Abstract
Cse4p is an essential histone H3 variant in Saccharomyces cerevisiae that defines centromere identity and is required for proper segregation of chromosomes. In this study, we investigated phenotypic consequences of Cse4p mislocalization and increased dosage of histone H3 and Cse4p, and established a direct link between histone stoichiometry, mislocalization of Cse4p, and chromosome segregation. Overexpression of the stable Cse4p mutant, cse4K16R, resulted in its mislocalization, increased association with chromatin, and a high rate of chromosome loss, all of which were suppressed by constitutive expression of histone H3 (Δ16H3). We determined that Δ16H3 did not lead to increased chromosome loss; however, increasing the dosage of histone H3 (GALH3) resulted in significant chromosome loss due to reduced levels of centromere (CEN)-associated Cse4p and synthetic dosage lethality (SDL) in kinetochore mutants. These phenotypes were suppressed by GALCSE4. We conclude that the chromosome missegregation of GALcse4K16R and GALH3 strains is due to mislocalization and a functionally compromised kinetochore, respectively. Suppression of these phenotypes by histone Δ16H3 and GALCSE4 supports the conclusion that proper stoichiometry affects the localization of histone H3 and Cse4p and is thus essential for accurate chromosome segregation.
THE term “chromosome cycle” describes the maintenance, replication, and segregation of chromosomal DNA, which constitutes a fundamental aspect of the cell division cycle. Coordination of multiple gene products that encode structural components for the kinetochore (centromere DNA and associated proteins), telomeres, spindle pole bodies, microtubules, and codensins, as well as regulatory components that establish checkpoints, is essential for maintenance of ploidy (Skibbens and Hieter 1998). The kinetochore is required for maintaining cohesion between sister chromatids in the vicinity of CEN DNA, attachment of chromosomes to the mitotic spindle, and their subsequent movement to the spindle poles (Ekwall 2007).
In contrast to the complex centromeric (CEN) structure of other eukaryotes, the CEN DNA sequence in Saccharomyces cerevisiae is relatively short (125 bp) with three conserved elements: CDEI, CDEII, and CDEIII (Hyman and Sorger 1995; Cheeseman et al. 2002; Cleveland et al. 2003). The integrity of CEN DNA and associated proteins is of pivotal importance to faithful chromosome transmission. The ≥70 kinetochore proteins identified to date are classified on the basis of their interaction with CEN DNA (inner kinetochore), with spindle microtubules (outer kinetochore), or between the inner and outer kinetochore components (central kinetochore) (McAinsh et al. 2003; Westermann et al. 2003; Meraldi et al. 2006). Binding of CBF3, a multiprotein complex containing Ctf13p, Ndc10p, Cep3p, and Skp1p, to CDEIII is essential and constitutes one of the first steps in kinetochore assembly (Westermann et al. 2003).
In addition to CEN DNA-binding proteins, higher-order chromatin structure plays an important role in chromosome segregation. In S. cerevisiae, the kinetochore chromatin domain is delimited on each side by strong nuclease-hypersensitive sites and is flanked by arrays of phased nucleosomes (Bloom and Carbon 1982; Funk et al. 1989; Schulman and Bloom 1991; Glowczewski et al. 2000). In contrast to conventional nucleosomes containing histones H2A, H2B, H3, and H4, a distinguishing feature of centromeric chromatin is the replacement of histone H3 by a histone H3 variant (CenH3) (Meluh et al. 1998; Henikoff and Dalal 2005). CenH3 homologs (Cse4p in S. cerevisiae, CENP-A in humans, CID in Drosophila melanogaster, HTR12 in Arabidopsis thaliana, and Cnp1 in Schizosaccharomyces pombe) are essential in all organisms for determining centromere identity and kinetochore function (Stoler et al. 1995; Smith et al. 1996; Meluh et al. 1998; Buchwitz et al. 1999; Blower and Karpen 2001; Chen et al. 2003). CenH3 proteins have two structural domains, a divergent N terminus (NT) and a conserved C-terminal histone fold domain (HFD) that is highly homologous to histone H3 (Malik and Henikoff 2003). S. cerevisiae Cse4p was identified in two independent genetic screens for mutants defective in chromosome segregation (Smith et al. 1996; Baker et al. 1998). Recent studies have also identified an essential role for Cse4p in the segregation of yeast 2μ plasmids (Hajra et al. 2006) and an essential N-terminal domain (END) of 33 amino acids within the first 130 amino acids that is required for cell viability. The HFD of Cse4p has been proposed to be sufficient for CEN targeting and propagation of active centromeres (Morey et al. 2004). Additionally, characterization of kinetochore subcomplexes has established that components of the Ctf19p, Okp1, Mcm21p, and Ame1p (COMA) and Ctf3p, Mcm16p, and Mcm22p (Ctf3p) complexes exhibit genetic and physical interactions with Cse4p (Ortiz et al. 1999; Measday et al. 2002).
Despite the essential role of CenH3, mechanisms for its deposition and maintenance at the centromere are not well understood. In humans and flies, centromere identity and propagation seem to be determined epigenetically, with preexisting CenH3-containing nucleosomes providing the epigenetic mark (Karpen and Allshire 1997). Studies with mammalian cells have shown that CENP-A is recruited in late mitosis/early G1 phase of the cell cycle in contrast to conventional histone H3 recruitment, which is primarily concomitant with DNA replication (Jansen et al. 2007). A CenH3 chaperone, RbAp48, has been identified on the basis of in vitro studies with D. melanogaster (Furuyama et al. 2006) and is required for CenH3 localization in S. pombe and humans (Hayashi et al. 2004). It has been proposed that the point centromeres of S. cerevisiae consist of a single nucleosome containing Cse4p (Furuyama and Biggins 2007). Recent studies have led to the identification of a novel inner kinetochore protein suppressor of chromosome missegregation (Scm)3p, which is required for the recruitment and establishment of Cse4p-containing chromatin (Camahort et al. 2007; Mizuguchi et al. 2007; Stoler et al. 2007; Zhang et al. 2007). It has been proposed that Scm3p substitutes for histones H2A–H2B in the nucleosome specialized for kinetochore assembly (Mizuguchi et al. 2007). Characterization of stable mutant forms of Cse4p, such as cse4K16R and cse4-351p, has led to the conclusion that ubiquitin-mediated proteolysis contributes to the exclusive centromeric localization of Cse4p (Collins et al. 2004).
Localization of CenH3 to the kinetochore is critical for high-fidelity chromosome segregation (Smith 2002). Overexpression and mislocalization of CenH3 have been observed in colorectal cell lines (Tomonaga et al. 2003), and aneuploidy and ectopic kinetochore formation were shown to be associated with the overexpression and mislocalization of CenH3 in D. melanogaster (Heun et al. 2006). To further elucidate the relationship between CenH3 and chromosome segregation, we used S. cerevisiae to investigate the consequences of Cse4p mislocalization. Unlike results from mammalian and Drosophila systems, overexpression of S. cerevisiae Cse4p does not lead to increased chromosome loss (Crotti and Basrai 2004). However, mislocalization of Cse4p has been reported in S. cerevisiae cac1Δhir1Δ and spt4Δ strains. These strains exhibit defects in chromosome segregation and heterochromatic gene silencing, as well as other phenotypes (Sharp et al. 2002; Crotti and Basrai 2004). An important biological question derived from these data is whether mislocalization of Cse4p leads to chromosome segregation defects.
Given the fact that Cse4p localization to centromeres is essential for genome stability and that Cse4p has a high degree of homology to histone H3, we investigated phenotypes of mislocalized Cse4p and altered histone dosage. We show that mislocalization of a stable mutant form of Cse4p, cse4K16R, to noncentromeric loci correlates with defects in chromosome transmission fidelity (ctf). This conclusion is supported by the suppression of these phenotypes by constitutive expression of histone H3 (Δ16H3). We also determined that overexpression of histone H3 (GALH3) resulted in a high rate of chromosome loss, which was due to reduced levels of Cse4p at CEN DNA. Additionally, the absence of Ctf19p, Mcm21p, Okp1p, Ame1p, Ctf3p, Mcm22p, and Mcm16p, which are kinetochore components of the COMA and Ctf3p complexes, sensitizes cells to increased histone H3 dosage. Our results establish that proper stoichiometry and localization of histone H3 and Cse4p are essential for high-fidelity chromosome segregation.
MATERIALS AND METHODS
Strains and plasmids used:
Strains used are listed in Table 1. pLG41 (GAL1/10-HHT1, 2μ, URA3) and pLG39 (GAL1/10-HHF1, 2μ, URA3) were gifts from M. M. Smith. pMB1159 (GAL1/10-HHT1, 2μ, TRP1) and pMB1158 (GAL1/10-HHF1, 2μ, TRP1) were derived from subcloning the PvuI fragments of pLG41 and pLG39, respectively, into pRS424. pSB500 (MYC-CSE4∷LEU2 integrating vector), pSB816 (GAL1/10-MYC-CSE4, 2μ, URA3), and pSB817 (GAL1/10-MYC-cse4K16R, 2μ, URA3) were gifts from S. Biggins. pMB1147 (GAL1/10-MYC-CSE4, 2μ, LEU2) was constructed by subcloning the BglI fragment of pSB816 into pRS425 cut with the same restriction enzyme. Integrating plasmids pAB95 (Δ16H2A-H2B∷URA3), pAB157 (Δ16H3-H4∷LEU2), pAB203 (Δ16H4∷LEU2), pAB204 (Δ16H3∷LEU2), pAB221 (Δ16H3N*H4∷LEU2), pAB224 (Δ16H3C*H4∷LEU2), and pCC65 (H3-H4, 2μ, URA3) were gifts from M. A. Osley. pMB824 (GAL1/10-CSE4, CEN, LEU2) was from the Basrai lab.
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| Y5563 | MATα his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 lyp1Δ can1Δ∷MFA1pr-HIS3 | Tong and Boone (2006) |
| YMB372 | ctf14-42 | This study |
| YMB3467 | MATα ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 (pSB816-pGAL1/10-MYC-CSE4, 2μ, URA3) CFIII (CEN3L.YPH278) HIS3 SUP11 | This study |
| YMB3468 | MATα ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 (pSB817-pGAL1/10-MYC-cse4K16R, 2μ, URA3) CFIII (CEN3L.YPH278) HIS3 SUP11 | This study |
| YMB3477 | MATα ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 (pSB816-pGAL1/10-MYC-CSE4, 2μ, URA3) [HHT1-hhf1Δ]Δ16′∷LEU2 CFIII (CEN3L.YPH278) HIS3 SUP11 | This study |
| YMB3478 | MATα ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 (pSB816-pGAL1/10-MYC-cse4K16R, 2μ, URA3) [HHT1-hhf1Δ]Δ16′∷LEU2 CFIII (CEN3L.YPH278) HIS3 SUP11 | This study |
| YMB3482 | MATα ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 (pRS426, 2μ, URA3) CFIII (CEN3L.YPH278) HIS3 SUP11 | This study |
| YMB6003 | MATα ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 (pRS426, 2μ, URA3) CFIII (CEN3L.YPH278) HIS3 SUP11 | This study |
| YMB6005 | MATα ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 (pLG41-GAL1/10-HHT1, 2μ, URA3) CFIII (CEN3L.YPH278) HIS3 SUP11 | This study |
| YMB6108 | MATα ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 (pRS424-GAL1/10, 2μ, TRP1) CFIII (CEN3L.YPH278) HIS3 SUP11 | This study |
| YMB6110 | MATα ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 (pSB817-pGAL1/10-MYC-cse4K16R, 2μ, TRP1) CFIII (CEN3L.YPH278) HIS3 SUP11 | This study |
| YMB6125 | MATα ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 (pSB817-GAL1/10-MYC-cse4K16R, 2μ, TRP1) (pRS316, CEN, URA3) CFIII (CEN3L.YPH278) HIS3 SUP11 | |
| YMB6132 | MATα ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 (pRS426-GAL1/10, 2μ, URA3) (pRS424-GAL1/10, 2μ, TRP1) MYC-CSE4∷LEU2 CFIII (CEN3L.YPH278) HIS3 SUP11 | This study |
| YMB6133 | MATα ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 (pRS426-GAL1/10, 2μ, URA3) (pMB1159-GAL1/10-HHT1, 2μ, TRP1) MYC-CSE4∷LEU2 CFIII (CEN3L.YPH278) HIS3 SUP11 | This study |
| YMB6136 | MATα ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 (pSB816-GAL1/10-MYC-CSE4, 2μ, URA3) (pMB1159-GAL1/10-HHT1, 2μ, TRP1) MYC-CSE4∷LEU2 CFIII (CEN3L.YPH278) HIS3 SUP11 | This study |
| YMB6138 | MATα ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 (pCC65-HHT1-HHF1, 2μ, URA3) CFIII (CEN3L.YPH278) HIS3 SUP11 | This study |
| YMB6146 | MATα ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 MYC-CSE4∷TRP1 [HHT1-hhf1Δ]Δ16′∷LEU2 CFIII (CEN3L.YPH278) HIS3 SUP11 | This study |
| YMB6147 | MATα ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 MYC-CSE4∷TRP1 (pRS315, CEN, LEU2) CFIII (CEN3L.YPH278) HIS3 SUP11 | This study |
| YMB6257 | MATα ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 (pLG41-GAL1/10-HHT1, 2μ, URA3) (pMB1158-GAL1/10-HHF1, 2μ, TRP1) CFIII (CEN3L.YPH278) HIS3 SUP11 | This study |
| YMB6267 | MATa/α ura3-52/ura3-52 lys2-801/lys2-801 ade2-101/ade2-101 trp1-Δ63/trp1-Δ63 his3-Δ200/his3-Δ200 leu2-Δ1/leu2-Δ1 cse4Δ∷KanMX/CSE4 CFIII (CEN3L) HIS3 SUP11 | This study |
| YMB6310 | MATα ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 (pSB816-pGAL1/10-MYC-cse4K16R, 2μ, URA3) [HHT1-hhf1Δ]Δ16′∷LEU2 cse4Δ∷KanMX CFIII (CEN3L.YPH278) HIS3 SUP11 | This study |
| YMB6398 | MATaura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 CSE4-MYC∷LEU2 CFIII (CEN3L.YPH278) HIS3 SUP11 | This study |
| YMB6426 | MATa/α lys2-801/lys2-801 ade2-101/ade2-101 trp1-Δ63/trp1-Δ63 his3-Δ200/his3-Δ200 leu2-Δ1/leu2-Δ1 cse4Δ∷KanMX/CSE4 cse4K16R∷URA3/ura3-52 CFIII (CEN3L) HIS3 SUP11 | This study |
| YMB6427 | MATα ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 cse4Δ∷KanMX cse4K16R∷URA3 CFIII (CEN3L) HIS3 SUP11 | This study |
| YMB6467 | MATaura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 CFIII (CEN3L) HIS3 SUP11 | This study |
| YPH1018 | MATα ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 CFIII (CEN3L.YPH278) HIS3 SUP11 | P. Hieter |
| YPH1676 | MATaame1-4:TRP1 | Pot et al. (2005) |
| YPH1678 | MATaokp1-5:TRP1 | Ortiz et al. (1999) |
| YPH1712 | MATamfa1Δ∷MFa1pr-LEU2 can1Δ∷MFA1pr-HIS3 ura3Δ0 leu2Δ0 his3Δ1 lys2Δ0 ctf3Δ∷natR | Measday et al. (2005) |
| YPH1713 | MATamfa1Δ∷MFa1pr-LEU2 can1Δ∷MFA1pr-HIS3 ura3Δ0 leu2Δ0 his3Δ1 lys2Δ0 ctf19Δ∷natR | Measday et al. (2005) |
| YPH1714 | MATamfa1Δ∷MFa1pr-LEU2 can1Δ∷MFA1pr-HIS3 ura3Δ0 leu2Δ0 his3Δ1 lys2Δ0 mcm16Δ∷natR | Measday et al. (2005) |
| YPH1715 | MATamfa1Δ∷MFa1pr-LEU2 can1Δ∷MFA1pr-HIS3 ura3Δ0 leu2Δ0 his3Δ1 lys2Δ0 mcm21Δ∷natR | Measday et al. (2005) |
| YPH1716 | MATamfa1Δ∷MFa1pr-LEU2 can1Δ∷MFA1pr-HIS3 ura3Δ0 leu2Δ0 his3Δ1 lys2Δ0 mcm16Δ∷natR | Measday et al. (2005) |
Chromosome transmission fidelity and viability assays:
Assays for the loss of the nonessential chromosome fragment (CF) were done as previously reported (Basrai et al. 1996). Briefly, reporter strains containing the nonessential CF were grown to logarithmic phase in minimal media selecting for the CF and any plasmids. Cultures were diluted and plated on SC medium to maintain plasmid selection with limiting adenine to enhance red sectors. At least 2000 colonies were assayed from three individual transformants for each strain. Chromosome loss was provided as the percentage of colonies that were at least half red, which indicates the loss of the CF during the first cell division after plating. Viability assays were conducted by plating cells on selective media with 2% glucose or 2% galactose and 2% raffinose and incubating them for 5–7 days at 30°. Viability was determined by the ratio of the number of colonies obtained on galactose/raffinose vs. glucose. At least three transformants from each strain were tested and at least 900 colonies were counted.
Protein stability and chromatin fractionation:
Cultures were grown to logarithmic phase in glucose media, washed, and grown in medium containing raffinose (2%) plus galactose (2%) for 6 hr. Protein samples were prepared at various time points after addition of the protein synthesis inhibitor cycloheximide at 10 μg/ml. The protein extracts were prepared using TCA methods as described previously (Kastenmayer et al. 2006). The protein concentration was determined by the Bio-Rad (Hercules, CA) DC protein assay (catalog no. 500-0113). Equal amounts of protein from each sample were resolved on a 4–12% Tris–Glycine gel (Invitrogen, San Diego) for Western blot analysis. Primary antibody against c-Myc for detection of epitope-tagged Cse4p and cse4K16R was purchased from Santa Cruz Biotechnology (Santa Cruz, CA) (Z-5; sc-789). Rabbit polyclonal antibodies against histones H3 and H2B were purchased from Abcam, while antibodies against Tub2 were custom made by Covance. Quantification was done using Eagle Eye software (Stratagene, La Jolla, CA).
Whole-cell and chromatin fractions were prepared as previously described (Liang and Stillman 1997) with minor modifications. Logarithmic phase cultures were grown in medium containing raffinose (2%) and galactose (2%) for 16 hr. Before harvesting, cultures were treated with NaN3 at 0.1% for 5 min at 30°. Equal quantities of cells were pelleted and 10% of the pellet was subjected to TCA protein extraction for the whole-cell extract (WCE), while the remaining 90% of the pellet was resuspended in spheroplast buffer (0.1 m KPO4, pH 7.4, 0.5 mm MgCl2, 1.2 m sorbitol). Following Zymolyase digestion, the resulting spheroplast pellets were resuspended in elution buffer (EB) with protease inhibitor cocktails (Sigma, St. Louis) and Triton X-100 was then added to 0.25% final concentration. The extracts were overlaid onto 30% sucrose and centrifuged. The upper phase was collected as the soluble fraction, while the insoluble pellet (chromatin) was further purified by washing in EBX followed by centrifugation.
Chromatin immunoprecipitation:
Chromatin immunoprecipitation (ChIP) was carried out as described previously (Orlando et al. 1997; Kuras and Struhl 1999) with some modifications. Cultures grown to logarithmic phase in media containing raffinose (2%) and galactose (2%) were treated with formaldehyde (1% final concentration) for cross-linking at room temperature for 30 min. Glycine was added to quench the reaction. The cell pellet was resuspended in breaking buffer (0.1 m Tris–HCl, pH 8, 20% glycerol, 1 mm PMSF) and subjected to bead beating to lyse the cells. The slurry was centrifuged to obtain the insoluble chromatin pellets followed by vigorous sonication. The resulting soluble portion was denoted as input and used for chromatin immunoprecipitation with α-c-Myc- or α-GST-coated protein A magnetic beads (Invitrogen) to precipitate Myc-Cse4p cross-linked chromatin or as a negative control, respectively. The immunoprecipitated chromatin–protein products were washed extensively in a series of buffers and finally resuspended in elution buffer. Reversal of cross-linking was done at 65° for at least 16 hr, followed by proteinase K treatment for 4 hr at 55°. The immunoprecipitated DNA was then purified, precipitated, and resuspended in water as a template for PCR amplification, using specific primer pairs. PCR was performed using HotstarTaq polymerase master mix (QIAGEN, Valencia, CA), with annealing at 55° for 1 min and extension at 68° for 45 sec. The linear amplification condition was determined by various cycles of amplification and by serial dilution of the input DNA. Twenty-two to 26 cycles of amplification were determined to reflect linear quantitative difference of input DNA. Sequences of primers are available upon request.
RESULTS
Overexpression of cse4K16R leads to defects in chromosome transmission fidelity in S. cerevisiae:
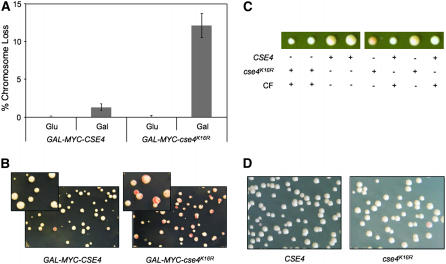
To determine the physiological consequence of mislocalized Cse4p, we utilized cse4K16R, a mutant form of Cse4p in which substitution of all 16 lysine residues to arginine (K16R) renders cse4K16R resistant to ubiquitin-mediated proteolysis (Collins et al. 2004). Overexpressed cse4K16R (GAL-MYC-cse4K16R) is relatively stable, is highly enriched in chromatin, and exhibits a diffuse nuclear localization pattern that is different from the discrete Cse4p foci characteristic of kinetochore localization (Collins et al. 2004). Using an assay based on the loss of a nonessential chromosome fragment that gives rise to red sectors in a white colony (Spencer et al. 1990), we determined that GAL-MYC-cse4K16R leads to defects in ctf in a wild-type strain. Specifically, the ctf phenotype was 10-fold higher in cells expressing GAL-MYC-cse4K16R than in those overexpressing CSE4 (GAL-MYC-CSE4) on galactose media (Figure 1, A and B). We conclude that the ctf phenotype is due to the overexpression of cse4K16R, as no measurable ctf phenotype was observed in strains expressing cse4K16R from its native promoter in wild-type or cse4Δ strain backgrounds (Figure 1, C and D). Also, since the Myc epitope-tagged versions of cse4K16R and Cse4p are able to complement a cse4Δ strain, it suggests that the essential function of these proteins is retained in the presence of an epitope tag.
Figure 1.—
Expression of GAL-MYC-cse4K16R leads to defects in chromosome transmission fidelity. (A) GAL-MYC-cse4K16R expression leads to a ctf phenotype. YMB3467 and YMB3468 were transformed with GAL-MYC-CSE4 (pSB816) and GAL-MYC-cse4K16R (pSB817) plasmids and plated on SC −URA with limiting adenine plus 2% glucose or 2% galactose/raffinose. The ctf phenotype was quantified by counting the number of colonies that were at least half red for each strain, using three independent transformants. (B) Red sectors in white colonies show the loss of the nonessential chromosome fragment. The inset displays a higher magnification of the colonies of YMB3467 and YMB3468 on 2% galactose/raffinose-containing media. (C) cse4K16R can complement a cse4Δ mutant. cse4K16R∷URA3, in which cse4K16R is expressed from a native CSE4 promoter, was integrated into the cse4Δ/CSE4 heterozygous diploid YMB6267 to make YMB6426, which was sporulated and dissected. The size difference in spores is due to the presence of the reporter chromosome fragment, which causes a slight growth delay. CF, chromosome fragment [CFIII (CEN3L) HIS3 SUP11]. (D) cse4K16R under a native CSE4 promoter does not result in a ctf phenotype. Strains derived from spores (C) containing a wild-type copy of CSE4 or cse4K16R under the endogenous CSE4 promoter (YMB6427) were diluted and plated on SC limiting adenine with 2% glucose.
Constitutive expression of histone H3 (Δ16H3) suppresses the ctf phenotype of overexpressed cse4K16R:
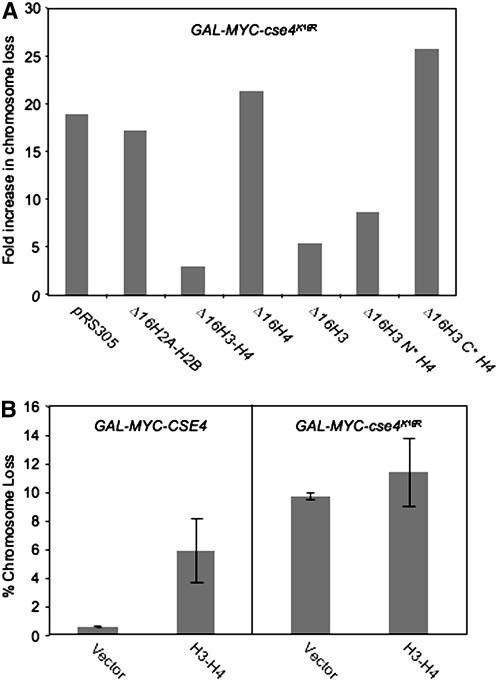
On the basis of previous data (Collins et al. 2004) and our results, we hypothesized that the mislocalization of cse4K16R to noncentromeric regions results in its observed ctf phenotype. We reasoned that increased H3 should favor [H3-H4]2 tetramer formation and compete with cse4K16R for H4, thus suppressing the mislocalization and ctf phenotype of cse4K16R. Since transcription of histones is tightly regulated during the cell cycle and peaks during S phase (Osley 1991), we examined the suppression of overexpressed cse4K16R by constitutively expressing the histones H2A, H2B, H3, and H4 from a mutant HTA1-HTB1 promoter lacking a 16-bp negative regulatory element (Δ16) (Bortvin and Winston 1996). Consistent with previous results (Stoler et al. 1995), we observed a 2.4-fold increase of soluble histone H3 in cells expressing integrated Δ16H3 (supplemental Figure 1A). Expression of GAL-MYC-cse4K16R resulted in a 19-fold increase in chromosome loss compared to vector alone in wild-type cells. When we assayed strains expressing the various integrated histone constructs, we determined that Δ16HHT1-HHF1 (Δ16H3-H4), but not Δ16HTA1-HTB1 (Δ16H2A-H2B) or vector, suppressed the GAL-MYC-cse4K16R ctf phenotype (Figure 2A). Further analysis showed that the suppression was observed with Δ16H3 and not Δ16H4. Expression of Δ16H3 did not lead to increased chromosome loss in a wild-type strain (data not shown).
Figure 2.—
Constitutive expression of histone H3 from a cell-cycle-independent promoter (Δ16H3) suppresses the GAL-MYC-cse4K16R ctf phenotype. (A) Wild-type strains containing a GAL vector (YMB6108) or expressing GAL-MYC-cse4K16R (YMB6110) were transformed with vector or histone constructs H2A-H2B (pAB95), H3-H4 (pAB157), H4 (pAB203), H3 (pAB204), Δ16H3N*H4 (pAB221), or Δ16H3C*H4 (pAB224) (integrated in the genome at the LEU2 locus), which constitutively express histones from a mutant promoter (Δ16), and were plated on SC limiting adenine with 2% galactose/raffinose. Fold increase in the percentage of colonies that are least half red for GAL-MYC-cse4K16R relative to a wild-type strain with the same integrated histone construct is plotted. The results represent an average of two independent experiments with variation of <5%. (B) The GAL-MYC-cse4K16R ctf phenotype is not suppressed by 2μ H3-H4. A reporter strain expressing 2μ H3-H4 (YMB6138) was transformed with a GAL vector, GAL-MYC-CSE4, or GAL-MYC-cse4K16R and plated on SC −URA with limiting adenine and 2% galactose/raffinose or 2% glucose. The ctf phenotype was quantified as described in Figure 1A. Values represent the average and standard deviation of chromosome loss for three transformants.
Evolutionarily conserved Cse4p shares a high degree of homology within the C-terminal histone fold domain of H3 (Keith et al. 1999; Morey et al. 2004). To further explore the mechanism of Δ16H3-mediated suppression, we utilized two mutant forms of constitutively expressed histone H3. The N-terminal mutant Δ16H3(Δ4-30)-H4 (Δ16H3N*H4), which interacts with H4 and contains an intact histone fold domain, suppressed the GAL-MYC-cse4K16R ctf phenotype. This suppression was not observed with the C-terminal mutant Δ16H3(Thr199Ile)-H4 (Δ16H3C*H4), which fails to assemble the histone octamer (Figure 2A) (Bortvin and Winston 1996). To determine if the suppression by Δ16H3 is mediated by the presence of endogenous CSE4, we examined the phenotype in a cse4Δ strain expressing GAL-MYC-cse4K16R. Our results showed that endogenous CSE4 was not required for suppression of the GAL-MYC-cse4K16R ctf phenotype by Δ16H3 (supplemental Figure 1B).
To ascertain whether the observed suppression by Δ16H3 was due to constitutive expression, we examined if expression of histones H3 and H4 from their native divergent promoter on a multicopy 2μ plasmid could suppress the GAL-MYC-cse4K16R ctf phenotype. Our data confirmed previous results (Meeks-Wagner and Hartwell 1986) that overexpression of H3-H4 leads to increased chromosome loss in GAL-MYC-CSE4 strains (6% vs. 0.7% half-sectored colonies, respectively) and failed to suppress the GAL-MYC-cse4K16R ctf phenotype (11.5% vs. 9.7%, respectively) (Figure 2B). We therefore conclude that constitutive expression of Δ16H3 suppresses the GAL-MYC-cse4K16R ctf phenotype.
Δ16H3 reduces the stability and chromatin enrichment of cse4K16R:
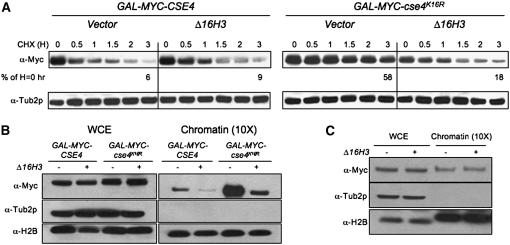
The suppression of the GAL-MYC-cse4K16R ctf phenotype by Δ16H3 prompted us to examine if expression of Δ16H3 altered any properties of cse4K16R, such as its increased stability and enrichment in chromatin (Collins et al. 2004). We hypothesized that mislocalization of cse4K16R and its increased stability are a result of increased chromatin association. To test this, the stabilities of Cse4p and cse4K16R were examined in whole-cell extracts from strains at various time points after a 6-hr induction in galactose media followed by a switch to glucose media with the protein synthesis inhibitor cycloheximide (CHX). We found that cse4K16R is much more stable than Cse4p (Collins et al. 2004) and that Δ16H3 reduced the stability of cse4K16R, but not Cse4p or Tub2p (Figure 3A). Specifically, the half-life of cse4K16R was reduced by >2 hr in the presence of Δ16H3 (data not shown).
Figure 3.—
Expression of Δ16H3 reduces the stability and chromatin association of cse4K16R. (A) Increased stability of cse4K16R is reduced by expression of Δ16H3. Western blot analysis was performed with protein extracts of GAL-MYC-CSE4 or GAL-MYC-cse4K16R strains with vector (YMB3467 and YMB3477, respectively) or Δ16H3 (YMB3468 and YMB3478, respectively). Protein samples were prepared from cultures grown in SC −URA with 2% galactose/raffinose medium for 6 hr and then shifted to 2% glucose and treated with cycloheximide (CHX) (10 μg/ml) for various times (H, hours). Blots were probed with α-Myc antibody to detect the epitope-tagged Cse4p or cse4K16R and α-Tub2p as a loading control. Blots were quantified by normalizing to Tub2p and setting the level in the untreated sample to 100%. (B) Expression of Δ16H3 reduces the level of chromatin-bound cse4K16R. Western blot analysis of whole-cell extract (WCE) or chromatin fractions of strains used in A were prepared after overnight growth in SC −URA with 2% galactose/raffinose. The blot was probed with α-Myc to detect epitope-tagged Cse4p and cse4K16R, and α-Tub2p and α-H2B as loading controls, for the WCE and chromatin, respectively. The chromatin fraction was prepared from 10-fold more cells than were used to prepare the WCE. (C) Expression of Δ16H3 does not result in altered levels of chromatin-bound Cse4p. Western blot analysis of WCE and chromatin fraction was carried out with wild-type strains expressing MYC-CSE4 from its native promoter with either vector (YMB6147) or integrated Δ16H3 (YMB6146) grown to logarithmic phase in YPD. The blot was probed with α-Myc to detect epitope-tagged Cse4p, and α-Tub2p and α-H2B as loading controls, for the WCE and chromatin, respectively.
These results next prompted us to examine if the reduced stability of cse4K16R correlated with its reduced enrichment in chromatin. Therefore, we analyzed the effect of Δ16H3 on levels of Cse4p or cse4K16R in WCEs and chromatin fractions from strains grown continuously in galactose media. Tub2p and H2B served as loading controls for WCEs and chromatin fractions, respectively. While Tub2p and H2B remained relatively constant, we observed enrichment of cse4K16R in the chromatin fraction as reported previously (Collins et al. 2004) (Figure 3B). Consistent with the protein stability data, expression of Δ16H3 reduced the high levels of chromatin-bound cse4K16R (Figure 3B). Additionally, we found that Δ16H3 reduced the levels of chromatin-bound Cse4p in GAL-MYC-CSE4 strains, but did not affect the level of chromatin-bound Cse4p expressed from its native promoter, suggesting a low level of mislocalization of Cse4p when overexpressed (Figure 3C). Unlike the result in Figure 3A, we did not observe an equivalent decrease in Cse4p and cse4K16R levels in the presence of Δ16H3 from the WCEs, which may be a result of the continuous and saturating expression of these proteins in galactose medium. Consistent with our hypothesis, the reduced stability of cse4K16R in the presence of Δ16H3 may be due to its replacement by canonical histone H3 at noncentromeric loci.
cse4K16R is mislocalized to noncentromeric DNA:
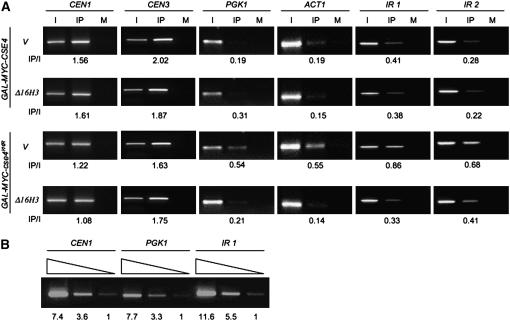
To determine the association of cse4K16R with chromosomal DNA, we used ChIP with strains expressing GAL-MYC-CSE4 or GAL-MYC-cse4K16R in the presence or absence of Δ16H3. We analyzed the association of these proteins at CEN DNA (CEN1 and CEN3), the actively transcribed genes PGK1 and ACT1, and AT-rich intergenic regions (IR1, IR2) of the genome. The latter regions were chosen because Cse4p is predicted to associate with the 78- to 86-bp central element of S. cerevisiae centromeres (CDEII), which is AT rich (Fitzgerald-Hayes et al. 1982). Serial dilutions of the input (I) DNA were tested to amplify three independent loci (CEN1, PGK1, and IR1), which confirmed that the PCR conditions used for the ChIP were semiquantitative (Figure 4B). The relative enhancement of binding is expressed as the ratio of signal strength of IP vs. I. Our results showed that the association of cse4K16R with CEN1 and CEN3 was similar to that observed for Cse4p (Figure 4A). Expression of Δ16H3 did not alter the CEN association of either cse4K16R or Cse4p. Compared to Cse4p, cse4K16R displayed a higher level of association with noncentromeric DNA (PGK1, ACT1, IR1, and IR2), which was reduced in the presence of Δ16H3. The slightly higher association of Cse4p observed at the noncentromeric regions IR1 and IR2 may reflect preferential association of Cse4p with AT-rich regions. Taken together, our data support the hypothesis that mislocalization of cse4K16R correlates with its defect in chromosome segregation.
Figure 4.—
cse4K16R mislocalizes to noncentromeric DNA and Δ16H3 suppresses this phenotype. (A) ChIP analysis was done using chromatin extracted from strains expressing GAL-MYC-CSE4 or GAL-MYC-cse4K16R with vector (YMB3467 and YMB3477, respectively) or Δ16H3 (YMB3468 and YMB3478, respectively) grown overnight in SC −URA with 2% galactose/raffinose. PCR analysis was done using input (I) and either α-Myc (IP) or α-GST (mock, M) immunoprecipitated DNA as a template for amplifying CEN1, CEN3, PGK1, ACT1, and two AT-rich intergenic regions IR1 (chromosome V, 2,227,624–2,227,965) and IR2 (chromosome III, 163,364–163,695). Fivefold more DNA was used for IP and M samples. PCR conditions using serially diluted input DNA as template were determined for linear amplification. The values for the IP/I are as shown below each row. (B) Verification of the linear range of PCR amplification for ChIP analysis. Input DNA from YMB3467 was fivefold serially diluted and used for PCR amplification of CEN1, PGK1, and IR1 DNA regions. The relative amount of PCR products normalized to the lowest input (designated as 1.0) is shown.
Overexpression of histone H3 (GALH3) results in defects in chromosome segregation and reduced levels of Cse4p at CEN DNA:
The high degree of homology between the HFD of Cse4p and histone H3 presents a challenge for their specific localization. Regulatory mechanisms in the cell must ensure the preferential incorporation of Cse4p in the centric nucleosome. In S. cerevisiae, overexpression of the HFD of Cse4p can complement a cse4Δ strain and is sufficient for association with CEN DNA (Morey et al. 2004). However, histone H3, when overexpressed, cannot substitute for Cse4p (our unpublished results). Our finding that Δ16H3 can suppress GAL-MYC-cse4K16R-mediated phenotypes suggests that faithful chromosome segregation is sensitive to perturbations in the expression and localization of Cse4p and histone H3. Therefore, we investigated the possibility that cell-cycle-independent overexpression of histone H3 affects chromosome segregation.
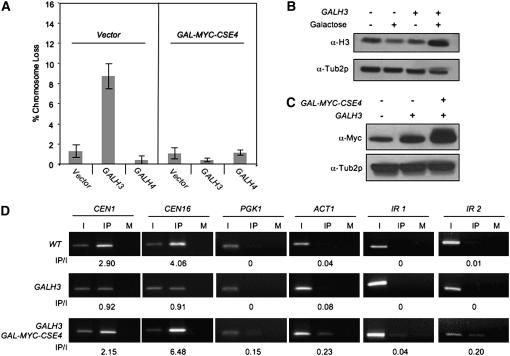
As shown in Figure 5A, expression of GALH3, but not GALH4, resulted in a severe ctf phenotype. On the basis of these data, we hypothesized that overexpressed H3 competes with Cse4p for association with CEN DNA and thereby leads to defective chromosome segregation. If this is true, then the following two predictions can be made. First, the expression of GAL-MYC-CSE4 should suppress the ctf phenotype of GALH3 in cells coexpressing both plasmids. Our results supported this prediction as GAL-MYC-CSE4 suppressed the ctf phenotype of GALH3 strains (Figure 5A). Second, strains expressing GALH3 should show reduced levels of Cse4p at CEN DNA. Since we used strains overexpressing H3 from a GAL promoter for chromosome loss and ChIP experiments, we confirmed that H3 levels were elevated in GALH3 strains upon galactose induction (Figure 5B) and that these levels did not affect the level of Myc-Cse4p (Figure 5C). To test for reduced CEN association of Cse4p, we assayed Myc-Cse4p expressed from its own promoter integrated at the LEU2 locus in wild-type and GALH3 strains. Consistent with our hypothesis, ChIP experiments showed that the association of Myc-Cse4p with CEN DNA was reduced in GALH3 strains (Figure 5D). When compared to wild-type strains, GALH3 strains showed 2- to 4-fold less binding of Myc-Cse4p at CEN1 and CEN16. Expression of GALH3 did not lead to association of Myc-Cse4p to noncentromeric loci such as PGK1, ACT1, IR1, and IR2. In parallel with our previous observations (Figure 4), expression of GAL-MYC-CSE4 resulted in low levels of Myc-Cse4p association to noncentromeric regions. The reduction in CEN association of Myc-Cse4p in GALH3 strains was suppressed by expression of GAL-MYC-CSE4. Taken together, these results support the hypothesis that the ctf phenotype of overexpressed histone H3 is a result of lower levels of CEN-associated Cse4p.
Figure 5.—
Phenotypes of overexpressed histone H3 (GALH3) are suppressed by GAL-MYC-CSE4. (A) Overexpression of histone H3 results in a ctf phenotype. Wild-type (YMB3482) or GALCSE4 (YMB3467) strains were transformed with vector (pRS424GAL1), GALH3 (pMB1159), or GALH4 (pMB1158) and plated on SC −URA limiting adenine with 2% galactose/raffinose. The ctf phenotype was quantified by counting the number of colonies that were at least half red from three independent transformants. (B) Histone H3 is expressed at a higher level in a GALH3 strain. Western blot analysis was carried out to detect histone H3 levels in the WCEs of WT cells containing empty GAL vector (YMB6132) or expressing GALH3 (YMB6133) in the presence of 2% glucose or 2% galactose/raffinose from overnight cultures. Tub2p is used as a loading control. (C) Overexpression of histone H3 does not alter Myc-Cse4p levels. Western blot analysis was done using whole-cell extracts with strains containing vector (YMB6132), GALH3 (YMB6133), or GALH3 and GALCSE4 (YMB6136) expressing epitope-tagged Cse4p (MYC-CSE4) from its native promoter in a wild-type strain. The strains were grown to logarithmic phase in SC medium with 2% galactose/raffinose. The blot was probed with α-Myc to detect epitope-tagged Cse4p and α-Tub2p as a loading control. (D) GALH3 leads to reduction in CEN-associated Cse4p. ChIP analysis was carried out using strains described in B. The strains were grown to logarithmic phase in SC with 2% galactose/raffinose. Input (I), immunoprecipitated (IP), and mock (M) samples were used for PCR amplification using primers specific for the loci indicated. PCR conditions identical to Figure 4B were used to obtain linear amplification of targeted chromosomal loci. Values of IP/I are shown below each row.
Overexpression of histone H3 (GALH3) is lethal in kinetochore mutants:
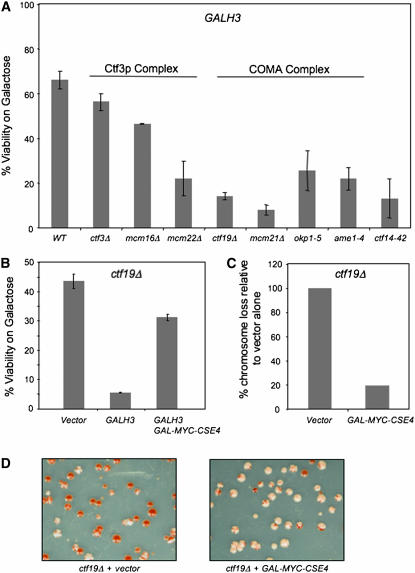
To further understand the mechanism by which GALH3 leads to a defect in chromosome segregation, we employed a genetic approach. We reasoned that, if overexpressed H3 associates with the centromere, the phenotype would be further exacerbated in mutants of kinetochore proteins that interact with Cse4p. Components of the COMA and Ctf3p complexes exhibit genetic and physical interactions with Cse4p (Ortiz et al. 1999; Measday et al. 2002). We proposed that kinetochore proteins, such as those of the COMA and Ctf3p complexes, contribute to the functional integrity of the Cse4p nucleosome and that the absence of these proteins may further enhance the phenotype of GALH3 strains. To test this, we examined whether GALH3 displayed lethality COMA and Ctf3p complex mutants. Viability was determined by comparing growth of strains on galactose vs. glucose media. GALH3 displayed maximum lethality in deletions of the COMA complex, specifically ctf19Δ and mcm21Δ strains, and to a lesser extent in okp1-5 and ame1-4 strains (Figure 6A). GALH3 also led to reduced viability in deletions of the Ctf3p complex, namely, ctf3Δ, mcm16Δ, and mcm22Δ. We conclude that deletions for components of the COMA and Ctf3p complexes are sensitive to overexpression of histone H3 because of a disruption in kinetochore–Cse4p interactions. If this is true, then GALCSE4 should suppress the ctf phenotype and the GALH3 synthetic dosage lethality (SDL) of a ctf19Δ strain. The results supported our hypothesis as GALCSE4 did indeed suppress the SDL of GALH3 (Figure 6B) and the chromosome loss phenotype (Figure 6, C and D) of ctf19Δ strains.
Figure 6.—
Expression of GALH3 is lethal in kinetochore mutants. (A) A wild-type strain (Y5563) and kinetochore mutants ctf3Δ (YPH1712), mcm16Δ (YPH1714), mcm22Δ (YPH1716), ctf14-42 (YMB372), ctf19Δ (YPH1713), mcm21Δ (YPH1715), okp1-5 (YPH1678), and ame1-4 (YPH1676) were transformed with 2μ GALH3 (pLG41) and plated on SC −URA with either 2% glucose or 2% galactose/raffinose. Percentage of viability was calculated by the ratio of viable colonies on galactose/raffinose media vs. glucose media for a least three independent transformants. (B) GAL-MYC-CSE4 suppresses the synthetic lethality of GALH3 in ctf19Δ strains. ctf19Δ containing vector (pRS425GAL1), GALH3 (pLG41), or GALH3 GAL-MYC-CSE4 (pMB1147) were plated on SC −TRP −URA with 2% glucose or 2% galactose/raffinose. Percentage of viability was determined as described in A. (C) GAL-MYC-CSE4 suppresses the chromosome loss defect of GALH3 in ctf19Δ strains. ctf19Δ containing vector (pRS315) or GAL-MYC-CSE4 (pMB1147) were plated on SC −LEU with 2% glucose or 2% galactose/raffinose media. Chromosome loss was determined as described in Figure 1A. Values plotted are based on setting the chromosome loss phenotype of the ctf19Δ strain with vector alone to 100%. (D) Representative results of the colony-sectoring assay using strains described in C.
DISCUSSION
Despite the lack of similarity between the structure of the simple point centromeres of S. cerevisiae and the complex regional centromeres in other systems, there is a high degree of conservation in the function of kinetochore components and regulatory mechanisms that ensure faithful chromosome segregation. Among the conserved components, the centromeric histone H3 variant CenH3 is essential for kinetochore function in all systems examined to date (Stoler et al. 1995; Smith et al. 1996; Meluh et al. 1998; Buchwitz et al. 1999; Blower and Karpen 2001; Chen et al. 2003). Overexpression and mislocalization of CenH3 have been observed in colorectal cell lines (Tomonaga et al. 2003), and aneuploidy and ectopic kinetochore formation were shown to be associated with the overexpression and mislocalization of CenH3 in D. melanogaster (Heun et al. 2006). To elucidate the relationship between CenH3, histone H3, and chromosome segregation, we utilized the simple point centromere of S. cerevisiae to investigate phenotypes associated with mislocalization of Cse4p and altered histone dosage. Our studies have established that overexpression of cse4K16R correlates with its mislocalization and defective chromosome segregation and that increased dosage of histone H3 leads to defects in kinetochore integrity. Our results suggest that proper stoichiometry of histone H3 and Cse4p is critical for ensuring their specific localization and is thus essential for high-fidelity chromosome segregation.
We used the stable cse4 mutant cse4K16R to ask the following questions: (a) Does overexpression of cse4K16R lead to a chromosome loss phenotype?, (b) Can constitutive expression of histone H3 suppress the overexpressed cse4K16R phenotypes such as chromosome loss, chromatin enrichment, and increased stability?, And (c) does overexpressed cse4K16R associate with centromeric DNA and/or noncentromeric regions? Our results have shown that overexpression of cse4K16R leads to increased chromosome loss, which we hypothesized may be due to defective kinetochore function and/or association of cse4K16R with noncentromeric DNA. Several independent approaches were used to examine these possibilities. ChIP experiments determined that there was no defect in centromere association of cse4K16R. Furthermore, cse4K16R expressed from a CSE4 promoter can complement a cse4Δ strain and does not exhibit a ctf phenotype (Figure 1, C and D). Therefore, in a wild-type strain, it is the overexpression of cse4K16R that leads to increased chromosome loss.
The suppression of the ctf phenotype, enrichment in chromatin, and increased stability of overexpressed cse4K16R by constitutive H3 expression, combined with data from ChIP experiments, support the latter possibility that it is the mislocalization of cse4K16R that results in its observed phenotype. Transcription of histones is cell-cycle regulated and histones are rapidly deposited into newly replicated DNA by nucleosome assembly factors (Gunjan and Verreault 2003). The lack of suppression of cse4K16R phenotypes by 2μ H3-H4 may be due to cell-cycle-regulated expression of H3-H4. However, it is also possible that altered levels of H3-H4 from the 2μ plasmid lead to incorporation into centromeric chromatin. This hypothesis is supported by previous observations for increased chromosome loss due to 2μ H3-H4 (Meeks-Wagner and Hartwell 1986) and supports our observations for lack of suppression of cse4K16R phenotype by 2μ H3-H4. Expression of Δ16H3 did not lead to increased chromosome loss in wild-type strains, suggesting that constitutively expressed histone H3 is maintained at levels that do not affect chromosome segregation. These results are consistent with observations that regulated proteolysis prevents accumulation of excess histones, which may serve to maintain a critical threshold level of H3 expressed in Δ16H3 strains. On the basis of our results, we propose that Δ16H3 expression increases the ratio of [H3-H4]2 tetramers compared to [cse4K16R-H4]2, thus competing with cse4K16R for H4 and incorporation into chromatin. Two predictions follow from this logic. The first is that H3 mutants that fail to interact with H4 should not suppress the ctf phenotype of overexpressed cse4K16R. Our results support this hypothesis as the C-terminal mutant Δ16H3C*H4, which fails to form a histone octamer, does not suppress this ctf phenotype (Figure 2A) (Bortvin and Winston 1996). The second prediction is that overexpression of cse4K16R and H4 should increase the proportion of [cse4K16R-H4]2 tetramers, resulting in increased mislocalization of cse4K16R and enhancement of the ctf phenotype. Our data are consistent with this prediction as expression of GALH4 resulted in increased chromosome loss when cse4K16R is overexpressed (data not shown). Recent studies have shown that downregulation of histone H4 leads to an increase in sister chromatid separation and an increased spindle length (Bouck and Bloom 2007). Taken together, the results show that proper stoichiometry of histones and the histone H3 variant, Cse4p, is essential for chromosome transmission fidelity.
As expected, we observed that expression of Δ16H3 reduced the stability and chromatin enrichment of cse4K16R. We propose that the increased stability and chromatin enrichment of cse4K16R are due to its incorporation into noncentromeric regions. Data from mammalian systems suggest that incorporation of CenH3 into the centric nucleosome requires exit from mitosis (Jansen et al. 2007). Since the suppression of overexpressed cse4K16R phenotypes correlated with constitutive expression of H3, our results raise the possibility that, as in other systems, incorporation of Cse4p into chromatin must not be concurrent with synthesis and incorporation of bulk histones into nucleosomes (Blower et al. 2006; Foltz et al. 2006). It should also be noted that a slight increase in Cse4p stability was observed in the presence of Δ16H3; however, no increase in ctf phenotype was observed in strains expressing Δ16H3, and hence an understanding of the physiological significance of this observation was not further pursued.
ChIP experiments showed that cse4K16R associates with CEN DNA at levels comparable to those observed for the association of Cse4p. However, we observed a higher level of association of cse4K16R at noncentromeric regions such as ACT1, PGK1, and two AT-rich intergenic regions (IR1 and IR2). Both overexpressed Cse4p and cse4K16R show enrichment at the AT-rich regions relative to ACT1 or PGK1. These results are not surprising as overexpressed Cse4p may have a preference to associate with AT-rich regions, since the central DNA element (CDE)II of S. cerevisiae centromeres is significantly AT rich (93%). Also, increased chromosome loss has been observed when cse4 mutants were combined with mutations in CDEI and CDEII, but not CDEIII (Keith and Fitzgerald-Hayes 2000; Smith 2002). Unlike cse4K16R, the low levels of mislocalization of overexpressed Cse4p to noncentromeric regions are not sufficient to result in increased chromosome loss, as we have not observed a severe ctf phenotype in GAL-CSE4 strains (Crotti and Basrai 2004).
Consistent with the suppression of the ctf phenotype associated with overexpressed cse4K16R, expression of Δ16H3 reduced levels of cse4K16R association at noncentromeric regions. It is possible that association of cse4K16R with noncentromeric regions recruits other kinetochore proteins and forms ectopic kinetochores. In contrast to studies of overexpression of CID in flies (Heun et al. 2006), functional kinetochores and multicentric chromosomes are unlikely formed in budding yeast at sites where cse4K16R is mislocalized, since overexpression of cse4K16R is not lethal. Additionally, if the levels of one or more proteins, such as kinetochore proteins or chromatin assembly factors, become rate limiting in the cell, this would result in a defective kinetochore. We used two independent approaches but failed to identify kinetochore proteins that are titrated away from the centromere in cse4K16R strains. The first approach utilized synthetic genetic array (SGA) analysis (Tong et al. 2001) to identify nonessential genes that exhibit synthetic lethality with overexpressed cse4K16R (data not shown). However, no candidates were found, which may be because the target gene is essential and not included in the screen or that a combination of deletions is required for manifesting a lethal phenotype. Additionally, the assay may not be sensitive enough for positive identification of candidate genes in the SGA screen. In the second approach, we performed ChIP on Ctf19p, which has been shown to physically associate with Cse4p in the kinetochore architecture (Ortiz et al. 1999) and is thus an ideal candidate for titration. However, we did not detect a difference in association of Ctf19p to centromeric and noncentromeric regions (IR1, ACT1) in strains overexpressing cse4K16R as compared to wild-type strains (data not shown).
In S. cerevisiae, overexpression of the HFD of Cse4p can complement a cse4Δ strain and is sufficient for association with CEN DNA (Morey et al. 2004). However, histone H3, when overexpressed, cannot substitute for Cse4p (our unpublished results). Even though a chimeric protein that contains H3 and the centromere targeting domain (CATD) of CenH3 can rescue CenH3 depletion in mammalian cells, such chimeras fail to rescue a cse4Δ strain (Black et al. 2007). Several studies suggest the presence of stringent regulatory and/or structural mechanisms to exclude histone H3 from centric nucleosomes. This may be facilitated by differences between H3 and Cse4p with respect to their (a) timing of synthesis and steady-state levels during the cell cycle, (b) timing of incorporation into nucleosomes, or (c) chaperones that mediate their association and/or elimination from specific chromosomal loci. We designed experimental strategies to investigate the first possibility: Does high-level constitutive expression of histone H3 affect chromosome segregation?
Several mechanisms in the cell influence the timing of expression as well as the steady-state levels of histone H3. Unlike expression of histone H3, which peaks in S phase, our Western blot results showed that levels of Cse4p are fairly constant throughout the cell cycle (supplemental Figure 3A). However, a genomewide cell-cycle analysis showed that the transcription of CSE4 is cell-cycle regulated (Pramila et al. 2006). In addition to regulated proteolysis, a Rad53p surveillance mechanism monitors excess free histones and triggers their degradation in S. cerevisiae (Gunjan and Verreault 2003). However, unlike canonical histones, Rad53p does not mediate the degradation of ectopic Cse4p (Collins et al. 2004). It is possible either that histone H3 is incorporated into centric nucleosomes and then replaced by Cse4p just before the onset of mitosis or that histone H3 is generally excluded from the centromere throughout the cell cycle. Experimental evidence so far has not conclusively established if histone H3 can associate with CEN DNA under physiological conditions in S. cerevisiae. Camahort et al. (2007) have shown low levels of H3 association within a 2-kb region surrounding CEN3 in nocodazole-arrested cells that is increased eightfold in strains depleted of Scm3p. Results from Mizuguchi et al. (2007) have shown exclusion of histone H3 from CEN DNA in logarithmically growing strains. Recent studies with S. pombe have shown that excess histone H3 can compete with Cnp1 and associate with the central domain of the centromere (Castillo et al. 2007).
We have shown that overexpression of histone H3 leads to increased chromosome loss (Figure 5A) and reduced viability in wild-type strains (data not shown). Hence, altered stoichiometry of [H3-H4]2 relative to [Cse4p-H4]2 tetramers affects kinetochore structure and function. Association of overexpressed histone H3 with CEN DNA results in reduced levels of CEN-associated Cse4p. Several pieces of experimental evidence support this conclusion, as we have observed (a) reduced levels of centromere-associated Cse4p in GALH3 strains, (b) suppression of GALH3-induced chromosome loss and reduced levels of CEN-associated Cse4p by GALCSE4, (c) synthetic dosage lethality interactions between mutants of the COMA and Ctf3p kinetochore complexes and GALH3, and (d) suppression of the ctf phenotype and GALH3 lethality in ctf19Δ strains by GALCSE4. Additional support comes from a recent observation in which rapid exchange of histone H3 at the centric nucleosome was detected in strains transiently overexpressing both GALH3 and GALH4 (Dion et al. 2007). As an extension of this, we have shown that coexpression of GALH3 and GALH4 results in lethality in wild-type strains (supplemental Figure 2).
We determined that overexpression of histone H3 is particularly detrimental in mutant strains for components of the COMA and Ctf3p complexes. Since proteins of the COMA and Ctf3p complexes interact with Cse4p, we hypothesize that the absence of these proteins affects the formation and/or maintenance of a stable and functional kinetochore. We propose that a functional kinetochore may require (1) hierarchical assembly of kinetochore proteins and (2) molecular interactions between the kinetochore proteins, histones, and the underlying DNA sequence that serve to stabilize and maintain a functional kinetochore. Since overexpression of CSE4 suppresses the ctf phenotype and synthetic dosage lethality of GALH3 in a ctf19Δ strain, we speculate that the chromosome loss phenotype of ctf19Δ strains may at least in part be due to a defect in stabilization of the Cse4p nucleosome as described above. This conclusion is in agreement with previous observations that Ctf19p is not required for the association of Cse4p with centromeric DNA (Measday et al. 2002). It is also possible that Cse4p and the COMA and Ctf3p complexes act cooperatively to prevent mislocalization of histone H3 to the centric nucleosome. Our data are consistent with results from Glowczewski et al. (2000) in which GALH3 exacerbates the temperature-sensitive phenotype of cse4 mutant strains. Taken together, these data support our hypothesis that both kinetochore assembly and maintenance are essential for its function and that defects in either one lead to increased chromosome loss.
Recent studies with a modified regional centromere of fission yeast show that CenH3 can associate with a reporter gene inserted within the central domain of the centromere (Castillo et al. 2007). These results are similar to those from D. melanogaster and human cells in which CenH3 can associate with noncentromeric DNA (Warburton et al. 1997; Williams et al. 1998; Heun et al. 2006). In S. cerevisiae, Cse4p localization has been observed only at centromeric DNA and the partitioning locus STB of the 2μ plasmid (Hajra et al. 2006). Also, in contrast to the association of CenH3 with several nucleosomes at regional centromeres, only a single Cse4p-containing nucleosome is reported at the point centromeres of budding yeast (Furuyama and Biggins 2007). Despite these differences, our results with the native centromere in S. cerevisiae are consistent with those of a modified centromere in S. pombe and establish that altered ratios between H3, H4, and CenH3 lead to chromosome loss (Castillo et al. 2007). Hence, the underlying mechanisms that regulate the assembly and maintenance of centric nucleosome(s) containing CenH3 seem to be evolutionarily conserved. Thus, future studies in S. cerevisiae that elucidate the molecular mechanisms for preventing the mislocalization of Cse4p and histone H3 may help us establish a possible link between mislocalization of CENP-A and aneuploidy in cancer cells (Cleveland et al. 2003; Tomonaga et al. 2003).
Acknowledgments
The authors thank Sue Biggins, Mitch Smith, and Vivian Measday for strains and plasmids. We are grateful to Oliver Kerscher, Duncan Clarke, Vivian Measday, Carole Carter, Robert Skibbens, and members of the Basrai lab for discussions and comments on the manuscript. This research was supported by the Intramural Research Program of the National Cancer Institute, National Institutes of Health.
References
- Baker, R. E., K. Harris and K. Zhang, 1998. Mutations synthetically lethal with cep1 target S. cerevisiae kinetochore components. Genetics 149 73–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basrai, M. A., J. Kingsbury, D. Koshland, F. Spencer and P. Hieter, 1996. Faithful chromosome transmission requires Spt4p, a putative regulator of chromatin structure in Saccharomyces cerevisiae. Mol. Cell. Biol. 16 2838–2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black, B. E., M. A. Brock, S. Bedard, V. L. Woods, Jr. and D. W. Cleveland, 2007. An epigenetic mark generated by the incorporation of CENP-A into centromeric nucleosomes. Proc. Natl. Acad. Sci. USA 104 5008–5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom, K. S., and J. Carbon, 1982. Yeast centromere DNA is in a unique and highly ordered structure in chromosomes and small circular minichromosomes. Cell 29 305–317. [DOI] [PubMed] [Google Scholar]
- Blower, M. D., and G. H. Karpen, 2001. The role of Drosophila CID in kinetochore formation, cell-cycle progression and heterochromatin interactions. Nat. Cell Biol. 3 730–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower, M. D., T. Daigle, T. Kaufman and G. H. Karpen, 2006. Drosophila CENP-A mutations cause a BubR1-dependent early mitotic delay without normal localization of kinetochore components. PLoS Genet. 2 e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortvin, A., and F. Winston, 1996. Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science 272 1473–1476. [DOI] [PubMed] [Google Scholar]
- Bouck, D. C., and K. Bloom, 2007. Pericentric chromatin is an elastic component of the mitotic spindle. Curr. Biol. 17 741–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchwitz, B. J., K. Ahmad, L. L. Moore, M. B. Roth and S. Henikoff, 1999. A histone-H3-like protein in C. elegans. Nature 401 547–548. [DOI] [PubMed] [Google Scholar]
- Camahort, R., B. Li, L. Florens, S. K. Swanson, M. P. Washburn et al., 2007. Scm3 is essential to recruit the Histone H3 variant Cse4 to centromeres and to maintain a functional kinetochore. Mol. Cell 26 853–865. [DOI] [PubMed] [Google Scholar]
- Castillo, A. G., B. G. Mellone, J. F. Partridge, W. Richardson, G. L. Hamilton et al., 2007. Plasticity of fission yeast CENP-A chromatin driven by relative levels of Histone H3 and H4. PLoS Genet. 3 e121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman, I. M., D. G. Drubin and G. Barnes, 2002. Simple centromere, complex kinetochore: linking spindle microtubules and centromeric DNA in budding yeast. J. Cell Biol. 157 199–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, E. S., S. Saitoh, M. Yanagida and K. Takahashi, 2003. A cell cycle-regulated GATA factor promotes centromeric localization of CENP-A in fission yeast. Mol. Cell 11 175–187. [DOI] [PubMed] [Google Scholar]
- Cleveland, D. W., Y. Mao and K. F. Sullivan, 2003. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112 407–421. [DOI] [PubMed] [Google Scholar]
- Collins, K. A., S. Furuyama and S. Biggins, 2004. Proteolysis contributes to the exclusive centromere localization of the yeast Cse4/CENP-A histone H3 variant. Curr. Biol. 14 1968–1972. [DOI] [PubMed] [Google Scholar]
- Crotti, L. B., and M. A. Basrai, 2004. Functional roles for evolutionarily conserved Spt4p at centromeres and heterochromatin in Saccharomyces cerevisiae. EMBO J. 23 1804–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion, M. F., T. Kaplan, M. Kim, S. Buratowski, N. Friedman et al., 2007. Dynamics of replication-independent histone turnover in budding yeast. Science 315 1405–1408. [DOI] [PubMed] [Google Scholar]
- Ekwall, K., 2007. Epigenetic control of centromere behavior. Annu. Rev. Genet. 41 63–81. [DOI] [PubMed] [Google Scholar]
- Fitzgerald-Hayes, M., L. Clarke and J. Carbon, 1982. Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell 29 235–244. [DOI] [PubMed] [Google Scholar]
- Foltz, D. R., L. E. Jansen, B. E. Black, A. O. Bailey, J. R. Yates, 3rd et al., 2006. The human CENP-A centromeric nucleosome-associated complex. Nat. Cell Biol. 8 458–469. [DOI] [PubMed] [Google Scholar]
- Funk, M., J. H. Hegemann and P. Philippsen, 1989. Chromatin digestion with restriction endonucleases reveals 150–160 bp of protected DNA in the centromere of chromosome XIV in Saccharomyces cerevisiae. Mol. Gen. Genet. 219 153–160. [DOI] [PubMed] [Google Scholar]
- Furuyama, S., and S. Biggins, 2007. Centromere identity is specified by a single centromeric nucleosome in budding yeast. Proc. Natl. Acad. Sci. USA 104 14706–14711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuyama, T., Y. Dalal and S. Henikoff, 2006. Chaperone-mediated assembly of centromeric chromatin in vitro. Proc. Natl. Acad. Sci. USA 103 6172–6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glowczewski, L., P. Yang, T. Kalashnikova, M. S. Santisteban and M. M. Smith, 2000. Histone-histone interactions and centromere function. Mol. Cell. Biol. 20 5700–5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunjan, A., and A. Verreault, 2003. A Rad53 kinase-dependent surveillance mechanism that regulates histone protein levels in S. cerevisiae. Cell 115 537–549. [DOI] [PubMed] [Google Scholar]
- Hajra, S., S. K. Ghosh and M. Jayaram, 2006. The centromere-specific histone variant Cse4p (CENP-A) is essential for functional chromatin architecture at the yeast 2-micron circle partitioning locus and promotes equal plasmid segregation. J. Cell Biol. 174 779–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi, T., Y. Fujita, O. Iwasaki, Y. Adachi, K. Takahashi et al., 2004. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell 118 715–729. [DOI] [PubMed] [Google Scholar]
- Henikoff, S., and Y. Dalal, 2005. Centromeric chromatin: What makes it unique? Curr. Opin. Genet. Dev. 15 177–184. [DOI] [PubMed] [Google Scholar]
- Heun, P., S. Erhardt, M. D. Blower, S. Weiss, A. D. Skora et al., 2006. Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores. Dev. Cell 10 303–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman, A. A., and P. K. Sorger, 1995. Structure and function of kinetochores in budding yeast. Annu. Rev. Cell Dev. Biol. 11 471–495. [DOI] [PubMed] [Google Scholar]
- Jansen, L. E., B. E. Black, D. R. Foltz and D. W. Cleveland, 2007. Propagation of centromeric chromatin requires exit from mitosis. J. Cell Biol. 176 795–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpen, G. H., and R. C. Allshire, 1997. The case for epigenetic effects on centromere identity and function. Trends Genet. 13 489–496. [DOI] [PubMed] [Google Scholar]
- Kastenmayer, J. P., L. Ni, A. Chu, L. E. Kitchen, W. C. Au et al., 2006. Functional genomics of genes with small open reading frames (sORFs) in S. cerevisiae. Genome Res. 16 365–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith, K. C., and M. Fitzgerald-Hayes, 2000. CSE4 genetically interacts with the Saccharomyces cerevisiae centromere DNA elements CDE I and CDE II but not CDE III. Implications for the path of the centromere DNA around a cse4p variant nucleosome. Genetics 156 973–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith, K. C., R. E. Baker, Y. Chen, K. Harris, S. Stoler et al., 1999. Analysis of primary structural determinants that distinguish the centromere-specific function of histone variant Cse4p from histone H3. Mol. Cell. Biol. 19 6130–6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuras, L., and K. Struhl, 1999. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature 399 609–613. [DOI] [PubMed] [Google Scholar]
- Liang, C., and B. Stillman, 1997. Persistent initiation of DNA replication and chromatin-bound MCM proteins during the cell cycle in cdc6 mutants. Genes Dev. 11 3375–3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik, H. S., and S. Henikoff, 2003. Phylogenomics of the nucleosome. Nat. Struct. Biol. 10 882–891. [DOI] [PubMed] [Google Scholar]
- McAinsh, A. D., J. D. Tytell and P. K. Sorger, 2003. Structure, function, and regulation of budding yeast kinetochores. Annu. Rev. Cell Dev. Biol. 19 519–539. [DOI] [PubMed] [Google Scholar]
- Measday, V., D. W. Hailey, I. Pot, S. A. Givan, K. M. Hyland et al., 2002. Ctf3p, the Mis6 budding yeast homolog, interacts with Mcm22p and Mcm16p at the yeast outer kinetochore. Genes Dev. 16 101–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Measday, V., K. Baetz, J. Guzzo, K. Yuen, T. Kwok et al., 2005. Systematic yeast synthetic lethal and synthetic dosage lethal screens identify genes required for chromosome segregation. Proc. Natl. Acad. Sci. USA 102 13956–13961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeks-Wagner, D., and L. H. Hartwell, 1986. Normal stoichiometry of histone dimer sets is necessary for high fidelity of mitotic chromosome transmission. Cell 44 43–52. [DOI] [PubMed] [Google Scholar]
- Meluh, P. B., P. Yang, L. Glowczewski, D. Koshland and M. M. Smith, 1998. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell 94 607–613. [DOI] [PubMed] [Google Scholar]
- Meraldi, P., A. D. McAinsh, E. Rheinbay and P. K. Sorger, 2006. Phylogenetic and structural analysis of centromeric DNA and kinetochore proteins. Genome Biol. 7 R23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi, G., H. Xiao, J. Wisniewski, M. M. Smith and C. Wu, 2007. Nonhistone Scm3 and histones CenH3–H4 assemble the core of centromere-specific nucleosomes. Cell 129 1153–1164. [DOI] [PubMed] [Google Scholar]
- Morey, L., K. Barnes, Y. Chen, M. Fitzgerald-Hayes and R. E. Baker, 2004. The histone fold domain of Cse4 is sufficient for CEN targeting and propagation of active centromeres in budding yeast. Eukaryot. Cell 3 1533–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando, V., H. Strutt and R. Paro, 1997. Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods 11 205–214. [DOI] [PubMed] [Google Scholar]
- Ortiz, J., O. Stemmann, S. Rank and J. Lechner, 1999. A putative protein complex consisting of Ctf19, Mcm21, and Okp1 represents a missing link in the budding yeast kinetochore. Genes Dev. 13 1140–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osley, M. A., 1991. The regulation of histone synthesis in the cell cycle. Annu. Rev. Biochem. 60 827–861. [DOI] [PubMed] [Google Scholar]
- Pot, I., J. Knockleby, V. Aneliunas, T. Nguyen, S. Ah-Kye et al., 2005. Spindle checkpoint maintenance requires Ame1 and Okp1. Cell Cycle 4 1448–1456. [DOI] [PubMed] [Google Scholar]
- Pramila, T., W. Wu, S. Miles, W. S. Noble and L. L. Breeden, 2006. The Forkhead transcription factor Hcm1 regulates chromosome segregation genes and fills the S-phase gap in the transcriptional circuitry of the cell cycle. Genes Dev. 20 2266–2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman, I., and K. S. Bloom, 1991. Centromeres: an integrated protein/DNA complex required for chromosome movement. Annu. Rev. Cell Biol. 7 311–336. [DOI] [PubMed] [Google Scholar]
- Sharp, J. A., A. A. Franco, M. A. Osley and P. D. Kaufman, 2002. Chromatin assembly factor I and Hir proteins contribute to building functional kinetochores in S. cerevisiae. Genes Dev. 16 85–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbens, R. V., and P. Hieter, 1998. Kinetochores and the checkpoint mechanism that monitors for defects in the chromosome segregation machinery. Annu. Rev. Genet. 32 307–337. [DOI] [PubMed] [Google Scholar]
- Smith, M. M., 2002. Centromeres and variant histones: What, where, when and why? Curr. Opin. Cell Biol. 14 279–285. [DOI] [PubMed] [Google Scholar]
- Smith, M. M., P. Yang, M. S. Santisteban, P. W. Boone, A. T. Goldstein et al., 1996. A novel histone H4 mutant defective in nuclear division and mitotic chromosome transmission. Mol. Cell. Biol. 16 1017–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer, F., S. L. Gerring, C. Connelly and P. Hieter, 1990. Mitotic chromosome transmission fidelity mutants in Saccharomyces cerevisiae. Genetics 124 237–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoler, S., K. C. Keith, K. E. Curnick and M. Fitzgerald-Hayes, 1995. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 9 573–586. [DOI] [PubMed] [Google Scholar]
- Stoler, S., K. Rogers, S. Weitze, L. Morey, M. Fitzgerald-Hayes et al., 2007. Scm3, an essential Saccharomyces cerevisiae centromere protein required for G2/M progression and Cse4 localization. Proc. Natl. Acad. Sci. USA 104 10571–10576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomonaga, T., K. Matsushita, S. Yamaguchi, T. Oohashi, H. Shimada et al., 2003. Overexpression and mistargeting of centromere protein-A in human primary colorectal cancer. Cancer Res. 63 3511–3516. [PubMed] [Google Scholar]
- Tong, A. H., and C. Boone, 2006. Synthetic genetic array analysis in Saccharomyces cerevisiae. Methods Mol. Biol. 313 171–192. [DOI] [PubMed] [Google Scholar]
- Tong, A. H., M. Evangelista, A. B. Parsons, H. Xu, G. D. Bader et al., 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294 2364–2368. [DOI] [PubMed] [Google Scholar]
- Warburton, P. E., C. A. Cooke, S. Bourassa, O. Vafa, B. A. Sullivan et al., 1997. Immunolocalization of CENP-A suggests a distinct nucleosome structure at the inner kinetochore plate of active centromeres. Curr. Biol. 7 901–904. [DOI] [PubMed] [Google Scholar]
- Westermann, S., I. M. Cheeseman, S. Anderson, J. R. Yates, 3rd, D. G. Drubin et al., 2003. Architecture of the budding yeast kinetochore reveals a conserved molecular core. J. Cell Biol. 163 215–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, B. C., T. D. Murphy, M. L. Goldberg and G. H. Karpen, 1998. Neocentromere activity of structurally acentric mini-chromosomes in Drosophila. Nat. Genet. 18 30–37. [DOI] [PubMed] [Google Scholar]
- Zhang, W., B. G. Mellone and G. H. Karpen, 2007. A specialized nucleosome has a “point” to make. Cell 129 1047–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]