Abstract
The Hsp90/Hsp70 chaperone machine is an essential regulator of cell growth and division. It is required for activation of select client proteins, chiefly protein kinases and transcription activators and thus plays a major role in regulating intracellular signaling and gene expression. This report demonstrates, in vivo, the association of the Saccharomyces cerevisiae maltose-responsive transcription activator Mal63 (MAL-activator) with the yeast Hsp70 (Ssa1), Hsp90 (Hsp82), and Hop (Sti1) homologs, using a collection of inducible, constitutive, and noninducible alleles. Each class of mutant activator forms a distinctly different stable multichaperone complex in the absence of maltose. Inducible Mal63p associates with Ssa1, Hsp82, and Sti1 and is released in the presence of maltose. Noninducible mal63 mutant proteins bind to Ssa1 alone and do not stably associate with Hsp82 or Sti1. Constitutive MAL-activators bind well to Hsp82 and poorly to Ssa1 and Sti1, but deletion of STI1 restores Ssa1 binding. Taken together, Mal63p regulation requires the formation of Hsp90/Hsp70 subcomplexes comparable to, yet distinct from those observed with previously characterized Hsp90 clients including glucocorticoid receptor and yeast Hap1p. Thus, comparative studies of different client proteins highlight functional diversity in the operation of the Hsp90/Hsp70 chaperone machine.
HSP70 and Hsp90 are essential molecular chaperones that work together as a “relay team” in the activation, folding, subcellular localization, and maturation of selected, so-called client proteins. This process is referred to as the Hsp70/Hsp90 chaperone machine (reviewed in Pratt and Toft 1997, 2003; Wegele et al. 2004; Bukau et al. 2006; Picard 2006; Caplan et al. 2007). Hsp90 client proteins fall chiefly into two categories, kinases and DNA-binding transcription activators. Thus, Hsp90 plays a major role in regulating intracellular signaling and gene expression. The chaperone activity of Hsp90 is unique in that it acts on nascent, partially folded client proteins that are recruited by the sequential activity of Hsp40 and Hsp70. Hsp70/Hsp90 chaperones function in the cytosol, endoplasmic reticulum, and mitochondrion. Compartment-specific isoforms of Hsp70 and Hsp40 have been characterized in eukaryote species and, in Saccharomyces, 14 genes encode various members of the Hsp70 family. In addition to Hsp70 and Hsp90, a number of cochaperones facilitate aspects of the process including stimulation of chaperone ATPase activity, nucleotide exchange, or client protein binding. Genetic methods have been particularly useful in identifying Hsp90 cochaperones and, while the function of some cochaperones is well understood, the role of others remains unclear. The results presented here lay the groundwork for studies of cochaperone function in vivo using a native client protein.
Perhaps the most extensively studied Hsp90 client is the glucocorticoid receptor, a DNA-binding transcription activator and member of the steroid receptor family (reviewed in Pratt et al. 1996, 2004, 2006; Pratt and Toft 2003; Wegele et al. 2006). Minimal requirements for the sequential Hsp70/Hsp90 chaperone activation of glucocorticoid receptor have been defined, largely on the basis of in vitro studies with purified proteins, but significant insights have come from studies of other steroid receptors. Nascent receptor is first recognized by Hsp40, which mediates receptor association with Hsp70 to form the “early complex.” Transfer to Hsp90 occurs in the “intermediate complex” consisting of receptor, Hsp90, Hsp70, Hsp40, and Hop, a scaffold protein that facilitates intermediate complex formation by binding both Hsp90 and Hsp70. Finally, p23 and an immunophilin replace Hop and Hsp70 to form the so-called “final complex.” Glucocorticoid receptor is held in the final complex in an inactive conformation capable of binding hormone with high affinity. Hormone binding causes release of the activated receptor and dissociation of the final complex components. Thus, Hsp90 is both required for and a repressor of glucocorticoid receptor activation. In vivo studies of glucocorticoid receptor and other mammalian clients such as v-Src are frequently carried out in Saccharomyces, a heterologous host that may exhibit significant differences compared to the native mammalian host cell, such as diversity in the classes and function of the encoded cochaperones.
In vivo studies of Hsp90/Hsp70 chaperone regulation of Hap1p, the Saccharomyces heme-responsive transcription activator, revealed significant differences compared to the glucocorticoid receptor. Hap1 associates with Hsp70 in cells grown in both the presence and absence of the inducer heme while Hsp90 binding is transient and enhanced only in the presence of heme (Lan et al. 2004). Hsp70, as opposed to Hsp90, is the negative regulator of Hap1p. Moreover, Hap1p binds DNA while still bound to Hsp70 (Lan et al. 2004; Xin et al. 2007). When compared with the studies of the glucocorticoid receptor, these findings reveal an unexpected plasticity in the fundamental regulatory roles of the chaperones with regard to these two transcription activators and underscores the importance of extending studies of the Hsp90/Hsp70 chaperone machine to other transcription activator clients in vivo in the native host cell. To this end, this report investigates the pathway by which Mal63p, a maltose inducible Saccharomyces MAL-activator, is regulated by the Hsp90/Hsp70 chaperone machine.
MAL63 encodes the MAL-activator from MAL6 (Hu et al. 1999). Mal63p is a DNA-binding transcription activator required for the maltose-dependent induction of the MAL structural genes encoding maltose permease and maltase. Mal63 protein is an Hsp90 client protein (Bali et al. 2003). Mal63p is sensitive to degradation in cells depleted of Hsp90 and binds to Hsp90 in vivo, both hallmarks of Hsp90 chaperone client proteins. A wide variety of mutant MAL-activator alleles is available with different regulatory phenotypes including inducible, noninducible, superinducible, and constitutive MAL-activator alleles. This situation is unique among Hsp90 client proteins and provides a rich resource with which to explore the mechanism by which Hsp90/Hsp70 chaperone regulates another member of the transcription activator class of client proteins in the native host.
The analysis reported here is similar to a substrate-dependent epistasis analysis except that the various alleles of the MAL-activator substrate progress differently through the Hsp90 chaperone pathway forming distinct chaperone complexes. These findings allow us to propose the pathway for inducible MAL-activator activation via the Hsp90/Hsp70 chaperone cycle. Activation of the MAL-activator appears to parallel that observed in vitro for the glucocorticoid receptor, at least with regard to the major Saccharomyces chaperone components Ssa1 (cytosol Hsp70 homolog), Sti1 (Hop homolog), and Hsp82 (Hsp90 homolog). In the absence of inducer maltose, inducible Mal63p forms a stable complex with Ssa1, Hsp82, and Sti1. Results reported here and in Bali et al. (2003) show that optimal maltose induction is dependent on all three chaperone complex components indicating that all play a positive role in Mal63p induction. However, we find that MAL-activator constitutivity is correlated with a loss of Ssa1 binding and reduced dependency on Ssa1 suggesting that, in contrast to glucocorticoid receptor but similar to Hap1p, Ssa1 has a second role as the negative regulator of inducible Mal63 MAL-activator. This study highlights the need to study a variety of different transcription activator clients of the Hsp90/Hsp70 chaperone machine. Moreover, it lays the foundation for future in vivo analyses of the component composition of different client protein-containing complexes and provides the reagents needed to explore the role of the cochaperones in client protein activation in vivo.
MATERIALS AND METHODS
Yeast strains and plasmids:
Table 1 lists the Saccharomyces strains used in this study. Strain W303 (MATa leu2-3,112 ura3-1 trp1-1 his3-11,15 can1-100 GAL SUC2) carries naturally occurring defective copies of the MAL1 and MAL3 loci (Han et al. 1995). Both loci contain functional maltose permease and maltase genes, referred to as MAL11 (also known as AGT1) and MAL12, respectively, at MAL1; and MAL31 and MAL32, respectively, at MAL3. Sequences homologous to the MAL63 MAL-activator gene are found at both MAL1 and MAL3, referred to as mal13 and mal33, respectively, but these genes are nonfunctional. Thus, strain W303 and its derivatives do not ferment maltose and require a plasmid-borne copy of the MAL-activator gene for the expression of the MAL structural genes.
TABLE 1.
List of strains
| Strain | Genotype | Source |
|---|---|---|
| W303 | MATaleu2-3,112 ura3-1 trp1-1 his3-11,15can1-100 GAL SUC2 | S. Lindquist |
| hsc82Δ | Isogenic to W303 except hsc82Δ∷LEU2 | A. Duina |
| CMY8015 | Isogenic to W303 except STI1/Myc | This study |
| CMY1200 | Isogenic to hsc82Δ except HSP82/Myc | This study |
| CMY8001 | Isogenic to CMY1200 except sti1Δ∷HygR | This study |
| S153 | MATaleu2-3,112 ura3-1 trp1-1 ade2-1 can1-100GAL SUC hsc82Δ∷LEU2 hsp82Δ∷LEU2pGPD-hsp82-T1011 | D. Nathan |
| cpr7Δ | Isogenic to W303 except cpr7Δ∷HIS3 | A. Duina |
| hsc82Δ cpr7Δ | Isogenic to W303 except hsc82Δ∷LEU2cpr7Δ∷TRP1 | A. Duina |
| 5CG2 | MATα ura3-52 lys2-801 ade2-101 trp1-63 his3-200 leu2-1 hsc82∷URA3 hsp82∷GAL1-HSP82∷LEU2 | S. Lindquist |
| JN516 | MATaura3-52 leu2-3,112 his3-11 trp1Δ1 lys2 SSA1ssa2∷LEU2 ssa3∷TRP1 ssa4∷LYS2 hem1-Δ100 | E. Craig and L. Zhang |
| CMY1300 | Isogenic to JN516 except SSA1/Myc | This study |
| CMY8002 | Isogenic to JN516 except sti1Δ∷HygR | This study |
| 5B6 | MATaura3-52 leu2-3,112 his3-11 trp1Δ1 lys2ssa1∷HIS3 ssa2∷LEU2 SSA3 ssa4∷LYS2 hem1-Δ100 pGAL1-SSA1 | E. Craig and L. Zhang |
Strain JN516 (MATa ura3-52 leu2-3,112 his3-11 trp1Δ1 lys2 ssa2∷LEU2 ssa3∷TRP1 ssa4∷LYS2 hem1-Δ100) (from Elizabeth Craig and as modified by Li Zhang) contains deletions in SSA2, SSA3, and SSA4 providing SSA1 as the sole source of this class of Hsp70 (Becker et al. 1996; Lan et al. 2004). Strain 5B6 (MATα ura3-52 leu2-3,112 his3-11 trp1Δ1 lys2 ssa1∷HIS3 ssa2∷LEU2 SSA3 ssa4∷LYS2 hem1-Δ100 pGAL1-SSA1) (from Elizabeth Craig and as modified by Li Zhang) contains deletions in SSA1, SSA2, and SSA4 providing GAL1-dependent SSA1 expression from a URA3 plasmid vector (Becker et al. 1996; Lan et al. 2004). The SSA3 gene of strain 5B6 by itself is not capable of supporting growth and this strain is viable only on galactose medium in the absence of glucose. As described above for W303, strains JN516 and 5B6 carry only nonfunctional MAL-activator genes, mal13 and mal33, and a plasmid-borne copy of the MAL-activator gene is required for the expression of the MAL structural genes.
Plasmids YCp50-MAL63 and YCp50-MAL43-C carry the inducible MAL63 and constitutive MAL-activator gene MAL43-C, respectively, in vector YCp50 (Gibson et al. 1997).
Plasmid p416GPD from the Mumberg et al. (1995) series contains URA3 as the selectable marker but is otherwise the same as p414 (Bali et al. 2003). The 2-kb SacI–KpnI fragment containing the entire GPD promoter-MAL63/HA3-tagged fusion gene was subcloned into plasmid p416 to create plasmid p416GPD-MAL63/HA3. An untagged version, referred to as p416GPD-MAL63, was created by an in-frame deletion of the NotI fragment encoding the sequence triple-HA. W303 transformants carrying this plasmid induced maltase expression to normal levels and fermented maltose in 1 day.
Construction of the triple-HA-tagged mutant MAL-activators:
A PCR-based method was used to construct triple-HA-tagged versions of mal63 mutant alleles expressed from the GPD promoter. The fragment encoding the mutant 3′ half of the mal63 ORF was amplified using an upstream primer (5′-GGGGAATTCCTTCCCTTCGGTGAACAA-3′), which anneals to the EcoRI site at the codon 215/216, and a downstream primer (5′-GGGGTCGACCCCGGGATCGATGTGAACAATAAA-3′), which inserts ClaI and SmaI sites immediately following the mal63 termination codon and just before the SalI cutting site, respectively. The EcoRI cutting site in the upstream primer and the SalI site in the downstream primer are in boldface type. The amplified 0.7-kb product was digested with EcoRI and SalI and used to replace the 0.7-kb EcoRI and SalI fragment containing the 3′ end of the wild-type MAL63 gene in plasmid p416GPD-MAL63/HA3. The ClaI and SmaI (underlined) sites were inserted for the purpose of diagnostic digestion. The constructs were confirmed by sequencing the full open reading frame.
Construction of a triple-HA-tagged MAL63(1-283)-T247A:
The upstream primer (5′-GGGGAATTCCTTCCCTTCGGTGAACAA-3′), which contains an EcoRI site (in boldface) at codons 215/216 of MAL63, and the downstream primer (5′-CCCCGTCGACTTACTTTCCTGGTATAGTGAA-3′), which anneals to codons 278–283 of MAL63 but creates translation stop codon at 284 (underlined) followed by a SalI site (in boldface), were used to amplify a 200-bp fragment using the constitutive mutant MAL63-NS284-C (Gibson et al. 1997). The amplified product was digested with EcoRI and SalI and inserted into the vector fragment of EcoRI- and SalI-digested p416GPD-MAL63/HA3 to produce p416GPD-MAL63(1-283)-T247A/HA3.
Construction of 6His-tagged SSA1 plasmids:
The upstream primer (5′-GGG GGATCCATGCATCATCATCATCATCATGGGGGGTCAAAAGCTGTCGGTATTGAT-3′) contains a BamHI site (in boldface) and anneals to the 5′ end of the SSA1 open reading frame. It places six histidine codons (underlined) and two glycine codons at the 5′ end of SSA1 creating a 6His tag at the N-terminal end of Ssa1 protein. The downstream primer (5′-GGGCTCGAGTTAATCAACTTCTTCAACGGT-3′) contains an XhoI site (in boldface) and anneals to the 3′ end of the SSA1 open reading frame. The stop codon is underlined. This primer pair was used to amplify the SSA1 open reading frame from the pGAL1-SSA1 plasmid carried by strain 5B6. The amplified product was digested with BamHI and XhoI and subcloned into BamHI- and XhoI-digested p414GPD and p414TEF, both CEN vectors that contain TRP1, to create p414GPD-SSA1/6His and p414TEF-SSA1/6His (Mumberg et al. 1995).
A 6His-tagged allele of SSA1 expressed from its native promoter was constructed as follows. Upstream primer (5′-GGGGGATCCATGCATCATCATCATCATCATGGGGGGTCAAAAGCTGTCGGTATTGAT-3′) contains a BamHI site (in boldface) and anneals to the 5′ end of the SSA1 open reading frame. It places six histidine codons (underlined) and two glycine codons in frame at the 5′ end of SSA1, creating a 6His tag at the N-terminal end of Ssa1 protein. The downstream primer (5′-GGGGCATGCGTTAGCGATAATCAAGAAGTG-3′) contains a SphI site (in boldface) and anneals to the 3′ end of the SSA1 open reading frame. The stop codon is underlined. The SSA1 promoter was amplified from the pGAL1-SSA1 plasmid carried by strain 5B6 using the following primer pair. Primer (5′-GGGGAGCTCCAAAGGCTCGGTTGTCGACAA-3′) contains a SacI site (in boldface) and anneals to a site ∼600 bp upstream of the SSA1 open reading frame. Primer (5′-GGGGGATCCATTATCTGTTATTTACTTGAA-3′) contains a BamHI site (in boldface) and anneals to a site immediately upstream of the SSA1 open reading frame. The SSA1 promoter was amplified using this PRO primer pair from the genome of S288C. The ORF amplified product was digested with BamHI and SphI and the PRO amplified product was digested with BamHI and SacI and the two digested products cloned into SphI–SacI-digested pUN30 (Elledge and Davis 1988). This created plasmid pUN30-SSA1/6His, a TRP1 CEN plasmid.
Addition of a Myc epitope tag to HSP82, SSA1, and STI1:
The Myc epitope consists of 10 amino acids (EQKLISEEDL) derived from the protein sequence of the human proto-oncogene p62-Myc. The sequence encoding one copy of the Myc tag was inserted at the 3′ end of the ORF of the genomic copy of HSP82, SSA1, and STI1 by a PCR-based technique described below thereby creating a C-terminal Myc-tagged allele expressed from the native promoter. Despite the fact that each gene is the sole copy of the chaperone or cochaperone present in the strain, the Myc-tagged strains do not exhibit any growth defects or defects in maltase expression (see data below) indicating that tags do not significantly alter the function of the tagged gene.
To construct HSP82/Myc, the kanR gene cassette was amplified using the following primers. The upstream primer contains the following sequences in the order presented: 45 bp upstream of the TAA stop codon of HSP82, 2 glycine codons, 30 bp of sequence encoding the Myc epitope, a stop codon, and an 18-bp sequence complementary to the 5′ end of the kanR cassette (5′- CCGGTTGAAGAGGTTCCAGCTGACACCGAAATGGAAGAGGTAGAT GGGGGGAACAAAAACTTATTTCTGAAGAAGATCTGTAGCAGCTGAAGCTTCGTACG-3′). The sequence encoding the Myc tag is underlined; bases homologous to the template are in boldface. The downstream primer contains 45 bp from the 3′ end of HSP82 followed by 18 bp of sequence from the 3′ end of the kanR cassette (5′-CATTGTAATGTTTTACCCAGTTATTTC CATGCAGATGCCCTATTTACGCATAGGCCACTAGTGGA-3′). Bases homologous to the template are in boldface. Plasmid pFA6-kanMX2 was used as a template to amplify the kanamycin (G418) resistance gene (Wach et al. 1994). The PCR product was transformed directly into strain W303 hsc82Δ HSP82 and transformants were selected on YPD supplemented with 200 μg/ml geneticin (catalog no. 10131-035; Invitrogen Life Technologies, Carlsbad, CA) (Guldener et al. 1996). Epitope tagging of HSP82 was confirmed by PCR and Western blotting. When transformed with pUN30-MAL63, maltose-induced expression of maltase in the tagged strain, CMY1200, was not significantly different from that in the untagged parental strain (368 ± 38 units vs. 367 ± 57 units, respectively).
SSA1 and STI1 were Myc tagged by a similar procedure. For SSA1, the upstream primer (5′-TCCAGCTCCAGAGGCTGAAGGTCCAACCGTTGAAGAAGTTGATGGGGGGGAACAAAAACTTATTTCTGAAGAAGATCTGTAACAGCTGAAGCTTCGTACG-3′) and downstream primer (5′-TTCCTCATTATACCCAGATCATTAAAAGACATTTTCGTTATTAT CAATTGCGCATAGGCCACTAGTGGA-3′) were used to amplify the kanR cassette. The PCR product was transformed directly into strain JN516 (from Elizabeth Craig), which contains deletions of SSA2, SSA3, and SSA4 leaving SSA1 as the only Hsp70 gene. When transformed with pUN30-MAL63, maltose-induced expression of maltase in the tagged strain, CMY1300, was not significantly different from that in the untagged parental strain (1201 ± 177 units vs. 1313 ± 203 units, respectively).
For STI1, the upstream primer (5′-CAGACGTTGATCGCTGCTGGTATCATCCGGACTGGCCGCGGGGGGGAACAAAAACTTATTATTTCTGAAGAAGATCTGTAACAGCTGAAGCTTCGTACG -3′) and downstream primer (5′-TTTCCCTCGCCCAAGAAACTTATTATCTTCAAGTTCCGATTTCTCAAAGCATAGGCCACTAGTGGA-3′) were used to amplify the kanR cassette. The PCR product was transformed directly into strain W303. When transformed with pUN30-MAL63, maltose-induced expression of maltase in the tagged strain, CMY8015, was not significantly different from that in the untagged parental strain (1215 ± 190 units vs. 1240 ± 239 units, respectively).
Construction of sti1Δ∷HygR strains:
Deletion of STI1 in strains CMY1200 and CMY1300 carrying Myc-tagged HSP82/Myc and SSA1/Myc, respectively, was complicated by the presence of the kanR cassette at each of these tagged genes. One-step gene disruption to replace STI1 with the hygromycin resistance gene hygR was accomplished by replacing the open reading frame of STI1 with the coding region of hygR without including homologous sequences derived from the kanR cassette. The upstream primer (5′-GCCCAAAAGTCTGCTCCCAAA TTCCTCACTGTAGCTACTAAAACAACCTATACGCAAGAAAGATGGGTAAAAAGCCTGAACTCA-3′) consists of 62 bp of the STI1 promoter sequence immediately upstream of the start codon (ATG) of STI1 followed by 22 bp of the hygR open reading frame (boldface). The downstream primer (5′-ATGTATGAAAAAGCAGTAAAAAAAGAATTCAAGATAATAAAGTTATATTTCGTATTATTTTTATTCCTTTGCCCTCGGACG-3′) consists of 60 bp of sequence homologous to the region immediately downstream of STI1 and 21 bp homologous to the 3′ end of the hygR open reading frame (boldface) including the stop codon. This primer pair was used to amplify the hygR open reading frame from the template plasmid pAG32 (Goldstein and McCusker 1999). The PCR product was transformed directly into strains CMY1200 and CMY1300 and hygromycin-resistant transformants were selected on YPD supplemented with 200 μg/ml hygromycin (catalog no. 10687-010, Invitrogen Life Technologies). Constructs were confirmed by PCR and Western blotting.
Maltase activity:
Extracts were prepared and maltase activity was measured in whole cell extracts as described in Dubin et al. (1985) and Hu et al. (1999). Activity is expressed as nanomoles of p-nitrophenol-α-glucopyranoside (PNPG) hydrolyzed per minute per milligram of protein.
Preparation of cell extracts, immunoblot analysis, and co-immunoprecipitation:
Strains were grown in the appropriate selective minimal medium to midlogarithmic phase (OD600 of 0.2–0.5). Denaturing cell extracts were prepared as described in Bali et al. (2003). Western blot analysis was carried out using standard methods and the proteins detected using the Vistra ECF kit (Amersham, Piscataway, NJ) in which the secondary antibody is conjugated to a fluorescent dye. The signal was visualized using a Molecular Dynamics Storm 860 and quantified using software provided by the manufacturer. Anti-Myc antibody was obtained from Roche Applied Science (Indianapolis). PGK (phosphoglycerol kinase) was detected by anti-PGK antibody from Molecular Probes (Eugene, OR).
Nondenaturing total cell extracts were prepared as follows. An aliquot of the culture containing ∼20 OD units of cells was harvested by filtration, washed with 50 mm KPO4 buffer pH 7.4 plus 2% sodium azide, and frozen while still on the filter paper at −80° for at least 20 min. Cells were resuspended in 1 ml of a nondenaturing extraction buffer containing 50 mm sodium molybdate, 20 mm HEPES, pH 7.5, 150 mm NaCl, 10% glycerol, 1 mm EDTA plus a cocktail of protease inhibitors [Roche, complete, mini, EDTA-free protease inhibitor tablets (catalog no. 1836170), yeast protease inhibitor cocktail (catalog no. P8215, Sigma, St. Louis), and a toothpick tip-full of sodium bisulfite], and flash-frozen in liquid nitrogen. Protein extracts were made via glass bead lysis. Protein concentration of the cell extract was determined by the Lowry assay method and extraction buffer added to equalize protein concentration and volume. A sample was removed for Western blot analysis (referred to as extract) as described above.
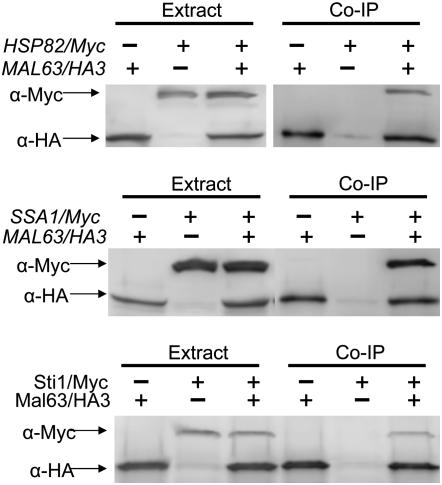
Immunoprecipitation was carried out as follows. Approximately 200 μl of lysate were then combined with 30–40 μl of anti-HA agarose slurry (Roche anti-HA affinity matrix, catalog no. 11 815 01 6001) and incubated with gentle mixing at 4° overnight. The beads were washed three times with 300 μl of cold extraction buffer, 20 μl of cold extraction buffer, and 40 μl of 2× sample buffer were added to the beads, and the suspension incubated at 75° in a heating block for 15 min to extract the bead-bound protein. Samples were analyzed by Western blot analysis probed with anti-HA antibody and anti-Myc antibody (Roche) and the signals quantified as described above. The results in Figure 1 demonstrate that the Myc-tagged proteins encoded by SSA1/Myc, HSP82/Myc, and STI1/Myc do not bind to the anti-HA agarose beads in the absence of HA-tagged MAL-activator.
Figure 1.—
Myc-tagged Hsp82, Ssa1, and Sti1 do not immunoprecipitate in the absence of HA-tagged Mal63. Strains carrying the genomic Myc-tagged alleles and the parental strains lacking the Myc-tagged HSP82 (strains hsc82Δ and CMY1200), SSA1 (strains JN516 and CMY1300), and STI1 (strains W303 and CMY8015) were transformed with plasmid p416GPD-MAL63/3HA carrying the triple-HA-tagged MAL63 or p416GPD-MAL63 carrying the untagged MAL63. Transformants were grown to midlog at 30° in selective synthetic medium lacking uracil and containing 3% glycerol, 2% lactate. Nondenaturing protein extracts were prepared and incubated with anti-HA bound agarose beads as described in materials and methods. Co-immunoprecipitation (Co-IP) samples and the total cell extracts from which they were prepared were analyzed by Western blotting using anti-HA and anti-Myc antibodies. Experiments were done at least in triplicate and representative blots are shown.
RESULTS
Strains with reduced Hsp70 expression exhibit defects in maltase expression:
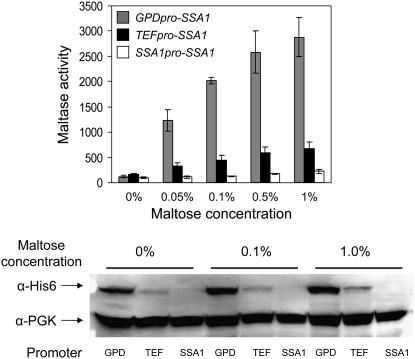
Bali et al. (2003) reported that very low concentrations of maltose restricted growth of strains defective for Hsp90 molecular chaperone complex, including the temperature-sensitive strain hsc82Δ hsp82-T101I and the double-null strain hsc82Δ cpr7Δ. Induction of maltase expression in these strains requires higher maltose concentrations and does not achieve wild-type induced levels (data not shown). Figure 2 explores the role of Ssa1 in MAL-activator regulation. SSA1-4 encoding Saccharomyces Hsp70 are essential (Becker et al. 1996). A series of strains expressing varying levels of Ssa1 protein were constructed that express a plasmid-borne N-terminal-tagged SSA1/6His gene fused to the GPD, TEF, and native SSA1 promoters in a strain lacking genomic Ssa1, -2, -4 expression (ssa1Δ ssa2Δ SSA3 ssa4Δ). The impact of reduced Ssa1 levels on MAL63-dependent maltase expression was assayed (Figure 2). The turnover rate and expression level of Mal63/HA3p were determined in the strains carrying the GPD- and TEF-expressed SSA1 and no effect of the reduced level of Hsp70 was observed (data not shown).
Figure 2.—
Maltase expression is dependent on Ssa1 chaperone. Plasmid series p414GPD-SSA1/6His, p414TEF-SSA1/6His, and pUN30-SSA1/6His, expressing 6His-tagged SSA1/6His from the GPD, TEF, and native SSA1 promoters were used to replace the pGAL1-SSA1 plasmid of strain 5B6 (ssa1∷HIS3 ssa2∷LEU2 SSA3 ssa4∷LYS2 pGAL1-SSA1). This strain series was transformed with plasmid pUN90-MAL63/HA. Transformants were grown at 30° in selective synthetic medium lacking uracil and tryptophan and containing 3% glycerol and 2% lactate (v/v) plus the indicated concentration of maltose. Maltase activity was assayed as described in materials and methods and is expressed as nmoles PNPG (p-nitrophenol-α-glucopyranoside) produced per milligram of protein per minute (top). Assays were carried out on at least three independent transformants. The error bars indicate standard deviation from three independent experiments done in duplicate. Western blot analysis was carried out as described in materials and methods on total cell extracts prepared from transformants grown at the indicated maltose concentration (bottom). Blots were probed with anti-6His antibody and anti-PGK antibody was used as a loading control. Analysis was carried out on three independent transformants; a representative blot is shown.
Maximal SSA1 expression is observed from the GPD promoter and expression from the SSA1 promoter is the lowest of the three constructs (bottom), although growth rates are not particularly affected (data not shown). Decreased expression of Ssa1p severely affects maltase expression at all inducer concentrations and maltase activity correlates with the level of Ssa1p expression (top). While SSA3 expression may be upregulated in response to the extremely low levels of SSA1 expression from the construct with its native promoter, it clearly is not sufficient to complement the defect in SSA1 expression, a finding consistent with the fact that strains carrying only SSA3 are inviable (Becker et al. 1996). We conclude that Ssa1 is required for MAL-activator activation and plays a positive role in Mal63 MAL-activator activation.
Binding of inducible Mal63/3HA MAL-activator to Hsp90 and Hsp70 is regulated by maltose:
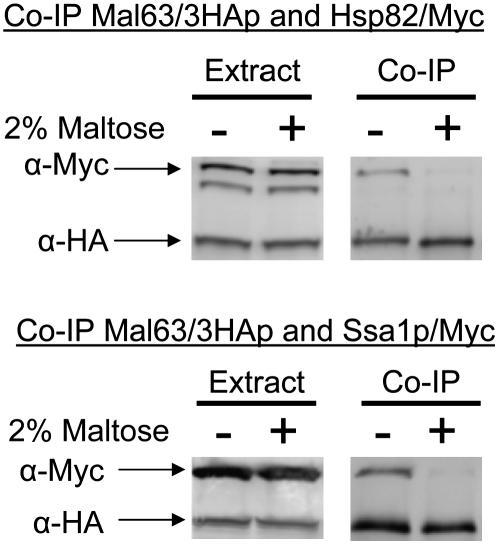
The results in Figure 2 predict that the MAL-activator should be found in association with Hsp70. Bali et al. (2003) demonstrated that Mal63/3HA MAL-activator coprecipitates with Hsp90 but in this study the His-tagged HSP82/6His was overexpressed from the GPD promoter and was carried on a multicopy plasmid. We decided to explore MAL-activator interaction in strains expressing Ssa1 and Hsp82 at normal levels from their native promoters. For this, the genomic copy of SSA1 and HSP82 was tagged in strains JN516 and hsc82Δ, respectively. The tag was placed at the C terminus of the open reading frame by a one-step gene insertion. The SSA1/Myc and HSP82/Myc gene in each strain is the sole source of the essential chaperone and appears by several measures to be fully functional. Both Myc-tagged strains grow normally and express maltase at rates comparable to the untagged parent (see materials and methods). Additionally, glucocorticoid receptor activation is unaffected in the strain containing SSA1/Myc as the sole source of this Hsp70 (K. Morano, unpublished results). The C-terminal TPR-binding EEVD domains of Hsp90 and Hsp70 are essential for interaction with Sti1 (Scheufler et al. 2000; Brinker et al. 2002; Odunuga et al. 2003). Our results indicate that, despite the proximity of the Myc epitope to the EEVD domain and its location at the C terminus, the Myc-tag does not interfere in any measurable way with Sti1 interaction. Moreover, because HSP82/Myc and SSA1/Myc are carried in the genome and expressed from their native promoter, expression levels should be relatively constant from cell to cell. Thus, we are able to detect differences in the amount of Mal63/3HA activator bound with Myc-tagged chaperone by quantifying the amount of chaperone that coprecipitates with the Mal63/3HA activator and normalizing the intensity of the Myc signal to the HA signal. Control experiments shown in Figure 1 demonstrate that neither Myc-tagged Hsp82/Myc nor Ssa1/Myc proteins bind to the HA-sepharose beads in the absence of HA-tagged Mal63 MAL-activator.
Plasmid-borne MAL63/3HA was coexpressed in strains containing the genomic Myc-tagged HSP82/Myc or SSA1/Myc and binding of Mal63 protein to the chaperones determined by co-immunoprecipitation of the chaperone with the triple-HA-tagged Mal63p. As can be seen in Figure 3, Mal63/3HA MAL-activator binds to Ssa1 and Hsp82 when cells are grown under uninduced conditions. It should be noted that expression of Mal63/3HA and the chaperones does not appear to be affected by growth in the presence or absence of maltose. In contrast to the results with cells grown under uninduced conditions, association of the chaperones with Mal63/3HA is dramatically reduced by growth in the presence of 2% maltose. This result is consistent with our finding that Ssa1 plays a positive role in MAL-activator activation. Moreover, it indicates that growth under induced conditions leads to the release of the activator from its association with both Ssa1 and Hsp82 chaperones.
Figure 3.—
Co-immunoprecipitation of inducible Mal63/3HA MAL-activator protein with Ssa1 and Hsp82. Strains CMY1200 (HSP82/Myc hsc82Δ) (top) and CMY1300 (SSA1/Myc ssa2Δ ssa3Δ ssa4Δ) (bottom) were transformed with the plasmid p416GPD-MAL63/3HA harboring the triple-HA-tagged inducible MAL63 MAL-activator gene. Transformants were grown to midlog at 30° in selective synthetic medium lacking uracil and containing 3% glycerol, 2% lactate, with or without 2% maltose. Nondenaturing protein extracts were prepared as described in materials and methods. Co-immunoprecipitation (Co-IP) samples and the total cell extracts from which they were prepared were analyzed by Western blotting using anti-HA and anti-Myc antibodies. Experiments were done at least in triplicate and representative blots are shown.
Noninducible Mal63/3HA mutant MAL-activator proteins are defective in their ability to bind to Hsp90:
Given these results for inducible Mal63 MAL-activator, we asked if mutant MAL-activators exhibited differences in their interactions with Hsp70, Hsp90, and other components of the chaperone complex and if these differences were reflected in their mutant phenotype. For this, we used a variety of noninducible and constitutive MAL-activator mutants characterized by Gibson et al. (1997) and Danzi et al. (2000, 2003). Table 2 lists the phenotype and the mutant alterations of each compared to inducible Mal63 MAL-activator. Danzi et al. (2003) isolated the following noninducible MAL-activator alleles by charged cluster-to-alanine mutagenesis: mal63-283, mal63-331, mal63-364, mal63-401, and mal63-467A9N. Noninducible mal63-467S9V was obtained by random mutagenesis of the C-terminal codons of MAL63 (Danzi et al. 2003).
TABLE 2.
MAL-activator mutant alleles used in this study
| MAL-activator allele | Mutant alterations compared to Mal63p | Reference |
|---|---|---|
| Inducible | ||
| MAL63 | None | Kim and Michels (1988) |
| Noninducible | ||
| mal63-283 | K283A, D287A | Danzi et al. (2000) |
| mal63-331 | R331A, R335A | Danzi et al. (2000) |
| mal63-364 | E364A, R367A | Danzi et al. (2000) |
| mal63-401 | D401A, K405A | Danzi et al. (2000) |
| mal63-467A9N | D467A, I469N | Danzi et al. (2000) |
| mal63-467S9V | D467S, I469V | Danzi et al. (2000) |
| Constitutive | ||
| MAL43-C | 31 alterations compared to Mal63p, 27 of which lie in residues 238–461 | Gibson et al. (1997) |
| MAL63/43-C | 27 alterations compared to Mal63p mapping to residues 238–461 | Gibson et al. (1997) |
| MAL63(1-283)-T247A | C284NS, T247A | Gibson et al. (1997) |
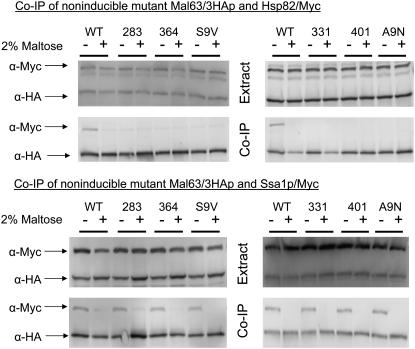
Triple-HA-tagged alleles of each of the noninducible mal63 mutants were coexpressed in strains carrying genomic Myc-tagged HSP82/Myc and SSA1/Myc and interaction between the mutant mal63 proteins and the chaperones determined. The results in Figure 4 demonstrate that the noninducible MAL-activator mutant proteins are expressed at levels comparable to inducible Mal63p but exhibit severe defects in their ability to bind Hsp90. The noninducible MAL-activator mutant proteins form complexes with Ssa1 chaperone apparently normally. That is, the noninducible mutant proteins bind to Ssa1/Myc chaperone in cells grown under uninduced but not under induced conditions. In contrast, the noninducible mutant activators do not form a complex with Hsp82/Myc even in cells grown under uninduced conditions. It should be noted that the noninducible mutants used in this study map to sites spanning nearly the full length of the regulatory region of Mal63p from residues 283 to 467 and each mutation alters two to three residues within a five-residue window. Thus, it is unlikely that this lack of Hsp82 binding could be explained by a nonspecific phenomenon such as the gross misfolding of the entire region. We conclude, consistent with the findings of Bali et al. (2003), that association with Hsp90 is required for MAL-activator activation and that, for the mutants studied here, the noninducible phenotype results from an inability to form the intermediate complex and bind Hsp90 chaperone. Additionally, this result indicates that binding of the MAL-activator to Hsp70 occurs independent of stable Hsp90 binding and prior to association with Hsp90 in an intermediate complex.
Figure 4.—
Co-immunoprecipitation of non-inducible MAL-activator mutant alleles with Ssa1 and Hsp82. Strains CMY1200 (HSP82/Myc hsc82Δ) (top) and CMY1300 (SSA1/Myc ssa2Δ ssa3Δ ssa4Δ) (bottom) were transformed with plasmids p416GPD-mal63-283, p416GPD-mal63-331, p416GPD-mal63-364, p416GPD-mal63-401, p416GPD-mal63-467S9V, and p416GPD-mal63-467A9N harboring the triple-HA-tagged alleles of the noninducible MAL-activator mutant genes (Danzi et al. 2003). Transformants were grown as described in Figure 3. Preparation of nondenaturing extracts, co-immunoprecipitation, and Western blot analysis were carried out as described for Figure 3.
Constitutive MAL-activators require Hsp90 but not Hsp70:
Strains defective for Hsp90 and Hsp70 were used to determine whether constitutive MAL-activator alleles are dependent on Hsp90 and Hsp70, as is seen for inducible MAL63. Constitutive MAL-activator mutants activate high levels of MAL gene expression in the absence of inducer maltose, even in the presence of glucose (Gibson et al. 1997). MAL63/43-C is a hybrid MAL-activator containing the first 215 codons of MAL63 fused to codons 216–470 of MAL43-C (Gibson et al. 1997). Mal63p and Mal43-Cp encode 470 residue proteins that differ at 31 residues, 27 of which are in the C-terminal region of the Mal63/43-C fusion protein from residues 238 to 461. Both MAL43-C and MAL63/43-C exhibit similar, if not identical, phenotypes. Of these 27 alterations, 10 and 8, respectively, are found clustered into two putative negative regulatory domains in residues 343–359 and 419–461 and either cluster of alterations is sufficient to produce the constitutive phenotype when introduced into inducible Mal63p (Danzi et al. 2000). MAL63(1-283)-T247A was isolated by random mutagenesis of the noninducible nonsense mutation mal63/NS284 and contains a single base change, T247A, that converts it to a constitutive activator (Gibson et al. 1997).
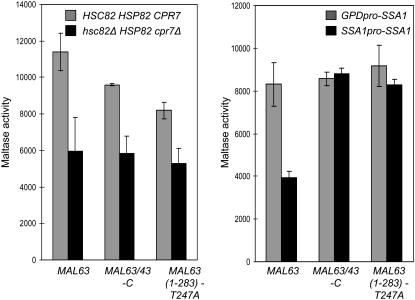
To test dependence on Hsp90, plasmid constructs carrying MAL63 or the two constitutive mutant genes expressed from the high-level GPD promoter were introduced into the strains W303 (HSC82 HSP82) and hsc82Δ cpr7Δ (hsc82Δ HSP82 cpr7Δ) and the affect on maltase expression was determined (Figure 5, left). All three transformed strains expressed very high levels of maltase ∼5- to 8-fold higher than the induced levels of maltase expression in strains carrying the inducible MAL63 expressed from its native promoter. This is likely due to the high level of MAL-activator expression from the GPD promoter. Nonetheless, maltase expression is decreased ∼30–50% in the double-deletion strain hsc82Δ cpr7Δ carrying either MAL63 or the constitutive mutant MAL-activator alleles. Comparable results were obtained using the MAL63 or MAL43-C genes expressed from their native promoters in this Hsp90 defective strain (data not shown). Thus, MAL gene activation by both the inducible and constitutive MAL-activators is similarly dependent on Hsp90 chaperone activity.
Figure 5.—
Maltase expression in strains carrying constitutive MAL-activators is dependent on Hsp90 but not Hsp70. Strains W303 (HSC82 HSP82) and hsc82Δ cpr7Δ (hsc82Δ HSP82 cpr7Δ) (left) were transformed with plasmids p416GPD-MAL63/3HA, carrying the triple-HA-tagged inducible MAL63 or p416GPD-MAL63/43-C and p416GPD-MAL63(1-283)-T247A, carrying the triple-HA-tagged constitutive MAL-activator mutant genes MAL63/43-C and MAL63(1-283)-T247A, respectively (Gibson et al. 1997; Danzi et al. 2000). The strains expressing SSA1 from the GPD and native SSA1 promoters described above in Figure 2 were transformed with plasmids p416GPD-MAL63/3HA, p416GPD-MAL63/43-C, and p416GPD-MAL63(1-283)-T247A (right). Transformants were grown at 30° in selective synthetic medium lacking the appropriate nutrients for plasmid selection and containing 3% glycerol and 2% lactate (v/v) with 1% maltose added to transformants carrying MAL63/3HA. Maltase activity was assayed on at least three independent transformants as described in Figure 2.
Dependency of the constitutive MAL-activators on Hsp70 was tested using the strain series expressing different levels of SSA1/His described above. Much to our surprise, maltase expression in the strains carrying the constitutive MAL-activator alleles is not affected by reduced Ssa1 expression while inducible maltase expression is ∼50% reduced in the strain with low levels of Ssa1. A more pronounced effect on maltase expression is observed in Figure 2 where Mal63p is expressed at a lower level from its native promoter suggesting that the apparent independence from Hsp70 of the constitutive alleles may be buffered somewhat by high MAL-activator expression but similar results were obtained with the native MAL43-C carried on a CEN plasmid (data not shown). Nonetheless, these results suggest that the constitutive MAL-activators are significantly less dependent than inducible Mal63p on Hsp70 for activation/maturation. On the basis of the findings shown in Figure 5, we investigated constitutive MAL-activator binding to Hsp70 and Hsp90.
Altered binding of constitutive MAL-activator mutant proteins to Hsp70:
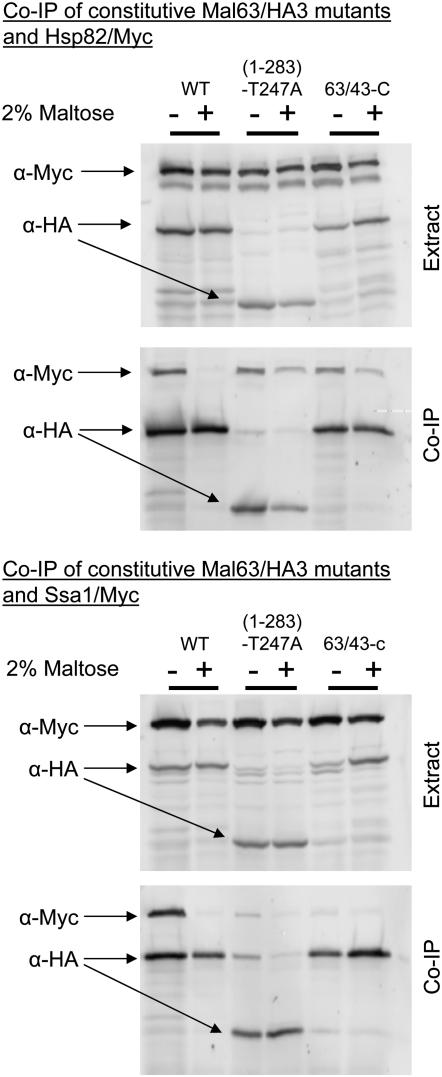
The constitutive MAL-activators encoded by MAL43/63-C and MAL63(1-283)-T247A were tested for their ability to associate with Hsp70 and Hsp90. The results are shown in Figure 6. Consistent with their dependency on Hsp90, both of the constitutive MAL-activator proteins bind Hsp82/Myc. The finding that the truncated Mal63(1-283)-T247A protein binds well to Hsp90 indicates that the full-length protein is involved in this interaction not just the C-terminal regulatory region. Some residual binding of Hsp82/Myc with the constitutive MAL-activator proteins is sometimes observed even in cells grown in maltose, although this is usually quite modest (see also Figure 8).
Figure 6.—
Co-immunoprecipitation of constitutive MAL-activator mutant proteins with Ssa1 and Hsp82. Strains CMY1200 (HSP82/Myc hsc82Δ) (top) and CMY1300 (SSA1/Myc ssa2Δ ssa3Δ ssa4Δ) (bottom) were transformed with the plasmids p416GPD-MAL63/43-C and p416GPD-MAL63(1-283)-T247A harboring the triple-HA-tagged constitutive MAL-activator mutant genes MAL63/43-C and MAL63(1-283)-T247A. Transformants were grown as described in Figure 3. Preparation of nondenaturing extracts, co-immunoprecipitation, and Western blot analysis were carried out as described for Figure 3.
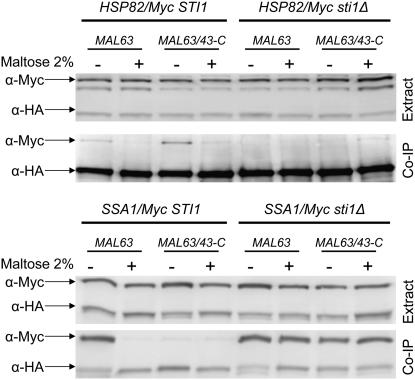
Figure 8.—
Effect of sti1Δ on the co-immunoprecipitation of inducible Mal63/3HA MAL-activator with Ssa1 and Hsp82. Strains CMY1200 (STI1 HSP82/Myc hsc82Δ), CMY8001 (sti1Δ HSP82/Myc hsc82Δ), CMY1300 (STI1 SSA1/Myc ssa2Δ ssa3Δ ssa4Δ), and CMY8002 (sti1Δ SSA1/Myc ssa2Δ ssa3Δ ssa4Δ) were transformed with plasmids p416GPD-MAL63/3HA or p416GPD-MAL63/43-C carrying triple-HA-tagged MAL63 and MAL63/43-C, respectively. Transformants were grown as described in Figure 3. Preparation of nondenaturing extracts, co-immunoprecipitation, and Western blot analysis were carried out as described for Figure 3.
The results for Hsp70 binding are quite different but again consistent with the apparent lack of dependency on Hsp70 reported in Figure 5. Binding of the constitutive MAL-activator proteins to Ssa1/Myc is very weak or nonexistent, even in extracts prepared from cells grown under uninduced conditions, and addition of maltose to the growth medium further reduces association of the constitutive activators to Ssa1/Myc Hsp70 (see also Figure 8). Therefore, in the absence of inducer, constitutive activators form a stable complex containing Hsp82 but not Ssa1.
Constitutive MAL-activator is dependent on STI1:
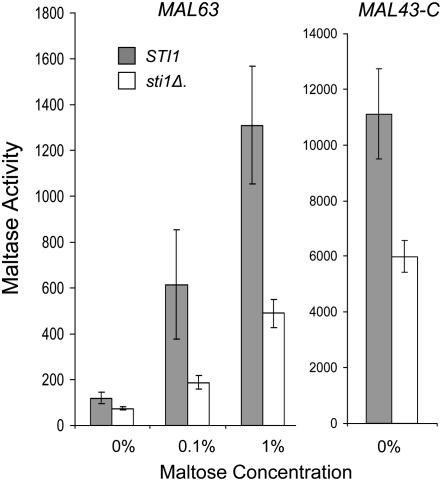
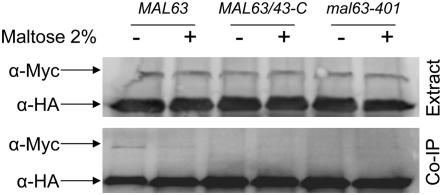
The finding that constitutive MAL-activators exhibit very modest levels of binding to Ssa1 (Figure 6) and are independent of Ssa1 levels (Figure 5) suggests the possibility that constitutive mutant proteins might bypass Hsp70 and, instead, bind to Hsp90 without the participation of Hsp70 or the formation of complexes containing cochaperones such as Sti1. We tested this possibility by investigating the effect of the loss of STI1 on MAL-activator chaperone binding. STI1 encodes the Saccharomyces Hop/p60 homolog (Chang et al. 1997). Sti1 functions as a scaffold protein that binds to Hsp70 and Hsp90 promoting the formation of the so-called intermediate complex and stimulating transfer of the client protein from Hsp70 to Hsp90 (reviewed in Pratt and Toft 2003; Wegele et al. 2004). Null mutations of STI1 are viable but loss of Sti1 reduces glucocorticoid receptor activity in Saccharomyces and increases its binding to Hsp40 (Ydj1) (Chang et al. 1997). STI1 was deleted in the strain carrying HSP82/Myc and maltase expression assayed in transformants carrying plasmid-borne inducible MAL63 or constitutive MAL43-C. Loss of Sti1 causes an ∼50% decrease in maltase activity in transformants carrying either allele, suggesting that both the inducible and the constitutive activators require Sti1 (Figure 7).
Figure 7.—
Maltase expression is dependent on STI1. Strains CMY1200 (STI1 HSP82/Myc hsc82Δ) and CMY8001 (sti1Δ HSP82/Myc hsc82Δ) were transformed with plasmid YCp50-MAL63 or YCp50-MAL43-C carrying the inducible MAL63 and constitutive MAL43-C MAL-activator alleles, respectively. Transformants were grown at 30° in selective synthetic medium lacking uracil and containing 3% glycerol and 2% lactate (v/v) plus the indicated concentration of maltose. Maltase activity was assayed in three independent transformants as described in Figure 2.
HSP82/Myc and SSA1/Myc strains containing sti1Δ were transformed with triple-HA-tagged MAL63 or MAL63/43-C and binding of the chaperones to the HA-tagged MAL-activator determined. The results, shown in Figure 8, are consistent with our finding that maltase expression in the constitutive mutant is dependent on Sti1. Binding of both inducible Mal63 and constitutive Mal63/43-C MAL-activators to Hsp82/Myc protein is severely reduced, almost absent, in the sti1Δ strain even when grown in the absence of maltose, the condition under which maximal Hsp82/Myc binding is observed in the STI1 strain. On the other hand, loss of Sti1 dramatically increases binding of both inducible Mal63 and constitutive Mal63/43-C MAL-activators to Ssa1 to comparable levels and binding is observed in cells grown under either uninduced or induced conditions. These results indicate that the constitutive MAL-activators bind with Hsp70 during Hsp90 chaperone activation but only transiently. We propose that the alterations present in the constitutive activators weaken binding to Hsp70 and this results in the premature dissociation of the intermediate complex formed by constitutive activators. The weakened Ssa1 binding also appears to result in an apparent loss of dependency on Hsp70.
The results reported in Figures 4 and 6 predict that neither the noninducible nor the constitutive mutant activators would be expected to stably bind Sti1. To investigate this, plasmid-borne inducible MAL63/3HA, constitutive MAL63/43-C/3HA, and noninducible mal63-401/3HA were coexpressed in a strain containing the C-terminal Myc-tagged STI1/Myc and binding of the various MAL-activators to Sti1/Myc determined by co-immunoprecipitation. Control experiments shown in Figure 1 demonstrate that Sti1/Myc protein does not bind to the HA-sepharose beads in the absence of HA-tagged Mal63 MAL-activator. Because STI1 expression is low, the Myc signal is weak. Nonetheless, as is shown in Figure 9, inducible Mal63 MAL-activator binds Sti1/Myc protein in cells grown under uninduced but not induced (maltose) conditions. Along with the results reported in Figure 3, this supports our proposal that inducible Mal63 activator forms a stable intermediate complex in the absence of inducer. Neither the constitutive nor the noninducible mutant MAL-activators binds significant levels of Sti1/Myc, even in cells grown in the absence of inducer, suggesting that neither of these mutant activator proteins is found in the so-called intermediate complex.
Figure 9.—
Co-immunoprecipitation of inducible, constitutive, and noninducible MAL-activators with Sti1. Strain CMY8015 (STI1/Myc) was transformed with plasmids p416GPD-MAL63/3HA, p416GPD-MAL63/43-C, and p416-mal63-401 carrying the triple-HA-tagged alleles of inducible MAL63, constitutive MAL63/43-C, and noninducible mal63-401. Transformants were grown as described in Figure 3. Preparation of nondenaturing extracts, co-immunoprecipitation, and Western blot analysis were carried out as described for Figure 3.
DISCUSSION
This report investigates the roles of Hsp90, Hsp70, and Sti1 in the regulation of the Saccharomyces MAL-activator, a DNA-binding transcription activator. Inducible MAL63 and several noninducible and constitutive MAL-activator mutant alleles from our collection are utilized (Charron and Michels 1987; Needleman 1991; Gibson et al. 1997; Danzi et al. 2000, 2003). The availability of these MAL-activator mutants is unique among Hsp90 client proteins and provides a novel tool to investigate regulation of a specific native client protein in vivo. The results demonstrate the formation of distinct stable chaperone complexes with the different classes of MAL-activator proteins (inducible, noninducible, and constitutive). Our analysis allows us to propose the in vivo pathway of MAL-activator regulation by the Hsp70/Hsp90 chaperone machine.
Activation of inducible Mal63 MAL-activator by the Hsp70/Hsp90 chaperone machine exhibits significant similarities to the activation process described for other clients but with a few important distinctions. As for other clients, Hsp70 is clearly required for Mal63 MAL-activator activation (Figure 2). However, in the absence of inducer, the stable chaperone complex formed by Mal63 is different from others that have been described in the literature. Under uninduced conditions, inducible Mal63 MAL-activator forms a stable complex with Ssa1, Hsp82, and Sti1 (Figures 3 and 9) in what appears to be an intermediate complex. This contrasts with glucocorticoid receptor which, in the absence of hormone, forms a stable so-called final complex with Hsp90, the cochaperone p23, and immunophilin (reviewed in Pratt and Toft 2003; Lan et al. 2004; Wegele et al. 2004, 2006). Alternately, the Saccharomyces heme-responsive transcription activator Hap1p is bound to Hsp70 in both the absence and presence of heme but is only transiently associated with Hsp90, although Hsp90 binding is enhanced in the presence of heme (Lan et al. 2004). Thus, the stable chaperone complexes formed by these three transcription activators are distinct with regard to their component composition. We also found that growth under maltose-induced conditions leads to a dramatic reduction in the relative binding of Mal63p to both chaperones and Sti1. Similarly, hormone releases glucocorticoid receptor from Hsp90 and allows it to enter the nucleus and bind DNA. In the case of Hap1p, Hsp70 but not Hsp90 is reported to bind to Hap1p-dependent promoters along with Hap1p (Xin et al. 2007). Thus, the interactions of MAL-activator with the chaperones are similar to those reported for other transcription activator clients but each has unique variations on the common theme. Given that only a small number of transcription activator clients have been studied in detail, our findings suggest that there is a greater variability in the functioning of the components of Hsp90/Hsp70 chaperone machine and that the roles played by different chaperone and cochaperones with different clients may be more flexible than has been anticipated.
Support for the hypothesis that MAL-activator association with Ssa1 and Hsp82 is sequential and in this order comes from studies with the various MAL-activator mutants. Noninducible mutants all exhibit normal binding to Ssa1, that is, they bind under uninduced growth conditions and not under induced growth conditions (Figure 4). On the other hand, binding to Hsp82 or Sti1 is not observed under any growth condition (Figures 4 and 9). This suggests that the noninducible mutant proteins are capable of forming the so-called early complex and of being released from Hsp70 in response to inducer but are not able to progress beyond the early complex to form a stable Hsp90-containing intermediate complex. It would appear that sequences required for recognition of the MAL-activator by Hsp70 are still functional in the noninducible mutant proteins but sequences required for recognition as an Hsp90 client are altered or unavailable.
The suggestion that inducible Mal63 MAL-activator is transferred from the early complex to the intermediate complex comes from the results in Figure 8. These results show that loss of Sti1 leads to Mal63 and Hsp70 binding in the presence of inducer, most likely by inhibiting the formation of the intermediate complex and thereby blocking the normal activation pathway. Additionally, deletion of STI1 significantly enhances binding of the constitutive MAL-activators to Ssa1. This finding, taken together with the dramatic loss of the Hsp82-constitutive MAL-activator interaction observed in the sti1Δ strain, clearly suggests that the constitutive activator mutants utilize the same chaperone activation pathway as inducible Mal63p. That is, constitutive activators initially bind to Ssa1 and then form an intermediate-like complex with Hsp82, Ssa1, and Sti1, albeit transiently. Rather than being retained in this intermediate complex, Ssa1 and Sti1 are released thereby allowing the constitutive activators to form a stable complex with Hsp82 alone. Whether this Hsp82-constitutive activator complex is a final complex similar to that formed by glucocorticoid receptor remains to be determined and will require the analysis of the complex for the presence of other components, such as the cochaperones Sba1 (p23) and the immunophilin Cpr7.
We propose that Ssa1 binding is required to retain the inducible activator in the intermediate complex in the absence of inducer. Moreover, we suggest that the alterations in the constitutive activators weaken Ssa1 binding and facilitate disruption of the intermediate complex formed by constitutive activators without the requirement for inducer. The correlation between decreased Ssa1 binding and constitutivity indicates that, in addition to its positive role in MAL-activator regulation, Ssa1 also serves as a repressor of inducible Mal63 MAL-activator in the absence of maltose. It is important to note that the constitutive activators bind Hsp82 in cells grown in the absence of inducer, that growth condition in which, by definition, these constitutive activator alleles are active. It is not apparent from our data whether all of the constitutive activator protein is bound to Hsp82 or whether the Hsp82-bound form is capable of DNA binding, as is seen for Hap1p and Hsp70 (Xin et al. 2007).
The results above have several interesting implications. Hsp70-bound MAL-activator is capable of maltose sensing even when bound in the early complex. This is consistent with the mutation analysis of Danzi et al. (2003), which suggests that the maltose-sensing domain of Mal63p is in residues 244–316. All of the noninducible mutants tested here except for mal63-283 map outside of this region. Additionally, this finding suggests that Sti1 alone is not sufficient to promote formation of the intermediate complex and raises the possibility that other cochaperones or sequences within the client protein itself are also involved (Chang et al. 1997). Cdc37p is required in the transfer of kinase clients to Hsp90. CDC37 overexpression suppresses defects in v-Src kinase activation in a sti1Δ strain (Lee et al. 2002) and the Cdk4 kinase–Cdc37p–Hsp90 complex has been well characterized by X-ray crystallography (Vaughan et al. 2006). The cochaperones that play this role for other classes of Hsp90 clients remain to be identified, although Aha1p has been suggested as a possible candidate (Hawle et al. 2006).
Three negative regulatory domains were identified by Danzi et al. (2000). Interestingly, these map in close proximity to sites at which alterations cause a noninducible phenotype and inhibit Hsp90 binding (Figure 4). Additionally, Danzi et al. (2000) found that very slight changes, even single-residue variations, in the C-terminal regulatory domain of Mal63p can convert a constitutive mutant into an inducible allele. These results indicate that multiple, noncontiguous sites in the MAL-activator interact with Hsp70 and Hsp90. It appears that alterations in a few critical residues can disrupt or restore binding to the chaperones and affect phenotypes in very complex ways. These findings are reminiscent of studies reported for the Src-family kinase Lck, which appears to bind to Hsp90 via multiple secondary structure sequences (Prince and Matts 2004). In contrast, the Hsp90-binding site of the glucocorticoid receptor is localized to a relatively defined region of the hormone-binding domain (Howard et al. 1990; Dalman et al. 1991).
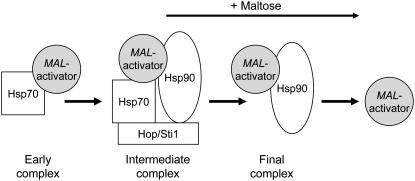
Figure 10 summarizes our current model for the regulation of the MAL-activator by the Hsp90/Hsp70 chaperone cycle. Binding to Hsp70 is the first step in MAL-activator activation but, under uninduced growth conditions, inducible Mal63 MAL-activator quickly binds with Hsp90 and Sti1 to form a stable intermediate complex. Binding to Hsp70 in this intermediate complex represses the MAL-activator in a conformation that maintains its ability to respond to maltose. Thus, Hsp70 is both required for and is a repressor of MAL-activator activation, the role played by Hsp90 in glucocorticoid activation. Addition of maltose causes release of inducible MAL-activator in a chaperone-free DNA-binding competent form. While we have no evidence that maltose binds directly to the MAL-activator, some of our findings are consistent with this possibility. In summary, while the key players in the chaperone cycle for the MAL-activator are the same as those for glucocorticoid receptor and Hap1p, their regulatory function and the components present in the stable complexes formed under the uninduced condition differ substantially. We are currently extending the in vivo analysis of the MAL-activator chaperone cycle described here to include other Saccharomyces cochaperones.
Figure 10.—
Model of MAL-activator regulation by the Hsp90/Hsp70 chaperone cycle.
Acknowledgments
We thank Susan Lindquist, Richard Gaber, and Li Zhang for providing various strains and Kevin A. Morano for helpful in-depth discussions. Western blot visualizations were done in the Core Facilities for Imaging, Cell, and Molecular Biology of Queens College, City University of New York (CUNY). We greatly appreciate the assistance of Areti Tsiola for the same. This work was carried out in partial fulfillment of the requirements of the Ph.D. degree from the Graduate School of CUNY (M.B. and F.R.) and was supported by a grant from the National Institutes of Health (GM-28216) to C.A.M.
References
- Bali, M., B. Zhang, K. A. Morano and C. A. Michels, 2003. The Hsp90 molecular chaperone complex regulates maltose induction and stability of the Saccharomyces MAL gene transcription activator Mal63p. J. Biol. Chem. 278 47441–47448. [DOI] [PubMed] [Google Scholar]
- Becker, J., W. Walter, W. Yan and E. A. Craig, 1996. Functional interaction of cytosolic hsp70 and a DnaJ-related protein, Ydj1p, in protein translocation in vivo. Mol. Cell. Biol. 16 4378–4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinker, A., C. Scheufler, F. Von Der Mulbe, B. Fleckenstein, C. Herrmann et al., 2002. Ligand discrimination by TPR domains. Relevance and selectivity of EEVD-recognition in Hsp70 × Hop × Hsp90 complexes. J. Biol. Chem. 277 19265–19275. [DOI] [PubMed] [Google Scholar]
- Bukau, B., J. Weissman and A. Horwich, 2006. Molecular chaperones and protein quality control. Cell 125 443–451. [DOI] [PubMed] [Google Scholar]
- Caplan, A. J., A. K. Mandal and M. A. Theodoraki, 2007. Molecular chaperones and protein kinase quality control. Trends Cell Biol. 17 87–92. [DOI] [PubMed] [Google Scholar]
- Chang, H. C., D. F. Nathan and S. Lindquist, 1997. In vivo analysis of the Hsp90 cochaperone Sti1 (p60). Mol. Cell. Biol. 17 318–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charron, M. J., and C. A. Michels, 1987. The constitutive, glucose-repression-insensitive mutation of the yeast MAL4 locus is an alteration of the MAL43 gene. Genetics 116 23–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalman, F. C., L. C. Scherrer, L. P. Taylor, H. Akil and W. B. Pratt, 1991. Localization of the 90-kDa heat shock protein-binding site within the hormone-binding domain of the glucocorticoid receptor by peptide competition. J. Biol. Chem. 266 3482–3490. [PubMed] [Google Scholar]
- Danzi, S. E., B. Zhang and C. A. Michels, 2000. Alterations in the Saccharomyces MAL-activator cause constitutivity but can be suppressed by intragenic mutations. Curr. Genet. 38 233–240. [DOI] [PubMed] [Google Scholar]
- Danzi, S. E., M. Bali and C. A. Michels, 2003. Clustered-charge to alanine scanning mutagenesis of the Mal63 MAL-activator C-terminal regulatory domain. Curr. Genet. 44 173–183. [DOI] [PubMed] [Google Scholar]
- Dubin, R. A., R. B. Needleman, D. Gossett and C. A. Michels, 1985. Identification of the structural gene encoding maltase within the MAL6 locus of Saccharomyces carlsbergensis. J. Bacteriol. 164 605–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge, S. J., and R. W. Davis, 1988. A family of versatile centromeric vectors designed for use in the sectoring-shuffle mutagenesis assay in Saccharomyces cerevisiae. Gene 70 303–312. [DOI] [PubMed] [Google Scholar]
- Gibson, A. W., L. A. Wojciechowicz, S. E. Danzi, B. Zhang, J. H. Kim et al., 1997. Constitutive mutations of the Saccharomyces cerevisiae MAL-activator genes MAL23, MAL43, MAL63, and mal64. Genetics 146 1287–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein, A. L., and J. H. McCusker, 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15 1541–1553. [DOI] [PubMed] [Google Scholar]
- Guldener, U., S. Heck, T. Fielder, J. Beinhauer and J. H. Hegemann, 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24 2519–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, E. K., F. Cotty, C. Sottas, H. Jiang and C. A. Michels, 1995. Characterization of AGT1 encoding a general alpha-glucoside transporter from Saccharomyces. Mol. Microbiol. 17 1093–1107. [DOI] [PubMed] [Google Scholar]
- Hawle, P., M. Siepmann, A. Harst, M. Siderius, H. P. Reusch et al., 2006. The middle domain of Hsp90 acts as a discriminator between different types of client proteins. Mol. Cell. Biol. 26 8385–8395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard, K. J., S. J. Holley, K. R. Yamamoto and C. W. Distelhorst, 1990. Mapping the HSP90 binding region of the glucocorticoid receptor. J. Biol. Chem. 265 11928–11935. [PubMed] [Google Scholar]
- Hu, Z., A. W. Gibson, J. H. Kim, L. A. Wojciechowicz, B. Zhang et al., 1999. Functional domain analysis of the Saccharomyces MAL-activator. Curr. Genet. 36 1–12. [DOI] [PubMed] [Google Scholar]
- Kim, J., and C. A. Michels, 1988. The MAL63 gene of Saccharomyces encodes a cysteine-zinc finger protein. Curr. Genet. 14 319–323. [DOI] [PubMed] [Google Scholar]
- Lan, C., H. C. Lee, S. Tang and L. Zhang, 2004. A novel mode of chaperone action: heme activation of Hap1 by enhanced association of Hsp90 with the repressed Hsp70-Hap1 complex. J. Biol. Chem. 279 27607–27612. [DOI] [PubMed] [Google Scholar]
- Lee, P., J. Rao, A. Fliss, E. Yang, S. Garrett et al., 2002. The Cdc37 protein kinase-binding domain is sufficient for protein kinase activity and cell viability. J. Cell Biol. 159 1051–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg, D., R. Muller and M. Funk, 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156 119–122. [DOI] [PubMed] [Google Scholar]
- Needleman, R., 1991. Control of maltase synthesis in yeast. Mol. Microbiol. 5 2079–2084. [DOI] [PubMed] [Google Scholar]
- Odunuga, O. O., J. A. Hornby, C. Bies, R. Zimmermann, D. J. Pugh et al., 2003. Tetratricopeptide repeat motif-mediated Hsc70-mSTI1 interaction. Molecular characterization of the critical contacts for successful binding and specificity. J. Biol. Chem. 278 6896–6904. [DOI] [PubMed] [Google Scholar]
- Picard, D., 2006. Chaperoning steroid hormone action. Trends Endocrinol. Metab. 17 229–235. [DOI] [PubMed] [Google Scholar]
- Pratt, W. B., U. Gehring and D. O. Toft, 1996. Molecular chaperoning of steroid hormone receptors. EXS 77 79–95. [DOI] [PubMed] [Google Scholar]
- Pratt, W. B., M. D. Galigniana, Y. Morishima and P. J. Murphy, 2004. Role of molecular chaperones in steroid receptor action. Essays Biochem. 40 41–58. [DOI] [PubMed] [Google Scholar]
- Pratt, W. B., Y. Morishima, M. Murphy and M. Harrell, 2006. Chaperoning of glucocorticoid receptors. Handb. Exp. Pharmacol., 111–138. [DOI] [PubMed]
- Pratt, W. B., and D. O. Toft, 1997. Steroid receptor interactions with heat shock protein and immunophilin chaperones. Endocr. Rev. 18 306–360. [DOI] [PubMed] [Google Scholar]
- Pratt, W. B., and D. O. Toft, 2003. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp. Biol. Med. 228 111–133. [DOI] [PubMed] [Google Scholar]
- Prince, T., and R. L. Matts, 2004. Definition of protein kinase sequence motifs that trigger high affinity binding of Hsp90 and Cdc37. J. Biol. Chem. 279 39975–39981. [DOI] [PubMed] [Google Scholar]
- Scheufler, C., A. Brinker, G. Bourenkov, S. Pegoraro, L. Moroder et al., 2000. Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell 101 199–210. [DOI] [PubMed] [Google Scholar]
- Vaughan, C. K., U. Gohlke, F. Sobott, V. M. Good, M. M. Ali et al., 2006. Structure of an Hsp90-Cdc37-Cdk4 complex. Mol. Cell 23 697–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach, A., A. Brachat, R. Pohlmann and P. Philippsen, 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10 1793–1808. [DOI] [PubMed] [Google Scholar]
- Wegele, H., L. Muller and J. Buchner, 2004. Hsp70 and Hsp90–a relay team for protein folding. Rev. Physiol. Biochem. Pharmacol. 151 1–44. [DOI] [PubMed] [Google Scholar]
- Wegele, H., S. K. Wandinger, A. B. Schmid, J. Reinstein and J. Buchner, 2006. Substrate transfer from the chaperone Hsp70 to Hsp90. J. Mol. Biol. 356 802–811. [DOI] [PubMed] [Google Scholar]
- Xin, X., C. Lan, H. C. Lee and L. Zhang, 2007. Regulation of the HAP1 gene involves positive actions of histone deacetylases. Biochem. Biophys. Res. Commun. 362 120–125. [DOI] [PMC free article] [PubMed] [Google Scholar]