Abstract
DNA double-strand breaks (DSB) are repaired through two different pathways, homologous recombination (HR) and nonhomologous end joining (NHEJ). Yeast Xrs2, a homolog of human Nbs1, is a component of the Mre11-Rad50-Xrs2 (MRX) complex required for both HR and NHEJ. Previous studies showed that the N-terminal forkhead-associated (FHA) domain of Xrs2/Nbs1 in yeast is not involved in HR, but is likely to be in NHEJ. In this study, we showed that the FHA domain of Xrs2 plays a critical role in efficient DSB repair by NHEJ. The FHA domain of Xrs2 specifically interacts with Lif1, a component of the ligase IV complex, Dnl4-Nej1-Lif1 (DNL). Lif1, which is phosphorylated in vivo, contains two Xrs2-binding regions. Serine 383 of Lif1 plays an important role in the interaction with Xrs2 as well as in NHEJ. Interestingly, the phospho-mimetic substitutions of serine 383 enhance the NHEJ activity of Lif1. Our results suggest that the phosphorylation of Lif1 at serine 383 is recognized by the Xrs2 FHA domain, which in turn may promote recruitment of the DNL complex to DSB for NHEJ. The interaction between Xrs2 and Lif1 through the FHA domain is conserved in humans; the FHA domain Nbs1 interacts with Xrcc4, a Lif1 homolog of human.
DNA double-strand breaks (DSBs) are repaired mainly through two distinct pathways, homologous recombination (HR) and nonhomologous end joining (NHEJ). In the NHEJ process, two DSB ends are protected from massive degradation, are held together, and are rejoined to recover the original junction or to create a new junction. On the basis of the differences in junction sequences and in genetic requirement, several pathways for NHEJ have been defined. NHEJ pathways require the HDF1/YKU70, HDF2/YKU80, LIF1, NEJ1, DNL4, MRE11, RAD50, and XRS2 genes and, in addition, the POL4 gene might be involved in microhomology-dependent NHEJ (Daley et al. 2005a,b). All of the pathways require the Dnl4-Lif1-Nej1 (DNL) complex, which functions as a DNA ligase in the rejoining step of the DSB ends. Dnl4, a homolog of human ligase IV, is a catalytic subunit, which contains DNA-binding and adenylation domains and oligonucleotide binding (OB)-fold. Dnl4 (ligase IV in humans) is a core component, which binds to both Lif1 and Nej1 (Xrcc4 and XLF in humans, respectively). In the DNL complex, Lif1 and Nej1 contribute to stabilization and activation of Dnl4/ligase IV protein (Grawunder et al. 1997; Herrmann et al. 1998; Valencia et al. 2001). Recruitment of the DLN complex to DSB ends is considered to be a critical step in various NHEJ pathways. However, how the DNL complex is recruited to the DSB sites remains largely unknown.
In the budding yeast Saccharomyces cerevisiae, the Mre11-Rad50-Xrs2 (MRX) complex is required not only for HR, but also for NHEJ (Moore and Haber 1996). Mre11 and Rad50, homologs of bacterial sbcC and sbcD, respectively, are well conserved from bacteria to mammals (Sharples and Leach 1995). The MRX complex is required for the formation of DSBs and the processing of DBS ends in both meiotic recombination (Johzuka and Ogawa 1995; Sharples and Leach 1995) and mitotic repair of a subset of irradiation-induced DSBs (Llorente and Symington 2004). Mre11 has a phosphodiesterase motif while Rad50, an SMC-like protein, functions in bridging of DSB ends in HR and NHEJ (Chen et al. 2001; Wiltzius et al. 2005). The third subunit, Xrs2, is a homolog of human Nbs1. The Xrs2/Nbs1 homolog is found only in eukaryotes (Connelly and Leach 2002). Xrs2 consists of three domains: forkhead-associated (FHA), Mre11-binding, and Tel1-binding domains (Nakada et al. 2003; Shima et al. 2005). Like Rad50 and Mre11, Xrs2 protein is involved in DNA repair, telomere maintenance, and damage checkpoint, possibly as a mediator protein for the recruitment of Mre11 (/Rad50) as a component of the MRX complex and of Tel1 to either DSB sites or the telomere. These three domains are conserved even in human Nbs1 protein whose dysfunction results in the Nijmigen breakage syndrome (NBS), an autosomal recessive disorder with a high risk of lymphoid cancers and immunodeficiency (Weemaes et al. 1981). Importantly, cells from NBS patients express Nbs1 proteins lacking a N-terminal region containing FHA domain as well as less-conserved BRCA1 C-terminal (BRCT) domain (Carney et al. 1998; Matsuura et al. 1998; Varon et al. 1998). This suggests that the function of the FHA domain in Xrs2/Nbs1 is important for genome stability and differentiation of immune cells. The FHA domain, known as a phospho-protein recognition/interaction domain, is found in various proteins involved in DNA repair and checkpoint pathways (Sun et al. 1998). However, the exact role of the FHA domain of Xrs2/Nbs1, including which protein(s) binds to the FHA domain of Nbs1, is controversial.
While many studies reveal functions of the MRX complex in HR at a molecular level, molecular function of the complex in NHEJ is still unknown. Our previous study showed that xrs2 mutations in the FHA domain do not confer a significant defect in repair of DNA damage, telomere maintenance, and meiotic recombination (Shima et al. 2005). Recently, Wilson and his colleagues revealed that the FHA domain of Xrs2 is involved in NHEJ (Palmbos et al. 2005). Particularly, they showed that the NHEJ defect in the xrs2 mutant lacking the FHA domain is largely found in the yku80 mutant background. Here we confirmed and extended their results. We found that the FHA domain of Xrs2 plays a critical role in NHEJ even in the presence of Ku function. Particularly, our results indicate that the FHA domain of Xrs2 is required for a rapid NHEJ pathway in yeast through the interaction with Lif1. Furthermore, the FHA-mediated interaction between Xrs2 and the Lif1 is also conserved between human Nbs1 and Xrcc4, suggesting that Nbs1 may promote a NHEJ pathway through the interaction with the ligase IV complex in human cells.
MATERIALS AND METHODS
Yeast strains and plasmids:
All plasmids, yeast strains, and their genotypes are listed in supplemental Table S1. The dnl4Δ and lif1Δ allele were constructed by PCR-mediated gene disruption. The LIF1-HA was constructed by the PCR-based tagging methodology (De Antoni et al. 2002). All of the primer sets for PCR-mediated gene disruption or tagging are described in supplemental Table S2.
Plasmid-rejoining assay:
After the digestion of plasmid pRS313 (ARS-CEN, HIS3) with BamHI or plasmid pRS315 (Sikorski and Hieter 1989) (ARS-CEN, LEU2) with PstI (NEB), the plasmids were gel purified and resuspended in the TE buffer at a concentration of 100 ng/μl. Yeast cells were transformed with 100 ng of the linear plasmid as well as with 10 ng of undigested plasmid as a control. Then cells harboring plasmid pRS313 or pRS315 were selected on SD−His plate or SD−Leu plate, respectively. After 3 days incubation at 30°, or at 23° in the case of the hdf1 mutant, colonies grown on the plates were counted and a plasmid-rejoining frequency was calculated as a ratio of the number of transformants with digested DNA to that with undigested DNA. Experiments were carried out more than three times.
Plasmid DNAs were recovered from the yeast transformants and reintroduced into Escherichia coli DH5α. The plasmids were prepared from E. coli cells and digested with a restriction enzyme to check the presence of an original junction.
Detection of HO-induced DSBs by Southern blotting:
Log-phase cells (JKM139 background) were arrested in G1 by treatment with α-factor (6 μg/ml; Bachem) for 2.5 hr in YP–raffinose medium. After the treatment, galactose was added to induce homothallic (HO) endonuclease as described previously (Miyazaki et al. 2004). The cells were harvested at each time point for preparation of genomic DNAs. Purified genomic DNA was digested with HindIII, separated by electrophoresis with a 0.7% agarose gel in 0.5× TBE buffer, and then transferred onto membrane (Hybond-N; GE Healthcare) after the treatment with HCl and then with NaCl/NaOH as described previously (Shinohara et al. 1997). A PCR fragment amplified from the yeast genome using a specific primer set was labeled in the presence of [α-32P]dATP using a random-labeling kit (Mega Prime labeling kit; GE Healthcare) and was used as a probe for the hybridization. DNA bands were visualized using the BAS2000 phosphorimager (Fuji) and quantified using an image-analyzing software (Image Gauge, Fuji). The percentage of bands is calculated by dividing the intensity of a given band (either parent or DSB bands) by the sum of intensities of both types of bands (parent and DSB bands). Primer sequences used for the probe are as follows: HO-proxi-Hind probe-f, 5′-CTTTGCAGCAAACGCACACCATTTCCTACT-3′, and HO-proxi Hind-probe-r, 5′-CCGCCTTTACCGTAGTTTTGCTGCACCTT-3′.
Survival after induction of HO endonuclease:
JKM179 and its derivatives were used for the analysis as described previously (Valencia et al. 2001). Early log-phase cells at ∼5 × 105 cells/ml were regrown in YP–raffinose at 30° for 12 hr as preculture. Then galactose was added to the culture in a final concentration of 2% to induce DSBs. To measure the survival ratio, aliquots of cell culture at 4 hr after galactose induction were withdrawn and plated onto YPAD plates after serial dilution. Cells were incubated for 3 days and colonies were counted. Survival was expressed as a ratio of number of cells at 4 hr after the galactose induction to those before the induction (0-hr time point).
Homologous recombination between intrachromosomal direct repeat:
Recombination rates were calculated using the median method (Lea and Coulson 1948). The analysis was performed using the strain NKY1068 (Bishop et al. 1992) and its derivatives as described previously (Shinohara et al. 1997).
Two-hybrid analysis:
PCR-amplified fragments from a given gene were cloned into both pAS2-1 and pACT2 (Clontech Laboratories) as described previously (Shima et al. 2005). Plasmids for the analysis are described in supplemental Table S1. Fragments of human NBS1, hMRE11, and XRCC4 genes were prepared from cDNAs from HeLa S3 cells. The Nbs1-SH gene was constructed by the PCR-based site-directed mutagenesis. Yeast cells, AH109 (Clontech Laboratories), were cotransformed with a pair of plasmids: pAS2-1 and pACT2 derivatives. The transformants selected on −WL (SD−Trp−Leu) plates were cultured in liquid SD–WL medium overnight and then were spotted onto −WL or −WLH (SD−Trp−Leu−His) plates. For the β-galactosidase assay, at least three independent colonies were grown to log phase in liquid −WL medium. The cell extracts were prepared using a beads beater, and protein concentration of each extract was determined by Bradford method (Bio-Rad, Hercules, CA). Cell extracts were incubated with ONPG (4 mg/ml) and OD420 was measured using the spectrophotometer. β-Galactosidase activity was calculated as follows: 1 unit = 1.7× OD420/0.0045(t × V × P), where t is time of incubation in minutes, V is volume of cells in milliliters, and P is the protein concentration in cell extracts (milligrams/milliliter).
Immunoprecipitation assay and Western blotting:
Immunoprecipitation was essentially carried out as described previously (Hayase et al. 2004), with minor change in the composition of the lysis buffer (50 mm Tris–HCl, pH 7.5, 100 mm KCl, 0.1 m NaF, 1 mm Na2VO4, 1 mm DTT, 20% glycerol, 0.3% Tween-20). In the Western blot for Figure 4, B and C, one membrane after the transfer was cut into two pieces and then incubated with different first antibodies to compare the relative amount of various proteins in immunoprecipitation (IP) fractions. The antibodies against recombinant Lif1 and recombinant Xrs2-314M (Shima et al. 2005) protein were raised in rats and in both guinea pigs (MBL) and rabbits (Kitayama LABES), respectively. For detection of tagged proteins, anti-FLAG M2 (Sigma) and Anti-HA 16B12 (Babco) antibodies were used. Antitubulin (Serotec) antibody was used for detection of α-tubulin in yeast. Proteins on the blots were detected with a BCIP/NBT kit (Nacalai) and secondary antibodies conjugated with alkaline phosphatase (Promega, Madison, WI) or by using the Odyssey infrared imaging system (LI-COR) after staining with secondary antibodies conjugated with Alexa-680 (Molecular Probes, Eugene, OR) or IR-Dye-800 (Rockland).
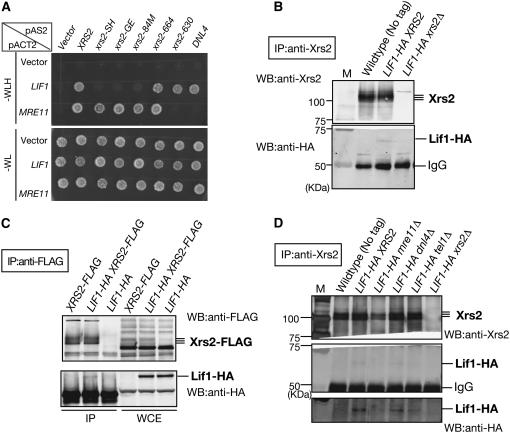
Figure 4.—
Interaction between Xrs2 and Lif1 protein. (A) Interactions of Lif1 or Mre11 with various Xrs2 mutant proteins were studied using two-hybrid analysis as described in materials and methods. (Top and bottom) Growth on SD–WLH selective and SD–WL nonselective plates, respectively. (B) In vivo complex formation of Xrs2 and Lif1. Complex formation of Xrs2 and Lif1 proteins were studied by IP using wild-type (W303-1A), LIF1-HA (MSY2644), and LIF1-HA xrs2Δ (KMY063) cells. Anti-Xrs2 (rabbit) was used for the precipitation. Xrs2 and Lif1-HA proteins were detected on immunoblots using anti-Xrs2 (guinea pig) and anti-HA antibodies, respectively. (C) Complex formation of Xrs2 and Lif1 proteins were studied by IP in another way, using XRS2-FLAG (KMY293), LIF1-HA XRS2-FLAG (KMY305), and LIF1-HA (MSY2644) cells. Anti-FLAG was used for the precipitation. Xrs2-FLAG and Lif1-HA proteins were detected on immunoblots using anti-FLAG and anti-HA antibodies, respectively. (D) Xrs2–Lif1 interaction was examined by immunoprecipitation in wild-type (W303-1A), LIF1-HA (MSY2644), LIF1-HA mre11Δ (MSY2665), LIF1-HA dnl4Δ (KMY168), LIF1-HA tel1Δ (KMY176), and LIF1-HA xrs2Δ (KMY063) cells. IP was carried out as described in B. (Bottom) Overexposed image of middle blots with anti-HA.
Phosphatase treatment of Lif1 protein:
Immunoprecipitation using anti-Lif1 antibody was performed with extracts from log-phase cells cultured in YPAD treated with phleomycin (20 μg/ml) for 2 hr. The precipitates were resuspended in the CIP buffer (100 mm NaCl, 10 mm Tris–HCl, pH 7.9, 1 mm DTT) and were divided into two aliquots. One fraction was incubated with 100 units of CIP (alkaline phosphatase, calf intestinal; NEB) at 30° for 30 min and the other was incubated in the absence of CIP. Lif1 protein was detected by Western blotting using anti-Lif1 antibody.
RESULTS
The FHA domain of Xrs2 is involved in NHEJ:
In budding yeast, Xrs2, a homolog of human Nbs1, is involved not only in telomere length control and DNA damage checkpoint, but also in DSB repair, including both HR and NHEJ. Previously, we dissected roles of various domains in Xrs2 in different DNA damage tolerance pathways (Shima et al. 2005). In the budding yeast, HR plays a major role in DSB repair, which may mask the effect of xrs2 mutations on NHEJ when DNA damage sensitivity was examined only for the single mutants. We therefore evaluated the effect of various xrs2 mutations on NHEJ by directly analyzing NHEJ reactions.
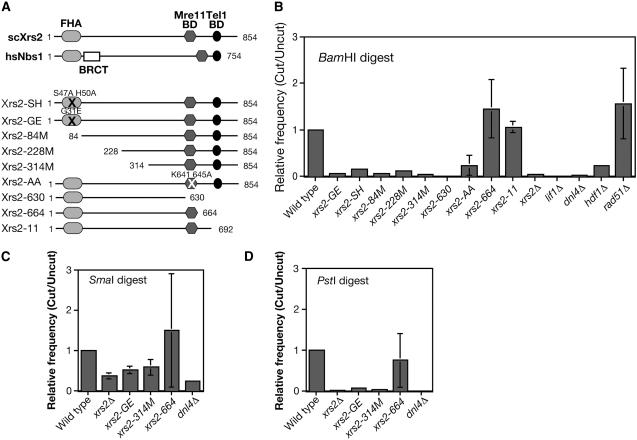
Our previous study identified three classes of xrs2 mutants (Shima et al. 2005) (Figure 1A). The first class has mutations in an N-terminal region including the FHA domain: xrs2-GE (G31E), -SH (S47A, H50A), -84M (Δ1-83), -228M (Δ1-227), and -314M (Δ1-313). The second class lacks the Mre11-binding domain, which is located in the C terminus: xrs2-630 (Δ631-854) and -AA (K641A, K645A). This mutant class shows almost similar phenotypes to the xrs2 null mutant. The third class lacks the C-terminal Tel1-binding domain: xrs2-664 (Δ665-854) and xrs2-11 (Nakada et al. 2003). The mutant has a defect in recruitment of Tel1 to DSB sites and telomere ends (Nakada et al. 2003; Shima et al. 2005).
Figure 1.—
xrs2-FHA mutants show a defect in the NHEJ. (A) The functional domains of human Nbs1 and yeast Xrs2 and the constructions of various Xrs2 mutant proteins used in this study. (B and C) Plasmid-rejoining assay was performed using plasmid digested with BamHI (B) (5′-overhang), (C) SmaI (blunt end), and (D) PstI (3′-overhang) in various mutant cells as described in materials and methods. Relative ratios compared to a wild-type ratio in each experiment are calculated and the average value with error bars (standard deviation) is shown for each experiment using different plasmids. At least three experiments were carried out to obtain SD. The following mutants were used: wild type (W303-1A), xrs2-GE (MSY2187), xrs2-SH (MSY2199), xrs2-84M (MSY2171), xrs2-228M (MSY2207), xrs2-314M (MSY2183), xrs2-630 (MSY2195), xrs2-AA (MSY2191), xrs2-664 (MSY2273), xrs2-11 (MSY2152), xrs2Δ (MSY2140), lif1Δ (KMY128), dnl4Δ (MSY2469), hdf1Δ (MSY2475), and rad51Δ (MSY2398).
Initially, we employed the plasmid-rejoining assay to assess the role of various Xrs2 domains in NHEJ. After the digestion of an autonomous plasmid with the HIS3 marker by a restriction enzyme, such as BamHI, the linearized plasmid was introduced in various strains and selected for the marker (Figure1B). Rejoining frequency is indicative of NHEJ activity, since the restriction site of the plasmid is located in a region without any homology to the yeast genome. We also analyzed mutants of the LIF1, DNL4, HDF1, and RAD51 genes as a control. As previously reported (Milne et al. 1996; Moore and Haber 1996; Wilson et al. 1997; Herrmann et al. 1998), as with lif1Δ, dnl4Δ, and hdf1Δ mutants, the xrs2Δ mutant is defective in plasmid recircularization. In contrast, the rad51Δ mutant showed a slightly higher frequency of recircularization than wild type. As expected, the xrs2-630 mutant reduced the NHEJ activity to a background level that is comparable to levels in the lif1Δ, dnl4Δ, and xrs2Δ cells. The xrs2-AA mutant shows a little higher frequency relative to the xrs2 null mutant, consistent with the fact that xrs2-AA mutant is partially active (Shima et al. 2005). In addition, the xrs2-11 and -664 mutants, lacking a C-terminal Tel1-binding domain, did not show any defect in rejoining of the plasmid.
Interestingly, three N-terminal deletion mutants, xrs2-84M, -228M, and -314M, show a strong defect in the circularization of the plasmid—an 8- to 10-fold decrease. This is consistent with the report by Palmbos et al. (2005), although they observed only a weak defect in the rejoining of the linearized plasmids for their xrs2 mutant (xrs2-FHA; ∼2.5-fold), which lacks N-terminal 125-amino-acid regions, including the FHA domain. The xrs2-84M mutant lacks only a FHA domain, which is conserved from yeast to humans. These confirmed previous results and showed that the 1- to 84-aa region of Xrs2 FHA is critical for NHEJ. Indeed, when the residues in the Xrs2, which are conserved among the FHA domains, are mutated, the mutant cells are defective in NHEJ. The xrs2-GE and -SH mutants almost abolished the rejoining of the linear plasmid by 8- to 10-fold.
To determine the pathways involved in NHEJ, the plasmids were recovered from transformants and the restoration of the original junction was confirmed by restriction enzyme digestion. Wild type restored the original junction (BamHI site) at a frequency of 100% (20/20) (data not shown). Importantly, all of the above xrs2 mutants showed complete restoration of the BamHI site on the plasmid.
We performed the same experiments using plasmids cleaved with SmaI (blunt end) or PstI (3′-overhang end) and received almost the same results on the PstI-digested plasmids as in the case of the BamHI (5′-overhang) digest (Figure 1, C and D) while various xrs2 mutants exhibited a weak defect in the rejoining of the plasmids with a blunt end (Figure 1C). As with wild type, the plasmid purified from the xrs2 mutants recovered the original junctions (PstI and SmaI).
The FHA domain of Xrs2 is required for repair of HO-induced DSBs:
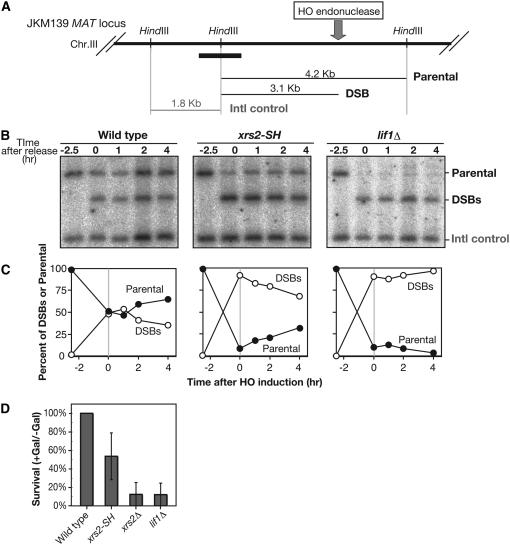
We next analyzed the NHEJ directly by Southern blotting for the DSB repair process. For this purpose, we studied the fate of DSB at the MAT locus on chromosome III in a strain lacking the donor loci the HML and HMR (JKM139) (Figure 2A). In this strain, HO-induced DSB cannot be repaired by HR due to the lack of donor sequence, but rather by either NHEJ or single-strand annealing (SSA) (Valencia et al. 2001). The expression of HO endonuclease is under the control of the GAL1-10 promoter. Cells were synchronized at G1 in the presence of α-factor and then were cultured in the presence of galacatose for 2 hr to induce a DSB at the MAT locus. After a 2-hr incubation, glucose was added to shut off continuous cleavage by the nuclease. The repair process was analyzed in the presence of α-factor to prevent repair between sister chromatids. At 0 hr, when glucose was added, wild-type cells showed both DSB and parental bands with almost same amount (∼50%; Figure 2, B and C). The presence of the parental DNAs at this time point is not due to inefficiency in DSB formation, but rather due to rapid repair of the break, since lif1 mutant cells can convert most of the parental DNA into DSBs. In wild type, after the shutoff of cleavage, the amount of DSB gradually decreases, while amounts of parental DNA increases in parallel (Figure 2C). These results suggest the presence of two types of DSB repair—rapid and slow processes—under such conditions. On the other hand, the lif1 cells completely abolish DSB repair under these conditions; there is very little recovery of the parental band. Thus, Lif1 is essential for two DSB repair processes. In the xrs2-SH mutant, the recovery of parental bands is very slow. After a 4-hr incubation, only 25% of DSBs are converted into the product. This indicates that the xrs2-SH mutant is defective in the rapid repair of DSB, but is proficient in the slow repair of the NHEJ. Consistent with this result, after the induction of DSBs, the xrs2-SH mutant forms more viable colonies than the lif1 mutant, but still shows reduced viability relative to wild type (Figure 2D). Taken together, these observations strongly suggest that the FHA domain of Xrs2 protein plays a critical role in NHEJ.
Figure 2.—
Physical analysis of repair of HO-induced DSBs in G1 phase. (A) A schematic of the MAT locus on yeast chromosome III. Sites for a diagnostic HindIII restriction enzyme site and sizes of DNA fragments for DSBs and parental and internal control DNAs are shown. The thick solid bar indicates a probe for Southern blotting analysis. (B and C) Physical analysis of NHEJ. In G1-arrested cells, DSBs are introduced by incubation with galactose for 2.5 hr. At 0 hr, glucose was added to shut off DSB induction. DNAs at indicated times were analyzed by Southern blotting. (C) Quantification, percentage of DSBs, and parental fragment in total DSBs and parental control. •, parental DNA, ○, DSBs. Wild type (JKM139), xrs2-SH (MSY3412), lif1Δ (KMY200). (D) Survival after induction of HO-endonuclease in various cells was examined as described in materials and methods. Error bars indicate SD for at least three independent experiments. Wild type (JKM179), xrs2-SH (MSY2998), xrs2Δ (KMY102), and lif1Δ (KMY082).
Xrs2 FHA domain is dispensable in single-strand annealing:
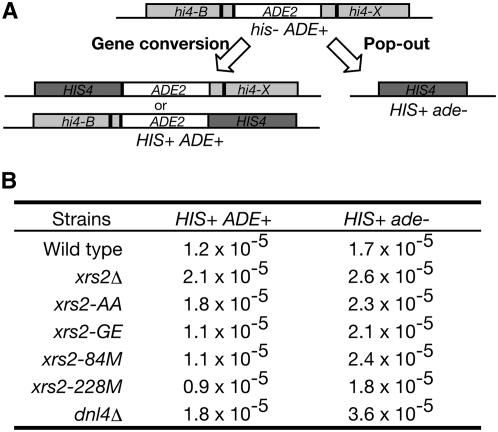
We examined the requirement of the Xrs2 and/or the Xrs2 FHA domain for intrachromosomal recombination between direct repeats using the his4B-ADE2-his4X allele on chromosome III (Figure 3A). In this construct, the His+ Ade+ prototrophs are generated mainly through gene conversion. On other hand, His+ Ade− prototrophs arise by either intrachromosome pop-out or unequal sister-chromatid exchange. Pop-out product is predominantly formed by SSA rather than by intrachromosomal crossing over. As reported previously (Johzuka and Ogawa 1995), an xrs2 null mutant generates His+ Ade+ prototrophs at a higher frequency than wild type. In this assay, a slight increase of recombination rate per generation was observed in the xrs2 null mutant (1.75-fold; Figure 3B) and the same is true for the xrs2-AA mutant. On the other hand, various xrs2 N-terminal mutants show a wild-type level of His+ Ade+ prototroph formation. Furthermore, these mutants significantly increase His+ Ade− prototroph formation more than wild type. A greater increase in the His+ Ade− prototroph is shown in the dnl4 mutant. These results suggest that the FHA domain of Xrs2 is dispensable for both gene conversion and SSA.
Figure 3.—
Intrachromosomal recombination between intrachromosomal direct repeats in the xrs2-FHA mutants. (A) Structure of the parental and recombinant DNAs. (B) Intrachromosomal recombination was assayed between a tandem repeat of his4 hetero-alleles flanking the ADE2 gene. Either His+Ade+ or HIS+Ade− prototrophs were selected on plates. Eleven independent colonies were assayed for each mutant. Recombination rate per generation was calculated using median values in the 11 given recombination frequencies. Wild type (NKY1068), xrs2Δ (MSY1668), xrs2-AA (MSY1722), xrs2-GE (MSY1730), xrs2-84M (MSY1699), xrs2-228M (MSY1701), and dnl4Δ (MSY2503).
The FHA domain is required for the Lif1–Xrs2 interaction:
Previous work showed the interaction between the Xrs2 and Lif1 proteins in vitro (Chen et al. 2001) or in vivo (Palmbos et al. 2005), prompting us to examine the role of the Xrs2 FHA domain in the Xrs2–Lif1 interaction. First, we performed yeast two-hybrid analysis (Figure 4A). As reported previously (Palmbos et al. 2005), we confirmed that specific interactions between Xrs2 and Mre11, Dnl4 and Lif1, and Xrs2 and Lif1 were detected by the assay. Truncation and substitution(s) of the FHA domain in Xrs2 (Xrs2-84M, -GE, and -SH) abolished the Xrs2–Lif1 interaction. However, these FHA mutations do not affect the binding of Xrs2 to Mre11. In the Xrs2-630 construct, which loses the Mre11-binding activity, a specific interaction with Lif1 was still intact. These results indicate a critical role of the FHA domain of Xrs2 in the binding to Lif1.
Next, we examined complex formation between Xrs2 and Lif1 by immunoprecipitation (Figure 4, B and C). For this purpose, we introduced the HA tag in the C terminus of the Lif1 open reading frame on yeast genome. The addition of the tag did not affect functions of Lif1 in vivo (data not shown). Importantly, we tried to detect the interaction between proteins under physiological conditions. Indeed, Lif1-HA protein was recovered in immunoprecipitates using the anti-Xrs2 antibody (Figure 4B). This precipitation depends on the presence of wild-type Xrs2 and also on the presence of HA tag on the Lif1 protein. However, the amount of Lif1 protein in the precipitates was <10% of the total amount of Lif1 in whole-cell extract. To refine this experiment, we tagged both Xrs2 and Lif1 with FLAG and HA tags, respectively. The double-tagged strain does not show any DNA repair defects (K. Matsuzaki, unpublished results). However, a small fraction of Lif1-HA still was recovered in the precipitates by using anti-FLAG antibody to immunoprecipitate Xrs2 protein in the LIF1-HA XRS2-FLAG double-tagged cells (Figure 4C). These results of polyclonal antibody against Xrs2 did not affect the interaction with Lif1 protein. The treatment of cell extracts with DNAaseI did not change the complex formation. This result suggests that only the small fraction of Lif1 protein can form the complex with Xrs2 in vivo. The complex formation of Xrs2 and Lif1 does not depend on Tel1 or the Lif1 partner, ligase VI (Figure 4D). Interestingly, the absence of the Xrs2 partner, Mre11, seemed to reduce the formation of Xrs2-Lif1 complex.
Lif1 is phosphorylated in vivo:
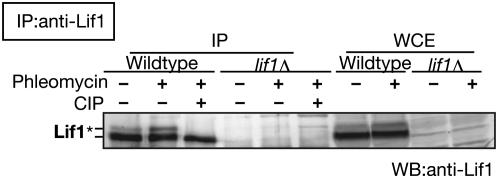
The FHA domain is known as a phospho-protein recognition domain and functions in protein–protein interaction (Sun et al. 1998; Durocher et al. 1999). During our analysis of Lif1 protein by Western blot using anti-Lif1 antibody, we noted that Lif1 migrates on gels as at least two distinct bands (Figure 5). To check possible Lif1 phosphorylation, we performed immunoprecipitation with anti-Lif1 antibody to detect Lif1 protein in yeast cells treated with or without phleomycin, a DSB-inducing agent (Figure 5). In wild-type cells, Lif1 protein was detected as two distinct bands, and after phleomycin treatment, a relative amount of top band to that of the bottom band was slightly increased. Importantly, the top band disappeared after the treatment of precipitates with CIP, a phosphatase. These results indicate that Lif1 is phosphorylated in yeast cells and that phosphorylation of Lif1 is induced by DNA damage.
Figure 5.—
Phosphorylation of Lif1 protein. In vivo phosphorylataion of Lif1. Lif1 protein from wild type (W303-1A) and lif1 null mutant cells (KMY128) with or without phleomycin treatment was immunoprecipitated using anti-Lif1 antisera and was treated with CIP as described in materials and methods. Lif1 protein in the precipitate and whole-cell extract (WCE) was detected using anti-Lif1 antibody. Wild-type (W303-1A) and lif1Δ (KMY128) cells with or without phleomycin treatment were analyzed as described in materials and methods. Anti-Lif1 antibody was used for precipitation and detection. The asterisk indicates the bands of phosphorylated Lif1 protein.
Two separated regions of Lif1 protein are required for the Lif1–Xrs2 interaction:
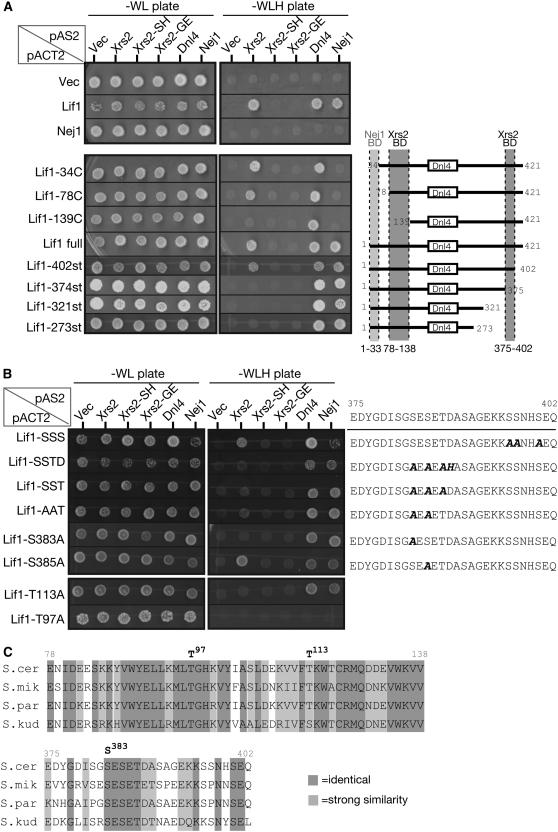
To dissect functional domains of Lif1, we constructed several truncated versions of Lif1 and examined the ability of the constructs to bind to Dnl4 and Nej1 as well as to Xrs2 by two-hybrid analysis. It is known that a highly conserved region among the orthologs of Lif1 (204–233 aa) is required for interaction with Dnl4 (Dore et al. 2006). We confirmed the binding of Lif1 to Dnl4. It is known that Nej1 protein also forms a complex with Lif1 and that a C-terminal region of Nej1 is important for the interaction (Frank-Vaillant and Marcand 2001). In our two-hybrid analysis, Lif1-34C, -78C, and -139C, which lack the N-terminal region of Lif1, did not interact with Nej1 (Figure 6A). This result shows that the most N-terminal (1–33 aa) region of Lif1 protein is required for Nej1 binding.
Figure 6.—
Identification of the Xrs2-binding domain in Lif1 protein. (A) Interaction among various Lif1 mutant proteins as determined by two-hybrid analysis. (Left) Results for the spot assay on selective SD–WLH plates and on nonselective SD–WL plates are shown. (Bottom) Interactions of Lif1 derivatives with a schematic of Lif1 deletion constructs. Dnl4-binding domain (open box) and binding domains for Nej1 (light shading) and for Xrs2 (dark shading) are also indicated. (B) Two-hybrid analysis of Lif1 proteins with substitution(s) in the Xrs2-binding domains. The analysis was carried out as in A. Amino acid sequence of the identified C-terminal Xrs2-binding domain and the positions of substitution (in boldface italics) are shown together with results of the two-hybrid spot assay. (C) Comparison of amino acid sequences of the two isolated Xrs2-binding domains among the Saccharomyces species. Light shading and dark shading highlight identical and similar amino acids, respectively. Similarity grouping was determined using the Gonnet Pam250 matrix (Gonnet et al. 1992), referenced from Saccharomyces Genome Database. S. cer (S. cerevisiae), S. mik (Saccharomyces mikatae), S. par (Saccharomyces paradoxus), and S. kud (Saccharomyces kudriavzevii).
Next we looked for the Xrs2 interaction site in Lif1 protein (Figure 6A). While the deletion of the N-terminal 78 aa did not affect the binding of Lif1 to Xrs2, the deletion of N-terminal 138 aa reduced the binding. This result indicates that the region of 78–138 aa of Lif1 is important for Lif1–Xrs2 interaction. In addition, Lif1-402st, which lacks the C-terminal 20 aa, can interact with Xrs2 but Lif1-374st, which deletes the C-terminal 48 aa, cannot interact with Xrs2. These results suggest that two regions of Lif1 protein, 78–138 and 375–402 aa, are necessary for binding with Xrs2. More importantly, mutations in the FHA domain of Xrs2 abolished interaction with the two regions of Lif1.
Given that the FHA domain generally recognizes a phosphorylated serine/threonine on a partner protein (Durocher et al. 1999), we assumed that the FHA domain of Xrs2 can specifically recognize a phosphorylated serine/threonine residue(s) in Lif1 protein. We looked for candidate residues of Lif1 and focused on serine/threonine residue(s) in these two Xrs2-binding regions, which are well conserved among Lif1 homologs found in Saccharomyces relatives (Figure 6C). In the 78–138-aa region, there are two conserved threonine residues, T97 and T113. We introduced lif1-T97A and -T113A mutations into the LIF1 gene and analyzed the effect of the mutations on the interaction with Xrs2. In lif1-T113A, interactions with Dnl4 or Nej1 were detected, but specific interaction with Xrs2 was lost (Figure 6B). In the case of the lif1-T97A mutation, all of the interaction with Xrs2, Dnl4, and Nej1 was abolished, suggesting that this region or threonine 97 might be important for a fundamental function of Lif1, such as protein structure or stability.
In the C terminal 375- to 402-aa region of Lif1, there are six well-conserved serine/threonine residues (Figure 6C). Interestingly, this region is rich in acidic amino acids as well as serine and threonine. To narrow down the interaction site for Xrs2, we divided the region into two parts and changed several conserved serine or threonine residues in each region. Three serine residues in the 375- to 402-aa region (396, 397, and 400) are dispensable for the Xrs2 binding. When we substituted all three conserved residues, S383, S385, and T387, to alanine (lif1-SST), these substitutions abolished the binding of Lif1 to Xrs2 (Figure 6B), but not to Dnl4 or Nej1. Among the three residues, at least S383 is critical for binding to Xrs2, since single substitution of S383 with alanine disrupted the interaction between Lif1 and Xrs2, but substitution of S385 with alanine did not.
These results suggest that there are two regions of Lif1 for Xrs2 interaction in Lif1 protein and that the C-terminal region includes an essential serine residue (S383). This serine residue is a potential candidate for phosphorylation (see discussion).
Xrs2-interaction regions of Lif1 is critical for NHEJ:
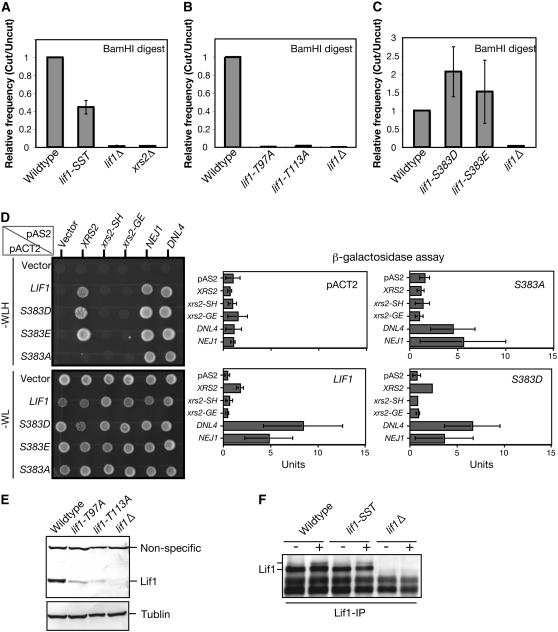
To determine the role of the Xrs2-binding domain of Lif1 in NHEJ, we characterized recircularization of linear plasmids in the lif1-SST (S383A, S385A, T387A), lif1-T97A, and lif1-T113A mutants (Figure 7, A and B). As expected, the lif1-SST mutation, which lacks an ability to bind to Xrs2, significantly decreases the rejoining efficiency of the plasmid compared to wild type. However, the efficiency in the lif1-SST mutant is significantly higher than those of the lif1 null mutant. This partial defect may be explained by the fact that the Lif1-SST mutant protein still remains the second Xrs2-interaction site.
Figure 7.—
Analysis of mutation at serine 383 of Lif1 in Xrs2 interaction and NHEJ; (A and B) Plasmid-rejoining assay was performed using plasmid cut with BamHI as described in materials and methods. Wild-type (W303-1A), lif1-SST (MSY3398), lif1-T97A (MSY3042), lif1-T113A (MSY3048), lif1Δ (KMY128), and xrs2Δ (MSY2140) cells were used for the assay. (C) Plasmid-rejoining assay was performed using plasmid cut with BamHI. Wild type (W303-1A), lif1-S383D (MSY3416), lif1-S383E (MSY3420), and lif1Δ (KMY128) cells. (D) Effects of amino acid substitution at serine 383 of Lif1 on interaction with Xrs2; Nej1 and Dnl4 were examined by two-hybrid assay. (Left, top and bottom) Result using selective (SD–WLH) and nonselective (SD–WL) plates, respectively. (Right) Results of β-garactosidase assay performed as described in materials and methods. Units for β-galactosidase are calculated and shown with error bars for three independent experiments. (E) The expression level of Lif1 proteins were compared among the wild-type (W303-1A), lif1-T97A (MSY3048), lif1-T113A (MSY3042), and lif1Δ (KMY128) cells. Lif1 proteins in whole-cell extract were detected using anti-Lif1 antibody and α-tubulin was detected using antitubulin as an internal control. (F) Effect of mutations on Lif1 phosphorylation was analyzed by immunoprecipitation using wild-type (W303-1A), lif1-SST (MSY3398), and lif1Δ (KMY128) cells with or without treatment with phleomycin. Lif1 proteins in the IP product were detected using anti-Lif1 antibody.
The lif1-T97A and lif1-T113A mutations show a defect in the recircularization of the plasmid similar to the lif1Δ mutant although this is merely speculative at this point since we do not know the linearity of plasmid rejoining relative to Lif1 abundance. However, in these two mutant cells, the expression levels of mutant Lif1 proteins are much lower relative wild type (Figure 7E). It is possible that the defects in plasmid rejoining and Xrs2 interaction of these two mutations, T97A and T113A, are caused by low expression or the combined defects of both dissociation of the complex and an expression problem. Especially in the case of lif1-T113A, specific dissociation of the Lif1-Xrs2 complex might cause a defect in the recircularization of the plasmid.
If the serine at 383 is phosphorylated in vivo, phospho-mimetic substitution might alter the functions of Lif1 in respect to the NHEJ activity. We constructed two phospho-mimetic substations of Lif1, S383E and S383D, which are the substitutions of serine at 383 with glutamate and aspartate, respectively. Both Lif1-S383E and -S383D proteins can bind to Xrs2, but not to Xrs2-FHA mutant protein, when analyzed by two-hybrid analysis (Figure 7D). Interestingly, both lif1-S383E and lif1-S383D significantly increased the efficiency of recircularization of linear plasmids (Figure 7C). This suggests that the phosphorylation of serine 383 plays a positive role in Lif1-mediated NHEJ activity.
In addition, we examined the mobility of mutant Lif1-SST protein on SDS–PAGE gels (Figure 7F). We found that Lif1-SST protein is as stable as wild-type Lif1. However, we could not detect change in the mobility of Lif1-SST protein on a gel. Therefore, we conclude that possible candidate serine residues are not responsible for the formation of the top band of Lif1.
The FHA-mediated interaction is conserved in human Nbs1-Xrcc4:
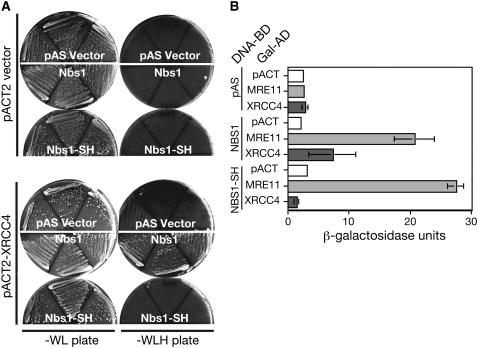
The FHA domain is conserved for the human Xrs2 ortholog, Nbs1 protein. However, it is still unknown whether the FHA domain is functionally conserved or not. We next examined interaction between human Nbs1 and Xrcc4, a homolog of Lif1, by the two-hybrid assay (Figure 8, A and B). We constructed NBS1-SH mutation (S42A H45A), as in yeast Xrs2-SH, in the FHA domain of Nbs1 to analyze the interaction with hMre11 or Xrcc4. While both wild-type Nbs1 and Nbs1-SH mutant proteins bind to hMre11, Nbs1-SH mutant protein does not interact with Xrcc4. This result indicates that Nbs1 interacts with Xrcc4 in a FHA domain-dependent manner as yeast Xrs2. Therefore, it is likely that Nbs1 binds to Xrcc4 in human cells and plays a role in NHEJ.
Figure 8.—
Interaction between Nbs1 and Xrcc4. (A) The interaction between human Nbs1 and Xrcc4 was examined by the two-hybrid plate assay. (Left and right) Results of nonselective (SD–WL) and selective (SD–WLH) plates, respectively. (B) β-Galactosidase assay for the interaction of Nbs1 proteins with Mre11 or Xrcc4 was performed as described in materials and methods. Units for β-galactosidase are calculated and shown with error bars for three independent experiments.
DISCUSSION
FHA domain of Xrs2 plays a critical role in NHEJ:
In budding yeast, it was known that the MRX complex is required for both NHEJ and HR pathways. Previous results reported that the deletion of the FHA domain in XRS2 causes a weak defect in NHEJ (Palmbos et al. 2005). We confirmed this finding using various deletions for the FHA domain in the XRS2 gene and extended the conclusion by constructing point mutations in the Xrs2 FHA domain (xrs2-SH and xrs2-GE). Both groups found that the xrs2-FHA mutants are deficient in the recircularization of plasmids, although the defect of the xrs2-FHA mutants in the previous report is weak. Furthermore, the Wilson group found that the xrs2-FHA mutants showed little defect in the repair of HO-induced DSB using a suicide substrate, but showed a defect when combined with a weak hypomorphic allele of ku70. On the other hand, we observed impairment in the repair of HO-induced DSBs in xrs2-FHA single mutants (Figure 2). The variation is due to differences in the strain background. Alternatively, we particularly analyzed the repair of HO-induced DSB in G1-arrested cells, while the Wilson group used asynchronous cultures for their assay. Since NHEJ seems to be regulated in a cell cycle (Daley et al. 2005b), this could explain the difference between the results.
As with lif1 or dnl4 mutants, xrs2 null mutant cells showed severe defects in plasmid religation and reduced survival after the formation of a single DSB by HO-endonuclease under conditions in which HR is suppressed. On the other hand, the xrs2-FHA mutant cells showed milder defects than the null mutant (Figure 1, B–D, and Figure 2D). The budding yeast has at least three pathways for DSB repair pathway in the absence of HR: classical NHEJ, nonclassical NHEJ (or microhomology dependent), and SSA. Our results suggest that xrs2-FHA mutants impair one or two of these pathways. Physical analysis of the repair of HO-induced DSBs in G1 phase revealed that there are two modes of DSB repair: rapid and slow repairs (Figure 2, B and C). While the lif1 null mutant is defective in both repairs, the xrs2-FHA mutant cells are deficient only in the rapid repair pathway. At this point we do not know the exact nature of the two pathways. The rapid pathway could be a simple religation of two DSB ends without any loss of DNA information: error-free NHEJ. The rapid pathway depends on the Xrs2 FHA function. Given that the MRX complex is recruited to DSB sites very early, the MRX might recruit the DNL complex efficiently through the interaction of the Xrs2 FHA with Lif1.
The slow repair might involve the resection of DSB ends and include ligation accompanied with microhomology, error-prone NHEJ, or SSA. This slow repair requires Lif1 but not FHA function of Xrs2, suggesting that there should be another recruiting mechanism for the DNL complex in this pathway. In the case of microhomology-dependent NHEJ, Pol4 interacts with Dnl4 directly (Tseng and Tomkinson 2002).
Xrs2 interacts with Lif1 through the FHA domain:
Previous work reported that purified Xrs2 protein binds to Lif1 protein directly in vitro and this interaction activates the Dnl4 ligase activity (Chen et al. 2001). Wilson and his colleagues (Palmbos et al. 2005) indicated that, when overproduced from the ADH1 promoter, Xrs2 and Lif1 proteins were able to interact with each other in yeast cells. In this article, we carried out an IP experiment under physiological conditions without any overproduction of proteins and found that Xrs2 forms a complex with Lif1 protein in vivo. Furthermore, our two-hybrid analysis revealed that the interaction is mediated through the FHA domain of Xrs2, but not through Mre11-binding and Tel1-binding domains. A similar result of two-hybrid analysis was reported by Palmbos et al. (2005). Substitution of amino acid residues in Xrs2, which are conserved among FHA domains, abolishes the interaction between Xrs2 and Lif1, confirming the importance of FHA domain in the interaction. Taken together, our results strongly indicate that the FHA domain of Xrs2 plays a role in NHEJ through the interaction with a critical NHEJ regulator, Lif1.
The two-hybrid analysis identified two regions on the Lif1 protein critical for binding to Xrs2 FHA: the N terminus and the C terminus. The importance of the C-terminal domain of Lif1 was verified by the mutational analysis. The mutation in conserved serines abolishes not only the interaction with Xrs2 FHA, but also NHEJ activity, without affecting the protein stability. This region contains possible phosphorylation sites (see below), which in turn may facilitate the NHEJ activity of Lif1. However, the functional significance of the N-terminal region is still obscured since the substitution of conserved threonine with alanine decreases the stability of Lif1 protein in cells. Further studies are necessary to determine the role of this region in the interaction with Xrs2.
Importantly, we can detect only a subset of Lif1 protein that forms a stable complex with a small fraction of Xrs2 under physiological conditions. Thus, it is likely that the interaction is very weak and/or transient and somehow could be regulated in a cell (see below).
The formation of the Xrs2-Lif1 complex in vivo depends upon the presence of Mre11 (Figure 4D). Mre11 has a nuclear localization signal that is essential for nuclear transport of Xrs2 protein (Tsukamoto et al. 2005). It is possible to consider that only Xrs2 protein in nuclei can interact with Lif1. However, this is unlikely since a nuclear transport signal was fused to Xrs2 in our two-hybrid assay. Another Xrs2-interacting factor, Tel1, may contribute little to the Xrs2–Lif1 interaction. Absence of Dnl4, a Lif1 partner, does not affect the Xrs2–Lif1 interaction.
Recruitment of the MRX complex to a DSB site is one of the earliest steps in the response of cells to DSBs (Lisby and Rothstein 2004). It is likely that the MRX complex recruits the DNL complex through the Xrs2–Lif1 interaction and facilitates rejoining DSB in an efficient way, if the DSB ends are clean enough to rejoin directly. Interestingly, Xrs2–Lif1 interaction is well conserved for human counterparts. Nbs1 plays an essential role in cell viability in mammalian cells, since knockout cells of the NBS1 gene in mice are lethal (Demuth et al. 2004). NBS is caused by partial dysfunction of Nbs1 protein. All known NBS patients express Nbs1 protein lacking the most N-terminal region, including FHA and BRCT domains. Many studies of NBS cells revealed that NBS cells have a defect in both HR and DNA damage checkpoint, as well as telomere maintenance, which might explain the cancer-prone phenotypes of such patients (Carney et al. 1998; Matsuura et al. 1998; Varon et al. 1998). However, these observations cannot fully explain the immunodeficiency of NBS patients. Our finding of an interaction between Nbs1 and Xrcc4 raises the interesting possibility that dysfunction of the NHEJ pathway, which may play a role in V(D)J recombination or class switching, may cause immunodeficiency in the NBS patients. This is supported by the fact that the ligase IV gene has been identified as a NBS variant gene (O'Driscoll et al. 2001). There is still not solid evidence to show that Nbs1 plays a direct role in NHEJ in mammals. However, there is some evidence for the presence of a NHEJ pathway, which is independent of Ku and DNA-PKcs, both of which are a major determinant of NHEJ activity in mammalian cells (Wang et al. 2003; Kuhfittig-Kulle et al. 2007). It is likely that Nbs1 promotes a minor “unidentified” NHEJ pathway in humans.
Casein kinase II could regulate the interaction between Xrs2 and Lif1:
The FHA domain is known as a phospho-protein recognition/interaction domain among several proteins (Durocher et al. 1999). In this article, we have shown that Lif1 protein is phosphorylated in vivo. This suggests that a phosphorylated region of Lif1 is responsible for binding to the Xrs2 FHA domain. We looked for a candidate site for phosphorylation on Lif1 protein, and serine at position 383 is a possible candidate site for the phosphorylation. When this serine is changed to alanine (lif1-S383A and lif1-SST mutant), it reduces not only the binding to Xrs2, but also NHEJ. Furthermore, when this serine is substituted with either aspartate or glutamate as phospho-mimic substitution, the substitutions do not affect binding to Xrs2 but stimulate NHEJ activity significantly. These results suggest that serine 383 of Lif1 could be phosphorylated in vivo and that phosphorylated Lif1 could be an active form for NHEJ. Phosphorylation at serine 383 might be directly recognized by the FHA domain of Xrs2, which in turn may promote the recruitment of the DNL complex to DSB ends.
We found that the amino acid sequence around the S383 and S385 (S-E-S-E-T-D) fits with a phosphorylation motif (S/T-D/E-X-D/E) for casein kinase II (CK2) (Meggio and Pinna 2003). CK2 is a conserved kinase among eukaryotes and plays a role in various metabolisms in a cell. In budding yeast, CK2 consists of a catalytic subunit (Cka1 or Cka2) and a regulatory subunit (Ckb1 or Ckb2). Loss of both genes (e.g., cka1 cka2 double) leads to loss of viability, suggesting an essential role of this protein kinase in cell growth and proliferation. Recent work showed that CK2 is involved in histone H4 modification at DSB sites in yeast (Cheung et al. 2005). Furthermore, an interaction between Cka2 and Lif1 has already been reported by high-throughput mass spectrometry analysis on yeast proteins (Ho et al. 2002). From these observations, it is possible to consider that CK2 is recruited to a DSB site through an unknown mechanism and may phosphorylate Lif1 protein at DSBs to stabilize the interaction between the MRX and DNL complexes. Alternatively, it is possible to think that CK2 is recruited to DSB through interaction with Lif1.
We are currently engaged in studies to show that CK2 can indeed phosphorylate Lif1 at serine 383. However, it is very hard to prove this, since Lif1 protein contains at least 13 potential candidates for phosphorylation by CK2. In addition, we raised an antibody specific to a peptide containing phosphorylated S383, but this phospho-specific antibody does not work (M. Shinohara, unpublished results). The detection by the antibody might be hampered by multiple phosphorylation around this region (see above).
On Western blots, Lif1 proteins were detected as a mixture of bands with differing mobility. The lif1-SST mutation did not induce a significant change in the mobility of Lif1 protein and many shift bands were still observed. This result suggests that there are some other phosphorylation(s) on Lif1 protein. Indeed, Lif1 contains various potential phosphorylation sites by other kinases, for example, one site for CDK and two sites for ATM/ATR. Lif1 protein might be regulated through phosphorylation at multiple sites, including serine 383 phosphorylation.
Acknowledgments
We are grateful to J. E. Haber and all members of the Shinohara Lab for critical discussions. We thank A. Murakami for excellent technical assistance. And we also thank M. Ito, M. Suzuki, and H. Shima for contribution in the early phase of this study. K. M. was supported by the Institute for Protein Research as a research fellow. This work was supported by special coordination funds for promoting science and technology from the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT) to M.S. and a grant-in-aid from MEXT to M.S. and A.S.
References
- Bishop, D. K., D. Park, L. Xu and N. Kleckner, 1992. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell 69 439–456. [DOI] [PubMed] [Google Scholar]
- Carney, J. P., R. S. Maser, H. Olivares, E. M. Davis, M. Le Beau et al., 1998. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell 93 477–486. [DOI] [PubMed] [Google Scholar]
- Chen, L., K. Trujillo, W. Ramos, P. Sung and A. E. Tomkinson, 2001. Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol. Cell 8 1105–1115. [DOI] [PubMed] [Google Scholar]
- Cheung, W. L., F. B. Turner, T. Krishnamoorthy, B. Wolner, S. H. Ahn et al., 2005. Phosphorylation of histone H4 serine 1 during DNA damage requires casein kinase II in S. cerevisiae. Curr. Biol. 15 656–660. [DOI] [PubMed] [Google Scholar]
- Connelly, J. C., and D. R. Leach, 2002. Tethering on the brink: the evolutionarily conserved Mre11-Rad50 complex. Trends Biochem. Sci. 27 410–418. [DOI] [PubMed] [Google Scholar]
- Daley, J. M., R. L. Laan, A. Suresh and T. E. Wilson, 2005. a DNA joint dependence of pol X family polymerase action in nonhomologous end joining. J. Biol. Chem. 280 29030–29037. [DOI] [PubMed] [Google Scholar]
- Daley, J. M., P. L. Palmbos, D. Wu and T. E. Wilson, 2005. b Nonhomologous end joining in yeast. Annu. Rev. Genet. 39 431–451. [DOI] [PubMed] [Google Scholar]
- De Antoni, A., J. Schmitzova, H. H. Trepte, D. Gallwitz and S. Albert, 2002. Significance of GTP hydrolysis in Ypt1p-regulated endoplasmic reticulum to Golgi transport revealed by the analysis of two novel Ypt1-GAPs. J. Biol. Chem. 277 41023–41031. [DOI] [PubMed] [Google Scholar]
- Demuth, I., P. O. Frappart, G. Hildebrand, A. Melchers, S. Lobitz et al., 2004. An inducible null mutant murine model of Nijmegen breakage syndrome proves the essential function of NBS1 in chromosomal stability and cell viability. Hum. Mol. Genet. 13 2385–2397. [DOI] [PubMed] [Google Scholar]
- Dore, A. S., N. Furnham, O. R. Davies, B. L. Sibanda, D. Y. Chirgadze et al., 2006. Structure of an Xrcc4-DNA ligase IV yeast ortholog complex reveals a novel BRCT interaction mode. DNA Repair 5 362–368. [DOI] [PubMed] [Google Scholar]
- Durocher, D., J. Henckel, A. R. Fersht and S. P. Jackson, 1999. The FHA domain is a modular phosphopeptide recognition motif. Mol. Cell 4 387–394. [DOI] [PubMed] [Google Scholar]
- Frank-Vaillant, M., and S. Marcand, 2001. NHEJ regulation by mating type is exercised through a novel protein, Lif2p, essential to the ligase IV pathway. Genes Dev. 15 3005–3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonnet, G. H., M. A. Cohen and S. A. Benner, 1992. Exhaustive matching of the entire protein sequence database. Science 256 1443–1445. [DOI] [PubMed] [Google Scholar]
- Grawunder, U., M. Wilm, X. Wu, P. Kulesza, T. E. Wilson et al., 1997. Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature 388 492–495. [DOI] [PubMed] [Google Scholar]
- Hayase, A., M. Takagi, T. Miyazaki, H. Oshiumi, M. Shinohara et al., 2004. A protein complex containing Mei5 and Sae3 promotes the assembly of the meiosis-specific RecA homolog Dmc1. Cell 119 927–940. [DOI] [PubMed] [Google Scholar]
- Herrmann, G., T. Lindahl and P. Schar, 1998. Saccharomyces cerevisiae LIF1: a function involved in DNA double-strand break repair related to mammalian XRCC4. EMBO J. 17 4188–4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, Y., A. Gruhler, A. Heilbut, G. D. Bader, L. Moore et al., 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415 180–183. [DOI] [PubMed] [Google Scholar]
- Johzuka, K., and H. Ogawa, 1995. Interaction of Mre11 and Rad50: two proteins required for DNA repair and meiosis-specific double-strand break formation in Saccharomyces cerevisiae. Genetics 139 1521–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhfittig-Kulle, S., E. Feldmann, A. Odersky, A. Kuliczkowska, W. Goedecke et al., 2007. The mutagenic potential of non-homologous end joining in the absence of the NHEJ core factors Ku70/80, DNA-PKcs and XRCC4-LigIV. Mutagenesis 22 217–233. [DOI] [PubMed] [Google Scholar]
- Lea, D. E., and C. A. Coulson, 1948. The distribution of the number of mutants in bacterial populations. J. Genet. 49 264–284. [DOI] [PubMed] [Google Scholar]
- Lisby, M., and R. Rothstein, 2004. Localization of checkpoint and repair proteins in eukaryotes. Biochimie 87 579–589. [DOI] [PubMed] [Google Scholar]
- Llorente, B., and L. S. Symington, 2004. The Mre11 nuclease is not required for 5′ to 3′ resection at multiple HO-induced double-strand breaks. Mol. Cell. Biol. 24 9682–9694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura, S., H. Tauchi, A. Nakamura, N. Kondo, S. Sakamoto et al., 1998. Positional cloning of the gene for Nijmegen breakage syndrome. Nat. Genet. 19 179–181. [DOI] [PubMed] [Google Scholar]
- Meggio, F., and L. A. Pinna, 2003. One-thousand-and-one substrates of protein kinase CK2? FASEB J. 17 349–368. [DOI] [PubMed] [Google Scholar]
- Milne, G. T., S. Jin, K. B. Shannon and D. T. Weaver, 1996. Mutations in two Ku homologs define a DNA end-joining repair pathway in Saccharomyces cerevisiae. Mol. Cell. Biol. 16 4189–4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki, T., D. A. Bressan, M. Shinohara, J. E. Haber and A. Shinohara, 2004. In vivo assembly and disassembly of Rad51 and Rad52 complexes during double-strand break repair. EMBO J. 23 939–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, J. K., and J. E. Haber, 1996. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol. Cell. Biol. 16 2164–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada, D., K. Matsumoto and K. Sugimoto, 2003. ATM-related Tel1 associates with double-strand breaks through an Xrs2-dependent mechanism. Genes Dev. 17 1957–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Driscoll, M., K. M. Cerosaletti, P. M. Girard, Y. Dai, M. Stumm et al., 2001. DNA ligase IV mutations identified in patients exhibiting developmental delay and immunodeficiency. Mol. Cell 8 1175–1185. [DOI] [PubMed] [Google Scholar]
- Palmbos, P. L., J. M. Daley and T. E. Wilson, 2005. Mutations of the Yku80 C terminus and Xrs2 FHA domain specifically block yeast nonhomologous end joining. Mol. Cell. Biol. 25 10782–10790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharples, G. J., and D. R. Leach, 1995. Structural and functional similarities between the SbcCD proteins of Escherichia coli and the RAD50 and MRE11 (RAD32) recombination and repair proteins of yeast. Mol. Microbiol. 17 1215–1217. [DOI] [PubMed] [Google Scholar]
- Shima, H., M. Suzuki and M. Shinohara, 2005. Isolation and characterization of novel xrs2 mutations in Saccharomyces cerevisiae. Genetics 170 71–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara, M., E. Shita-Yamaguchi, J. M. Buerstedde, H. Shinagawa, H. Ogawa et al., 1997. Characterization of the roles of the Saccharomyces cerevisiae RAD54 gene and a homologue of RAD54, RDH54/TID1, in mitosis and meiosis. Genetics 147 1545–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski, R. S., and P. Hieter, 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Z., J. Hsiao, D. S. Fay and D. F. Stern, 1998. Rad53 FHA domain associated with phosphorylated Rad9 in the DNA damage checkpoint. Science 281 272–274. [DOI] [PubMed] [Google Scholar]
- Tseng, H. M., and A. E. Tomkinson, 2002. A physical and functional interaction between yeast Pol4 and Dnl4-Lif1 links DNA synthesis and ligation in nonhomologous end joining. J. Biol. Chem. 277 45630–45637. [DOI] [PubMed] [Google Scholar]
- Tsukamoto, Y., C. Mitsuoka, M. Terasawa, H. Ogawa and T. Ogawa, 2005. Xrs2p regulates Mre11p translocation to the nucleus and plays a role in telomere elongation and meiotic recombination. Mol. Biol. Cell 16 597–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia, M., M. Bentele, M. B. Vaze, G. Herrmann, E. Kraus et al., 2001. NEJ1 controls non-homologous end joining in Saccharomyces cerevisiae. Nature 414 666–669. [DOI] [PubMed] [Google Scholar]
- Varon, R., C. Vissinga, M. Platzer, K. M. Cerosaletti, K. H. Chrzanowska et al., 1998. Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell 93 467–476. [DOI] [PubMed] [Google Scholar]
- Wang, H., A. R. Perrault, Y. Takeda, W. Qin, H. Wang et al., 2003. Biochemical evidence for Ku-independent backup pathways of NHEJ. Nucleic Acids Res. 31 5377–5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weemaes, C. M., T. W. Hustinx, J. M. Scheres, P. J. van Munster, J. A. Bakkeren et al., 1981. A new chromosomal instability disorder: the Nijmegen breakage syndrome. Acta Paediatr. Scand. 70 557–564. [DOI] [PubMed] [Google Scholar]
- Wilson, T. E., U. Grawunder and M. R. Lieber, 1997. Yeast DNA ligase IV mediates non-homologous DNA end joining. Nature 388 495–498. [DOI] [PubMed] [Google Scholar]
- Wiltzius, J. J., M. Hohl, J. C. Fleming and J. H. Petrini, 2005. The Rad50 hook domain is a critical determinant of Mre11 complex functions. Nat. Struct. Mol. Biol. 12 403–407. [DOI] [PubMed] [Google Scholar]