Abstract
Histone acetylation levels are regulated through the opposing activities of histone acetyltransferases (HATs) and deacetylases (HDACs). While much is known about gene-specific control of histone acetylation, little is understood about how total or cellular levels of histone acetylation are regulated. To identify regulators of cellular levels of histone acetylation, we developed an immunofluorescence-based approach to screen the single-gene deletion library of Saccharomyces cerevisiae for strains with significant reductions in cellular histone acetylation levels. Of the 4848 mutants screened, we identified 63 strains with considerable cellular hypoacetylation of N-terminal lysines in histones H3 and H4. The cellular hypoacetylation was validated for subsets of the identified strains through secondary screens including mass spectrometric analysis of individual lysines and chromatin immunoprecipitation of specific genomic loci. Among the identified mutants were several members of the Ccr4-Not complex, V-type ATPases, and vacuolar protein-sorting complexes as well as genes with unknown functions. We show that Gcn5, a major HAT in yeast, has diminished histone acetyltransferase activity in particular mutants, providing a plausible explanation for reduction of cellular acetylation levels in vivo. Our findings have revealed unexpected and novel links between histone acetylation, Gcn5 HAT activity, and diverse processes such as transcription, cellular ion homeostasis, and protein transport.
ACETYLATION (Ac) of histones has been widely demonstrated as an important post-translational modification that can regulate many DNA-based processes including gene expression (Struhl 1998; Kurdistani and Grunstein 2003a). There are numerous lysines (K) in both the amino termini and the globular domains of the core histones including K9, K14, K18, K23, K27, and K56 in histone H3 and K5, K8, K12, and K16 in H4 as well as other lysines in histones H2A and H2B. These lysines can be acetylated or deacetylated by various histone acetyltransferases (HATs) or deacetylases (HDACs) that show specificity toward histone subtypes as well as individual residues within a given histone. For instance, Gcn5, a member of the Spt-Ada-Gcn5-acetyltransferase (SAGA) complex in Saccharomyces cerevisiae, is the main HAT for lysine residues in the amino terminus of histone H3 (Kuo et al. 1996; Grant et al. 1999) whereas Rtt109 specifically acetylates K56, a lysine in the globular domain of H3 (Han et al. 2007a). HDACs also show similar specificities. For example, Rpd3 deacetylates essentially all lysines in the four core histones with the exception of H4K16 (Suka et al. 2001).
Since acetylation is a reversible process, the level of acetylation at a particular lysine residue is maintained through the balance of HAT and HDAC activities at both individual chromatin loci such as promoters and at the whole-cell level. The mechanisms by which various histone-modifying enzymes affect histones in chromatin involve targeted recruitment to specific genomic loci as well as global activities toward most histones throughout the genome, including nonpromoter sequences (Kuo et al. 2000; Vogelauer et al. 2000; Kurdistani et al. 2002). Targeted recruitment occurs when a specific DNA-binding protein such as a transcription factor directs the histone modifiers to its target genes. This results in modification of nearby histones with consequent effects on gene expression. The mechanism of global activities is less understood but may partly involve direct binding to specifically modified histones themselves. For instance, the Rpd3S HDAC complex deacetylates histones in the coding regions of genes during gene activity by binding to methylated H3K36. The binding is mediated in part by the chromodomain of Eaf3, a member of the Rpd3S complex, and is required for suppression of aberrant transcription initiation (Carrozza et al. 2005; Keogh et al. 2005). Therefore, the opposing actions of different histone modifiers through targeted and/or global mechanisms may establish specific patterns of acetylated and deacetylated lysine residues at different locations throughout the genome (Kurdistani et al. 2004).
Significant effort has been expended in the past few years, in model organisms as well as in human cells, to determine how various HATs and HDACs may be recruited to promoters and coding regions and the potential downstream effects on expression of nearby genes. However, little is known about how the activities of the histone-modifying enzymes or the complexes in which they reside may be regulated. While “modification-by-recruitment” may affect local histone modification patterns, regulation of enzyme activity could significantly change the total levels of a modified lysine throughout the genome. For instance, the Akt kinase can phosphorylate several histone-modifying enzymes such as the p300 HAT and the H3K27 methyltransferase Ezh2. Phosphorylation of p300 increases its intrinsic HAT activity (Huang and Chen 2005) whereas that of Ezh2 decreases its affinity for histone H3, leading to lower cellular levels of H3K27 methylation (Cha et al. 2005). Drastic changes in total or cellular levels of histone modifications may have broad effects in both normal biology of a cell and disease processes. In fact, our laboratory has shown that differences in total levels of specific histone modifications in cancer cells generate a novel “cellular epigenetic heterogeneity” that can predict clinical outcome of prostate cancer patients (Seligson et al. 2005). Unexpectedly, the increased prevalence of cells with lower cellular levels of specific histone modifications, especially of H3K18 acetylation, was associated with poorer clinical outcome. The biological basis for such association remains to be determined but it is conceivable that aberrant regulation of histone-modifying enzyme activity may in part underlie the cell–cell differences in total levels of specific histone modifications.
The yeast S. cerevisiae as a model organism has been extraordinarily useful in providing important insights for understanding epigenetic processes. For instance, a link between histone acetylation and transcriptional activation was shown for the first time for the yeast Gcn5 HAT (Kuo et al. 1996). The histones, their modified residues, and the histone-modifying enzymes also show a great degree of sequence and functional conservation among eukaryotes, including humans (Wang et al. 1997; Doyon and Cote 2004). Therefore, further understanding of epigenetic mechanisms in yeast should shed light on similar mechanisms in human cells. Due to its powerful genetics, we thus decided to use yeast to identify regulators of histone acetylation, expecting not only to discern epigenetic regulatory mechanisms in yeast but also to generate testable hypotheses for how cancer cells may regulate cellular levels of acetylation.
To identify genes that may potentially regulate the total cellular levels of H3K18 acetylation, we developed a 96-well plate format immunofluorescence (IF)-based assay to screen the single-gene deletion library of the yeast S. cerevisiae for strains that have lower levels of H3K18ac compared to wild-type cells. We reasoned that strains with significantly low levels of H3K18ac should be detectable when stained at the whole-cell level with an antibody against H3K18ac, similar to the findings in primary cancer tissues. We further reasoned that a subset of the identified genes may result in significant reductions in total levels of H3K18ac, directly or indirectly, through modulation of SAGA/Gcn5 HAT activity. Identification of such genes may eventually lead to elucidation of potential regulatory pathways that control H3K18ac levels in a cell, with potentially important implications for prostate cancer biology (Seligson et al. 2005). Of the 4848 single-gene deletion mutants screened, we identified 63 strains with significantly decreased cellular H3K18ac levels including known HATs and their cofactors. For subsets of the candidate mutants, we validated the reduction in histone acetylation through Western blotting and mass spectrometry of histones, determined their effects on the HAT activity of Gcn5, and mapped the decrease in total levels of acetylation to specific regions of the genome. Our results suggest that diverse cellular pathways operating in both nucleus and cytoplasm may have roles in regulation of cellular histone acetylation levels in part through modulation of Gcn5 HAT activity.
MATERIALS AND METHODS
Yeast strains, deletion library, plasmids, and media:
Wild-type BY4741 (MATa his3Δ1 leu2Δ0 ura3Δ0 met15Δ0) and RMY200 (MATa ade2-101 his3-200 lys2-801 trp1-901 ura3-52, hht1, hhf1∷LEU2 hht2, hhf2∷HIS3, pRM200) were used as isogenic wild-type controls when appropriate. The point mutants H3K18R and H4K16R were generated using RMY200. The MATa S. cerevisiae single-gene deletion library (Winzeler et al. 1999) was obtained from Research Genetics (Birmingham, AL). All strains were grown in YPD media with 100 μg/ml ampicillin, with 200 μg/ml G418 added for library. For reintroduction of wild-type copy of genes into the corresponding mutants, vector BG1805-derived plasmids containing the genes of interest (Open Biosystems, Huntsville, AL) under a GAL-inducible promoter were used for transformation according to standard protocols. Synthetic dextrose (SD −Ura) or galactose media lacking uracil (SG −Ura) were used to repress or induce the expression of plasmid-borne genes, respectively.
Yeast IF:
The yeast strains from each library plate were cultured in sterile 96-well plates without agitation for 1 or 2 days at 30° till saturation. A suitable volume of the saturated culture was then used to inoculate 600 μl media per well in new sterile 96-deep-well plates (BD Falcon, San Diego) for an initial A600 ∼ 0.2 and grown till A600 ∼ 0.8–1.0. Cells were fixed with 37% w/v formaldehyde (for 4% final concentration) at room temperature for 1 hr with slow shaking. Cells were spun down, washed once with PBS and once with sorbitol buffer (1.2 m sorbitol, 0.1 m K2HPO4, 0.1 m KH2PO4, pH 6.5), and then resuspended in 400 μl sorbitol buffer. The fixed cells were immediately used for digestion or stored at 4° for use within several days. To digest the cell wall, 5 units zymolyase 100T (ICN Biochemicals, Irvine, CA) and 1 μl of 14 m β-mercaptoethanol (Sigma, St. Louis) were added into each well and incubated for 50 min at 30° with slow shaking. The subsequent spheroplasts in each well were washed twice with 200 μl sorbitol buffer, permeabilized by 50 μl 0.5% NP-40 for 10 min at 30°, followed by two washes with 200 μl PBS–BSA (0.04 m K2HPO4, 0.01 m KH2PO4, 0.15 m NaCl, pH 7.5, 5 mg/ml BSA), and then resuspended in 500 μl of PBS–BSA. A suitable amount of the cell suspension was added to 96-well glass-bottom black-frame detection plates (Whatman, Florham Park, NJ) precoated with 0.1% poly-l-lysine (Sigma). The plates were incubated at room temperature for 15 min to allow cells to sink to the bottom of the plates and get immobilized on the poly-l-lysine-coated surface. After the unattached cells were discarded, the plates were heated at 60° for 10 min to strengthen the cell immobilization and then cooled down to room temperature. For histone modification staining, 45 μl primary antibody (Suka et al. 2001) diluted with PBS–BSA (1:100 for anti-H3K18ac or 1:400 for anti-H4K16ac) were added to each well for overnight incubation at 4°. The plates were washed three times with 150 μl/well PBS–BSA after which 45 μl/well of the mixture of secondary antibody (1:1000 anti-rabbit IgG conjugated with FITC; Molecular Probes, Eugene, OR) and DAPI (1:1000 working dilution, Sigma) were added for 2 hr incubation in the dark at room temperature. The plates were washed twice with distilled H2O. The fluorescent intensity of each well was scored with an inverted fluorescence microscope (Carl Zeiss, Göttingen, Germany) with the 40× objective lens. Pictures were taken with the 100× oil objective lens. The strains that scored lower than wild-type were pooled and reanalyzed two to four times as described above. A similar procedure was used to check the localization of myc-tagged Gcn5 in mutants of interest. The monoclonal anti-myc antibody (9E10) (Roche, Indianapolis) (working dilution 1:800) was used accordingly.
Histone extraction:
For Western blotting, histones were extracted from 5 ml culture (A600 ∼ 0.8–1.0) by a fast trichloroacetic acid (TCA)-precipitation method. Briefly, cell pellets were washed with 20% TCA and quickly frozen at −80°. Pellets were resuspended in 20% TCA and lysed by vigorous vortexing with glass beads. Crude histones were precipitated with TCA (10% final), washed with 100% ethanol, and resuspended with 1 m Tris (pH 8.0). For mass spectrometry, crude histones were extracted and isolated from yeast as described (Xu et al. 2005). Briefly, histones were acid extracted from isolated nuclei from 1 liter of culture and precipitated using TCA. Yeast lytic enzyme (ICN Biomedicals, Aurora, OH) was used for cell wall digestion.
Mass spectrometry:
Quantification of histone acetylation at specific lysine residues was carried out as previously reported with some modification for processing on a QTOF instrument (Zhang et al. 2004; Smith 2005). Briefly, core histones were separated by reverse-phase HPLC on a C4 column into four fractions, each containing histone H2A, H2B, H3, or H4. Histones H3 and H4 were reacted with d6-acetyl anhydride in d4-acetic acid (5/50) for 6 hr at room temperature to acetylate in vitro all unacetylated lysine residues, resulting in the prevention of trypsin digestion at lysine residues that were either derivatized with d3-acetyl groups or naturally modified with acetyl groups. The reactant solutions were completely dried and digested by trypsin (limited to arginine residues in this case) in 25 mm ammonium bicarbonate buffer overnight and then submitted for liquid chromatography/mass spectrometry/mass spectrometry (LC/MS/MS) analyses. LC/MS/MS experiments were performed on a QTOF Ultima-Global (Micromass) instrument with operation conditions set as previously reported with a modification for quantification of acetylation (Zhang et al. 2004). Quantification of acetylation at a specific site was obtained from the relative abundance of acetyl-modified vs. d3-acetyl-modified fragmentation ions, along with the distribution of precursor ions. Quantification of histone methylation at H3K79 was done as described previously (Zhang et al. 2004: Shahbazian et al. 2005).
Western blotting and chromatin immunoprecipitation:
Histones were separated by 15% SDS–PAGE and probed with anti-β-actin (1:12,000; Abcam, Cambridge, MA), anti-H3 (1:6000, Abcam), anti-acetyl H3 polyclonal antibody (1:6000; Upstate Biotechnology, Lake Placid, NY), and anti-rabbit IgG secondary antibody linked with HRP (1:10,000) (Amersham Biosciences, Piscataway, NJ). Western Lightning Chemiluminescence Reagent Plus (Perkin-Elmer Life Sciences, Waltham, MA) was used for detection. Chromatin immunoprecipitation (ChIP) was carried out essentially as described (Suka et al. 2001; Kurdistani and Grunstein 2003b).
Liquid HAT activity assay:
The liquid HAT activity assay was performed according to standard protocols (Eberharter et al. 1998; Mizzen et al. 1999). Briefly, cells from log-phase cultures (OD600 ∼ 1.0) were harvested and resuspended in lysis buffer (50 mm HEPES/KOH, pH 7.5, 150 mm NaCl, 1% Triton X-100, 1 mm EDTA, 1× complete protease inhibitors) and lysed by vortexing with sterile glass beads. The crude protein concentration in lysates of each strain was measured using Bio-Rad (Hercules, CA) protein assay reagent and the amount of lysates of each strain used for Gcn5-myc immunoprecipitation was adjusted accordingly. The Myc-tagged Gcn5 was immunoprecipitated with anti-myc monoclonal antibody 9E10 (Roche) by overnight incubation at 4° and bound to 50 μl of protein A Sepharose CL-48 beads slurry (Amersham Biosciences, Uppsala, Sweden) by an additional 2 hr incubation at 4°. After five washes with lysis buffer, the beads were suspended in 50 μl reaction buffer (50 mm Tris-HCl, pH 8.0, 50 mm KCl, 10% glycerol, 10 mm sodium butyrate, 1 mm PMSF, and 1 mm dithiothreitol). Reactions were assembled on ice. Each reaction contained 20 μg of calf thymus total histones (type IIA, Sigma) or oligonucleosomes isolated from a gcn5 deletion strain. Oligonucleosomes were prepared as described (Gregory and Horz 1999; Mizzen et al. 1999). A total of 0.5 μCi of [3H]-acetyl-CoA (Perkin-Elmer, Norwalk, CT) was added to start the reaction, which was performed at 30° for 40 min. The reactions were stopped by spotting the reaction material onto a P-81 filter paper (Whatman, London) and the radioactivity was counted by a Tri-Carb 2800 TR liquid scintillation analyzer (Perkin-Elmer).
RESULTS
Systematic identification of potential regulators of cellular histone acetylation levels:
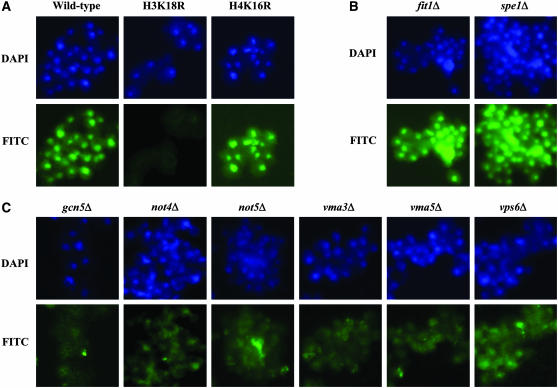
To identify genes that may be required for maintenance of cellular H3K18ac levels, we developed a 96-well plate, IF-based technique to screen the MATa single-gene deletion library of S. cerevisiae. We also examined the levels of H4K16ac. Before screening the yeast library, we first determined the feasibility of discerning the potential mutants with low levels of H3K18ac from wild-type cells. A highly specific antibody recognizing H3K18ac was used to detect cellular acetylation levels of wild-type RMY200 vs. H3K18R and H4K16R mutant cells (Suka et al. 2001). As expected, no IF signal was observed in the H3K18R compared to the wild-type or H4K16R mutant cells (Figure 1A), confirming the specificity of the antibody in IF staining. The specificity and feasibility of an anti-H4K16ac antibody was also verified using the same strains (data not shown). Therefore, IF staining of individual nuclei with highly specific antibodies can distinguish cells with low levels of individual sites of acetylation.
Figure 1.—
Ascertaining cellular levels of specific histone acetylation sites via immunofluorescence. Yeast cells were subjected to indirect immunofluorescence (IF) staining using highly specific histone acetylation antibodies. Shown are false color images for the nuclear stain (blue, DAPI) and immunostain (green, FITC). (A) The feasibility and specificity of an anti-H3K18ac in immunofluorescence staining was tested using strains with wild-type histones H3 and H4 (RMY200) and H3K18R or H4K16R mutations. As expected, H3K18 acetylation is undetectable in H3K18R but unaffected in H4K16R compared to wild type. (B and C) Representative strains immunostained with the anti-H3K18ac antibody from the single-gene deletion library are shown. Most deletion strains such as fit1Δ and spe1Δ showed no difference in H3K18ac levels compared to wild type but a subset had significantly lower cellular levels of H3K18ac such as gcn5Δ, not4Δ, not5Δ, vma3Δ, vma5Δ, and vps6Δ.
The protocol for the immunofluorescence-based screening of the library is described in detail in materials and methods. Briefly, cultures of yeast cells were grown overnight in 96-well plates and used to inoculate fresh media in a new plate and grown for 4–5 hr. Cells were harvested in log phase, fixed with formaldehyde, and digested to generate spheroplasts. Cells were then permeabilized and transferred to 96-well glass-bottom plates with black frames and precoated with 0.1% poly-l-lysine. The plates were incubated at room temperature for 15 min and then at 60° for 10 min to immobilize the cells on the glass surface. Immobilized cells were incubated with the primary antibody at 4° overnight and the secondary antibody for 2 hr at room temperature. The staining was visualized with an inverted fluorescence microscope (Zeiss) with the 40× objective. Semiquantitative scoring of IF intensity was performed for individual wells on a scale of 1–4, with 1 showing significant reduction and 4 having wild-type levels of acetylation. The acetylation levels in strains that were scored <4 were confirmed through independently repeated experiments. To eliminate observer bias, we were blinded to the identity of deletion mutants until after all scoring was completed.
We examined 4848 deletion strains in the library and identified 63 mutants that have lower than wild-type histone acetylation at H3K18 (Table 1). Thirty-one of the 63 mutants had lower levels of H4K16ac as well (Table 1, footnote b). We grouped the mutants according to their semiquantitative IF scores: of the 63 mutants, 19 scored 1, 15 scored 2, and 29 scored 3 for cellular levels of H3K18ac. We verified the identity of the mutants scored at 1 or 2 by amplification and sequencing of the identifying bar codes (data not shown). Figure 1, B and C, shows representative IF staining for H3K18ac in two randomly selected strains (fit1Δ and spe1Δ) that showed wild-type levels of acetylation and six deletion mutants that exhibited significant reduction in the cellular levels of H3K18ac. Among the identified mutants, there were six members of the SAGA complex including gcn5 that were identified blindly as part of the screen, validating our approach. We conclude that our approach is able to successfully identify deletion mutants with low cellular levels of histone modifications. As shown below, the effect of deletion mutants on lowering histone acetylation was not specific to H3K18 but affected other sites of modification as well with the exception of H3K56 (Figure 3 and supplemental Table 1). This is consistent with the broad specificities of HATs and HDACs for multiple sites of histone acetylation.
TABLE 1.
Genes affecting cellular levels of histone acetylation identified from screening of the single-gene deletion library of Saccharomyces cerevisiae
| Immunostain scorea | Genes identified from the library screen |
|---|---|
| 1 | ADA2b, APT2b, COG1b, CUP5/VMA3b, GAS1b, GCN5/ADA4b, HFI1/ADA1, MOT2/NOT4b, NGG1/ADA3, NOT5b, PRD1, RNR1b, SPT7, SPT10b, SPT20/ADA5, TFP3/VMA11b, VMA5b, VPS45b, YKL118Wb |
| 2 | ALF1, ANP1/MNN8, ASF1, CAX4b, CCR4b, DIA2b, ELM1, MRE11, NHX1/VPS44b, PEP12/VPS6b, PMT2b, RIB4b, SNF7/VPS32, VMA8b, YGL101W |
| 3 | BST1/PER17, CDC40b, CUL3, CYC8b, ERG28, ERP5, GDS1, GRX5, HUR1, KIN3/FUN52, KIP3, LAS21b, MNN10, OCH1, PPA1/VMA16b, ROG1, RPB4b, SEC28, SIC1, SPC72, THP1, TYR1, UBA4, VMA2b, VMA13b, VMA22b, VPS34, YDL177C, YPR099Cb |
The IF intensity was scored on a scale of 1–4, with 1 showing significant hypoacetylation and 4 having wild-type levels of acetylation.
Showed low acetylation on both H3K18 and H4K16.
Figure 3.—
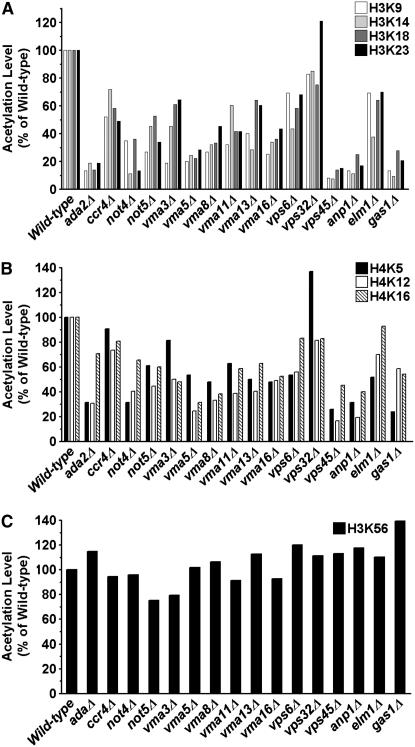
Mass spectrometry analysis of cellular levels of histone acetylation validates the identified hits. The acetylation levels of different lysine residues in histones H3 and H4 were measured by LC/MS/MS (see materials and methods) for wild type, ada2Δ (positive control), and 13 deletion mutants. Shown are the percentages of total histones modified in each mutant relative to wild type set at 100%. The absolute levels of acetylation levels of the indicated lysines residues in the deletion mutants are listed in supplemental Table 1. (A) H3K9ac, K14ac, K18ac, and K23ac; (B) H4K5ac, K12ac, and K16ac; (C) H3K56ac.
Identified regulators of histone acetylation are enriched in common functional groups:
Since we screened essentially all of the nonessential deletion mutants, we expected that a subset of the identified regulators would be involved in common cellular pathways. This is because deletion of some or most members of a given pathway or complex should have similar phenotypic effects. To determine whether the identified mutants were enriched in common functional groups, we mapped the 63 identified mutants to multiple classification databases, including MIPS, GO, MDS, Cellzome Proteomics Complexes, and Proteome Localization, using FunSpec (Functional Specifications at http://funspec.med.utoronto.ca). After correcting for multiple hypothesis testing (Bonferroni correction), statistical analysis revealed that the identified mutants were significantly enriched in common functional categories. As listed in Table 2, these categories include the SAGA complex (P = 6.0 × 10−9), the transcription factor complex (P = 6.4 × 10−7), vacuolar acidification (P = 4.5 × 10−14) and transport (P = 2.7 × 10−13), the V-type ATPase complex (P = 7.9 × 10−11), and protein modifications (P = 7.9 × 10−8). Considering that we selected the mutant strains solely on the basis of low levels of histone acetylation, enrichment of specific functional categories among the identified mutants further validates our methodology.
TABLE 2.
Enrichment of the identified deletion mutants in specific functional categories or cellular components
| Category | P-value | Genes identified from screen |
|---|---|---|
| Vacuolar acidification (GO: 0007035) | 4.54E-14 | VMA2NHX1 CUP5 VMA8 PPA1 VMA22 VMA5 TFP3 VMA13 |
| Vacuolar transport (GO: 0007034) | 2.67E-13 | VMA2 NHX1 CUP5 VMA8 PPA1 VMA22 VMA5 VPS34 TFP3 VMA13 |
| Vacuole organization and biogenesis (GO: 0007033) | 9.29E-09 | VMA2 NHX1 CUP5 VMA8 VPS45 PPA1 VMA22 VMA5 VPS34 TFP3 VMA13 |
| Hydrogen-translocating V-type ATPase complex (GO: 0016471) | 7.85E-11 | VMA2 CUP5 VMA8 PPA1 VMA5 TFP3 VMA13 |
| Hydrogen-transporting ATPase V1 domain (GO: 0000221) | 1.86E-06 | VMA2 VMA8 VMA5 VMA13 |
| Hydrogen-transporting ATPase V0 domain (GO: 0000220) | 1.90E-05 | CUP5 PPA1 TFP3 |
| Organelle organization and biogenesis (GO: 0006996) | 1.34E-07 | SPC72 VMA2 MNN10 CDC40 NHX1 CUP5 VMA8 BST1 VPS45 KIP3 PPA1 VMA22 VMA5 SNF7 VPS34 ALF1 THP1 TFP3 VMA13 |
| Protein modification (GO: 0006464) | 7.88E-08 | PMT2 SPT7 NGG1 MNN10 ADA2 ANP1 OCH1 CAX4 GCN5 UBA4 LAS21 ELM1 VPS34 SPT20 HFI1 |
| Protein amino acid acetylation (GO: 0006473) | 3.98E-07 | SPT7 NGG1 ADA2 GCN5 SPT20 HFI1 |
| SAGA complex (GO: 0000124) | 5.99E-09 | SPT7 NGG1 ADA2 GCN5 SPT20 HFI1 |
| Transcription factor complex (GO: 0005667) | 6.41E-07 | CCR4 SPT7 NGG1 ADA2 MOT2 GCN5 SPT20 HFI1 NOT5 |
Western blotting and mass spectrometric analysis of histone modifications validate the identified mutants:
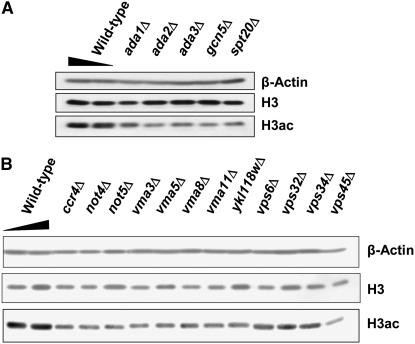
To further verify that the identified deletion mutants contained lower levels of histone acetylation than wild-type cells, we performed two sets of validation experiments. First, we extracted histones from a subset of the deletion mutants and subjected them to standard Western blotting with an anti-H3K9/K14ac antibody (note that the antibodies used for IF, which were originally tested for ChIP, did not work in Western blotting of yeast histones). All tested mutants reproducibly showed lower acetylation levels than the wild-type strain. Two representative blots are shown in Figure 2. The Western data indicate that cellular histone acetylation levels are indeed reduced in the identified mutants.
Figure 2.—
Western blotting for cellular levels of histone acetylation validates the identified hits. Histones extracted from a subset of identified deletion strains were subjected to standard Western blotting with anti-H3 and anti-H3K9/K14ac to detect total histone H3 and its acetylation levels, respectively. Anti-β-actin was used as additional loading control. Deletion mutants of (A) the SAGA components and (B) the Ccr4-Not complex, V-ATPase, and VPS proteins are shown.
Second, we isolated total histones from wild-type and mutant strains and subjected them to MS for quantitative determination of total levels of modified histones. We examined histone acetylation levels of wild type, ada2Δ (to serve as a positive control because of expected low acetylation levels due to disruption of the SAGA complex), and 13 deletion mutants that were selected to represent the various functional categories from Table 1. Briefly, histones H3 and H4 from nuclear extracts were separated by reverse-phase HPLC and the acetylation levels of K9, K14, K18, K23, and K27 in H3 and K5, K12, and K16 in H4 were measured by LC/MS/MS as described in materials and methods. The data were analyzed as described elsewhere (Zhang et al. 2004; Smith 2005). Supplemental Table 1 contains the percentage of total histones that were modified at the indicated lysines in the different strains. In Figure 3, we plotted these percentages as fractions of wild-type acetylation levels for each lysine for better visualization of the percentage of decreased histone acetylation in the identified mutants.
Of the lysine acetylation sties examined, H4K16 was the most abundant site of acetylation in wild-type cells (53.7% of H4), consistent with previous reports (Smith et al. 2003). Interestingly, H3K18ac was the least abundant with only 3.6% of wild-type H3 modified at this site. Other sites of acetylation showed varied levels of abundance, suggesting that the total histone acetylation levels may be regulated in a lysine-dependent manner. In the ada2Δ mutant, all H3 lysines with the exception of H3K56 showed significant reductions in acetylation, consistent with loss of SAGA HAT activity. Interestingly, both H4K5 and H4K12 also showed significant reductions in acetylation levels while H4K16ac showed only an ∼1.4-fold decrease. Examining the mutant strains, we found that, with the exception of H3K56, all the examined lysines showed lower levels of acetylation compared to wild type, albeit to different extents depending on the strain and the specific lysine (Figure 3). For instance, Ccr4, Not4, and Not5 are members of the Ccr4-Not transcriptional regulator but ccr4 deletion has a moderate effect on acetylation levels (50–90% of wild type) whereas not4 and not5 have more drastic effects, with acetylation levels reaching as low as 10% of wild type (Figure 3, A and B). All the V-type ATPases examined and two vacuolar sorting protein (VPS) mutants (vps6Δ and vps45Δ) also showed decreased acetylation levels on both H3 and H4 histones. (Note that the vps45Δ mutant from the commercial library may harbor additional mutations; see below for more details.) Deletions of ANP1, a type II membrane protein and subunit of the α-1,6-mannosyltransferase complex, and GAS1, a β-1,3-glucanosyltransferase that is required for cell wall assembly, also showed significant decreases in histones H3 and H4 acetylation levels (Figure 3, A and B). Notably, the cellular level of H3K56ac was not adversely affected in most mutants relative to wild type (Figure 3C). In the deletion mutants, H3K56ac ranged from 22.4 to 41.5% compared to wild type at 29.8%. The similar H3K56 acetylation levels in the mutants indicate that the decrease of histone acetylation on N-terminal lysines is unlikely due to a general lack of acetyl coenzyme A availability. They also suggest a potentially different regulatory mechanism for maintenance of acetylation levels on the N-terminal lysines vs. the globularly located H3K56. This is consistent with the fact that Rtt109, which specifically acetylates K56, shares little homology with other known HATs (Han et al. 2007a). Taken together, the quantitative MS data support the IF and Western findings that cellular histone acetylation levels of N-terminal lysines are adversely affected in the identified mutants.
To verify that the global decrease in histone acetylation was indeed caused by the deleted genes, we carried out complementation assays for two strains each from the three main functional groups plus the ada2 control strain. We reintroduced a plasmid (Open Biosystems) containing the wild-type copies of ADA2, VMA5, VMA8, NOT4, CCR4, VPS6, or VPS32 under the control of a galactose-inducible promoter (pGAL1) into each of their respective deletion strains. We then determined the levels of H3 acetylation by Western blotting under repressed (SD media) or induced (SG media) conditions (supplemental Figure 1). In all cases, compared to repressed conditions, expression of the exogenous wild-type allele resulted in increased H3ac, complementing the phenotype of the mutants.
Global decrease in histone acetylation maps to specific regions of the genome:
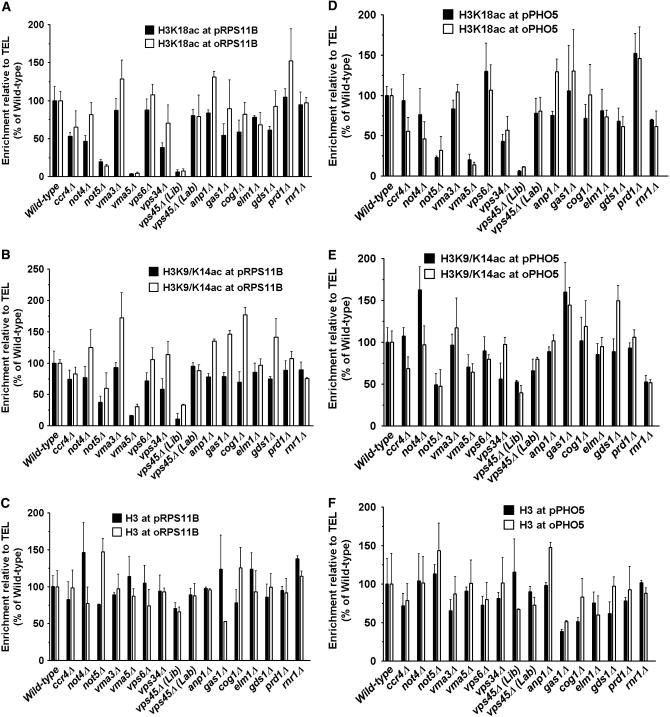
To determine how the decrease in cellular histone acetylation levels maps to individual genes and their promoters, we performed ChIP experiments using antibodies specifically against either H3K18ac or H3K9/K14ac to analyze local acetylation levels at selected gene loci including RPS11B (Figure 4, A and B) and PHO5 (Figure 4, D and E). These two genes are expressed and repressed, respectively, in rich media and are targets of the SAGA complex. The levels of H3 itself were also measured by ChIP to control for nucleosome content (Figure 4, C and F). At each gene, we analyzed the promoter (Figure 4, solid bars) and the open reading frame (ORF) (Figure 4, open bars) regions separately. We also examined histone acetylation levels within the ORFs of three additional randomly selected genes, MEC1, PMA1, and PYK1 (supplemental Figure 2). In general, we found that the cellular decrease in total levels of histone modifications in each mutant maps to specific regions of the genome, affecting some loci but not others. For instance, not5 deletion resulted in a significant decrease in acetylation at both RPS11B and PHO5 genes, but with only modest effects at the MEC1 gene. Compared to not5, ccr4 and not4 deletions had more limited effects on histone acetylation. While not5 deletion decreased H3K18ac levels at the PHO5 promoter and ORF, ccr4 and not4 deletions affected mainly the coding region with little to no effects on the promoter. For the vma strains, we did not detect significant changes in histone acetylation at the examined loci in the vma3 deletion strain. However, vma5 deletion resulted in significant loss of histone acetylation at most loci, with RPS11B, PHO5, PYK1, and MEC1 more adversely affected than PMA1. In these cases, there was little change in H3 distribution (Figure 4, C and F).
Figure 4.—
Reduction in cellular levels of histone acetylation maps to specific genomic loci. Levels of histone acetylation at H3K18 and H3K9/K14 and unmodified histone H3 were determined at promoter (p) and open reading frame (o) regions of RPS11B and PHO5 genes by standard chromatin immunoprecipitation (ChIP). A region 500 bp from the telomere of chromosome VI-R (TEL) was used as an internal control in each PCR reaction. The values reflect enrichment of ChIPed DNA relative to the TEL region and input DNA and are shown as percentage of wild type that was set to 100%. The data are the average of three independent experiments. (A) H3K18ac at RPS11B; (B) H3K9/K14ac at RPS11B; (C) H3 at RPS11B; (D) H3K18ac at PHO5; (E) H3K9/K14ac at PHO5; (F) H3 at PHO5.
Of the VPS genes, vps6 affected H3K9/K14ac only at the PYK1 gene but vps34 had a broader effect, causing diminished acetylation at both the PHO5 and RPS11B genes, affecting promoters more than ORFs. In the vps45 deletion, we detected only background ChIP signals at PHO5 and RPS11B, suggesting that little or no H3 acetylation exists at these loci. In addition, as shown below, Gcn5 had little HAT activity in this mutant. This raised our suspicion as to whether vps45 deletion by itself could result in such a drastic loss of acetylation. To address this issue, we generated a vps45 deletion strain (CTY135) in the same wild-type background as the library mutant, using a similar strategy to that described by the Saccharomyces Genome Deletion Project (http://www-sequence.stanford.edu/group/yeast_deletion_project/deletions3.html). In this article, we refer to the two strains as vps45Δ (Lib) and vps45Δ (Lab) to distinguish the library strain from the one made in our laboratory, respectively. Deletion of VPS45 was confirmed in both strains using multiple PCR strategies (data not shown). Comparing the two strains, we found that they differed in growth rate (supplemental Figure 3), histone H3 acetylation levels (supplemental Figure 4A), SAGA complex composition (supplemental Figure 4B), and Gcn5 in vitro HAT activity (Figure 5). Therefore, we conclude that the vps45Δ (Lib) strain must have additional mutations or defects to account for cellular loss of acetylation. Nonetheless, like other VPS deletion mutants, vps45Δ (Lab) showed a modest but consistently detectable decrease in histone acetylation both at the cellular level (supplemental Figure 4) and at specific loci (Figure 4 and supplemental Figure 2).
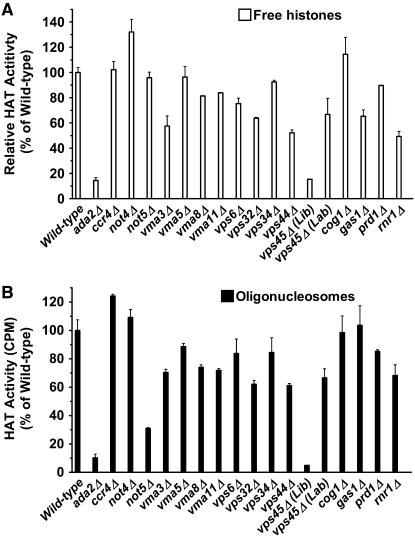
Figure 5.—
The Gcn5 in vitro HAT activity is reduced in a subset of the identified deletion mutants. The in vitro HAT activity of immunoprecipitated myc-tagged Gcn5 was determined for the wild type, ada2Δ (positive control), and 17 deletion mutants. Shown are the percentages of Gcn5-myc HAT activity on (A) free histones and (B) oligonucleosomes relative to wild type set at 100%. The data are averages of at least three independent experiments.
The remaining deletion mutants also showed variable effects on histone acetylation. Of note is the rnr1 deletion (Rnr1 is the large subunit of ribonucleotide-diphosphate reductase) that caused moderate to significant reduction in histone acetylation at RPS11B and PHO5 genes, respectively. Deletion of GAS1 did not result in decreased histone acetylation but, unlike other mutants, caused significant reduction in total H3 levels, especially at PHO5. Taken together, we conclude that the decrease in histone acetylation in each mutant at the cellular level maps to different genomic regions at the molecular level. It is important to note that, since different histone modifiers affect common regions of chromatin, the level of acetylation at individual loci reflects the balance of specific HATs and HDACs that are present at each locus. So, loss of acetylation at a given chromatin region may be due to decreased HAT activity and/or increased HDAC activity. Loss of acetylation could also be secondary to gene expression changes. Below, we show that a subset of mutants adversely affects acetylation levels in part through decreased Gcn5 HAT activity.
Global decrease in histone acetylation is in part due to decreased Gcn5 HAT activity:
To determine whether the decrease in the levels of histone acetylation in the various deletion mutants was due to decreased HAT activity, we determined the in vitro HAT activity of Gcn5 on both free histones and oligonucleosomes. We introduced a myc-tag in the chromosomal locus of GCN5 at its 3′ end in wild type and in each of the deletion mutant cells including ada2Δ as indicated in Figure 5. The Gcn5-myc protein was immunoprecipitated from cell lysates by an anti-myc monoclonal antibody and used in an in vitro liquid HAT assay in the presence of [3H]-acetyl-CoA on free histones or oligonucleosomes isolated from a gcn5Δ mutant (for lower baseline acetylation levels). Expression of Gcn5-myc was confirmed in all strains using Western blotting (data not shown) and equal protein concentrations were used in each assay as described in materials and methods. Figure 5, A and B, shows the average levels of Gcn5 HAT activity from three independent experiments for wild type (set to 100%), ada2Δ, and 17 deletion mutants on free histones or oligonucleosomes, respectively. We found that, as expected, deletion of ADA2 essentially abolished Gcn5 HAT activity on either substrate. While deletions of CCR4, NOT4, and NOT5 had no effect on the ability of Gcn5 to acetylate free histones, deletion of NOT5 reduced Gcn5 HAT activity toward oligonucleosomes to ∼30% of wild type. All V-ATPase mutants showed lower HAT activity on either substrate or both, ranging from 58 to 84% of wild-type activity levels. Deletions of VPS6, VPS32, VPS34, and VPS44, all identified in the screen (Table 1), had moderate effects, decreasing Gcn5 HAT activity to 52–85% of wild type depending on the substrate. The Gcn5 from the vps45Δ (Lib) strain showed essentially no activity above background whereas the Gcn5 from vps45Δ (Lab) retained 60–67% of wild-type HAT activity, further suggesting that the library strain may have additional mutations. Deletions of PRD1 (a metalloendopeptidase) and RNR1, both of which were identified in the screen, also resulted in decreased Gcn5 HAT activity. The effect of gas1Δ differed from all other mutants as it reduced Gcn5 HAT activity toward free histones (62% of wild type) but not on oligonucleosomes. Taken together, these results indicate that decreases in cellular histone acetylation levels may in part be due to modulation of HAT activity toward free or nucleosomal histones.
DISCUSSION
Understanding how histone-modifying enzymes maintain cellular levels of histone modifications requires elucidation of not only their recruitment to chromatin but also regulation of their activity. Here, we report identification of regulators of histone acetylation in the yeast S. cerevisiae. We developed a high-throughput immunofluorescence-based method to screen the yeast single-gene deletion library for strains with significantly lower cellular levels of histone acetylation than wild-type cells. A major advantage of this method is that histone acetylation is examined by indirect immunofluorescence of intact nuclei, avoiding potentially aberrant modification patterns that may occur if histones were to be extracted. Since cells are fixed with formaldehyde and the histones are assessed in their native location, this method also obviates the need for use of HDAC inhibitors that are usually included in histone extraction protocols. A disadvantage is the labor-intensive scoring of the IF intensity for each strain, which, however, can be overcome if combined with automated plate readers with image analysis software. Nonetheless, our approach was successful in identifying deletion strains with significantly decreased histone acetylation levels.
Blinded to the identity of the strains, we identified 63 deletion mutants, including members of the SAGA HAT complex, that showed significantly lower cellular levels of H3K18ac and/or H4K16ac compared to wild-type cells. We did not detect other HATs presumably because their deletions may not cause a significant decrease in H3 acetylation to be detected in our assay. The decrease in histone acetylation as detected by immunofluorescence was validated for subsets of the mutants through Western blotting as well as mass spectrometry. Although the mutants were selected initially for lower cellular levels of H3K18ac, Western and mass spectrometry analyses indicated lower histone acetylation levels at other acetylatable histone lysines as well. This suggests that regulation of H3K18ac levels is linked to other acetylation sites and is consistent with the ability of yeast HATs and HDACs to affect multiple histones and acetylation sites. In all the mutants tested, mass spectrometry revealed lower acetylation levels of N-terminal lysine in histones H3 and H4 compared to wild-type cells, albeit to different extents for the various lysine residues. Interestingly, H3K56ac levels showed little change in the tested mutants, suggesting a different regulatory mechanism for this lysine and excluding a generalized deficiency in acetyl CoA levels. In contrast, some of the deletion mutants affected the modification levels of another globularly located lysine, H3K79 methylation (supplemental Figure 5). We also showed that the reduction in histone acetylation at the cellular level maps to distinct regions of the genome at the molecular level. Finally, in certain mutants, lower cellular levels of histone acetylation could be due to decreased HAT activity of the SAGA complex. Despite the global decrease in histone acetylation levels, we did not detect a unified growth defect in the deletion mutants (supplemental Figure 3). Altogether, our data strongly implicate the identified mutants in regulation of global histone acetylation levels.
Among the mutants, we found three members of the Ccr4-Not transcriptional complex, a global regulator of transcription in yeast and other eukaryotes including humans (Collart 2003). In addition to gene expression, this complex also regulates many other cellular functions such as RNA degradation and acts as a regulatory platform for sensing nutrient level and stress (Collart 2003; Lenssen et al. 2005; Traven et al. 2005). While a role for this complex in regulating cellular histone acetylation levels was not known, an interaction between Not2 of the Ccr4-Not and Ada2 of the SAGA complexes has been reported (Benson et al. 1998). Several functional and genetic interactions between the Ccr4-Not complex and members of the SAGA complex in regulation of gene expression have also been identified (Collart 2003; Biswas et al. 2006; James et al. 2007). In addition, the Ccr4-Not complex is also required to maintain global levels of H3K4 trimethylation that has also been linked to proper acetylation of histone H3 (Bernstein et al. 2002; Laribee et al. 2007). Our findings now suggest a more widespread role for this complex in regulation of cellular levels of both histones H3 and H4 acetylation. The low histone acetylation in the Ccr4-Not mutants could be due to the disruption of interactions between Ccr4-Not components and the SAGA complex as well as direct effects on Gcn5 HAT activity.
The V-ATPases are a family of proton pumps that couple the energy of ATP hydrolysis to active proton transport across both intracellular and plasma membranes of eukaryotic cells. Each functional V-ATPase comprises a large peripheral domain (V1) responsible for ATP hydrolysis and a membrane integral domain (V0) for proton translocation (Nishi and Forgac 2002; Inoue et al. 2003). The V-ATPases are present in compartments such as vacuoles, endosomes, lysosomes, Golgi-derived and secretory vesicles, and even the plasma membrane in some eukaryotic cells, including certain tumor cells (Sennoune et al. 2004). By regulating intracellular pH, the V-ATPases play a variety of roles in protein sorting and transport, processing, degradation, and potentially post-translational modification, provide a driving force for uptake of ions, amino acids, and metabolites (Rothman et al. 1989; Nishi and Forgac 2002; Inoue et al. 2003). Of the VMA genes identified in this study, VMA2, VMA5, VMA8, and VMA13 are subunits of the V1 domain and VMA3, VMA11, and VMA16 are subunits of the V0 domain. The mechanism by which deletion of these genes affects histone acetylation may be through changes in intracellular pH that adversely affect the activity of HAT complexes. For instance, the glutamic acid 173 of Gcn5, an essential catalytic residue, must be deprotonated for proper HAT activity and the protonation state of Glu173 depends on the pH (Tanner et al. 1999). Consistent with this model, we found that deletion of VMA3, VMA5, or VMA8 genes results in reduction in HAT activity of Gcn5 on free histones and oligonucleosomes in vitro. Changes in pH may also lead to deregulated expression of many genes including HAT or HDAC complex members. However, at least for vma5 and vma8 mutants, we did not detect significant gene expression changes in known HATs, HDACs, or their associated factors (supplemental Figure 6), suggesting a more direct effect on Gcn5 HAT activity.
We also identified several genes involved in intracellular protein transport including VPS proteins. The VPS genes comprise a large family of >50 genes with known functions in protein transport within the intracellular membrane system and are necessary for correct sorting of proteins to various cellular compartments (Peterson and Emr 2001; Bowers and Stevens 2005). In our study, deletions of VPS6, VPS32, VPS34, VPS44, and VPS45 genes showed lower levels of histone acetylation (Table 1). Strikingly, Vps6, a target soluble N-ethylmaleimide sensitivity factor receptor (t-SNARE) protein, docks with Vps45, a vacuolar-SNARE (v-SNARE), for fusion of Golgi-derived vesicles with the prevacuolar compartment (Cowles et al. 1994; Bryant et al. 1998). Vps34 is a phosphatidylinositol 3-phosphate [PtdIns(3)P] kinase that helps form a membrane-associated signal transduction complex for proper protein sorting (Roth 2004). Many of the other identified genes in our screen are also involved in protein transport, including COG1, ANP1/MNN8, MNN10, ERP5, SEC28, and OCH1. Cog1 is an essential component of the conserved oligomeric Golgi (COG) complex that plays a key role in controlling Golgi-associated membrane trafficking and glycoconjugate processing (Oka et al. 2004, 2005; Fotso et al. 2005). Cog1 deficiency in human cells results in congenital disorder of glycosylation type II (Foulquier et al. 2006). Anp1/Mnn8 and Mnn10 are membrane protein subunits of the α-1,6 mannosyltransferase complex that is required for proper Golgi function in S. cerevisiae (Hashimoto and Yoda 1997; Jungmann et al. 1999). But how do these proteins affect histone acetylation? Disruption of intracellular trafficking could have indirect effects on histone acetylation but possible direct mechanisms could involve aberrant post-translational processing and/or inappropriate targeting of one or members of HAT complexes. However, we did not detect aberrant localization of Gcn5 in the vps6 deletion mutant (supplemental Figure 7). Although none of the Vps proteins identified in this study associates with known HAT complexes, it is noteworthy that the Rtt109 HAT is in a complex with the Vps75 protein, a NAP family histone chaperone with a proposed role in vacuolar protein sorting (Han et al. 2007b; Tsubota et al. 2007). Curiously, besides histone acetylation, disruption of the secretory/protein sorting pathway affects a variety of other nuclear events, such as nuclear morphology (Matynia et al. 2002; Teixeira et al. 2002), meiotic segregation (Marston et al. 2004), nuclear pore assembly and morphology (Bryant et al. 1998; Teixeira et al. 2002), and telomere length (Rog et al. 2005), as well as some other seemingly unrelated processes, such as ribosome biogenesis (Li and Warner 1996) and splicing (Vincent et al. 2003). The underlying mechanisms in all these cases remain unclear but changes in histone acetylation with subsequent effects on gene regulation may provide a common and testable explanation for the pervasive effects of the secretory pathway mutants on cellular processes.
Interestingly, orthologs of many of the genes identified in this study have been implicated in human and animal models of carcinogenesis. For instance, the human Ccr4-Not complex acts as a repressor of transcription by nuclear hormone receptors in breast cancer cell lines (Winkler et al. 2006). In colon cancer, a subunit of the Ccr4-Not complex, CNOT8, is overexpressed in primary and metastatic carcinomas relative to normal colon mucosa (Seiden-Long et al. 2006). Mice lacking Tob, a member of the antiproliferative gene family that interacts with the Ccr4-Not complex, are predisposed to cancer due to deregulated expression of cell cycle genes (Yoshida et al. 2003). The V-ATPases are important regulators of intracellular pH in tumor cells, which have acidic cytosols due to high glycolytic rate (Sennoune et al. 2004). The V-ATPases also contribute to the metastatic potential and drug resistance of tumor cells (Sennoune et al. 2004). The VPS genes are involved in a catabolic membrane-trafficking phenomenon termed autophagy that occurs in response to adverse environmental signals such as nutrient or hormonal deprivation and is linked to the carcinogenic potential (Abeliovich and Klionsky 2001). Finally, activity of ribonucleotide reductase including its large subunit, Rnr1 (identified in our screen), is crucial for rapidly dividing cells and is a target for anticancer and antiviral therapies (Xu et al. 2006). Taken together with our data, it is conceivable that the effect of these genes on cancer may be mediated partly through regulation of histone acetylation.
Our study has uncovered unexpected links between histone acetylation, Gcn5 HAT activity, and diverse cellular processes such as transcription, cellular ion homeostasis, and protein transport. As most of the proteins identified in this study, including the Ccr4-Not, V-ATPase, and VPS proteins, are highly conserved from fungi to humans, our data also provide useful information for understanding histone acetylation and gene regulation in human cells under both physiological and pathological conditions.
Acknowledgments
We thank Gregory Payne for providing the Saccharomyces cerevisiae library, Michael Grunstein for providing anti-H3K18ac and anti-H4K16ac antibodies, and Feng Xu for providing technical expertise with histone isolation. This research was supported by a Beckman Young Investigator Award to S.K.K.
References
- Abeliovich, H., and D. J. Klionsky, 2001. Autophagy in yeast: mechanistic insights and physiological function. Microbiol. Mol. Biol. Rev. 65 463–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson, J. D., M. Benson, P. M. Howley and K. Struhl, 1998. Association of distinct yeast Not2 functional domains with components of Gcn5 histone acetylase and Ccr4 transcriptional regulatory complexes. EMBO J. 17 6714–6722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein, B. E., E. L. Humphrey, R. L. Erlich, R. Schneider, P. Bouman et al., 2002. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc. Natl. Acad. Sci. USA 99 8695–8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas, D., Y. Yu, D. Mitra and D. J. Stillman, 2006. Genetic interactions between Nhp6 and Gcn5 with Mot1 and the Ccr4-Not complex that regulate binding of TATA-binding protein in Saccharomyces cerevisiae. Genetics 172 837–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers, K., and T. H. Stevens, 2005. Protein transport from the late Golgi to the vacuole in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1744 438–454. [DOI] [PubMed] [Google Scholar]
- Bryant, N. J., R. C. Piper, S. R. Gerrard and T. H. Stevens, 1998. Traffic into the prevacuolar/endosomal compartment of Saccharomyces cerevisiae: a VPS45-dependent intracellular route and a VPS45-independent, endocytic route. Eur. J. Cell Biol. 76 43–52. [DOI] [PubMed] [Google Scholar]
- Carrozza, M. J., B. Li, L. Florens, T. Suganuma, S. K. Swanson et al., 2005. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123 581–592. [DOI] [PubMed] [Google Scholar]
- Cha, T. L., B. P. Zhou, W. Xia, Y. Wu, C. C. Yang et al., 2005. Akt-mediated phosphorylation of EZH2 suppresses methylation of lysine 27 in histone H3. Science 310 306–310. [DOI] [PubMed] [Google Scholar]
- Collart, M. A., 2003. Global control of gene expression in yeast by the Ccr4-Not complex. Gene 313 1–16. [DOI] [PubMed] [Google Scholar]
- Cowles, C. R., S. D. Emr and B. F. Horazdovsky, 1994. Mutations in the VPS45 gene, a SEC1 homologue, result in vacuolar protein sorting defects and accumulation of membrane vesicles. J. Cell Sci. 107(12): 3449–3459. [DOI] [PubMed] [Google Scholar]
- Doyon, Y., and J. Cote, 2004. The highly conserved and multifunctional NuA4 HAT complex. Curr. Opin. Genet. Dev. 14 147–154. [DOI] [PubMed] [Google Scholar]
- Eberharter, A., S. John, P. A. Grant, R. T. Utley and J. L. Workman, 1998. Identification and analysis of yeast nucleosomal histone acetyltransferase complexes. Methods 15 315–321. [DOI] [PubMed] [Google Scholar]
- Fotso, P., Y. Koryakina, O. Pavliv, A. B. Tsiomenko and V. V. Lupashin, 2005. Cog1p plays a central role in the organization of the yeast conserved oligomeric Golgi complex. J. Biol. Chem. 280 27613–27623. [DOI] [PubMed] [Google Scholar]
- Foulquier, F., E. Vasile, E. Schollen, N. Callewaert, T. Raemaekers et al., 2006. Conserved oligomeric Golgi complex subunit 1 deficiency reveals a previously uncharacterized congenital disorder of glycosylation type II. Proc. Natl. Acad. Sci. USA 103 3764–3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant, P. A., A. Eberharter, S. John, R. G. Cook, B. M. Turner et al., 1999. Expanded lysine acetylation specificity of Gcn5 in native complexes. J. Biol. Chem. 274 5895–5900. [DOI] [PubMed] [Google Scholar]
- Gregory, P. D., and W. Horz, 1999. Mapping chromatin structure in yeast. Methods Enzymol. 304 365–376. [DOI] [PubMed] [Google Scholar]
- Han, J., H. Zhou, B. Horazdovsky, K. Zhang, R. M. Xu et al., 2007. a Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science 315 653–655. [DOI] [PubMed] [Google Scholar]
- Han, J., H. Zhou, Z. Li, R. M. Xu and Z. Zhang, 2007. b The Rtt109-Vps75 histone acetyltransferase complex acetylates non-nucleosomal histone H3. J. Biol. Chem. 282 14158–14164. [DOI] [PubMed] [Google Scholar]
- Hashimoto, H., and K. Yoda, 1997. Novel membrane protein complexes for protein glycosylation in the yeast Golgi apparatus. Biochem. Biophys. Res. Commun. 241 682–686. [DOI] [PubMed] [Google Scholar]
- Huang, W. C., and C. C. Chen, 2005. Akt phosphorylation of p300 at Ser-1834 is essential for its histone acetyltransferase and transcriptional activity. Mol. Cell. Biol. 25 6592–6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, T., S. Wilkens and M. Forgac, 2003. Subunit structure, function, and arrangement in the yeast and coated vesicle V-ATPases. J. Bioenerg. Biomembr. 35 291–299. [DOI] [PubMed] [Google Scholar]
- James, N., E. Landrieux and M. A. Collart, 2007. A SAGA-independent function of SPT3 mediates transcriptional deregulation in a mutant of the Ccr4-not complex in Saccharomyces cerevisiae. Genetics 177 123–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungmann, J., J. C. Rayner and S. Munro, 1999. The Saccharomyces cerevisiae protein Mnn10p/Bed1p is a subunit of a Golgi mannosyltransferase complex. J. Biol. Chem. 274 6579–6585. [DOI] [PubMed] [Google Scholar]
- Keogh, M. C., S. K. Kurdistani, S. A. Morris, S. H. Ahn, V. Podolny et al., 2005. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell 123 593–605. [DOI] [PubMed] [Google Scholar]
- Kuo, M. H., J. E. Brownell, R. E. Sobel, T. A. Ranalli, R. G. Cook et al., 1996. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature 383 269–272. [DOI] [PubMed] [Google Scholar]
- Kuo, M. H., E. vom Baur, K. Struhl and C. D. Allis, 2000. Gcn4 activator targets Gcn5 histone acetyltransferase to specific promoters independently of transcription. Mol. Cell 6 1309–1320. [DOI] [PubMed] [Google Scholar]
- Kurdistani, S. K., and M. Grunstein, 2003. a Histone acetylation and deacetylation in yeast. Nat. Rev. Mol. Cell. Biol. 4 276–284. [DOI] [PubMed] [Google Scholar]
- Kurdistani, S. K., and M. Grunstein, 2003. b In vivo protein-protein and protein-DNA crosslinking for genomewide binding microarray. Methods 31 90–95. [DOI] [PubMed] [Google Scholar]
- Kurdistani, S. K., D. Robyr, S. Tavazoie and M. Grunstein, 2002. Genome-wide binding map of the histone deacetylase Rpd3 in yeast. Nat. Genet. 31 248–254. [DOI] [PubMed] [Google Scholar]
- Kurdistani, S. K., S. Tavazoie and M. Grunstein, 2004. Mapping global histone acetylation patterns to gene expression. Cell 117 721–733. [DOI] [PubMed] [Google Scholar]
- Laribee, R. N., Y. Shibata, D. P. Mersman, S. R. Collins, P. Kemmeren et al., 2007. CCR4/NOT complex associates with the proteasome and regulates histone methylation. Proc. Natl. Acad. Sci. USA 104 5836–5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenssen, E., N. James, I. Pedruzzi, F. Dubouloz, E. Cameroni et al., 2005. The Ccr4-Not complex independently controls both Msn2-dependent transcriptional activation—via a newly identified Glc7/Bud14 type I protein phosphatase module—and TFIID promoter distribution. Mol. Cell. Biol. 25 488–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, B., and J. R. Warner, 1996. Mutation of the Rab6 homologue of Saccharomyces cerevisiae, YPT6, inhibits both early Golgi function and ribosome biosynthesis. J. Biol. Chem. 271 16813–16819. [DOI] [PubMed] [Google Scholar]
- Marston, A. L., W. H. Tham, H. Shah and A. Amon, 2004. A genome-wide screen identifies genes required for centromeric cohesion. Science 303 1367–1370. [DOI] [PubMed] [Google Scholar]
- Matynia, A., S. S. Salus and S. Sazer, 2002. Three proteins required for early steps in the protein secretory pathway also affect nuclear envelope structure and cell cycle progression in fission yeast. J. Cell Sci. 115 421–431. [DOI] [PubMed] [Google Scholar]
- Mizzen, C. A., J. E. Brownell, R. G. Cook and C. D. Allis, 1999. Histone acetyltransferases: preparation of substrates and assay procedures. Methods Enzymol. 304 675–696. [DOI] [PubMed] [Google Scholar]
- Nishi, T., and M. Forgac, 2002. The vacuolar (H+)-ATPases—nature's most versatile proton pumps. Nat. Rev. Mol. Cell. Biol. 3 94–103. [DOI] [PubMed] [Google Scholar]
- Oka, T., D. Ungar, F. M. Hughson and M. Krieger, 2004. The COG and COPI complexes interact to control the abundance of GEARs, a subset of Golgi integral membrane proteins. Mol. Biol. Cell 15 2423–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka, T., E. Vasile, M. Penman, C. D. Novina, D. M. Dykxhoorn et al., 2005. Genetic analysis of the subunit organization and function of the conserved oligomeric golgi (COG) complex: studies of COG5- and COG7-deficient mammalian cells. J. Biol. Chem. 280 32736–32745. [DOI] [PubMed] [Google Scholar]
- Peterson, M. R., and S. D. Emr, 2001. The class C Vps complex functions at multiple stages of the vacuolar transport pathway. Traffic 2 476–486. [DOI] [PubMed] [Google Scholar]
- Rog, O., S. Smolikov, A. Krauskopf and M. Kupiec, 2005. The yeast VPS genes affect telomere length regulation. Curr. Genet. 47 18–28. [DOI] [PubMed] [Google Scholar]
- Roth, M. G., 2004. Phosphoinositides in constitutive membrane traffic. Physiol. Rev. 84 699–730. [DOI] [PubMed] [Google Scholar]
- Rothman, J. H., C. T. Yamashiro, P. M. Kane and T. H. Stevens, 1989. Protein targeting to the yeast vacuole. Trends Biochem. Sci. 14 347–350. [DOI] [PubMed] [Google Scholar]
- Seiden-Long, I. M., K. R. Brown, W. Shih, D. A. Wigle, N. Radulovich et al., 2006. Transcriptional targets of hepatocyte growth factor signaling and Ki-ras oncogene activation in colorectal cancer. Oncogene 25 91–102. [DOI] [PubMed] [Google Scholar]
- Seligson, D. B., S. Horvath, T. Shi, H. Yu, S. Tze et al., 2005. Global histone modification patterns predict risk of prostate cancer recurrence. Nature 435 1262–1266. [DOI] [PubMed] [Google Scholar]
- Sennoune, S. R., K. Bakunts, G. M. Martinez, J. L. Chua-Tuan, Y. Kebir et al., 2004. Vacuolar H+-ATPase in human breast cancer cells with distinct metastatic potential: distribution and functional activity. Am. J. Physiol. Cell Physiol. 286 C1443–C1452. [DOI] [PubMed] [Google Scholar]
- Shahbazian, M. D., K. Zhang and M. Grunstein, 2005. Histone H2B ubiquitylation controls processive methylation but not monomethylation by Dot1 and Set1. Mol. Cell 19 271–277. [DOI] [PubMed] [Google Scholar]
- Smith, C. M., 2005. Quantification of acetylation at proximal lysine residues using isotopic labeling and tandem mass spectrometry. Methods 36 395–403. [DOI] [PubMed] [Google Scholar]
- Smith, C. M., P. R. Gafken, Z. Zhang, D. E. Gottschling, J. B. Smith et al., 2003. Mass spectrometric quantification of acetylation at specific lysines within the amino-terminal tail of histone H4. Anal. Biochem. 316 23–33. [DOI] [PubMed] [Google Scholar]
- Struhl, K., 1998. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 12 599–606. [DOI] [PubMed] [Google Scholar]
- Suka, N., Y. Suka, A. A. Carmen, J. Wu and M. Grunstein, 2001. Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol. Cell 8 473–479. [DOI] [PubMed] [Google Scholar]
- Tanner, K. G., R. C. Trievel, M. H. Kuo, R. M. Howard, S. L. Berger et al., 1999. Catalytic mechanism and function of invariant glutamic acid 173 from the histone acetyltransferase GCN5 transcriptional coactivator. J. Biol. Chem. 274 18157–18160. [DOI] [PubMed] [Google Scholar]
- Teixeira, M. T., B. Dujon and E. Fabre, 2002. Genome-wide nuclear morphology screen identifies novel genes involved in nuclear architecture and gene-silencing in Saccharomyces cerevisiae. J. Mol. Biol. 321 551–561. [DOI] [PubMed] [Google Scholar]
- Traven, A., A. Hammet, N. Tenis, C. L. Denis and J. Heierhorst, 2005. Ccr4-not complex mRNA deadenylase activity contributes to DNA damage responses in Saccharomyces cerevisiae. Genetics 169 65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubota, T., C. E. Berndsen, J. A. Erkmann, C. L. Smith, L. Yang et al., 2007. Histone H3–K56 acetylation is catalyzed by histone chaperone-dependent complexes. Mol. Cell 25 703–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent, K., Q. Wang, S. Jay, K. Hobbs and B. C. Rymond, 2003. Genetic interactions with CLF1 identify additional pre-mRNA splicing factors and a link between activators of yeast vesicular transport and splicing. Genetics 164 895–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelauer, M., J. Wu, N. Suka and M. Grunstein, 2000. Global histone acetylation and deacetylation in yeast. Nature 408 495–498. [DOI] [PubMed] [Google Scholar]
- Wang, L., C. Mizzen, C. Ying, R. Candau, N. Barlev et al., 1997. Histone acetyltransferase activity is conserved between yeast and human GCN5 and is required for complementation of growth and transcriptional activation. Mol. Cell. Biol. 17 519–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler, G. S., K. W. Mulder, V. J. Bardwell, E. Kalkhoven and H. T. Timmers, 2006. Human Ccr4-Not complex is a ligand-dependent repressor of nuclear receptor-mediated transcription. EMBO J. 25 3089–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson et al., 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285 901–906. [DOI] [PubMed] [Google Scholar]
- Xu, F., K. Zhang and M. Grunstein, 2005. Acetylation in histone H3 globular domain regulates gene expression in yeast. Cell 121 375–385. [DOI] [PubMed] [Google Scholar]
- Xu, H., C. Faber, T. Uchiki, J. Racca and C. Dealwis, 2006. Structures of eukaryotic ribonucleotide reductase I define gemcitabine diphosphate binding and subunit assembly. Proc. Natl. Acad. Sci. USA 103 4028–4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, Y., T. Nakamura, M. Komoda, H. Satoh, T. Suzuki et al., 2003. Mice lacking a transcriptional corepressor Tob are predisposed to cancer. Genes Dev. 17 1201–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, K., P. M. Yau, B. Chandrasekhar, R. New, R. Kondrat et al., 2004. Differentiation between peptides containing acetylated or tri-methylated lysines by mass spectrometry: an application for determining lysine 9 acetylation and methylation of histone H3. Proteomics 4 1–10. [DOI] [PubMed] [Google Scholar]