Abstract
The SQD1 enzyme is believed to be involved in the biosynthesis of the sulfoquinovosyl headgroup of plant sulfolipids, catalyzing the transfer of SO3− to UDP-glucose. We have determined the structure of the complex of SQD1 from Arabidopsis thaliana with NAD+ and the putative substrate UDP-glucose at 1.6-Å resolution. Both bound ligands are completely buried within the binding cleft, along with an internal solvent cavity which is the likely binding site for the, as yet, unidentified sulfur-donor substrate. SQD1 is a member of the short-chain dehydrogenase/reductase (SDR) family of enzymes, and its structure shows a conservation of the SDR catalytic residues. Among several highly conserved catalytic residues, Thr-145 forms unusually short hydrogen bonds with both susceptible hydroxyls of UDP-glucose. A His side chain may also be catalytically important in the sulfonation.
The sulfolipid sulfoquinovosyldiacylglycerol (SQDG) is common to all plants and many photosynthetic bacteria (1) and is found exclusively in the photosynthetic (thylakoid) membranes. SQDG-deficient null mutants from different photosynthetic bacteria exhibit a conditionally lethal phenotype under phosphate-limiting growth conditions (2, 3), suggesting that SQDG can substitute for phosphatidylglycerol under these conditions (1). This hypothesis also seems to be valid for plants such as Arabidopsis thaliana, for which concomitant changes in thylakoid membrane lipid composition and in the regulation of sulfolipid gene expression have been observed in response to phosphate starvation (4). Beyond its role in photosynthetic membranes, SQDG has been found to inhibit retroviral infections of mammalian cells and specifically the viral reverse transcriptase (5, 6).
Genes essential for the biosynthesis of the SQDG were isolated from photosynthetic bacteria by genetic complementation of mutants (7, 8). The amino acid sequence of the product of one of these genes, sqdB, is highly conserved among different organisms and a plant ortholog, SQD1, has been recently isolated from A. thaliana and expressed in Escherichia coli (4). The SQDB/SQD1 proteins show modest sequence similarity to sugar-nucleotide enzymes (4), and this similarity has been recently exploited by Essigmann and coworkers (9) to predict the structure of the A. thaliana SQD1 protein. As they also demonstrated that SQD1 binds NAD+ and contains characteristic conserved Y-XXX-K and glycine-rich (G-XX-G-XX-G) sequence patterns, SQD1 appears to be a member of the short-chain dehydrogenase/reductase (SDR) family (10–12).
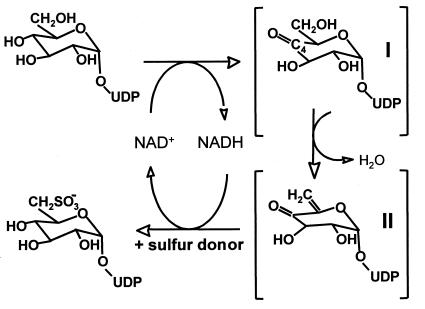
The sugar-nucleotide UDP-sulfoquinovose is thought to be the headgroup donor for SQDG biosynthesis (1), a conclusion supported by in situ labeling experiments with isolated chloroplasts and using synthetic UDP-sulfoquinovose (13, 14) and by the discovery of UDP-sulfoquinovose in bacterial sulfolipid mutants and other organisms (15, 16). Pugh et al. (17) proposed a metabolic pathway for SQDG biosynthesis in which UDP-glucose is first converted to a UDP-4-ketoglucose-5-ene intermediate, with subsequent addition of sulfite by some unknown donor. Because of their similarity to sugar nucleotide-modifying enzymes, the SQDB/SQD1 proteins, known to be essential for SQDG biosynthesis in all organisms analyzed thus far, are prime candidates for enzymes catalyzing these reactions. Essigmann and colleagues (9) have proposed that conversion of UDP-glucose to UDP-sulfoquinovose involves a single enzyme, SQD1, and its catalytic mechanism (Fig. 1) would be similar to that of UDP-galactose 4′-epimerase (GalE) (18), dTDP-glucose 4′,6′-dehydratase (dTGDH) (19), and CDP-glucose 4′,6′-dehydratase (CGDH) (20, 21), including its use of a tightly bound NAD+ cofactor. We describe here the crystal structure of SQD1 from A. thaliana, at 1.6-Å resolution, complexed with the putative substrate, UDP-glucose.
Figure 1.
A scheme of the SQD1 reaction mechanism derived from Essigmann et al. (9).
Materials and Methods
SQD1, lacking the 84-residue N-terminal thylakoid-targeting sequence, was overexpressed in E. coli and purified as previously described (4), although 0.3 M NaCl was added to all buffers. Purified protein was dialyzed against 25 mM Hepes (pH 7.5), with 0.3 M NaCl and concentrated to 3–4 mg/ml. Crystals were grown by sitting drop vapor diffusion: equal volumes of protein were mixed with buffer A, which contains 1.0 M ammonium sulfate, 0.1 M Mes (pH 6.5), 5 mM NAD+, and 5 mM UDP-glucose, and then equilibrated against reservoirs of 1.0–2.0 M ammonium sulfate, 0.1 M Mes (pH 6.5). The resulting bipyramidal crystals (0.35 × 0.35 × 0.20 mm) belong to the tetragonal space group I4122 (a = b = 159.6 Å, c = 98.9 Å). For data collection, crystals were transferred directly into buffer A containing 30% glycerol (wt/vol) as a cryoprotectant and flash-cooled in nitrogen gas at 100 K. X-ray diffraction data were measured by using Cu Kα radiation, monochromated and focused by Osmic (Troy, MI) Max-Flux multilayer mirrors, and a Siemens HI-STAR area detector; the data were then processed with saint version 5.00 (Bruker).
A low-resolution native data set (Native 2 in Table 1) was phased by multiple isomorphous replacement (MIR) by using solve version 1.10 (22). Five heavy-atom derivatives were prepared by soaking native crystals overnight in buffer A [with 30% glycerol (wt/vol)] containing the appropriate heavy atom compounds (Table 1). Although the phasing power (〈Fh〉/〈E〉 in Table 1) of the derivatives was low, the overall figure-of-merit was quite good: 0.65 for data between 20 and 3.0 Å and 0.58 for data in the highest shell. After solvent-flattening, the MIR phased electron density map was readily interpretable, and a model for the SQD1 protein was built with fragments taken from the homologous structures of dTGDH (PDB entry 1BXK, J. Thoden, A. Hegeman, P. Frey, and H. Holden, personal communication) and GalE [PDB entry 1XEL (23)]. x-plor (24) positional refinement and simulated annealing were performed using the Native 2 data at a 2σ cutoff level and 2.8-Å resolution. With an initial, partial protein model (R factor = 0.34, Rfree = 0.41), the refinement was continued using the high-resolution Native 1 data set, and the resolution was gradually extended from 2.8 Å to 1.6 Å (using data above 1σ and a free R test set of 5% of the total data). In later stages of refinement, individual B factor refinement and a bulk solvent contribution were included and water molecules were added to the model. The final refined model yields the R factor 0.17 (Rfree = 0.191) for 63,023 data from 30- to 1.6-Å resolution and includes 3,470 nonhydrogen atoms (residues 2–391, NAD+, UDP-glucose, and 344 water molecules); the rms deviations for bond lengths, bond angles, and dihedral angles are 0.005 Å, 1.4°, and 24.5°, respectively. The protein has an average thermal factor of 13 Å2, with no breaks or significantly disordered regions in the main chain. Density is not observed for the N-terminal His tag or for 16 surface protein side chains. The protein model conforms well to ideal geometry, as defined by procheck (25). All main-chain dihedral angles fall in allowed regions of the Ramachandran plot, with 90% within the most favored conformations.
Table 1.
Summary of data collection and phasing results
| Data set* | Resolution, Å | Unique reflections | Completeness,† % | Rsym† | Rmerge | 〈Fh〉/〈E〉‡ |
|---|---|---|---|---|---|---|
| Native 1 | 1.6 | 75,606 | 90.4 (60.3) | 0.055 (0.38) | — | — |
| Native 2 | 2.8 | 13,952 | 90.8 (92.6) | 0.07 (0.10) | — | — |
| AuCN (10 mM) | 3.0 | 12,621 | 97.2 (96.9) | 0.051 (0.075) | 0.145 | 0.78 |
| AuCN (20 mM) | 3.0 | 12,620 | 96.9 (94.8) | 0.048 (0.083) | 0.220 | 0.67 |
| EMTS (1 mM) | 3.0 | 12,711 | 97.6 (97.3) | 0.057 (0.089) | 0.126 | 1.00 |
| PHMBS (1 mM) | 3.0 | 12,601 | 96.8 (96.0) | 0.087 (0.141) | 0.222 | 1.01 |
| UOAc (5 mM) | 3.0 | 12,767 | 98.0 (96.2) | 0.077 (0.138) | 0.097 | 0.43 |
*Abbreviations: AuCN, KAu(CN)2; EMTS, ethylmercurithiosalicylate; PHMBS, p-hydroxymercuribenzenesulfonate; UOAc, uranyl acetate.
†Values in the highest shell of data are shown in parentheses.
‡〈Fh〉 is the rms of the heavy atom scattering factor and 〈E〉 is the rms of the lack-of-closure.
Results and Discussion
Overall Structure.
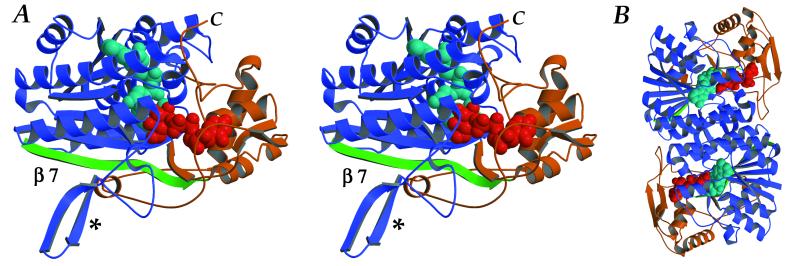
The SQD1 protein has a bidomain structure (Fig. 2A), with the catalytic site located in the cleft formed between the two lobes. The large N-terminal domain, which is responsible for NAD+ binding, consists predominantly of a seven-stranded parallel β-sheet with six connecting α-helices. The first six β-strands (β1–6) represent a classical Rossmann fold motif, commonly associated with dinucleotide binding (26). The C-terminal third of the protein (residues 300–391) and an extended loop inserted between the true Rossmann fold and added α/β unit (residues 211–267) form the small domain, which is responsible for UDP-glucose binding. SQD1 crystallizes as a C2-symmetric homodimer (Fig. 2B) where two long helices from the large domain in each monomer pack against one another to form an antiparallel four-helix bundle at the subunit interface.
Figure 2.
(A) Ribbon drawing of the SQD1 structure shown in stereoview. The large domain is shown in blue with additional 7th strand of Rossmann motif in green; small domain is gold; the C terminus is labeled C. Bound NAD (aqua) and UDP-glucose (red) are shown as space-filling models. Asterisk indicates β-hairpin insertion loop. (B) View of SQD1 homodimer along C2 dimer axis showing interaction of four-helix bundle. The figure was generated by using molscript (46) and raster3d (47).
SQD1 belongs to a large, structurally homologous class of SDR enzymes (10–12). These enzymes catalyze a diverse set of reactions, but all contain the modified “6+1” Rossmann fold and share a small number of conserved residues. SQD1 is structurally most similar to the sugar-nucleotide enzyme subfamily, for which only three crystal structures have been determined: GalE (23), dTGDH (Thoden et al., personal communication), and GDP-fucose synthetase (GFS) (27, 28). Despite very modest levels of amino acid sequence identity (19–30%), these structures represent a quite homologous subfamily of enzymes and are distinguished structurally from the broader class of SDR enzymes by the presence of a distinct second domain. Within this subfamily, the SQD1 structure is most similar to that of GalE, with an rms deviation of 2.5 Å for 249 corresponding Cα pairs when the analogous NAD(H)/UDP-glucose ternary complexes are optimally superimposed (1.7 Å for 196 Cα pairs of large domain only). Moreover, the dimer interface in SQD1 is also conserved within this subfamily (27, 29).
Unlike the ternary complexes of GalE (23, 29), in SQD1 an elongated C terminus folds over the sugar nucleotide-binding site along the interdomain cleft and effectively buries the entire active site. Moreover, residues Val-35–Gly-44 form an additional helical segment inserted between the β2 and α2 components of the Rossmann fold, which further buries NAD+ within the protein. Thus, the bound NAD+ and UDP-glucose ligands are almost completely inaccessible to bulk solvent (Fig. 3). Another distinctive feature of the SQD1 fold is a long β-ribbon with a hairpin turn (residues 161–172) inserted after β5 of the Rossmann fold (Fig. 2A). This β-ribbon extends out from each monomer along the dimer interface and partially wraps around its neighbor with a left-handed twist.
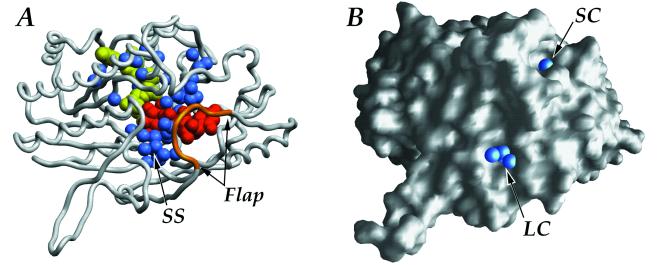
Figure 3.
(A) View of the buried ligands (NAD+ in yellow and UDP-glucose in red) and water molecules (blue) within the protein (SS is the presumptive sulfur donor site). The C-terminal flap, which may open to allow access to the sugar-dinucleotide-binding site, is shown in gold. (B) Surface representation highlighting the water molecules (blue spheres) at entrances of the large (LC) and small (SC) solvent channels leading into the SQD1 binding cleft.
NAD+ Binding.
NAD+ binds in an extended conformation and is essentially buried within the protein, consistent with the fact that it is tightly bound to the enzyme (9). On the basis of the proposed mechanisms for the related enzymes, GalE (30), dTGDH (19), and CGDH (21), the NAD+ would undergo a cycle of reduction and oxidation during catalysis without being released by the enzyme. In SQD1, NAD+ appears to be present as the oxidized form: electron density maps show a planar nicotinamide ring, and structure refinement gives no indication of the reduced form through elevated B factors or puckering of the ring when planar stereochemical restraints are released.
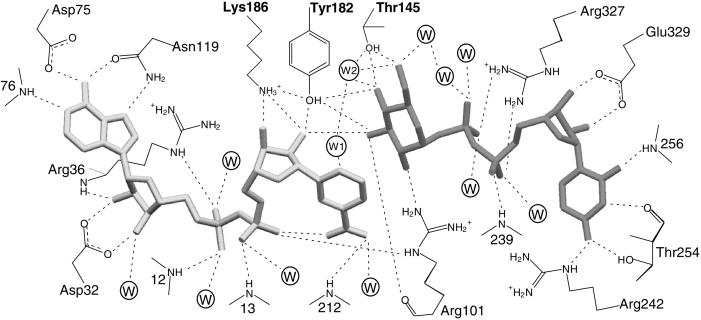
Interactions of NAD+ in the SQD1 binding cleft, shown in Fig. 4, are provided by residues near the C termini of the Rossmann fold β-strands. The NAD+ pyrophosphate binds at the N terminus of the α1 helix, with the phosphoryl groups hydrogen bonding to amide nitrogens of Tyr-12 and Cys-13 (Fig. 4), an interaction characteristic of Rossmann folds. Interestingly, the G-XXX-G-X-G (where X is any amino acid) sequence fingerprint of the Rossmann fold at the pyrophosphate site in monodomain SDR enzymes is replaced by a G-XX-G-XX-G sequence in SQD1 and the other bidomain enzymes. In SQD1, the NAD+ pyrophosphate is also hydrogen bonded by Arg-36 and Arg-101 side chains (Fig. 4). Although a hydrogen bond similar to that of Arg-101 is provided by a Lys side chain in GalE complexes (23, 29, 31), direct protein side-chain interactions with the pyrophosphate moiety of NAD+/NADP+ vary greatly among related enzymes, and are absent altogether in some cases.
Figure 4.
Schematic diagram of potential hydrogen-bonding interactions of bound NAD+ and UDP-glucose with the protein. All potential interactions (dashed lines) are within 3.2 Å except those involving Asn-119 Nδ (3.4 Å) and Arg-36 Nɛ (3.4 Å). Circles represent bound water molecules; W1 and W2 are waters Wat1 and Wat2, respectively.
Both hydroxyls of the NAD+ adenosyl ribose are liganded by the carboxyl oxygens of Asp-32 (Fig. 4), located at the base of the β2 strand. The amide group of adjacent Arg-36 is also within hydrogen bonding distance of the O2′ adenosyl ribose hydroxyl. Analogous interactions of a conserved Asp residue are observed for GalE and dTGDH, as well as some monodomain SDR enzymes that bind NAD+ rather than NADP+. Highly conserved interactions with the NAD+ adenosine moiety are also observed. The Asp-75 side chain hydrogen bonds to the adenosyl amino group, whereas the subsequent main-chain amide of Ile-76 interacts with the N1 ring nitrogen. An Asp (or more rarely an Asn) is found at this position in all SDR enzymes, and both possible hydrogen bonds are highly conserved. In SQD1, the Asn-119 Oδ and Nδ side-chain atoms make additional hydrogen bonds to the adenosyl N6 and N7 atoms, respectively. This residue is maintained in GalE, but is not widely conserved.
Around the nicotinamide ribose moiety in SQD1, the conserved Tyr-182 and Lys-186 side chains interact with the ribose hydroxyls, as expected in an enzyme from the SDR family. The NAD+ nicotinamide moiety adopts a syn conformation, with the carboxamide nitrogen atom within 2.8 Å of the nearest NAD+ phosphoryl oxygen. This conformation is consistent with the GalE complexes with NAD+ and NADH/UDP-glucose and is the proper orientation for B-side hydride transfer. In the SQD1 complex, the syn conformation may be further stabilized by an additional hydrogen bond between the carboxamide oxygen atom and the amide nitrogen of Val-212 (3.2 Å), an interaction precluded in GalE by substitution to Pro at this position. The carboxamide oxygen also makes a solvent-mediated interaction with the Val-212 carbonyl oxygen. Thus, the interactions of NAD+ in the SQD1 complex are generally very similar to those observed for GalE in the ternary complex with NAD+ and UDP (31) and in the abortive complex with NADH and UDP-glucose (23). However, the protein environment surrounding the nicotinamide moiety is not well conserved between SQD1 and GalE. The major difference appears to be the presence of adjacent bulky Tyr and Pro side chains in GalE, which crowd the nicotinamide ring toward the neighboring binding position of UDP-glucose; the smaller corresponding residues Gln-209 and Val-212 in SQD1 result in a relative translation of the nicotinamide ring by 1.2 Å.
NAD+ interactions in the SQD1 binding site also include five solvent-mediated hydrogen bonds with phosphoryl oxygen atoms and one adenosyl ribose hydroxyl oxygen (Fig. 4). Another interaction of note near the catalytic site involves a water molecule (Wat1) which is aligned with the plane of the nicotinamide ring and lies within 3.2 Å of the C6 atom. CH⋅⋅⋅O hydrogen bonds occur in proteins for carbon centers having slight acidic character (32), as would be expected in a positively charged nicotinamide ring. The C–O distance and CH–O angle of 163° (based on calculated ideal H positions) agree well with water molecules coordinated by CH hydrogen bonds in a survey of small molecule neutron crystal structures (33), where the typical C–O distance was found to be 3.4–3.5 Å. Although otherwise buried within the complex, Wat1 is also in good hydrogen-bonding distance of both Gln-209 amide nitrogen and Leu-207 carbonyl oxygen atoms, as well as another buried water molecule (Wat2).
UDP-Glucose Binding.
The binding of the UDP pyrophosphate and ribose moieties, also shown in Fig. 4, is reminiscent of the NAD+ interactions. The pyrophosphoryl group is positioned near the N terminus of an α-helix (239–249) from the small domain and makes a hydrogen bond to the amide nitrogen of Ala-239. Both phosphate groups interact with the Arg-327 side chain, and both make additional solvent-mediated interactions to SQD1 main-chain atoms. The UDP-ribose hydroxyls hydrogen bond to either oxygen atom of the Glu-329 carboxylate, an interaction mimicking that of the NAD+ adenosyl ribose with Asp-32. The uridine ring is hydrogen bonded by the side chain Oγ and main-chain carbonyl oxygen atoms of Thr-254, the Arg-242 Nɛ atom, as well as the amide nitrogen of Tyr-256. The binding of UDP-glucose may be further stabilized by the parallel stacking interaction of the uridine and Tyr-256 rings, with an interplanar distance of 3.5 Å. Both the main-chain hydrogen bonds and aromatic stacking interaction are also observed in the structures of the GalE abortive complex with NADH and UDP-glucose. In the GalE complex with UDP and NAD+, the aromatic stacking was reported to be disrupted by a second conformation of the corresponding Phe-218 side chain (31). However, steric interference by the surrounding side chains in SQD1 would prevent the occurrence of a similar conformational change in the Tyr-256 side chain.
UDP-glucose extends into the SQD1 cleft, such that the plane of the hexose ring is parallel to, and partially overlaps, the NAD+ nicotinamide, with a distance of approximately 3.6 Å between the two rings (Fig. 5A). The glucosyl 3′-hydroxyl is in hydrogen bonding distance (2.7 Å) of one hydroxyl from the NAD+ nicotinyl ribose. The glucosyl ring also abuts closely against the surface of the large domain and is well stabilized by protein interactions. The Oγ hydroxyl of Thr-145 makes two short hydrogen bonds (2.4 Å) with the O4′ and O6′ glucosyl hydroxyls (Fig. 5B). The hydroxyl of Tyr-182 is also within hydrogen bonding distance of both O4′ (2.5 Å) and O3′ (2.9 Å) glucosyl hydroxyls. Additionally, Arg-101 forms hydrogen bonds with the O2′ and O3′ glucosyl hydroxyls through the guanidinium nitrogen and main-chain carbonyl oxygen atoms, respectively. In the SQD1 and GalE ternary complexes, the relative positions of the glucose ring with respect to the nicotinamide ring and binding site residues differ significantly, because of the combined effects of conformational differences in their small domains and the differing binding conformations of the NAD+ nicotinamide moiety as well as UDP-glucose. However, as neither structure may accurately show a productive complex, the importance of this observation is unclear.
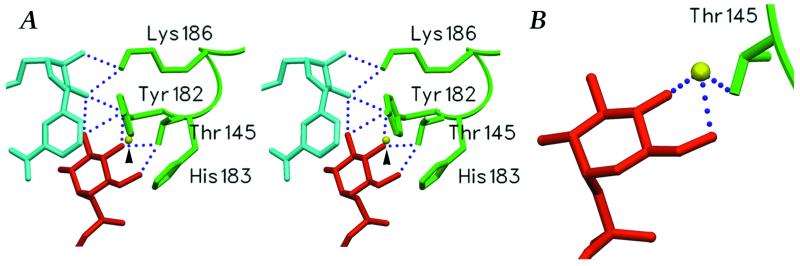
Figure 5.
The active center of SQD1. (A) Stereoview showing the bound NAD+ (aqua), UDP-glucose (red), and Wat2 (arrowhead) with respect to presumed catalytic residues of SQD1 active site. (B) A close-up view of the tightly bound water Wat2 and its interactions with Thr-145 and the hexose ring. All hydrogen bonds (dotted lines) involving Thr-145 and bound water are 2.4 Å. The figure was generated by using setor (48).
As the bound UDP-glucose ligand is completely buried within the SQD1 structure (Fig. 3), a significant conformational change of the protein must occur to allow entry and exit of substrate and product. Residues 323–330 form a C-terminal flap (Fig. 3) that covers the end of the binding cleft but is poorly stabilized with respect to the rest of the SQD1 tertiary structure: only one main-chain hydrogen bond (326 CO⋅⋅⋅HN 382) and one intraloop hydrogen bond (between Asn-325 and Glu-331) are made. Other side-chain interactions include only those of Arg-327 and Glu-329 with the bound UDP moiety, described above. As this flap is apparently stabilized by the presence of bound UDP-glucose, the absence of ligand may increase its flexibility and allow access to the binding site.
Sulfur-Donor Site.
The Arg-327 side chain on the flap also partitions off two distinct channels of buried solvent leading from the enzyme surface to the bound UDP-glucose (Fig. 3). The first is a small channel containing 8 buried water molecules that ends at the uridinyl and pyrophosphate moieties of UDP-glucose. The second, larger, channel ends at a much wider solvent cavity, occupied by 14 water molecules, near the O5′–C5′–C6′–O6′ edge of the UDP-glucose hexose ring. This cavity (9–10 Å in diameter) is the presumptive binding site for the, as yet, unidentified sulfur donor (Fig. 3). Arg-263 extends into the center of the cavity and is an obvious candidate for potential interaction with the sulfite-donating substrate. Suggested possibilities for the sulfur donor (1) have included free sulfite, adenosyl-5′-phosphosulfate, and 3′-phosphoadenosyl-5′-phosphosulfate, as well as a protein sulfur donor. Of these, only the last may be reasonably ruled out, as the reactive glucosyl C5′ atom of bound UDP-glucose is 12 Å from the protein surface. A smaller sulfur donor, however, may be able to diffuse into the closed binding cleft through the larger solvent channel, which has a diameter of 7–9 Å at the protein surface.
The Active Site and Mechanistic Implications.
Only a few residues are highly conserved throughout the entire SDR family; however among these are a characteristic Y-XXX-K motif and a Ser/Thr residue, which together form a catalytic triad (34). Mutation studies with GalE (35–37) have demonstrated the importance of these residues for catalysis, with Tyr being most critical. The corresponding residues in SQD1, Thr-145, Tyr-182, and Lys-186, are found in similar orientations and can be reasonably assumed to fulfill analogous mechanistic roles.
The first step of the proposed SQD1 mechanism involves proton abstraction from the UDP-glucose 4′-hydroxyl, with concomitant hydride transfer from C4′ to NAD+, to produce the 4′-keto intermediate I (Fig. 1). The conserved Tyr residue, in the form of a negatively charged tyrosinate, acts as the general base that abstracts the proton. The tyrosinate catalytic base hypothesis was originally suggested for Drosophila alcohol dehydrogenase (38) and is a generally accepted mechanistic principle among the monodomain SDR enzymes, as well as for GalE. Consistent with this, Tyr-182 makes a close hydrogen bond (2.5 Å) with the O4′ hydroxyl of bound UDP-glucose in the SQD1 complex (Figs. 4 and 5). In the SDR enzymes, the surrounding positive charges, particularly on the adjacent conserved Lys side chain and the oxidized nicotinamide ring, substantially lower the pKa of the catalytic Tyr (35). The interaction between the Tyr/Lys pair has always been assumed to be electrostatic in nature, since the OH–Nζ distance in all determined structures of related SDR enzymes has been greater than 4 Å, well beyond hydrogen-bonding range. In the SQD1 complex, in contrast, we observe the Tyr and Lys side chains within good hydrogen bonding distance at 2.9 Å (Fig. 5).
Whereas the catalytic Lys-Tyr pair is absolutely conserved in SDR sequences, the Thr/Ser appears to be less critical and is even replaced by other residues in some cases (39, 40). In SQD1, the corresponding Thr-145 conserves the side-chain hydroxyl in an orientation identical to the Ser or Thr Oγ seen in the homologous complexes of SDR enzymes. Thr-145 is located at the N terminus of a short segment of α-helix (residues Thr-145–Tyr-149), which puts the Thr-145 Oγ atom within hydrogen-bonding distance (3.0 Å) of the Gly-147 amide nitrogen atom. Interestingly, both the hydrogen bond and short α-helical segment are conserved in the three-dimensional structures of other SDR enzymes, suggesting that this interaction, perhaps enhanced by the helical dipole, may be important in maintaining the proper orientation of the catalytic Ser/Thr side chain.
For the closely related GalE enzyme, two mechanisms were originally suggested: one in which the conserved Ser side-chain hydroxyl serves as a proton shuttle to the Tyr catalytic base, and one in which the Tyr acts directly on the UDP-glucose 4′-hydroxyl, relegating the Ser side chain to a more secondary role (29). As a direct interaction between the catalytic Tyr and sugar dinucleotide was not observed in structures of ternary complexes of this enzyme, the former proposal was ultimately preferred by these researchers (37). In the SQD1 complex, however, the reactive 4′-hydroxyl of UDP-glucose clearly interacts directly with both Thr-145 and the catalytic base Tyr-182. This is consistent with the ternary complexes of many SDR enzymes (41–43), in which the susceptible oxygen atoms of bound inhibitors or reaction products also make analogous hydrogen bonds with both catalytic residues. Thus it appears that the conserved Thr side-chain hydroxyl is important in substrate binding and/or stabilization of reaction intermediates.
The Thr-145 side chain participates in a network of three unusually short hydrogen bonds (≈2.4 Å) with the glucosyl 4′-hydroxyl, glucosyl 6′-hydroxyl, as mentioned above, as well as a buried water molecule Wat2 (Figs. 4 and 5). The simultaneous interaction of Thr-145 with both glucosyl positions forces the 6′-hydroxyl to be held in the least sterically favored rotameric orientation and places the O6′ atom within 2.8 Å of O4′. Wat2, which is bound above the plane of these three atoms, not only makes short hydrogen bonds (≈2.4 Å) with the Thr-145 Oγ, but also with the O4′ and O6′ hydroxyls. Wat2 is additionally within hydrogen-bonding distance of Wat1, but is otherwise sequestered from additional solvent. Similar short or “low-barrier” hydrogen bonds (LBHBs) have been proposed to be important in the catalytic mechanisms of some enzymes by stabilizing reaction intermediates or transition states (44, 45). Distinguished structurally by hydrogen-bond distances less than 2.5 Å, these interactions are characterized by partial covalent contributions between the hydrogen and both heteroatoms and may occur when the proton affinities of these atoms are similar. The formation of LBHBs has been invoked specifically to facilitate the formation of enolic transition states (44, 45), as would occur during the conversion of the 4′-keto to the 4′-keto-glucose-5-ene intermediate II (Fig. 1). The unusual network observed at the reactive center of the SQD1 ternary complex suggests that LBHBs may be important in transition state stabilization and in promoting the removal of the O6′ hydroxyl.
A general base would also be required to abstract a proton from the acidic glucosyl C5′ atom during formation of intermediate II (Fig. 1). A likely candidate is His-183, which is located near the glucosyl ring such that the Nɛ atom is within 4.2 Å of the H5′ atom (Fig. 5). The orientation of the His side chain is stabilized by a hydrogen bond between the Nδ atom and the hydroxyl of Ser-180. In the subsequent step, the sulfur donor would then transfer SO3− to UDP-4′-keto-glucose-5-ene by electrophilic addition across the C5′⩵C6′ double bond, followed by reduction of the 4′-keto group and regeneration of NAD+. Interestingly, in the crystal structure of dTGDH, which should also form a 4-keto-glucose-5-ene intermediate during catalysis (19), the carboxylate side chain of Glu-136 occupies a nearly identical position within the catalytic site and may serve as the general base.
The proposed mechanism for SQD1 catalysis suggests that, in the absence of the sulfur donor, the initial reaction steps might proceed to the 4-keto-glucose-5-ene intermediate. However, we clearly observe UDP-glucose bound at the active site rather than either of the proposed reaction intermediates; the bound NAD+ also appears to be in the oxidized state. Thus, the UDP-glucose molecule appears poised, but unable to react. One obvious explanation may be that the binding of the sulfur donor is required to induce a productive arrangement of NAD+ and/or substrate. Indeed, the NAD+ and glucosyl rings are overlapped such that the nicotinamide C4 atom is poorly aligned for abstraction of hydride from the glucosyl C4′ position. The addition of the sulfonyl group at the C6′ may also be coordinated with NAD+ reduction, as the sulfur donor should be rather unstable. The buried active site and solvent channels thus suggest that SQD1 has adapted to deal with short-lived substrates in the absence of bulk water. It will be important to determine the structures of other SQD1–ligand complexes and to identify potential sulfur donors.
Acknowledgments
This work has been supported by the Michigan State University Foundation and grants from the National Science Foundation (MCB-9807993 to C.B.) and the Research Excellence Fund Center for Protein Structure, Function, and Design.
Abbreviations
- SQDG
sulfoquinovosyldiacylglycerol
- SDR
short-chain dehydrogenase/reductase
- GalE
UDP-glucose 4′-epimerase
- dTGDH
dTDP-glucose 4′,6′-dehydratase
- CGDH
CDP-glucose 4′,6′-dehydratase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1QRR).
References
- 1.Benning C. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:53–75. doi: 10.1146/annurev.arplant.49.1.53. [DOI] [PubMed] [Google Scholar]
- 2.Benning C, Beatty J T, Prince R C, Somerville C R. Proc Natl Acad Sci USA. 1993;90:1561–1565. doi: 10.1073/pnas.90.4.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Güler S, Seeliger A, Härtel H, Renger G, Benning C. J Biol Chem. 1996;271:7501–7507. doi: 10.1074/jbc.271.13.7501. [DOI] [PubMed] [Google Scholar]
- 4.Essigmann B, Güler S, Narang R A, Linke D, Benning C. Proc Natl Acad Sci USA. 1998;95:1950–1955. doi: 10.1073/pnas.95.4.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gustafson K R, Cardellina J H, Fuller R W, Weislow O S, Kiser R F, Snader K M, Patterson G M, Boyd M R. J Natl Cancer Inst. 1989;81:1254–1258. doi: 10.1093/jnci/81.16.1254. [DOI] [PubMed] [Google Scholar]
- 6.Loya S, Reshef V, Mizrachi E, Silberstein C, Rachamim Y, Carmeli S, Hizi A. J Nat Prod. 1998;61:891–895. doi: 10.1021/np970585j. [DOI] [PubMed] [Google Scholar]
- 7.Benning C, Somerville C R. J Bacteriol. 1992;174:2352–2360. doi: 10.1128/jb.174.7.2352-2360.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benning C, Somerville C R. J Bacteriol. 1992;174:6479–6487. doi: 10.1128/jb.174.20.6479-6487.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Essigmann B, Hespenheide B, Kuhn L, Benning C. Arch Biochem Biophys. 1999;369:30–41. doi: 10.1006/abbi.1999.1344. [DOI] [PubMed] [Google Scholar]
- 10.Baker M E, Blasco R. FEBS Lett. 1992;301:89–93. doi: 10.1016/0014-5793(92)80216-4. [DOI] [PubMed] [Google Scholar]
- 11.Jornvall H, Persson B, Krook M, Atrian S, Gonzalez-Duarte R, Jeffery J, Ghosh D. Biochemistry. 1995;34:6003–6013. doi: 10.1021/bi00018a001. [DOI] [PubMed] [Google Scholar]
- 12.Labesse G, Vidal-Cros A, Chomilier J, Gaudry M, Mornon J-P. Biochem J. 1994;304:95–99. doi: 10.1042/bj3040095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heinz E, Schmidt H, Hoch M, Jung K H, Binder H, Schmidt R R. Eur J Biochem. 1989;184:445–453. doi: 10.1111/j.1432-1033.1989.tb15037.x. [DOI] [PubMed] [Google Scholar]
- 14.Seifert U, Heinz E. Bot Acta. 1992;105:197–205. [Google Scholar]
- 15.Rossak M, Tietje C, Heinz E, Benning C. J Biol Chem. 1995;270:25792–25797. doi: 10.1074/jbc.270.43.25792. [DOI] [PubMed] [Google Scholar]
- 16.Tietje C, Heinz E. Planta. 1998;206:72–78. [Google Scholar]
- 17.Pugh C E, Roy A B, Hawkes T, Harwood J L. Biochem J. 1995;309:513–519. doi: 10.1042/bj3090513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nelsestuen G L, Kirkwood S. J Biol Chem. 1971;246:7533–7543. [PubMed] [Google Scholar]
- 19.Glaser L, Zarkowsky R. In: The Enzymes. Boyer P, editor. Vol. 5. New York: Academic; 1971. pp. 465–480. [Google Scholar]
- 20.Liu H-W, Thorson J S. Annu Rev Microbiol. 1994;48:223–256. doi: 10.1146/annurev.mi.48.100194.001255. [DOI] [PubMed] [Google Scholar]
- 21.He X, Thorson J, Liu H. Biochemistry. 1996;35:4721–4731. doi: 10.1021/bi952706p. [DOI] [PubMed] [Google Scholar]
- 22.Terwilliger T C, Berendzen J. Acta Crystallogr D. 1996;52:749–757. doi: 10.1107/S0907444996000832. [DOI] [PubMed] [Google Scholar]
- 23.Thoden J, Frey P, Holden H. Biochemistry. 1996;35:5137–5144. doi: 10.1021/bi9601114. [DOI] [PubMed] [Google Scholar]
- 24.Brunger, A. (1992) x-plor Manual (Yale Univ. Press, New Haven, CT), Version 3.1.
- 25.Laskowski R A, MacArthur M W, Moss D S, Thornton J M. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 26.Rossmann M G, Moras D, Olsen K W. Nature (London) 1974;250:194–199. doi: 10.1038/250194a0. [DOI] [PubMed] [Google Scholar]
- 27.Rizzi M, Tonetti M, Vigevani P, Sturla L, Bisso A, De Flora A, Bordo D, Bolognesi M. Structure. 1998;6:1453–1465. doi: 10.1016/s0969-2126(98)00144-0. [DOI] [PubMed] [Google Scholar]
- 28.Somers W, Stahl M, Sullivan F. Structure. 1998;6:1601–1612. doi: 10.1016/s0969-2126(98)00157-9. [DOI] [PubMed] [Google Scholar]
- 29.Thoden J, Frey P, Holden H. Protein Sci. 1996;5:2149–2161. doi: 10.1002/pro.5560051102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frey P A. FASEB J. 1996;10:461–470. [PubMed] [Google Scholar]
- 31.Thoden J, Frey P, Holden H. Biochemistry. 1996;35:2557–2566. doi: 10.1021/bi952715y. [DOI] [PubMed] [Google Scholar]
- 32.Derewenda Z S, Lee L, Derewenda U. J Mol Biol. 1995;252:248–262. doi: 10.1006/jmbi.1995.0492. [DOI] [PubMed] [Google Scholar]
- 33.Steiner T, Saenger W. J Am Chem Soc. 1993;115:4540–4547. [Google Scholar]
- 34.Oppermann U, Filling C, Berndt K, Persson B, Benach J, Ladenstein R, Jornvall H. Biochemistry. 1997;36:34–40. doi: 10.1021/bi961803v. [DOI] [PubMed] [Google Scholar]
- 35.Liu Y, Thoden J, Kim J, Berber E, Gulick A, Ruzicka F, Holden H, Frey P. Biochemistry. 1997;36:10675–10684. doi: 10.1021/bi970430a. [DOI] [PubMed] [Google Scholar]
- 36.Swanson B A, Frey P A. Biochemistry. 1993;32:13231–13236. doi: 10.1021/bi00211a035. [DOI] [PubMed] [Google Scholar]
- 37.Thoden J, Gulick A, Holden H. Biochemistry. 1997;36:10685–10695. doi: 10.1021/bi9704313. [DOI] [PubMed] [Google Scholar]
- 38.Chen Z, Jiang J C, Lin Z G, Lee W R, Baker M E, Chang S H. Biochemistry. 1993;32:3342–3346. doi: 10.1021/bi00064a017. [DOI] [PubMed] [Google Scholar]
- 39.Irie S, Doi S, Yorifuji T, Takagi M, Yano K. J Bacteriol. 1987;169:5174–5179. doi: 10.1128/jb.169.11.5174-5179.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shahbaz M, Hock J A, Trach K A, Hural J A, Webber S, Whiteley J M. J Biol Chem. 1987;262:16412–16416. [PubMed] [Google Scholar]
- 41.Tanaka N, Nonaka T, Nakanishi M, Deyashiki Y, Hara A, Mitsui Y. Structure. 1996;4:33–45. doi: 10.1016/s0969-2126(96)00007-x. [DOI] [PubMed] [Google Scholar]
- 42.Nakajima K, Yamashita A, Akama H, Nakatsu T, Kato H, Hashimoto T, Oda J, Yamada Y. Proc Natl Acad Sci USA. 1998;95:4876–4881. doi: 10.1073/pnas.95.9.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sawicki M W, Erman M, Puranen T, Vihko P, Ghosh D. Proc Natl Acad Sci USA. 1999;96:840–845. doi: 10.1073/pnas.96.3.840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gerlt J, Kreevoy M, Cleland W, Frey P. Chem Biol. 1997;4:259–267. doi: 10.1016/s1074-5521(97)90069-7. [DOI] [PubMed] [Google Scholar]
- 45.Cleland W W, Frey P A, Gerlt J A. J Biol Chem. 1998;273:25529–25532. doi: 10.1074/jbc.273.40.25529. [DOI] [PubMed] [Google Scholar]
- 46.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 47.Merritt E A, Murphy M E P. Acta Crystallogr D. 1994;50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- 48.Evans S V. J Mol Graphics. 1993;11:134–138. doi: 10.1016/0263-7855(93)87009-t. [DOI] [PubMed] [Google Scholar]