Jiang A. Molecular cloning and analysis of the human PCAN1 (GDEP) promoter. Cell Mol Biol Lett. 2007;12:482–492. doi: 10.2478/s11658-007-0016-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen C, Bubendorf L, Voeller HJ, Slack R, Willi N, Sauter G, Gasser TC, Koivisto P, Lack EE, Kononen J, Kallioniemi OP, Gelmann EP. Loss of NKX3.1 expression in human prostate cancers correlates with tumor progression. Cancer Res. 2000;60:6111–6115. [PubMed] [Google Scholar]
- Ju JH, Maeng JS, Zemedkun M, Ahronovitz N, Mack JW, Ferretti JA, Gelmann EP, Gruschus JM. Physical and functional interactions between the prostate suppressor homeoprotein NKX3.1 and serum response factor. J Mol Biol. 2006;360:989–999. doi: 10.1016/j.jmb.2006.05.064. [DOI] [PubMed] [Google Scholar]
- Simmons SO, Horowitz JM. Nkx3.1 binds and negatively regulates the transcriptional activity of Sp-family members in prostate-derived cells. Biochem J. 2006;393:397–409. doi: 10.1042/BJ20051030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Bieberich CJ. Structural and functional analysis of domains mediating interaction between NKX-3.1 and PDEF. J Cell Biochem. 2005;94:168–177. doi: 10.1002/jcb.20297. [DOI] [PubMed] [Google Scholar]
- Magee JA, Abdulkadir SA, Milbrandt J. Haploinsufficiency at the Nkx3.1 locus A paradigm for stochastic, dosage-sensitive gene regulation during tumor initiation. Cancer cell. 2003;3:273–283. doi: 10.1016/S1535-6108(03)00047-3. [DOI] [PubMed] [Google Scholar]
- Vasmatzis G, Essand M, Brinkmann U, Lee B, Pastan I. Discovery of three genes specifically expressed in human prostate by expressed sequence tag database analysis. Proc Natl Acad Sci USA. 1998;95:300–304. doi: 10.1073/pnas.95.1.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David JS, Domenica G, Edward PG. DNA-binding sequence of the human prostate-specific homeodomain protein NKX3.1. Nucleic Acids Res. 2000;28:2389–2395. doi: 10.1093/nar/28.12.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhr J, Andersson E, Persson M, Jessell TM, Ericson J. Groucho-mediated transcriptional repression establishes progenitor cell pattern and neuronal fatein the ventral neural tube. Cell. 2001;104:861–873. doi: 10.1016/S0092-8674(01)00283-5. [DOI] [PubMed] [Google Scholar]
- Choi CY, Kim YH, Kwon HJ, Kim Y. The homeodomain protein NK-3 recruits Groucho and a histone deacetylase complex to repress transcription. J Biol Chem. 1999;274:33194–33197. doi: 10.1074/jbc.274.47.33194. [DOI] [PubMed] [Google Scholar]
- Carson JA, Fillmore RA, Schwartz RJ, Zimmer WE. The smooth muscle gamma-actin gene promoter is a molecular target for the mouse bagpipe homologue, mNkx3-1, and serum response factor. J Biol Chem. 2000;275:39061–39072. doi: 10.1074/jbc.M006532200. [DOI] [PubMed] [Google Scholar]
- Nishida W, Nakamura M, Mori S, Takahashi M, Ohkawa Y, Tadokoro S, Yoshida K, Hiwada K, Hayashi K, Sobue K. A triad of serum response factor and the GATA and NK families governs the transcription of smooth and cardiac muscle genes. J Biol Chem. 2002;277:7308–7317. doi: 10.1074/jbc.M111824200. [DOI] [PubMed] [Google Scholar]