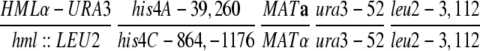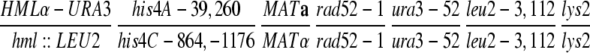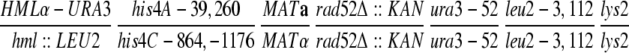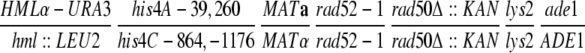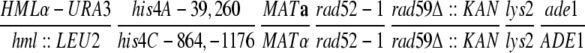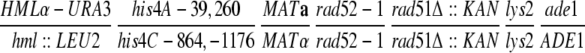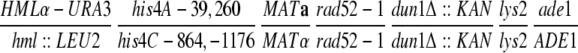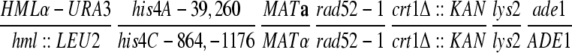Abstract
In wild-type diploid cells, heteroallelic recombination between his4A and his4C alleles leads mostly to His+ gene conversions that have a parental configuration of flanking markers, but ∼22% of recombinants have associated reciprocal crossovers. In rad52 strains, gene conversion is reduced 75-fold and the majority of His+ recombinants are crossover associated, with the largest class being half-crossovers in which the other participating chromatid is lost. We report that UV irradiating rad52 cells results in an increase in overall recombination frequency, comparable to increases induced in wild-type (WT) cells, and surprisingly results in a pattern of recombination products quite similar to RAD52 cells: gene conversion without exchange is favored, and the number of 2n − 1 events is markedly reduced. Both spontaneous and UV-induced RAD52-independent recombination depends strongly on Rad50, whereas rad50 has no effect in cells restored to RAD52. The high level of noncrossover gene conversion outcomes in UV-induced rad52 cells depends on Rad51, but not on Rad59. Those outcomes also rely on the UV-inducible kinase Dun1 and Dun1's target, the repressor Crt1, whereas gene conversion events arising spontaneously depend on Rad59 and Crt1. Thus, there are at least two Rad52-independent recombination pathways in budding yeast.
IN budding yeast, the Rad52 protein is required for efficient mitotic recombination (for a review see Krogh and Symington 2004). Deletion of RAD52 is epistatic to loss of any other gene implicated in homologous recombination, including the recombinase Rad51, Rad54, the Rad51 paralogs, Rad55 and Rad57, Mre11-Rad50-Xrs2 (MRX), and Rad59. There are at least two major Rad52-dependent pathways, one requiring Rad51, Rad55, Rad57, and Rad54 and the other independent of Rad51, but requiring Rad59 and MRX. Rad51-dependent and independent pathways have been identified among spontaneous recombination events, in the repair of eroding telomeres in cells lacking telomerase, as well as those induced by a double-strand break. Rad52's homolog, Rad22, is also a central protein in recombination in fission yeast; but curiously, in many higher eukaryotes, even though biochemical interactions between Rad52 and Rad51 appear to be preserved, Rad52 plays a surprisingly minor role in homologous recombination (Benson et al. 1998; Yamaguchi-Iwai et al. 1998). In fact, in Caenorhabditis, Rad52 does not appear to have been retained. In many metazoans, Rad52 function may have been taken over by BRCA2 (Wong et al. 1997; Chen et al. 1998; Yuan et al. 1999; Moynahan et al. 2001; Brough et al. 2007; Thorslund et al. 2007).
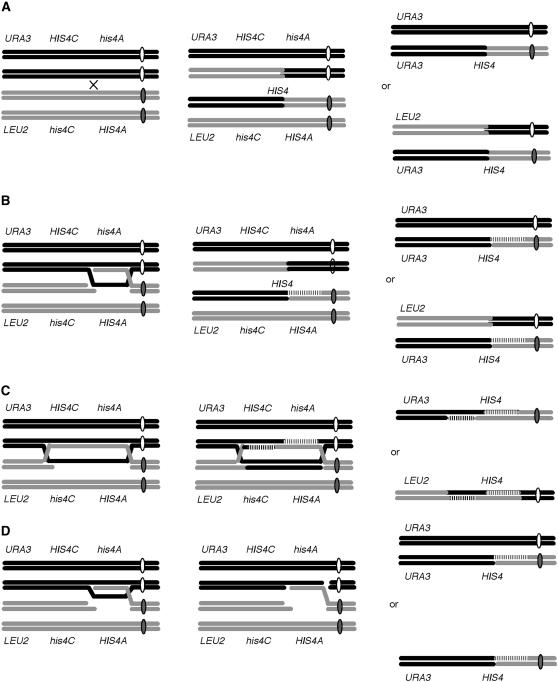
Even in Saccharomyces, there are several Rad52-independent mechanisms of double-strand break (DSB) repair. Single-strand annealing becomes Rad52 independent when homologous regions are several kilobases long, as between ribosomal rDNA repeats, although Rad52 is crucial when homologies flanking a DSB are <1 kb (Ozenberger and Roeder 1991; Sugawara et al. 2000). A rare class of survivors following loss of telomerase is also Rad52 independent (Maringele and Lydall 2004). In haploids and diploids selected for recombination between heteroalleles, the absence of Rad52 reduces the rate of recombination by 50- to 100-fold. Strikingly, the great majority of outcomes are crossover associated, in contrast to a relatively low rate of crossover seen in isogenic wild-type strains. Haber and Hearn (1985) analyzed mitotic recombination in rad52 diploids heteroallelic for pairs of nonreverting frameshift mutations in his4C and in his4A and found recombination was reduced to 1.3% of wild-type (WT) levels. Most crossovers resulted from events that are rarely seen in WT cells. Most rad52-independent His+ recombination events had undergone the loss of one of the participating chromosomes and the retained chromosome in the 2n − 1 diploid was nearly always a crossover recombinant (Figure 1D and Figure 2A). Second, in wild-type cells, only 11% of the HIS4 gene conversions were accompanied by loss of heterozygosity (LOH) of a distal marker, among which most (82%) became homozygous for URA3, as expected if gene conversions resulted either from a simple crossing over between his4 regions to join HIS4C and HIS4A or after heteroduplex formation or gap repair that did not cover both his4C and his4A (Figure 1, A and B). Heteroduplex formation could occur either in G1 or in G2, with crossing over completed after DNA replication (Esposito 1978). However, in rad52 diploids, among the 42% of His+ recombinants that present LOH, 74% unexpectedly became homozygous for the LEU2 marker distal to his4C (Haber and Hearn 1985) (see also Figure 2A). This outcome is most likely the result of heteroduplex formation covering both his4C and his4A followed by independent mismatch correction of each pair of mismatches (Figure 1C). Thus Rad52-independent recombination is not simply a marked reduction in a normal spectrum of events but the consequences of a different pathway of recombination.
Figure 1.—
Mechanisms of recombination between his4A and his4C alleles yielding His+ recombinants and loss of heterozygosity of a distal marker. (A) A simple crossover in the interval between HIS4C and HIS4A will lead either to a diploid homozygous for URA3 or to one with distal markers in reversed orientation, depending on which chromatids segregate together. (B) Strand invasion after a double-strand break that creates an asymmetric heteroduplex covering only his4A; this heteroduplex can be resolved by mismatch correction (vertical hatch marks) and reciprocal crossing over to yield either a HIS4 diploid with a reversed linkage of the distal URA3 and LEU2 markers or a diploid homozygous for URA3. Gene conversions without crossing over will retain a parental arrangement of markers. A similar outcome will prevail if the heteroduplex covers only his4C and similar outcomes are expected for spontaneous events initiated by single-stranded DNA lesions. (C) HIS4-containing recombinant chromosomes carrying LEU2 can be recovered if heteroduplex DNA covers both his4A and his4C and there is independent mismatch correction of these regions. Here only the HIS4-containing recombinant is shown, which can segregate with either sister chromatid of the opposite sister centromere. (D) Two outcomes of chromosome loss associated with recombination at HIS4.
Figure 2.—
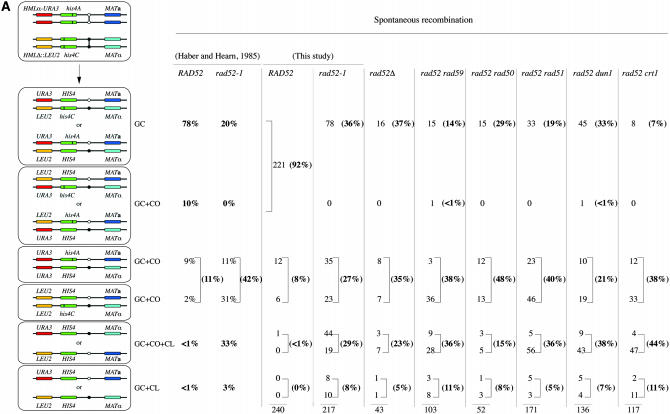
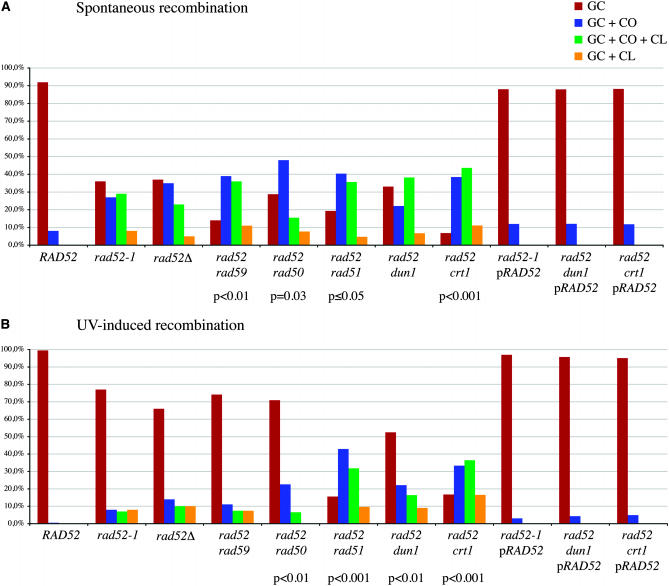
The distribution of recombination products in rad52 double mutants. (A) Spontaneous recombination. (B) UV-induced recombination. GC, gene conversion; GC + CO, gene conversion with associated crossing over; GC + CO + CL, gene conversion with associated crossing over and chromosome loss; GC + CL, gene conversion with associated chromosome loss.
UV irradiation is a potent inducer of recombination (Nakai and Mortimer 1969; Galli and Schiestl 1995). Since cells lacking Rad52 are hypersensitive to ionizing radiation, but show very little sensitivity to UV, we wondered if UV would stimulate Rad52-independent recombination. To our surprise, the events stimulated by UV in rad52 diploids were strikingly different from those arising spontaneously: the majority of recombinants had a pattern of recombination reminiscent of spontaneous (or UV-induced) WT recombination events. We have characterized these events and determined some of the genetic requirements of both spontaneous and UV-stimulated Rad52-independent recombination. A central role for the MRX protein Rad50 emerged for both spontaneous and UV-induced events. Both Rad51 and the UV-regulated Crt1 were important in promoting gene conversion spontaneously and after UV irradiation. Among spontaneous Rad52-independent recombinants, Rad59 also plays an important role.
MATERIALS AND METHODS
Strains and plasmids:
The relevant genotypes of the strains used are listed in Table 1. The construction of MH134 and MH160 was described previously (Haber and Hearn, 1985). Strains AL103 (rad50Δ), AL114 (rad59Δ), AL117 (rad51Δ), AL119 (dun1Δ), and TF122 (crt1Δ) were constructed by transforming the haploid strains U319 and U325 with an appropriate KAN-MX cassette. To overcome the homologous recombination defect of rad52 strains, these strains were first transformed with a HIS4-containing centromeric plasmid bearing a WT copy of RAD52 (pJH1035). The KAN-MX fragment was amplified from the Research Genetics (Birmingham, AL) deletion strains using primers also amplifying 500 bp upstream and downstream of the ORF to be deleted. Transformants were checked by colony PCR. For complementation analysis, a NAT-MX-containing centromeric plasmid bearing a WT copy of RAD52 (pTF01) was introduced into each double-mutant strain.
TABLE 1.
Yeast strains
| Strain | Genotype | Reference | |
|---|---|---|---|
| U319 | hml∷LEU2 his4C-864,-1176 MATα rad52-1 ura3-52 leu2-3,112 lys2 | Haber and Hearn (1985) | |
| U325 | HMLα-URA3 his4A-39,260 MATarad52-1 ura3-52 leu2-3,112 lys2 ade1 | Haber and Hearn (1985) | |
| MH134 |
|
Haber and Hearn (1985) | |
| MH160 |
|
Haber and Hearn (1985) | |
| ECY526-7 |
|
This study | |
| AL103 |
|
This study | |
| AL114 |
|
This study | |
| AL117 |
|
This study | |
| AL119 |
|
This study | |
| TF122 |
|
This study |
Selection of recombinants:
HIS4 (His+) recombinants were obtained by replica plating patches of diploid cells on YEPD to medium lacking histidine. After incubation for up to 5 days, a single His+ papillus from each patch was selected to ensure independence of the recombination event. UV-induced recombinants were selected by irradiating the replica-plated patches at 15 J/m2 prior to incubation.
Analysis of recombination:
Recombinant papillae were patched on medium lacking histidine and replica plated to SC −leu and SC −ura media. Recombinants were also tested for their mating type and for the linkage between the centromere-distal markers and MAT alleles by replica plating on minimum plates containing 200 μl of YEPD and a lawn of MATa or MATα tester strains that were either ura3 or leu2.
Determination of recombination rates and frequencies:
The spontaneous rate of His+ prototroph formation was determined by a fluctuation test (method of the median). Cultures were grown overnight to saturation, diluted in a large volume to a concentration of 102 cells/ml, and aliquoted to a minimum of nine tubes. These nine cultures were incubated at 30° to a concentration of 108 cells/ml and dilutions were plated on both YEPD and SC −his media. The rate of spontaneous His+ prototroph formation was determined analytically using the method of Lea and Coulson (1949).
UV sensitivity and the frequency of UV-induced His+ prototroph formation were determined by exposing strains on YEPD and on SC −his media to increasing doses of UV. Frequency of recombination was calculated after correcting for concentration and normalizing against survival on YEPD plates at the corresponding UV dose as well as spontaneous recombination.
Each experiment was repeated at least three times.
RESULTS
UV irradiation stimulates Rad52-independent recombinants different from those arising spontaneously:
We investigated whether UV irradiation would stimulate Rad52-independent recombination. Spontaneous recombination rates are very low in the rad52-1 strain: 6.9 His+/108 cells/generation compared to 440 His+/108 cells/generation in WT cells. As described previously (Prakash et al. 1980), we found that the rad52 homozygous diploid is only slightly sensitive to UV (Figure 3). In rad52 cells UV-induced recombination is reduced compared to WT almost to the same extent as spontaneous recombination (16- to 24-fold according to the dose of UV compared to 65-fold spontaneously, Figure 3), but recombination frequencies were still inducible: the frequency of Rad52-independent recombination is increased from 1.6 His+/108 survivors at 10 J/m2 to 12.9/108 at 30 J/m2. We presume that all of the His+ colonies arising in diploids heteroallelic for the his4A and his4C alleles arose by recombination since both alleles are double-frameshift mutations that show virtually no reversion (Jackson and Fink 1981).
Figure 3.—
Spontaneous rates and UV-induced frequencies of recombination in WT and rad52 strains. (A) Rate of spontaneous recombination in WT as compared to the rad52 single mutant. (B) Rate of spontaneous recombination in rad52 strains also deleted for recombination factors or UV-induced damage response factors. (C) UV sensitivity and UV-induced recombination in WT and rad52 strains. Rates in A–C were calculated for at least three independent experiments and the calculated standard error is represented by vertical bars. (D) UV sensitivity and UV-induced recombination in the rad52 strains studied. Error bars have been omitted for reasons of clarity. Only rad52 rad50Δ has been found to be significantly hyporecombinogenic and rad52 dun1Δ to be more sensitive to UV and hyperrecombinogenic compared with rad52.
In the study by Haber and Hearn (1985), the arrangement of markers along each of the two chromosomes after recombination in WT cells was analyzed through meiosis to separate the two chromosomes III and to determine without ambiguity the linkage between the HIS4 allele and the markers URA3, LEU2, and MAT. In rad52 cells, they took advantage of rare spontaneous chromosome loss observed in rad52 homozygous diploids (Mortimer et al. 1981) to analyze the configuration of alleles among recombinants. Here we analyzed spontaneous and UV-induced Rad52-independent recombinants in the same way (see materials and methods). Four recombination outcomes were observed (Figure 2):
Gene conversion without associated crossing over (GC) occurs when one of the his4 alleles is converted to the HIS4 allele, resulting in the parental phenotype of His+ Leu+ Ura+ and nonmating (i.e., MATa/MATα).
Gene conversions associated with chromosome loss (GC + CL) result in the conversion of one his4 allele to HIS4 and the concomitant loss of the homologous chromosome. GC + CL recombinants carry either only the LEU2 or the URA3 marker and show a mating-type phenotype corresponding to the parental configuration of alleles (LEU2 MATα or URA3 MATa).
Gene conversion may also be associated with a crossover (GC + CO) in which the flanking LEU2 and URA3 markers are exchanged. Depending on the mitotic segregation of the four chromatids at mitosis, three GC + CO phenotypes are possible. In two of these, the cells become homozygous either for LEU2 or for URA3 and are therefore auxotrophic for uracil or leucine, respectively, but they are still MATa/MATα nonmaters. The third GC + CO phenotype is phenotypically identical to the parental phenotype, although LEU2 is now linked to MATa instead of to MATα and URA3 is linked to MATα.
Gene conversion can also be associated with both crossing over and chromosome loss (GC + CO + CL). The recombinant mates strongly as either MATa or MATα and is auxotrophic for either uracil or leucine. What differentiates these recombinants from those in the GC + CO category is that they are 2n − 1 monosomic for chromosome III and have a strong MATa or MATα phenotype.
The analysis of 240 RAD52 and 217 rad52-1 spontaneous recombinants confirmed the results observed by Haber and Hearn (1985). In this study, WT recombinants were mostly the result of GC (78%). The rest were equally distributed between GC + CO with reverse linkage of the markers and GC + CO associated with LOH of the markers (Figures 2 and 4).
Figure 4.—
Proportions of recombinant classes. (A) Spontaneous recombination. (B) UV-induced recombination. Each bar represents the percentage of a given recombinant class from Figure 1. The probability (p) to have a distribution of classes of recombinants that is statistically different in the double mutants from rad52 through a chi-square calculation is shown when significant. Complementation experiments with a RAD52-bearing plasmid (pRAD52) in strains rad52-1, rad52-1 dun1Δ, and rad52-1 crt1Δ are also shown.
We confirmed that the rad52-1 homozygous diploid produces a very different spectrum of recombinants (Figures 2 and 4 and supplemental Table 1). Approximately 35% of the events were solely GC and 8% GC + CL. The remaining recombination events were associated with CO: 27% were GC + CO and another 29% were GC + CO + CL. Among the GCs + COs, none showed the simple reversed linkage arrangement expected if both participating chromatids segregated together. The inability to recover both participating chromatids may be the result of instability of the recombination intermediate and a high rate of CL. That GC + CO and GC + CO + CL are in the same proportions suggests that the two classes are the result of random segregations of the HIS4-bearing chromatid with either the chromatid of the homologous chromosome that participated in recombination (but was lost) or the chromatid that was not involved in the exchange. These results confirm that RAD52-independent recombination in vegetative cells occurs through a mechanism distinct from that in WT.
Surprisingly, UV-induced rad52 recombinants resembled WT recombinants (Figures 2 and 4). In WT cells, UV-induced recombinants were virtually all the result of GC. Only one event of GC + CO associated with LOH was detected among 207 recombinants. We did not analyze WT recombinants through meiosis because the very low proportion of GC + CO associated with LOH suggests that the proportion of GC + CO with a reverse linkage is also very low. In rad52 homozygous diploids, 78% of the recombination events were solely GC and only 15% were associated with a CO. Therefore, UV induces a very different mechanism of repair involving mostly GC.
Because the rad52-1 allele used in this study is a missense mutation with a null phenotype (Adzuma et al. 1984), we were concerned that UV irradiation could induce phenotypic suppression of the mutation. We therefore repeated these experiments in an isogenic diploid strain in which rad52-1 was replaced by a full deletion of the RAD52 coding sequence. The same results were obtained, both spontaneously and after UV irradiation (Figures 2 and 4).
The MRX protein Rad50 is required for both spontaneous and UV-induced Rad52-independent recombination pathways:
To investigate the genetic requirements of the two Rad52-independent pathways, we quantified spontaneous and UV-induced recombination in strains bearing deletions of three genes belonging to the RAD52 epistasis group but involved in different aspects of homologous recombination: RAD50, RAD51, and RAD59. Rad50 is a member of the MRX complex, involved in the 5′–3′ resection of DNA ends necessary to form invasive 3′ ssDNA tails (for review see Symington 2002; Krogh and Symington 2004; Williams et al. 2007). MRX has been implicated in an array of cellular responses including nonhomologous end joining, telomere maintenance, and cell-cycle checkpoints. Most recently the MRX complex has been shown to be required for the recruitment of cohesins and SMC5/6 complexes to the sites of DSBs and thus to facilitate sister-chromatid repair (Strom et al. 2004; Unal et al. 2004; De Piccoli et al. 2006; Lindroos et al. 2006; Strom and Sjogren 2007). This observation may explain why MRX mutants show an enhanced rate of heteroallelic recombination in diploids (Ivanov et al. 1992; Bressan et al. 1999), if lesions are less often repaired by sister chromatids.
Rad51 forms a nucleoprotein filament on single-stranded DNA that is active in searching for homology, in the formation of joint molecules, and in the exchange of DNA strands (Krogh and Symington 2004). Rad52 is required for Rad51 filament formation at DSBs (Sugawara et al. 2003; Wolner et al. 2003; Lisby et al. 2004). Rad59 has homology to the N terminus of Rad52 and promotes annealing of complementary DNA sequences. This protein is involved in both Rad51-dependent and -independent pathways (Krogh and Symington 2004). Of these three genes, only the deletion of RAD50 in the rad52-1 background shows a profound decrease both in spontaneous (0.19 HIS+/108 cells/generation, a 36-fold decrease compared to rad52-1; Figure 3) and in UV-induced recombination (0.15/108 survivors after 10 J/m2, a 10-fold decrease compared to rad52-1; Figure 3). Both rad52-1 rad51Δ and rad52-1 rad59Δ do not show a significant change compared to rad52-1.
The distribution of recombinants is statistically different in rad52-1 rad50Δ compared to rad52-1 (Figures 2 and 4 and supplemental Table 1). Among spontaneous recombinants, the class of GC + CO with LOH increased from 27% in rad52-1 to 48% in the double mutant, to the detriment of GC + CO + CL. We postulate that Rad50 facilitates the cosegregation of the two participating chromatids after recombination (although the second chromatid is not stable), thus leading to a high number of GC + CO + CL events; without Rad50, the HIS4 chromosome is recovered more often with the nonparticipating homologous chromatid. After UV irradiation, the same kind of distribution is found, but the GC + CL class disappears, consistent with the general finding that recombination in the presence of Rad50 occurs in ways that retain both participating chromatids. Alternatively, it is possible that without Rad50 the HIS4-containing chromosome itself is lost, so that these events are not recovered.
To establish that the effects of rad50Δ and other mutations discussed below are specific to recombination in the absence of Rad52, we introduced a centromeric plasmid carrying RAD52. Restoring RAD52 yielded outcomes that were very similar to WT cells, UV treated or not (data not shown). Therefore, the role of Rad50 described here is specific for Rad52-independent recombination.
Roles of Rad51 and Rad59 in Rad52-independent recombination:
Both rad52-1 rad51Δ and rad52-1 rad59Δ show a statistically significant decrease in the GC class and an increase of the two classes of CO among spontaneous recombinants, compared to rad52-1 (Figures 2 and 4 and supplemental Table 1). This suggests that in the absence of Rad52, GC not associated with CO results from a Rad51- and Rad59-dependent mechanism. Another mechanism that is Rad51 and Rad59 independent leads to the formation of half-COs. Since the recombination rates are unaffected in both double mutants, the lesions normally leading to GC in the presence of Rad51 or Rad59 can be directed efficiently to the half-CO pathway. After UV irradiation, the deletion of RAD51 in rad52-1 leads to a distribution of events similar to the one observed among spontaneous events. Therefore, the large increase in the UV-induced GC class is completely Rad51 dependent. Deletion of RAD59 did not change the distribution of recombinant types, suggesting that a specific Rad51-dependent pathway generates GC when rad52-1 cells are exposed to UV.
RAD52 complementation in both double mutants restored the distribution of recombination events seen in WT, both spontaneously and after UV irradiation (data not shown), indicating that the high proportion of half-COs observed in both rad52 rad51Δ and rad52 rad59Δ double mutants is linked to the absence of Rad52.
The UV-regulated genes CRT1 and DUN1 are involved in the UV-induced and spontaneous GC pathways:
One hypothesis to explain the UV-induced pathway of Rad52-independent recombination, with its quasi-WT spectrum of recombination, is that it requires an activator of gene conversion or a crossover suppressor that is induced by UV irradiation. We tested mutants of the UV-inducible kinase Dun1 and its downstream target, the transcriptional repressor Crt1. While the complete range of genes controlled by Crt1 is not yet known, it has been experimentally shown to repress the transcription of ribonucleotide reductase genes as well as the damage-induced gene HUG1 (Huang et al. 1998; Basrai et al. 1999). A simple hypothesis would be that UV exposure leads to derepression of Crt1-regulated genes.
The rad52 dun1Δ double mutant shows an additive sensitivity to UV irradiation, as previously noted by Fasullo et al. (1999), and a slight increase in UV-induced recombination. We found no modification of the spontaneous rate of recombination compared with rad52-1 (Figure 4). The double mutant, which might maintain Crt1 repression both spontaneously and after UV, would be expected to show low amounts of gene conversion without crossing over after UV induction; but, in fact, such gene conversions were the largest class (52% in Figures 2 and 4), as in the rad52-1 mutant.
If a UV-inducible gene under the control of the Crt1 repressor were responsible for the high proportion of GC, then we might expect to see a high level of GC even in the absence of UV irradiation in the rad52 crt1Δ mutant. This is not what we observed; GC dropped to 7% among spontaneous recombinants and to 17% after UV irradiation (Figures 2 and 4 and supplemental Table 1). This result indicates on the contrary that GC is somehow under the control of CRT1. The deletion of CRT1 does not increase UV sensitivity in rad52 mutants and recombination frequencies are not modified (Figure 3). However, spontaneous recombination shows a significant increase (threefold), but the rate is still very low compared to WT. Since the deletion of CRT1 does not reduce the rates of recombination and does not affect the incidence of half-COs, it seems that the lesions repaired by a CRT1-dependent GC pathway can alternatively be processed efficiently by a Crt1-independent pathway yielding half-COs.
Restoring RAD52 activity in the rad52 dun1Δ and rad52 crt1Δ double mutant leads to a wild-type distribution of spontaneous and UV-induced recombinants (Figure 4), arguing that the high proportion of half-COs depends on the absence of Rad52.
After UV treatment, half-crossovers are also more often associated with LEU2:
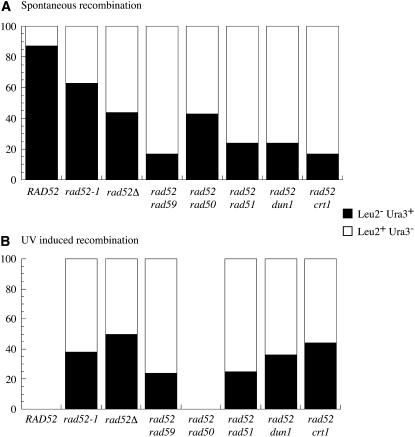
Haber and Hearn (1985) showed that there was a marked change in the proportion of His+ recombinants homozygous for each of the centromere-distal markers in the rad52 mutant. In wild-type diploids, there was a strong bias for the recovery of LOH events retaining the URA3 marker while in rad52 this bias was suppressed if not reversed (Figure 2).
UV-induced Rad52-independent recombinants associated with LOH are also more often associated with LEU2 (Figure 5). This indicates that the same pathway probably generates half-crossings over spontaneously and after UV irradiation. This bias is also observed in all the double mutants tested for spontaneous and UV-induced recombination.
Figure 5.—
Proportion of LOH retaining URA3 or LEU2: percentage of Ura3+ (solid bars) and Leu2+ (open bars) LOH among GCs + COs with LOH and GC + CO + CL compiled from Figure 1 in each mutant tested for (A) spontaneous and (B) UV-induced recombination. In B RAD52 and rad52 rad50 were not analyzed.
DISCUSSION
Most mitotic homologous recombination requires the action of the Rad52 protein, but there are significant Rad52-independent gene conversion mechanisms of both spontaneous and UV-induced recombination. Our previous study, confirmed here, demonstrated that Rad52-independent spontaneous recombination differs mechanistically from that seen in wild-type cells. First, the proportion of events involving a crossover is surprisingly high, in the present study accounting for 56% of all events being either 2n but homozygous for a distal marker or 2n − 1 with only a crossover chromosome remaining. Another 8% showed chromosome loss with a parental arrangement of flanking markers. In RAD52 cells, crossovers resulting in LOH accounted for only 8% of all gene conversions and events leading to chromosome loss were rare. If crossing over occurs in G2, then the two crossover classes are the expected outcomes if the HIS4-containing chromosome segregates at random with either the uninvolved homologous chromatid (i.e., 2n GC + CO) or the apparently unstable participating chromatid (i.e., 2n − 1 GC + CO + CL). We have suggested that Rad52-independent crossovers are in fact half-crossovers in which the second participating chromatid is still broken and/or unrepaired (Haber and Hearn 1985).
The rate of spontaneous gene conversion in rad52 diploids is strongly dependent on Rad50; in contrast, the rate of His+ recombinants was not affected by deleting Rad51 or Rad59. Given the many biochemical activities of MRX proteins (end tethering, control of 5′–3′ resection, strand annealing, strand unwinding, and damage-induced recruitment of cohesins and Smc5/6) (Paull and Gellert 1999; D'Amours and Jackson 2002; Strom et al. 2004; Unal et al. 2004; De Piccoli et al. 2006; Lindroos et al. 2006; Strom and Sjogren 2007; Williams et al. 2007), one can imagine many scenarios that might account for the role of MRX in facilitating recombination in the absence of the efficient Rad51 filament formation or the strand-annealing activities of Rad52. In spontaneous heteroallelic recombination, deleting Rad50 significantly increases prototroph formation, possibly because of its indirect role in recruiting cohesins or Smc5/6 or because reduced 5′–3′ resection would increase the proportion of heteroduplexes that would not result in coconversion. But here, in the absence of Rad52, deleting Rad50 nearly eliminates the recovery of HIS4 prototrophs. This suggests that the MRX complex must provide a means to carry out strand invasion—a conclusion that has also been reached in the analysis of Rad51-independent (but Rad52-dependent) DSB repair events (Chen et al. 2001; Signon et al. 2001; Ira and Haber 2002). Here MRX succeeds without Rad52. The fact that the absence of Rad50 also causes an increase in the proportion of GC + CO outcomes over GC + CO + CL suggests that the MRX complex might contribute to the preferential segregation of both participating chromatids.
The analysis of rad52 in combination with mutations in other important recombination genes showed some small, but often statistically significant, effects on the types of recombinants recovered. There was a significant reduction in noncrossover gene conversions in rad52 rad59Δ and rad52 rad51Δ compared to rad52. The most unexpected finding was that deleting the UV-regulated repressor protein Crt1 (Rfx1) had the most profound effect, such that 93% of the outcomes were crossover associated and/or exhibited chromosome loss, compared to 64% of rad52 cells. The basis of this effect is not understood. Until now, Crt1 has been known as a regulator of RNR genes; although it also affects other genes (Zaim et al. 2005), none have been implicated in recombinational repair. We note that deleting CRT1 in otherwise wild-type cells does not affect the types of outcomes that are recovered; CRT1 affects specifically a Rad52-independent recombination pathway. The role of CRT1 is not readily explained by assuming that Crt1 represses a gene that promotes noncrossover gene conversion; however, Crt1 could repress a downstream repressor so that a deletion would yield different results.
Another important discovery is that Rad51 must have a Rad52-independent role in recombination. Rad51 ensures that intermediates of gene conversion are resolved as noncrossovers. In the absence of Rad51, the spectrum of recombination outcomes for UV-induced His+ Rad52-independent recombinants resembles the profile seen in spontaneous rad52 rad51Δ cells; that is, there are relatively few noncrossover gene conversions and >85% of the events are LOH crossovers and/or chromosome losses. Again, the role of Rad51 in this pathway is distinct from what occurs in RAD52 cells; when the rad52 rad51Δ cells are complemented with RAD52, ∼90% of all His+ recombinants—either spontaneous or UV induced—are noncrossover gene conversions. The current biochemical consensus is that Rad51 acts after being loaded onto ssDNA by Rad52 (Sung et al. 2003). It is possible that other proteins (Rad55/Rad57 or the newly characterized Rad51D-like Shu2 protein) (Shor et al. 2005; Martin et al. 2006; Mankouri et al. 2007) might be able to promote at least limited successful filament formation without Rad52. Also, heteroduplex DNA has been detected physically in rad52 mutants during meiosis (Borts et al. 1986; Nag and Petes 1993). In any case, ∼60% of Rad52-independent UV-induced GC events are Rad51 dependent. It is possible that the anti-recombinogenic activity of Srs2 helicase in dismantling the Rad51 filament at the sites of damage leading to HIS4 recombinants (Krejci et al. 2003; Veaute et al. 2003) might be less active after UV induction.
On the other hand, after UV irradiation, the formation of single-stranded DNA by the nucleotide excision repair process could allow the formation of Rad51 filaments, which might not be eliminated by Srs2. Those filaments could engage in recombination through a mechanism that does not allow the formation of crossovers, such as synthesis-dependent strand annealing (SDSA), or by strand-assimilation mechanisms discussed below.
Possible mechanisms of Rad52-independent recombination:
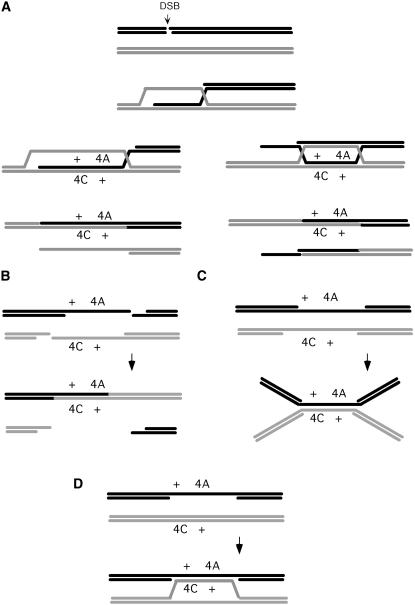
How Rad52-independent recombination events can form is unknown. Four possibilities are shown in Figure 6. In the first two, the initiating lesion is presumed to be a DSB. In the last two, the initiating lesion is imagined as a large single-stranded gap.
Figure 6.—
Possible mechanisms of Rad52-independent recombination. (A) A DSB leads to Rad51-strand invasion and strand assimilation that can either lead to a long heteroduplex flanked by one complete and one incomplete Holliday junction (HJ) or a pair of HJs. Resolution of the HJ will yield one crossover recombinant and incomplete fragments of a second chromosome. (B) Independent spontaneous DSBs on the two participating chromatids could yield long heteroduplex regions after SSA. The successful outcomes will have an associated crossover. (C) Large single-strand gaps could anneal and could produce one intact chromosome after nuclease cleavage, although in this case the outcomes need not be crossovers. (D) A single large single-strand gap might be filled by strand assimilation from an intact duplex. This association might be transient, allowing mismatch correction after which the gap would be filled by new DNA synthesis.
One possibility (Figure 6A) is that one end of a DSB is able to engage in strand invasion without Rad52 but that Rad52's strand-annealing activity would be needed to recruit the second end of a DSB (Sugiyama et al. 2006). Such events have been suggested by Neil Hunter (Lao et al. 2008), who has observed Rad52-independent single-end invasions in meiotic cells, where Dmc1 promotes strand invasion. Rad52-independent meiotic crossovers were first documented by Borts et al. (1986) and more recent analysis has suggested that many of these events may be nonreciprocal exchanges, with accompanying chromosome loss (A. Malkova and J. E. Haber, unpublished results). We note also that rare Rad52-independent recombinants were also recovered in mitotic diploids where one chromosome was cut by the HO endonuclease; consistent with the results discussed here, these rare outcomes were 2n − 1 diploids in which there had been a crossover (Malkova et al. 1996). In this scenario, an unstable strand-invasion intermediate could be stabilized by branch migration, to yield a more stable double Holliday junction whose resolution would yield one intact crossover chromosome and a chromosome fragment that would be lost. Whether such exchanges could occur without Rad51 (and when Dmc1 is not expressed) is not known, but our results in mitotic yeast cells argue that these events do occur without the only active strand-exchange protein, Rad51. As mentioned above, Rad51 may be able to form at least a short nucleoprotein filament without Rad52. This scenario could account for the other important difference in Rad52-independent recombination, the frequent recovery of diploids homozygous for the “wrong” distal marker. To obtain such diploids, the simplest mechanism would be to have a long region of heteroduplex DNA covering both the his4A and his4C markers, followed by independent mismatch repair. The single-end invasion model could generate such intermediates if the heteroduplex DNA between the two Holliday junctions were several kilobases long. Resolution of this intermediate could also generate gene conversions without an associated crossover.
A second mechanism that would account for both Rad52-independent exchanges and the frequent recovery of the wrong distal marker would be through Rad52-independent interhomolog single-strand annealing (SSA). In this scenario (Figure 6B), there would have to be sufficiently frequent spontaneous DSBs to have breaks on both participating chromatids. In a rad52Δ haploid, ∼10% of cell divisions produce one mother or daughter cell that grows into a colony and one cell that becomes arrested at G2/M, characteristic of a single unrepaired DSB (J. E. Haber, unpublished data). Thus there is ∼1 lesion—we assume it is a DSB that is not rejoined by DNA ligase4 and Ku-mediated end joining—that requires Rad52 for sister-chromatid repair in every five diploid cells. The DSB rate is 0.1/genome or 0.1 × 10−4/kb. The left arm of chromosome III is ∼100 kb, so the probability of a single DSB on the arm is 1 × 10−3. The probability of 2 DSBs on this chromosome arm in a diploid is 1 × 10−6. Rad52-independent HIS4 recombinants appear at a rate of ∼2 × 10−8. So ∼1/50 pairs of DSBs would have to result in HIS4. Resection can lead to SSA between sequences at least 50 kb apart and SSA can occur without Rad52 when the regions annealing are >10 kb (Ozenberger and Roeder 1991). Thus it is entirely possible that Rad52-independent events could arise from SSA-based repair of spontaneous DSBs. These considerations could equally be applied to other repair events, for example, damage leading to single-strand lesion.
A third possibility is that HIS4 recombinants could arise from the Rad52-independent annealing of two large single-strand gaps of opposite polarity on the two homologs (Figure 6C). There would be a long region of heteroduplex DNA that would be subjected to independent mismatch repair of his4A and his4C alleles. The postulated intermediate would be resolved either with or without crossing over although following UV the results would somehow be biased toward outcomes without crossing over. Moreover, after UV, there could be an increased use of another Rad51-dependent mechanism that involves strand invasion and that does not lead to CO, such as SDSA.
A variation on this idea would be strand assimilation of complementary sequences to provoke mismatch repair, following which the intact template might be reformed and the gap filled in (Figure 6D).
UV-induced Rad52-independent recombination: yet another pathway of recombination:
Although rad52 cells are mostly sensitive to ionizing radiation, they do have modest UV sensitivity. Nevertheless, UV irradiation of rad52 cells causes an induction of recombination. Measuring the frequency of these events after a brief exposure to UV is equivalent to determining the rate of recombination. At 10 J/m2 the rates of His+ recombinants in rad52 compared to spontaneous events increased 86-fold, compared to a very similar 94-fold increase in the same strain complemented by a RAD52 gene.
The significant increase in UV-stimulated events relative to spontaneous Rad52-independent events suggests that most of the His+ recombinants arose after UV irradiation rather than UV irradiation altering the outcomes of a small number of preexisting events. The most surprising aspect of the outcomes in UV-irradiated rad52-1 and rad52Δ diploids is that the proportion of noncrossover events rose from 36% among spontaneous events to 77% of the UV-irradiated cohort. These results suggest that UV either induces recombination proteins to carry out such events or creates DNA lesions that are intrinsically more likely to result in noncrossover outcomes. The UV-induced events apparently depend on an unidentified Dun1- and Crt1-dependent target. Earlier studies have suggested that ionizing radiation and UV are equally likely to result in mitotic crossovers leading to loss of heterozygosity of more distal markers (Nakai and Mortimer 1969), but we cannot specify the way that UV suppresses the nonreciprocal exchanges and loss of chromosomes seen for spontaneous events. UV irradiation would, both by excision repair and by replication fork stalling, create single-stranded regions that could be recombinogenic; this fact leads to the suggestion that the UV-induced Rad52-independent recombination events could be initiated in a different fashion from spontaneous events and consequently could have quite different outcomes. Repair apparently involves at least transient strand assimilation and mismatch correction and almost always occurs without a crossover. MRX must play a decisive role in this process.
Although deleting Rad51 does not significantly reduce the rate of His+ recombinants in the absence of Rad52, Rad51 plays an important Rad52-independent role in ensuring that intermediates of gene conversion are resolved as noncrossovers. In the absence of Rad51, the spectrum of recombination outcomes for UV-induced His+ recombinants resembles the profile seen in spontaneous rad52 and rad52 rad51Δ cells; that is, there are relatively few noncrossover gene conversions and >85% of the events are LOH crossovers and/or chromosome losses. Again, the role of Rad51 in this pathway is distinct from what occurs in RAD52 cells; when the rad52 rad51Δ cells are complemented with RAD52, ∼90% of all His+ recombinants—either spontaneous or UV induced—are noncrossover gene conversions. Possibly, there is a Rad52-independent, Rad51-dependent SDSA mechanism.
Acknowledgments
We are grateful to Michael Fasullo and Neil Hunter for their comments on the manuscript. E.C. was supported in part by grants from l'Association pour la Recherche sur le Cancer and the Philippe Foundation. Research was supported by National Institutes of Health grant GM20056.
References
- Adzuma, K., T. Ogawa and H. Ogawa, 1984. Primary structure of the RAD52 gene in Saccharomyces cerevisiae. Mol. Cell. Biol. 4 2735–2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basrai, M. A., V. Velculescu, K. Kinzler and P. Hieter, 1999. Norf5/Hug1 is a component of the Mec1-mediated checkpoint response to DNA damage and replication arrest in Saccharomyces cerevisiae. Mol. Cell. Biol. 19 7041–7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson, F., P. Baumann and S. West, 1998. Synergistic actions of Rad51 and Rad52 in recombination and DNA repair. Nature 391 401–404. [DOI] [PubMed] [Google Scholar]
- Borts, R., M. Lichten and J. Haber, 1986. Analysis of meiosis-defective mutations in yeast by physical monitoring of recombination. Genetics 113 551–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressan, D., B. K. Baxter and J. H. Petrini, 1999. The Mre11-Rad50-Xrs2 protein complex facilitates homologous recombination-based double-strand break repair in Saccharomyces cerevisiae. Mol. Cell. Biol. 19 7681–7687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brough, R., D. Wei, S. Leulier, C. Lord, Y. Rong et al., 2007. Functional analysis of Drosophila melanogaster BRCA2 in DNA repair. DNA Repair 7 10–19. [DOI] [PubMed] [Google Scholar]
- Chen, J., D. Silver, D. Walpita, S. Cantor, A. Gazdar et al., 1998. Stable interaction between the products of the BRCA1 and BRCA2 tumor suppressor genes in mitotic and meiotic cells. Mol. Cell 2 317–328. [DOI] [PubMed] [Google Scholar]
- Chen, Q., A. Ijpma and C. Greider, 2001. Two survivor pathways that allow growth in the absence of telomerase are generated by distinct telomere recombination events. Mol. Cell. Biol. 21 1819–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amours, D., and S. Jackson, 2002. The Mre11 complex: at the crossroads of DNA repair and checkpoint signaling. Nat. Rev. Mol. Cell. Biol. 3 317–327. [DOI] [PubMed] [Google Scholar]
- De Piccoli, G., F. Cortes-Ledesma, G. Ira, J. Torres-Rosell, S. Uhle et al., 2006. Smc5-Smc6 mediate DNA double-strand-break repair by promoting sister-chromatid recombination. Nat. Cell. Biol. 8 1032–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito, M., 1978. Evidence that spontaneous mitotic recombination occurs at the two-strand stage. Proc. Natl. Acad. Sci. USA 75 4436–4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasullo, M., J. Koudelik, P. Ahching, P. Giallanza and C. Cera, 1999. Radiosensitive and mitotic recombination phenotypes of the Saccharomyces cerevisiae dun1 mutant defective in DNA damage-inducible gene expression. Genetics 152 909–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli, A., and R. Schiestl, 1995. On the mechanism of UV and gamma-ray-induced intrachromosomal recombination in yeast cells synchronized in different stages of the cell cycle. Mol. Gen. Genet. 248 301–310. [DOI] [PubMed] [Google Scholar]
- Haber, J., and M. Hearn, 1985. Rad52-independent mitotic gene conversion in Saccharomyces cerevisiae frequently results in chromosomal loss. Genetics 111 7–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, M., Z. Zhou and S. J. Elledge, 1998. The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell 94 595–605. [DOI] [PubMed] [Google Scholar]
- Ira, G., and J. Haber, 2002. Characterization of RAD51-independent break-induced replication that acts preferentially with short homologous sequences. Mol. Cell. Biol. 22 6384–6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov, E., V. G. Korolev and F. Fabre, 1992. XRS2, a DNA repair gene of Saccharomyces cerevisiae, is needed for meiotic recombination. Genetics 132 651–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, J., and G. Fink, 1981. Gene conversion between duplicated genetic elements in yeast. Nature 292 306–311. [DOI] [PubMed] [Google Scholar]
- Krejci, L., S. Van Komen, Y. Li, J. Villemain, M. S. Reddy et al., 2003. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423 305–309. [DOI] [PubMed] [Google Scholar]
- Krogh, B., and L. Symington, 2004. Recombination proteins in yeast. Annu. Rev. Genet. 38 233–271. [DOI] [PubMed] [Google Scholar]
- Lao, J., S. Oh, M. Shinoara, A. Shinoara and N. Hunter, 2008. Rad52 promotes post-invasion steps of meiotic recombination. Mol. Cell 29 517–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea, D., and C. Coulson, 1949. The distribution of the number of mutants in bacterial population. J. Genet 49 264–285. [DOI] [PubMed] [Google Scholar]
- Lindroos, H., L. Strom, T. Itoh, Y. Katou, K. Shirahige et al., 2006. Chromosomal association of the Smc5/6 complex reveals that it functions in differently regulated pathways. Mol. Cell 22 755–767. [DOI] [PubMed] [Google Scholar]
- Lisby, M., J. Barlow, R. Burgess and R. Rothstein, 2004. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell 118 699–713. [DOI] [PubMed] [Google Scholar]
- Malkova, A., E. Ivanov and J. Haber, 1996. Double-strand break repair in the absence of RAD51 in yeast: a possible role for break-induced DNA replication. Proc. Natl. Acad. Sci. USA 93 7131–7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankouri, H., H. Ngo and I. D. Hickson, 2007. Shu proteins promote the formation of homologous recombination intermediates that are processed by Sgs1-Rmi1-Top3. Mol. Biol. Cell 18 4062–4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maringele, L., and D. Lydall, 2004. Telomerase- and recombination-independent immortalization of budding yeast. Genes Dev. 18 2663–2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, V., C. Chahwan, H. Gao, V. Blais, J. Wohlschlegel et al., 2006. Sws1 is a conserved regulator of homologous recombination in eukaryotic cells. EMBO J. 25 2564–2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer, R., R. Contopoulou and D. Schild, 1981. Mitotic chromosome loss in a radiation-sensitive strain of the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 78 5778–5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynahan, M., A. Pierce and M. Jasin, 2001. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol. Cell 7 263–272. [DOI] [PubMed] [Google Scholar]
- Nag, D., and T. Petes, 1993. Physical detection of heteroduplexes during meiotic recombination in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 13 2324–2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai, S., and R. Mortimer, 1969. Studies on the genetic mechanism of radiation-induced mitotic segregation in yeast. Mol. Gen. Genet. 103 329–338. [DOI] [PubMed] [Google Scholar]
- Ozenberger, B., and G. S. Roeder, 1991. A unique pathway of double-strand break repair operates in tandemly repeated genes. Mol. Cell. Biol. 11 1222–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull, T., and M. Gellert, 1999. Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev. 13 1276–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash, S., L. Prakash, W. Burke and B. Montelone, 1980. Effects of the RAD52 gene on recombination in Saccharomyces cerevisiae. Genetics 94 31–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shor, E., J. Weinstein and R. Rothstein, 2005. A genetic screen for top3 suppressors in Saccharomyces cerevisiae identifies SHU1, SHU2, PSY3 and CSM2: four genes involved in error-free DNA repair. Genetics 169 1275–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signon, L., A. Malkova, M. Naylor, H. Klein and J. Haber, 2001. Genetic requirements for RAD51- and RAD54-independent break-induced replication repair of a chromosomal double-strand break. Mol. Cell. Biol. 21 2048–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom, L., and C. Sjogren, 2007. Chromosome segregation and double-strand break repair: a complex connection. Curr. Opin. Cell Biol. 19 344–349. [DOI] [PubMed] [Google Scholar]
- Strom, L., H. Lindroos, K. Shirahige and C. Sjogren, 2004. Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol. Cell 16 1003–1015. [DOI] [PubMed] [Google Scholar]
- Sugawara, N., G. Ira and J. Haber, 2000. DNA length dependence of the single-strand annealing pathway and the role of Saccharomyces cerevisiae RAD59 in double-strand break repair. Mol. Cell. Biol. 20 5300–5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara, N., X. Wang and J. Haber, 2003. In vivo roles of Rad52, Rad54, and Rad55 proteins in Rad51-mediated recombination. Mol. Cell 12 209–219. [DOI] [PubMed] [Google Scholar]
- Sugiyama, T., N. Kantake, Y. Wu and S. Kowalczykowski, 2006. Rad52-mediated DNA annealing after Rad51-mediated DNA strand exchange promotes second ssDNA capture. EMBO J. 25 5539–5548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung, P., L. Krejci, S. Van Komen and M. Sehorn, 2003. Rad51 recombinase and recombination mediators. J. Biol. Chem. 278 42729–42732. [DOI] [PubMed] [Google Scholar]
- Symington, L., 2002. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 66 630–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorslund, T., F. Esashi and S. West, 2007. Interactions between human Brca2 protein and the meiosis-specific recombinase Dmc1. EMBO J. 26 2915–2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unal, E., A. Arbel-Eden, U. Sattler, R. Shroff, M. Lichten et al., 2004. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol. Cell 16 991–1002. [DOI] [PubMed] [Google Scholar]
- Veaute, X., J. Jeusset, C. Soustelle, S. C. Kowalczykowski, E. Le Cam et al., 2003. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature 423 309–312. [DOI] [PubMed] [Google Scholar]
- Williams, R., J. Williams and J. A. Tainer, 2007. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem. Cell Biol. 85 509–520. [DOI] [PubMed] [Google Scholar]
- Wolner, B., S. Van Komen, P. Sung and C. Peterson, 2003. Recruitment of the recombinational repair machinery to a DNA double-strand break in yeast. Mol. Cell 12 221–232. [DOI] [PubMed] [Google Scholar]
- Wong, A., R. Pero, P. A. Ormonde, S. Tavtigian and P. L. Bartel, 1997. RAD51 interacts with the evolutionarily conserved BRC motifs in the human breast cancer susceptibility gene BRCA2. J. Biol. Chem. 272 31941–31944. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Iwai, Y., E. Sonoda, J. Buerstedde, O. Bezzubova, C. Morrison et al., 1998. Homologous recombination, but not DNA repair, is reduced in vertebrate cells deficient in RAD52. Mol. Cell. Biol. 18 6430–6435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, S., S. Lee, G. Chen, M. Song, G. Tomlinson et al., 1999. BRCA2 is required for ionizing radiation-induced assembly of Rad51 complex in vivo. Cancer Res. 59 3547–3551. [PubMed] [Google Scholar]
- Zaim, J., E. Speina and A. Kierzek, 2005. Identification of new genes regulated by the Crt1 transcription factor, an effector of the DNA damage checkpoint pathway in Saccharomyces cerevisiae. J. Biol. Chem. 280 28–37. [DOI] [PubMed] [Google Scholar]