Abstract
Two types of cell death have been studied extensively in Caenorhabditis elegans, programmed cell death and necrosis. We describe a novel type of cell death that occurs in animals containing mutations in either of two genes, lin-24 and lin-33. Gain-of-function mutations in lin-24 and lin-33 cause the inappropriate deaths of many of the Pn.p hypodermal blast cells and prevent the surviving Pn.p cells from expressing their normal developmental fates. The abnormal Pn.p cells in lin-24 and lin-33 mutant animals are morphologically distinct from the dying cells characteristic of C. elegans programmed cell deaths and necrotic cell deaths. lin-24 encodes a protein with homology to bacterial toxins. lin-33 encodes a novel protein. The cytotoxicity caused by mutation of either gene requires the function of the other. An evolutionarily conserved set of genes required for the efficient engulfment and removal of both apoptotic and necrotic cell corpses is required for the full cell-killing effect of mutant lin-24 and lin-33 genes, suggesting that engulfment promotes these cytotoxic cell deaths.
DURING the development of the Caenorhabditis elegans hermaphrodite, 131 of the 1090 somatic cells generated are eliminated by the process of programmed cell death, also called apoptosis (Sulston and Horvitz 1977; Kimble and Hirsh 1979; Sulston et al. 1983). The identification of the C. elegans genes that function in the pathway for programmed cell death has provided valuable insights into this evolutionarily conserved process. For example, molecular identification of the caspase gene ced-3 defined the first biochemical mechanism of apoptosis (Yuan et al. 1993). Caspases are aspartate-specific cysteine proteases that act in apoptosis in all animals (Degterev et al. 2003). Similarly, studies of C. elegans ced-9 (Hengartner et al. 1992; Hengartner and Horvitz 1994; Shaham and Horvitz 1996) helped establish the function and pathway of action of its human homolog Bcl-2 and of the misregulation of apoptosis as a cause of human cancer (Vaux et al. 1988; Hockenbery et al. 1990; Vaux et al. 1992). At least 30 C. elegans genes that function in programmed cell death have been identified; many have homologs that function in mammalian apoptosis (Horvitz 1999; Lettre and Hengartner 2006).
Cells can die not only by apoptosis but also by autophagy and necrosis. In autophagy, cells consume themselves from within, for example, under situations of nutritional deprivation (Edinger and Thompson 2004). There are no known examples of autophagic deaths in C. elegans. Necrosis is a cellular death process defined largely by ultrastructure, such as the swelling of organelles and the loss of plasma membrane integrity (Zong and Thompson 2006). Necrosis can be caused by injury or physiological insult and contributes significantly to human disease. Some cell deaths that occur in C. elegans are considered to be necrotic (Hall et al. 1997). The best understood necrotic deaths in C. elegans are those caused by gain-of-function (gf) mutations that alter the mechanosensory sodium channel subunit MEC-4 (Driscoll and Chalfie 1991). The deaths caused by mec-4(gf) mutations and by mutations in several other genes, including deg-1 (which encodes a related channel subunit) (Chalfie and Wolinsky 1990), deg-3 (which encodes an acetylcholine-gated ion channel) (Treinin and Chalfie 1995), and activated transgenic Gαs (Korswagen et al. 1997; Berger et al. 1998) probably involve similar mechanisms. The mec-4(gf) mutations cause the hyperactivation of degenerin/epithelial Na+ channel (DEG/EnaC) channels and cause the deaths of the six touch-receptor neurons that express the MEC-4 subunit (Lai et al. 1996). These deaths seem to require release of calcium from the endoplasmic reticulum (Xu et al. 2001) and the activation of the calcium-activated calpain proteases and of specific cathepsin aspartyl proteases (Syntichaki et al. 2002). This death process is similar to that proposed for mammalian excitotoxic cell death (Driscoll and Gerstbrein 2003).
Here we describe a third type of cell death in C. elegans. We refer to this type as cytotoxic cell death. Cytotoxic cell death is distinguishable from programmed cell death and from necrotic cell death on the basis of morphological, genetic, and electron microscopic criteria. We observed the cytotoxic cell deaths of the C. elegans Pn.p hypodermal blast cells in animals containing gain-of-function mutations in either of two genes, lin-24 or lin-33. lin-24 and lin-33 appear to act together in causing the cytotoxicity that affects the fates and survival of the Pn.p cells.
MATERIALS AND METHODS
Strains and general techniques:
Strains were cultured as described by Brenner (1974) and grown at 20°. The Bristol strain N2 was used as the wild-type strain, except in multifactor mapping experiments that used the polymorphic wild-type strains N62 (Bergerac) or CB4856 (Williams et al. 1992; Wicks et al. 2001). The mutations used in the study are listed below and are described, unless otherwise noted, by Riddle et al. (1997):
LGI: ced-1(e1735) and ced-12(n3261) (Zhou et al. 2001).
LGIII: ced-9(n1950), ced-4(n1162), ced-6(n1813), and ced-7(n1996).
LGIV: lin-24(n432, n1057), lin-24(n1821, n2050) (S. G. Clark and H. R. Horvitz, unpublished results), lin-24(n2258, n2333, n4294Δ, n432 n1503) (this study), lin-33(n1043, n1044), lin-33(n1110) (M. K. Edwards and H. R. Horvitz, unpublished results), lin-33(n1302) (J. H. Thomas and H. R. Horvitz, unpublished results), lin-24(n1968, n2003, n4514Δ, n1043 n1502) (this study), ced-3(n717), ced-2(e1752), ced-5(n1812), ced-10(n1993), dpy-4(e1166), dpy-26(n199), unc-22(s7), unc-30(e191), unc-31(e169), unc-44(e362), and lntl-1(n4763Δ) (this study).
LGV: unc-76(e911) and egl-1(n1084 n3082) (Conradt and Horvitz 1998).
In addition, we used strains containing the following chromosomal aberrations: sDf21 (Clark and Baillie 1992), nDf41 (S. Kim and H. R. Horvitz, unpublished results), and yDp1 (Delong et al. 1987).
lin-24(n2258) was found in the strain CB362 unc-44(e362) and was originally identified during the mapping of lin-33. Briefly, we had constructed an unc-5(e53) unc-44(e362) IV strain with unc-44(e362) from strain CB362. We crossed lin-33(n1043) males with unc-5(e53) unc-44(e362) hermaphrodites and observed that the lin-33(n1043)/unc-5(e53) unc-44(e362) cross progeny hermaphrodites did not have a vulvaless phenotype. Given that lin-33(n1043)/+ hermaphrodites are 77% vulvaless, we postulated that, in the unc-5(e53) unc-44(e362) strain, there was also a loss-of-function lin-33 or lin-24 allele that suppressed the lin-33(n1043)/+ vulvaless phenotype. Additional experiments confirmed that our unc-5(e53) unc-44(e362) strain as well as CB362 unc-44(e362) were homozygous for a lin-24 allele, which we named n2258.
Assay for vulvaless animals:
All animals scored had been propagated through at least two generations without starvation. Five-centimeter petri plates were seeded 12–24 hr prior to the assay with 10 μl of an overnight culture of the Escherichia coli strain OP50 grown in B broth. Fourth-stage larval (L4) animals were placed singly on petri plates. After 18 hr, plates were examined for the presence of gravid adults. Only petri plates with gravid adults were scored for the presence or absence of eggs; animals on petri plates without any eggs were scored as vulvaless (Vul).
Percentage of suppression was calculated by dividing the penetrance of the vulvaless phenotype of lin(gf); ced strains by the penetrance of the vulvaless phenotype of the lin(gf) strain, subtracting that number from one and multiplying the result by 100.
To assay for the suppression of heterozygous lin-24(n432)- and lin-33(n1043)-induced death by loss-of-function alleles of genes that function in programmed cell death, males homozygous for both the lin(gf) mutation and the candidate suppressor [e.g., ced-5(n1812) lin-24(n432)] were generated by heat shock and mated with strains homozygous for the candidate suppressor and marked with unc-76(e911) [e.g., ced-5(1812); unc-76(e911)]; 3–5 days later cross-progeny (non-Unc) L4 hermaphrodites were recovered and assayed as described above.
The Vul assays for the dosage studies of lin-24(n1057) were performed as previously described (Ferguson and Horvitz 1985). To generate animals of the genotype lin-24(n1057)/lin-24(+); yDp1, we crossed lin-24(n1057) unc-31(e169)/lin-24(+) unc-31(+) males with dpy-26(n199) unc-31(e169); yDp1 hermaphrodites and picked the hermaphrodite progeny. We determined the Vul phenotype of each animal and by examining its progeny determined the genotype of each animal. Non-Dpy non-Unc animals that segregated Dpy Unc and Lin Unc animals had the genotype lin-24(n1057) unc-31(e169)/dpy-26(n199) unc-31(e169); yDp1. To generate animals with the genotype lin-24(n1057); yDp1 for analysis, we picked non-Dpy non-Unc progeny of lin-24(n1057) unc-31(e169)/dpy-26(n199) unc-31(e169); yDp1 animals. We determined which animals did not segregate Dpy Unc animals and assayed the non-Unc animals.
We used mDp4 in our attempted dosage studies for lin-33(n1043) but very frequent recombination between the free duplication and the chromosomal region under investigation precluded the isolation of strains with the genotypes required for this experiment.
Nomarski observation of Pn.p cells:
Pn.p cells in lin-24 or lin-33 mutant animals were observed using Nomarski differential interference contrast (DIC) microscopy at different times during development as previously described (Sulston and Horvitz 1977).
Electron microscopy:
Nomarski DIC microscopy was used to select a nematode with refractile Pn.p cells, and digital images were taken to note their positions. Mutant animals were recovered from the slide and fixed as previously described (Bargmann et al. 1993). The fixed, embedded animals were sectioned as previously described (Gumienny et al. 1999) and photographed using a JEOL 12000CX electron microscope at 80 kV. After finding the location of the anus or the gonad in the micrographs, nuclei were counted by examining micrographs of adjacent sections and the images corresponding to the Pn.p cell that had been identified as abnormal using Nomarski microscopy were examined.
Dominant suppression screen for revertant alleles of lin-24(n432) and lin-33(n1043):
We mutagenized L4 or early adult hermaphrodites homozygous for either lin-24(n432) or lin-33(n1043) using ethyl methanesulfonate (EMS) as previously described (Brenner 1974). After mutagenesis, three to four P0 hermaphrodites were placed on individual 5-cm petri plates. F1 self-progeny that were non-Vul were placed singly on petri plates. If >10% of F2 animals on a petri plate were non-Vul, individual non-Vul F2 animals were picked singly to petri plates and the next generation was assayed for suppression of the Vul phenotype by quantifying the penetrance of the egg-laying defect.
Cloning of lin-24 and lin-33:
Using standard three-point and polymorphism mapping, we localized lin-24(n1057) and lin-33(n1043) to ∼300-kb and 156-kb intervals, respectively. To clone the gene lin-24 we prepared cosmids T20H7, B0001, ZC197, and F20B10 found in the region containing lin-24 and tested these cosmids (10–50 μg/ml) for ability to suppress the Vul phenotype of lin-24(n2050) using germ-line transformation. This experiment was performed essentially as previously described (Mello et al. 1991) with the plasmid pRF4 (80 μg/ml) as the co-injection marker. To clone lin-33 we amplified DNA from the 156-kb region between a polymorphism at position 5412 of cosmid C10G6 and unc-44 from lin-33(n1302) animals using the Expand Long Template PCR System (Roche Applied Science, Indianapolis, IN) according to the manufacturer's instructions. This amplified DNA was then injected into unc-76(e911) animals, using the plasmid P76-16B (Bloom and Horvitz 1997) (50 μg/ml) as a co-injection marker, to identify DNA that caused the vulvaless phenotype.
Isolation of lin-24 cDNAs:
A 10.8-kb SmaI-NcoI fragment from cosmid T20H7 was used to screen >800,000 plaques of a λ-ZAP cDNA library derived from mixed-stage poly(A)-positive RNA collection as previously described (Barstead and Waterston 1989). Fourteen plaques were isolated, 6 of which hybridized to fragments of the 10.8-kb fragment sufficient to rescue the vulvaless phenotype of lin-24(n2050) animals.
Determination of mutant allele sequences:
We used PCR-amplified regions of genomic DNA to determine DNA sequences. For all alleles of lin-24 and lin-33, we determined the sequences of all exons and splice junctions. Sequences were determined using an AB1 373A Cycle Sequencer or ABI Prism 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA).
Isolation of deletion alleles:
Genomic DNA pools from EMS-mutagenized animals were screened for deletions using PCR as previously described (Jansen et al. 1997; Liu et al. 1999). Deletion mutants were recovered from frozen stocks and backcrossed to the wild type six times. lin-24(n4294Δ) removes nucleotides 2259–3253 of cosmid B0001 (GenBank accession no. Z69634). lin-33(n4514Δ) removes nucleotides 28,902–29,937 of fosmid H32C10 (GenBank accession no. AF125443). lntl-1(n4763Δ) removes nucleotides 27,211–28,463 of cosmid C31H1 (GenBank accession no. U42848).
RESULTS
Mutations in lin-24 and lin-33 cause the Pn.p cells to inappropriately undergo cell death and to adopt aberrant cell fates:
Four C. elegans strains with mutations in the gene lin-24 and five C. elegans strains with mutations in the gene lin-33 were isolated mostly from screens for animals with the vulvaless phenotype and for animals unable to lay eggs (Trent et al. 1983; Ferguson and Horvitz 1985). Previous observations of lin-24 and lin-33 mutants using Nomarski DIC microscopy suggested that the Pn.p cells (the posterior daughters of the 12 P cells P1–P12), some of which divide to generate descendants that form the vulva (Sulston and Horvitz 1977), frequently either die or fail to divide appropriately to form the vulva (Ferguson and Horvitz 1985; Ferguson et al. 1987). We confirmed and extended these observations. We observed that, in lin-24(n432) and lin-33(n1043) mutant hermaphrodites late in the first larval stage (L1) or early in the second larval stage (L2), the nuclei of the Pn.p cells increase in refractility and form noncircular often oval-shaped refractile bodies that can persist for several minutes to 3 hr (Figure 1). Once the refractility begins to decrease, within 2 hr, the nucleus becomes granular and then resolves and one of three outcomes occurs: the cell dies, the cell survives but the nucleus becomes abnormally small, or the cell survives and the nucleus regains a normal appearance. The refractile bodies seen in this process are distinct from the circular and more highly refractile corpses seen in programmed cell death (Sulston and Horvitz 1977). The refractile bodies are also distinct from the dying cells observed in C. elegans necrosis, in which dying cells swell to several times their original diameter (Chalfie and Sulston 1981; Chalfie and Wolinsky 1990). We observed 176 Pn.p cells (P1–P11.p) from the mid-L1 larval stage through the early L2 larval stage in 16 lin-24(n2050) animals and observed that 31 died (18%), 2 survived and had small nuclei, and 143 recovered the appearance of normal Pn.p cells (Table 1). Of the 99 Pn.p cells we observed in 9 lin-33(n2003) animals, 38 (38%) died inappropriately, 6 had small nuclei, and the remaining 55 recovered the appearance of normal Pn.p cells (Table 2). We discovered no spatial pattern as to which of the 11 cells examined (P1–P11.p) died. In particular, there appeared to be no preferential survival or death of the P3–P8.p cells, which have the potential to divide and differentiate to form the vulva (Sulston and Horvitz 1977; Sulston et al. 1980; Kimble 1981; Sternberg and Horvitz 1986).
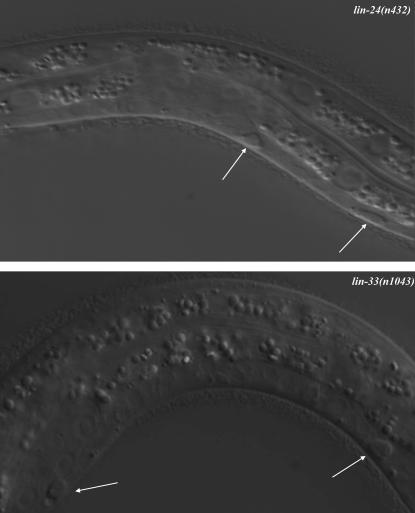
Figure 1.—
The Pn.p cells of lin-24(n432) and lin-33(n1043) mutants are morphologically abnormal. Nomarski photomicrographs of abnormal Pn.p cells in lin-24(n432) and lin-33(n1043) L1 larvae are shown. White arrows indicate Pn.p cells with an abnormal refractile appearance. Anterior, left; ventral, down.
TABLE 1.
lin-24(n2050) mutant Pn.p cell fate at early L2 larval stage
| Animal no. | P1.p | P2.p | P3.p | P4.p | P5.p | P6.p | P7.p | P8.p | P9.p | P10.p | P11.p | No. X, sm |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Wild type | + | + | + | + | + | + | + | + | + | + | + | 0, 0 |
| 1 | + | + | X | + | + | + | + | + | + | + | + | 1, 0 |
| 2 | X | + | + | + | + | + | + | + | X | + | + | 2, 0 |
| 3 | + | + | + | + | + | + | + | X | + | + | + | 1, 0 |
| 4 | + | X | + | X | + | X | + | + | X | + | + | 4, 0 |
| 5 | + | + | + | X | + | X | + | sm | + | + | + | 2, 1 |
| 6 | + | + | + | + | + | + | X | + | + | X | + | 2, 0 |
| 7 | + | + | + | + | X | + | + | + | + | + | + | 1, 0 |
| 8 | + | X | + | + | + | + | + | + | + | + | + | 1, 0 |
| 9 | X | + | + | + | X | + | + | + | X | + | + | 3, 0 |
| 10 | + | + | X | + | + | + | + | + | + | + | + | 1, 0 |
| 11 | + | X | X | + | X | + | + | + | + | + | + | 3, 0 |
| 12 | + | X | + | + | + | + | + | + | + | + | X | 2, 0 |
| 13 | + | + | + | + | + | X | + | + | + | + | + | 1, 0 |
| 14 | + | + | + | + | sm | X | + | X | + | + | + | 2, 1 |
| 15 | + | + | + | + | + | + | X | + | + | X | + | 2, 0 |
| 16 | X | + | X | + | + | + | + | + | + | + | X | 3, 0 |
| Total | 3, 0 | 4, 0 | 4, 0 | 2, 0 | 3, 1 | 4, 0 | 2, 0 | 2, 1 | 3, 0 | 2, 0 | 2, 0 | 31, 2 |
lin-24(n2050) animals were observed using Nomarski optics from the mid-L1 to the early L2 larval stage. In each box, the appearance of the corresponding Pn.p cell is noted: X, cell died; +, the nucleus looked normal; sm, the nucleus looked abnormally small. In the last column the number of Pn.p cells that died and the number of Pn.p cell nuclei that looked abnormally small are given, separated by a comma. In wild-type animals, all Pn.p cells looked normal.
TABLE 2.
lin-33(n2003) mutant Pn.p cell fate at early L2 larval stage
| Animal no. | P1.p | P2.p | P3.p | P4.p | P5.p | P6.p | P7.p | P8.p | P9.p | P10.p | P11.p | No. X, sm |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Wild type | + | + | + | + | + | + | + | + | + | + | + | 0, 0 |
| 1 | + | + | X | + | + | + | + | + | + | + | X | 2, 0 |
| 2 | X | + | X | + | + | + | + | X | sm | + | + | 3, 1 |
| 3 | + | + | + | X | X | + | X | + | + | X | + | 4, 0 |
| 4 | X | X | X | + | X | + | X | + | X | + | + | 6, 0 |
| 5 | + | X | + | sm | + | X | + | X | + | + | + | 3, 1 |
| 6 | + | X | X | sm | + | + | X | + | + | X | X | 5, 1 |
| 7 | X | sm | + | X | X | X | + | + | + | sm | sm | 4, 3 |
| 8 | + | X | + | X | + | + | sm | X | + | X | X | 5, 1 |
| 9 | X | + | X | + | + | X | X | + | X | + | X | 6, 0 |
| Total | 4, 0 | 4, 1 | 5, 0 | 3, 2 | 3, 0 | 3, 0 | 4, 1 | 3, 0 | 2, 1 | 3, 1 | 4, 1 | 38, 7 |
lin-33(n2003) animals were observed using Nomarski optics from the mid-L1 to the early L2 larval stage. In each box, the appearance of the corresponding Pn.p cell is noted: X, cell died; +, the nucleus looked normal; sm, the nucleus looked abnormally small. In the last column the number of Pn.p cells that died and the number of Pn.p cell nuclei that looked abnormally small are given, separated by a comma. In wild-type animals, all Pn.p cells looked normal.
P(5–7).p normally undergo three rounds of division to generate the cells that form the vulva (Sulston and Horvitz 1977). Three neighboring cells P(3, 4, and 8).p, although competent to make vulval cells, normally divide once to generate two descendants that fuse with the syncytial hypodermis (Sulston and Horvitz 1977; Sulston and White 1980). We examined the fates of P(3–8).p in lin-24 and lin-33 mutants and observed that these cells almost never divided. In lin-24(n2050) animals, of the 55 cells we tracked, all but 2 (96%) never divided (Table 3). Instead, these cells appeared to fuse with the hypodermal syncytium, a fate normally assumed by P(1, 2).p and P(9–12).p. Thus, lin-24 and lin-33 mutants are abnormal not only in Pn.p cell survival but also in Pn.p cell fate. The cell death of the Pn.p cells and the failure of surviving vulval equivalence group cells (P3–8.p) to divide in lin-24 and lin-33 mutant animals account for the vulvaless phenotype observed in these strains.
TABLE 3.
Fate of the Pn.p cells of the vulval equivalence group in lin-24(n2050) mutants
| Animal no. | P3.p | P4.p | P5.p | P6.p | P7.p | P8.p |
|---|---|---|---|---|---|---|
| Wild type | 3° | 3° | 2° | 1° | 2° | 3° |
| 1 | S | X | S | S | S | S |
| 2 | S | S | S | X | Sa | S |
| 3 | S | S | S | S | S | S |
| 4 | S | S | S | S | S | S |
| 5 | S | S | S | X | S | S |
| 6 | S | S | S | S | S | X |
| 7 | S | S | S | 1° | X | S |
| 8 | X | S | X | X | S | S |
| 9 | S | 3° | X | S | X | S |
| 10 | S | S | S | S | S | S |
| 11 | S | S | S | S | X | S |
lin-24(n2050) animals were picked in the early L3 larval stage and observed to the L3 molt. The fate of each cell is recorded: X, the Pn.p cell was not present and presumed dead; S, cell persisted into the L4 larval stage without dividing and probably joined the hypodermal syncytium; 3°, tertiary fate; 2°, secondary fate; 1°, primary fate (Sternberg and Horvitz 1986). The top row shows the normal fates of each of the P(3–8).p cells in wild-type hermaphrodites.
Nucleus was abnormally small.
We refer to the Pn.p cell deaths of lin-24 and lin-33 mutants as cytotoxic cell deaths to distinguish them from the previously studied programmed and necrotic cell deaths that are morphologically and genetically distinct (see below).
The cytotoxic cell deaths of lin-24 and lin-33 mutants are ultrastructurally distinct from programmed and necrotic cell deaths:
We used electron microscopy to examine the ultrastructure of the Pn.p cells of lin-24 and lin-33 mutants. Chromatin condensation and a shrinking of cell volume, which are hallmarks of apoptotic programmed cell death (Robertson and Thomson 1982), were not observed. Additionally, the nuclear crenellation and accumulation of single membrane-bound vacuoles observed in the nonapoptotic linker cell death were absent (Abraham et al. 2007). Finally, we did not observe the large vacuoles characteristic of the necrotic cell deaths induced by mec-4(gf) and deg-1(gf) mutations (Hall et al. 1997). We noted four ultrastructural abnormalities of the refractile Pn.p cells in lin-24(n423) animals, lin-33(n1043) animals, and lin-33(n1043) lin-24(n432) double-mutant animals: (1) few intact cristae and a nonelectron dense matrix in the mitochondria, (2) electron-dense puncta in the nuclei, (3) dilation of the lumen of the nuclear envelope, and (4) electron-dense membranous whorls in the cytoplasm (Figure 2 and supplemental Figure 2). Both the electron-dense membranous whorls and mitochondrial distortion are observed in deaths induced by mec-4(gf) and deg-1(gf) mutations (Hall et al. 1997). Nonetheless, together the four features observed in lin-24(n432) and lin-33(n1043) mutants distinguish their abnormal cells from the dying cells observed both in programmed cell death and in the necrotic death associated with gain-of-function mutations in the genes mec-4 and deg-1. Although the lin-24- and lin-33-induced cytotoxicity is distinct from the mec-4- and deg-1-induced necrotic deaths, the dilation of the nuclear envelope we observed is also observed in some mammalian necrotic deaths (Wyllie 1981).
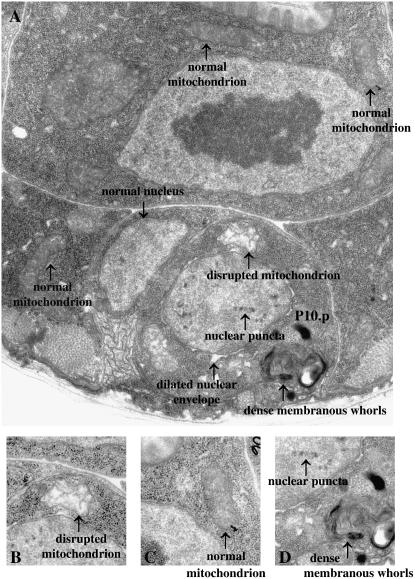
Figure 2.—
Ultrastructural characteristics of the abnormal Pn.p cells of lin-24(n432) and lin-33(n1043) mutants. (A) Electron micrograph of a refractile P10.p cell in a lin-33(n1043) mutant. Four ultrastructural characteristics of the abnormal Pn.p cells observed in lin-24(n432) and lin-33(n1043) animals are indicated: dilated mitochondrion with disrupted internal architecture, electron-dense puncta in the nucleus, dilation of the lumen of the nuclear envelope, and dense membranous whorls. Note that these four abnormal characteristics are not observed in the adjacent cells, which all have normal cellular architecture. For comparison, a normal nucleus and normal mitochondria in neighboring cells are indicated. (B) Increased magnification of A showing a disrupted mitochondrion. (C) Increased magnification of A showing a mitochondrion with normal architecture. (D) Increased magnification of A showing the electron-dense nuclear puncta and membranous whorls.
The lin-24 and lin-33 alleles that cause cytotoxic cell death and cell-fate defects are gain-of-function mutations:
To quantify the defects of different lin-24 and lin-33 mutants, we scored the vulvaless phenotype by determining the percentage of animals of each strain that failed to lay eggs. The penetrance of the egg-laying defect caused by eight of the nine lin-24 and lin-33 alleles ranged from 75 to 100% (Table 4). Some of the lin-24 and lin-33 alleles caused a semidominant vulvaless phenotype (Table 4). lin-24(n1057) was unique in that the penetrance of the vulvaless phenotype was greatest (41%; n = 74) when lin-24(n1057) was in trans to lin-24(+) but was only rarely observed in lin-24(n1057) homozygotes (2%; n = 90).
TABLE 4.
Penetrances of cytotoxic alleles of lin-24 and lin-33
| Genotype | % Vul (n) |
|---|---|
| Wild type | 0 (89) |
| lin-24(n432) | 92 (90) |
| lin-24(n432)/lin-24(+) | 66 (109) |
| lin-24(n1821) | 75 (89) |
| lin-24(n1821)/lin-24(+) | 0 (72) |
| lin-24(n2050) | 91 (87) |
| lin-24(n2050)/lin-24(+) | 0 (71) |
| lin-24(n1057) | 2 (90) |
| lin-24(n1057)/lin-24(+) | 41 (74) |
| lin-33(n1043) | 97 (90) |
| lin-33(n1043)/lin-33(+) | 77 (121) |
| lin-33(n1044) | 96 (90) |
| lin-33(n1110) | 92 (88) |
| lin-33(n1302) | 100 (90) |
| lin-33(n2003) | 99 (90) |
| lin-33(n1043) lin-24(n432) | 97 (89) |
| lin-33(n1043) lin-24(n432)/lin-24(+) lin-33(+) | 100 (49) |
The penetrance of the Vul phenotype (%) was determined as described in materials and methods. All genotypes were as indicated, except for the heterozygous animals, which also were heterozygous for unc-76(e911). n, number of animals.
We performed gene-dosage studies using the duplication yDp1 and counting animals that failed to properly lay eggs and as a result turned into “bags of worms.” We found penetrances of this bag-of-worms phenotype as follows: lin-24(n1057)/lin-24(n1057) (0% vulvaless, n > 300) = lin-24(n1057)/+/yDp1 (2% vulvaless, n = 153) = lin-24(n1057)/lin-24(n1057)/yDp1 (2% vulvaless, n = 141) < lin-24(n1057)/+ (33% vulvaless, n > 300). (The slight difference between the percentage of animals scored as vulvaless in the bag-of-worms assay and in the egg-laying assay (see materials and methods) is likely a result of the greater sensitivity of the egg-laying assay to defects in vulval development.) These data suggest that a balance between the dosages of the mutant and wild-type gene products determines the penetrance of the vulvaless phenotype, such that when the levels of the wild-type and mutant gene products are equal, the penetrance of the vulvaless phenotype is the highest. One simple interpretation is that the lin-24 protein forms a multimer, in a lin-24/+ heterozygote the mutant and wild-type proteins interact to form a cytotoxic complex, and having a greater dosage of either gene product results in a reduced level of the cytotoxic heteromeric protein complex.
The semidominance of some lin-24 and lin-33 alleles suggested that these alleles might not be loss-of-function mutations. To define the loss-of-function phenotype of these genes we sought to isolate mutations that reduce or eliminate lin-24 and lin-33 function. Quantification of the penetrances of the bag-of-worms phenotype of lin-24 alleles (n432, n1057, n1821, and n2050) or lin-33 alleles (n1043, n1044, n1110, n1302, or n2003) when in trans to the deficiencies sDf21 and nDf41, respectively, which remove either lin-24 or lin-33, suggested that lin-24(n432)/lin-24(null) and lin-33(n1043)/lin-33(null) animals were wild-type because no bag of worms were observed (supplemental Table 1). Thus, we predicted that we should be able to isolate lin-24 and lin-33 loss-of-function mutations as cis-dominant suppressors of the vulvaless phenotype of lin-24(n432) or lin-33(n1043) mutants in F1 reversion screens. In two separate screens (see materials and methods), we mutagenized strains that were homozygous for either lin-24(n432) or lin-33(n1043) and isolated non-Vul animals from the F1 self-progeny. One isolate from each of these two screens was a good candidate to be an intragenic suppressor mutation, on the basis of its map position, which in each case was <0.09 MU from the respective mutation [lin-24(n432 n1503) and lin-33(n1043 n1502)] (data not shown). Animals homozygous for either of the intragenic revertant alleles appeared wild type, suggesting that the loss-of-function phenotype of these genes is not vulvaless and that the previously isolated alleles of lin-24 and lin-33 that cause the vulvaless phenotype are gain-of-function alleles. We confirmed this hypothesis once we molecularly identified lin-24 and lin-33 and isolated deletion alleles of these genes (see below).
The alleles of lin-24 that cause a vulvaless phenotype fall into three categories: those that semidominantly cause a vulvaless phenotype (n432), those that recessively cause a vulvaless phenotype (n1821 and n2050), and those that cause a vulvaless phenotype only when in trans to a wild-type allele (n1057) (Table 4). Unlike the alleles of lin-33 that all cause a similar semidominant vulvaless phenotype, the differing natures of the lin-24 alleles suggest that the function of lin-24 can be altered in a variety of ways to be cytotoxic and cause the vulvaless phenotype.
Molecular identification of lin-24 and lin-33:
lin-24 had been previously mapped between unc-22 and dpy-26 on linkage group IV (Ferguson and Horvitz 1985). We further mapped lin-24 to an interval of ∼300 kb located to the left of an RFLP on cosmid C41D9 present in the C. elegans strain N62 (G. Garriga, personal communication) (Figure 3A). We cloned lin-24 using transformation rescue by injecting wild-type cosmid DNA into lin-24(n1821) mutant animals, which displayed a recessive vulvaless phenotype. Of the four cosmids that we tested corresponding to sequences within the defined interval between unc-22 and the RFLP on C41D9, two overlapping cosmids (T20H7 and B0001) rescued the recessive vulvaless phenotype of lin-24(n1821) animals in germ-line transformation experiments. A transgene carrying a 6.3-kb DNA sequence present in both T20H7 and B0001 and containing the single predicted gene B0001.1 was sufficient to rescue the recessive vulvaless phenotype of lin-24(n2050) animals. We determined the DNA sequences of the presumptive lin-24 gene in each of the four different lin-24 mutants and found that each mutant carried a different missense mutation in B0001.1 (Figure 3C). Additionally, as predicted, we found a second mutation in B0001.1 in lin-24(n432 n1503), which was isolated in the lin-24(n432) reversion screen (Figure 3C). On the basis of these mapping, rescue, and sequence results, we concluded that B0001.1 is lin-24. Six lin-24 cDNAs were isolated from a mixed-stage cDNA library; the longest appeared to be full length, as it contained an SL1 trans-spliced leader sequence. Using this cDNA, we established the gene structure of lin-24 shown in Figure 3B.
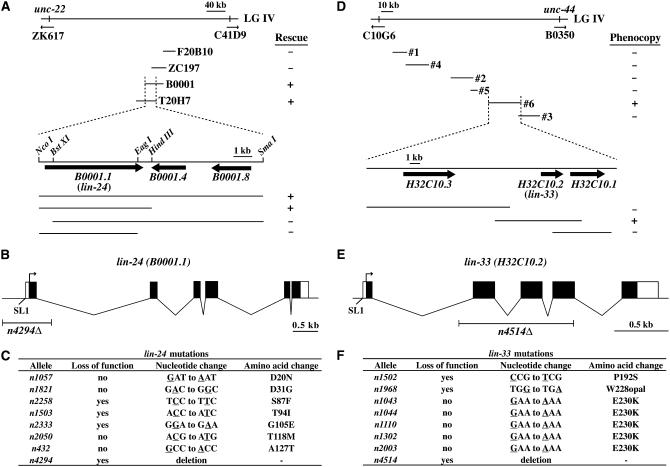
Figure 3.—
Molecular identification of lin-24 and lin-33. (A) Genetic and physical maps of the lin-24 region. lin-24 was mapped to the ∼300-kb interval between unc-22 and an RFLP on C41D9. The four cosmids shown were tested for their abilities to rescue the recessive vulvaless phenotype of lin-24 mutants. The overlapping cosmids B0001 and T20H7 and an NcoI-HindIII fragment containing the gene B0001.1 could rescue the Vulvaless phenotype. (B) The structure of the lin-24 gene as deduced from genomic and cDNA sequences. Exons (solid boxes), 5′ and 3′ untranslated regions (open boxes), the predicted transcriptional start, and the SL1 splice leader sequence are indicated. The location of the deletion lin-24(n4294Δ) and the scale are indicated by the labeled horizontal lines. (C) Sequences of lin-24 mutations. Mutated bases are underlined. Alleles were designated loss-of-function if they suppressed the semidominant vulvaless phenotype of lin-24(n432). (D) Genetic and physical maps of the lin-33 region. lin-33 was mapped to the ∼156-kb interval between a polymorphism on C10G6 and unc-44. PCR products generated from lin-33(n1302) genomic DNA spanning almost all of the shown interval were injected into lin-33(+) animals and tested for the ability to phenocopy the semidominant vulvaless phenotype of the lin-33(n1302) animals. The 26-kb PCR product no. 6 and a 9-kb PCR product containing only the gene H32C10.2 generated a vulvaless phenotype. (E) The structure of the lin-33 gene as deduced from genomic and cDNA sequences. Exons (solid boxes), 5′ and 3′ untranslated regions (open boxes), the predicted transcriptional start, and the SL1 splice leader sequence are indicated. The location of the deletion lin-33(n4514Δ) and the scale are indicated by the labeled horizontal lines. (F) Sequences of lin-33 mutations. Mutated bases are underlined. Alleles were designated loss-of-function if they suppressed the semidominant vulvaless phenotype of lin-33(n1043).
lin-33 was previously mapped between dpy-13 and unc-8 on linkage group IV (Ferguson and Horvitz 1985). We further mapped lin-33 to an interval located to the left of unc-44 and to the right of a polymorphism at position 5412 of cosmid C10G6 found in the C. elegans strain CB4856 (Figure 3D). We cloned lin-33 by injecting DNA amplified using PCR from lin-33(n1302) mutant animals [which display the lin-33(gf) semidominant vulvaless phenotype] into lin-33(+) animals and determining the minimal fragment of DNA sufficient to cause a vulvaless phenotype. Seven PCR products that had been amplified from lin-33(n1302) genomic DNA and that together almost completely covered the 156-kb interval were tested for their abilities to phenocopy the vulvaless phenotype of lin-33(n1302) animals as transgenes in a lin-33(+) background. One of the seven PCR products, predicted to contain three genes, caused a vulvaless phenotype in wild-type animals. A single 9-kb PCR product predicted to contain one of these three genes, H32C10.2, also caused a vulvaless phenotype when injected into lin-33(+) animals. We determined the DNA sequence of H32C10.2 from each of the five semidominant lin-33 mutants that cause Pn.p cell death and found that all five contain the identical missense mutation, predicted to change glutamic acid 230 to lysine (Figure 3F). Additionally, as predicted, we found a second mutation in H32C10.2 in lin-33(n1043 n1502), which was isolated in the lin-33(n1043) reversion screen (Figure 3F). On the basis of these mapping, rescue, and sequence results, we concluded that H32C10.2 is lin-33. Two lin-33 cDNAs have been isolated by Yuji Kohara and co-workers (Y. Kohara, personal communication). One, yk1122h12, appears to be full length, as it contains an SL1 trans-spliced leader sequence. The gene structure of lin-33 based on this cDNA is shown in Figure 3E.
LIN-24 contains a toxin domain, and LIN-33 is a novel protein:
BLAST and related searches with the predicted LIN-33 protein sequence did not identify any significant domains or homologies with other proteins. Similar searches using the LIN-24 protein sequence revealed that LIN-24 contains a domain found in many bacterial toxins. This domain, spanning amino acids 62–176, is most similar to the Bacillus sphaericus Mtx3 mosquitocidal toxin (Figure 4) (Liu et al. 1996). These toxins are thought to function by binding to eukaryotic cells and aggregating to form holes in the membrane, ultimately causing osmotic lysis by disrupting membrane permeability (Gilbert 2002). Five of the seven mutations in lin-24 are located in the toxin-related domain (Figure 4), including the three loss-of-function alleles, suggesting that this domain has an important function in the observed cytotoxicity of mutant LIN-24. All of the loss-of-function alleles affect conserved residues. It is tempting to speculate that LIN-24 causes cytotoxicity by a mechanism similar to that of these bacterial toxins.
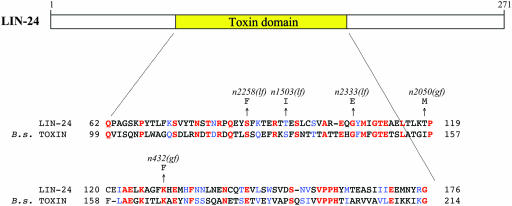
Figure 4.—
LIN-24 contains a domain similar to that in bacterial toxins. Sequence alignment of amino acids 62–176 of LIN-24 and the Mtx3 toxin domain of the 35.8-kDa mosquitocidal toxin from B. sphaericus (GenBank accession no. AAB36654). Numbers at the right and left indicate amino acid positions within the respective proteins. Identical residues are colored red, and similar residues are colored blue (29% identity and 45% similarity). Missense mutations found in lin-24 within the region of homology are indicated.
The lin-24(n432) mutation likely causes a novel gene activity:
We identified the deletion alleles lin-24(n4294Δ) and lin-33(n4514Δ) by screening a library of mutagenized animals (see Figure 3 and materials and methods). Animals homozygous for either deletion allele appeared wild type (Table 5), demonstrating that the null phenotype of these genes is not vulvaless. The lin-24(n432)/lin-24(+) and lin-33(n1043)/lin-33(+) strains have a more penetrant vulvaless phenotype (66 and 77% vulvaless, respectively) than lin-24(n432)/lin-24(Δ) and lin-33(n1043)/lin-33(Δ) strains (3 and 21% vulvaless, respectively), suggesting that the presence of a wild-type copy of the respective gene can increase the cytotoxicity of these alleles (Table 5). Using a duplication carrying a wild-type copy of the lin-24 locus, we scored the penetrance of the vulvaless phenotype of strains containing different combinations of lin-24 alleles. The relative penetrances of the vulvaless phenotype of the following lin-24(n432)-containing strains was lin-24(n432)/lin-24(+)/lin-24(+) (38% vulvaless) < lin-24(n432)/lin-24(+) (66% vulvaless) < lin-24(n432)/lin-24(n432)/lin-24(+) (81% vulvaless) < lin-24(n432)/lin-24(n432) (92% vulvaless) (Table 5), such that 0(null)/0 < m(mutant)/0 < m/+/+ < m/+ < m/m/+ < m/m. These results demonstrate that an increase in the dosage of the lin-24(+) gene product results in a decrease in the percentage of hermaphrodites that are vulvaless. These data indicate that the wild-type lin-24 gene product antagonizes the cytotoxicity of the semidominant mutation. Therefore, the lin-24(n432) mutation does not result in an increase in the wild-type function of the gene product, function as a dominant negative, or cause a loss of function. Instead, the lin-24(n432) mutation probably causes the generation of an abnormal cytotoxic gene product. Technical difficulties prevented similar investigations using the free duplication mDp4 of the effect of n1043 on lin-33 function (see materials and methods). The relative penetrances of the vulvaless phenotype of the following lin-33(n1043)-containing strains were lin-33(n1043)/lin-33(Δ) (21% vulvaless) < lin-33(n1043)/lin-33(+) (77% vulvaless) < lin-33(n1043)/lin-33(n1043) (97% vulvaless) (Table 5), such that 0/0 < m/0 < m/+ < m/m. These data suggest the semidominant n1043 mutation might cause either an increase in wild-type function or, like lin-24(n432), cause a novel abnormal function.
TABLE 5.
Gene dosage and gene interaction studies of lin-24 and lin-33 alleles
| Genotype | % Vul (n) |
|---|---|
| Wild typea | 0 (89) |
| lin-24(n432)/lin-24(n432)a | 92 (90) |
| lin-24(n4294Δ)/lin-24(n4294Δ) | 0 (102) |
| lin-24(n432)/lin-24(n4294Δ)b | 3 (89) |
| lin-24(n432)/lin-24(+)a | 66 (109) |
| lin-24(n432)/lin-24(+)/lin-24(+)c | 38 (56) |
| lin-24(n432)/lin-24(n432)/lin-24(+)d | 81 (117) |
| lin-33(n1043)/lin-33(n1043)a | 97 (90) |
| lin-33(n4514Δ)/lin-33(n4514Δ) | 0 (72) |
| lin-33(n1043)/lin-33(n4514Δ)e | 21 (77) |
| lin-33(n1043)/lin-33(+)a | 77 (121) |
| lin-33(n1043) lin-24(n432)a | 97 (89) |
| lin-33(n1043) lin-24(n4294Δ) | 0 (56) |
| lin-33(n4514Δ) lin-24(n432) | 0 (60) |
| lin-33(+) lin-24(n432)/lin-33(n4514Δ) lin-24(+)f | 0 (29) |
| lin-33(n1043) lin-24(+)/lin-33(+) lin-24(n4294Δ)g | 0 (12) |
The number of Vul animals was determined as described in materials and methods. n, number of animals.
These data are from Table 1.
lin-24(n432) homozygous males were mated with lin-24(n4294Δ) dpy-4(e1166) hermaphrodites, and non-Dpy cross progeny were scored.
This strain was of genotype unc-30(e191) dpy-4(e1166)/lin-24(n432) dpy-4(e1166); yDp1. Only non-Unc non-Dpy animals that segregated Unc Dpy and Unc Lin animals were scored.
This strain was of genotype lin-24(n432) dpy-4(e1166); yDp1. Only non-Dpy animals were scored.
lin-33(n1043) homozygous males were mated with lin-33(n4514Δ) unc-30(e191) hermaphrodites, and non-Unc cross progeny were scored.
lin-24(n432) homozygous males were mated with lin-33(n4514Δ) unc-30(e191) hermaphrodites, and non-Unc cross progeny were scored.
lin-33(n1043) homozygous males were mated with lin-24(n4294Δ) dpy-4(e1166) hermaphrodites, and non-Dpy cross progeny were scored.
Both lin-24(n432) and lin-33(n1043) show strikingly sharp gene-dosage effects. We suggest that the cytotoxic effects of both lin-24(n432) and lin-33(n1043) are highly sensitive to the level of mutant protein. For both mutants, the phenotype of lin/+ is intermediate between that of lin/deletion and lin/lin. This result indicates that the level of gene expression is critical for the mutant gene products to be cytotoxic.
We also isolated a deletion allele of a gene that encodes the only protein that is highly similar to LIN-24 (32% identical and 51% similar), lntl-1 [lin-24 (twenty-four) like]. Both the lntl-1(n4763Δ) single mutant and the lntl-1(n4763Δ) lin-24(n4294Δ) double mutant appeared wild type. The wild-type phenotype of the doubly mutant strain lntl-1(n4763Δ) lin-24(n4294Δ) suggests that these two genes do not function redundantly in a critical process in C. elegans. There are no C. elegans genes that encode proteins with similarity to LIN-33.
The cytotoxic effect of lin-24 and lin-33 each requires the function of the other gene:
In addition to isolating loss-of-function alleles of both lin-24 and lin-33 in their respective dominant suppressor screens, we also isolated alleles of the other gene. Specifically, we obtained one loss-of-function allele of lin-33, n1968, in the lin-24(n432) screen and one loss-of-function allele of lin-24, n2333, in the lin-33(n1043) screen. These results suggested a genetic interaction between lin-24 and lin-33, which we explored further using our deletion alleles. Strains of the genotypes lin-33(n1043) lin-24(n4294Δ) or lin-33(n4514Δ) lin-24(n432) were phenotypically wild type (Table 5). Additionally, as suggested by their isolation in F1 screens, loss-of-function alleles of lin-24 and lin-33 can each suppress the vulvaless phenotype caused by gain-of-function mutations in the other gene when the former are present only in one copy (data not shown). For example, animals of the genotype lin-33(n1043) lin-24(n4294Δ)/lin-33(n1043) lin-24(+) were not vulvaless, so a loss of one copy of lin-24 was able to suppress the vulvaless phenotype of lin-33(gf) mutant animals. Similarly, the vulvaless phenotype of lin-24(n432)/lin-24(+) animals was completely suppressed by the loss of one copy of lin-33, and the vulvaless phenotype of lin-33(n1043)/lin-33(+) animals was completely suppressed by the loss of one copy of lin-24 (Table 5). Together these results demonstrate that the cytotoxic forms of lin-24 and lin-33 each require the function of the other gene for their cytotoxicities.
lin-24- and lin-33-induced cytotoxicity is partially dependent on the genes that function to engulf and degrade dying cells:
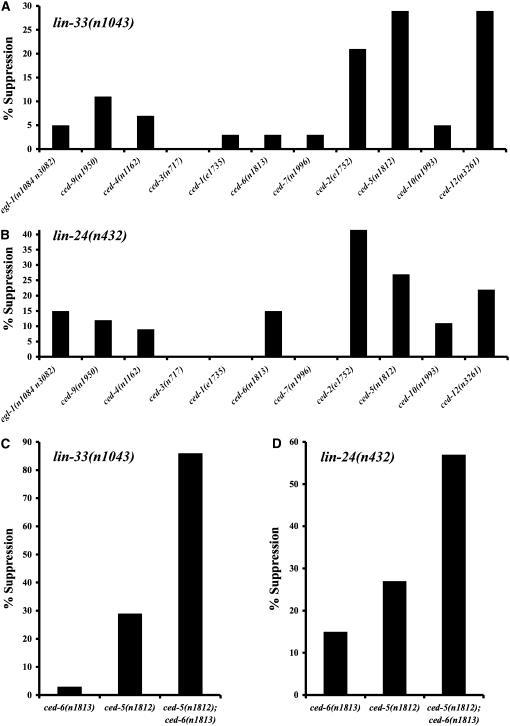
To determine whether genes involved in programmed cell deaths in C. elegans are also involved in the lin-24- or lin-33-induced cytotoxicity, we tested mutations in these genes for effects on the vulvaless phenotype of lin-24(n432) and lin-33(1043) animals. Loss-of-function mutations of genes that promote programmed cell death and a protective gain-of-function allele of ced-9 had a modest effect on the vulvaless phenotype (Figure 5, A and B). Notably, elimination of the caspase gene ced-3, the most downstream gene that functions in the killing step of all programmed cell deaths, had no effect on the vulvaless phenotype of lin-24(n432) and lin-33(n1043) animals. Loss-of-function alleles of the cell-death genes ced-2, ced-5, and ced-12 most strongly suppressed the vulvaless phenotype of lin-24(n432) and lin-33(n1043) animals (Figure 5, A and B). ced-2, ced-5, and ced-12 function together in one of two engulfment pathways involved in the removal of cell corpses during programmed cell death (Ellis et al. 1991; Gumienny et al. 2001; Wu et al. 2001; Zhou et al. 2001). ced-10 is also proposed to function together with ced-2, ced-5, and ced-12 (Ellis et al. 1991; Gumienny et al. 2001; Wu et al. 2001; Zhou et al. 2001). Since ced-10 is an essential gene, we used a partial loss-of-function allele, which might explain why we did not observe suppression of the vulvaless phenotype by this allele of ced-10. Loss-of-function alleles of genes that function together in the second engulfment pathway, ced-1, ced-6, and ced-7, did not suppress the vulvaless phenotype of lin-24(n432) and lin-33(n1043) animals (Figure 5, A and B).
Figure 5.—
The cytotoxicity of lin-24(n432) and lin-33(n1043) is partially dependent on genes that mediate the removal of apoptotic cell corpses. The penetrances of the vulvaless phenotypes of lin-24(n432); ced and lin-33(n1043); ced strains were analyzed and the percentage of suppression was calculated as described (see materials and methods). At least 56 animals were assayed for each genotype. (A) Mutations in some of the genes that act in programmed cell death partially suppress the vulvaless phenotype of lin-33(n1043) animals. Genes are organized in the following order along the x-axis: cell-killing genes in order of action (egl-1, ced-9, ced-4, and ced-3); one of two partially redundant engulfment pathways (ced-1, ced-6, and ced-7); and the other partially redundant engulfment pathway (ced-2, ced-5, ced-10, and ced-12). (B) Mutations in some of the genes required for programmed cell death partially suppress the vulvaless phenotype of lin-24(n432) animals. (C) A ced-5 and ced-6 double mutant was more strongly suppressed for the vulvaless phenotype caused by lin-33(n1043) than was either single mutant. (D) A ced-5 and ced-6 double mutant was more strongly suppressed for the vulvaless phenotype caused by lin-24(n432) than was either single mutant.
We also examined the ability of mutations disrupting genes that function in programmed cell death to suppress the vulvaless phenotype of animals heterozygous for lin-24(n432) or lin-33(n1043). Since the penetrances of the vulvaless phenotype caused by these alleles are weaker when heterozygous, this background might be sensitized such that it enables detection of roles in the cytotoxicity for additional genes. As expected from the results with lin-24(n432) and lin-33(n1043) homozygotes, loss-of-function mutations in ced-2, ced-5, and ced-12 strongly suppressed the vulvaless phenotype of animals heterozygous for the cytotoxic mutations (supplemental Figure 1). For example, ced-12 mutations almost completely restored the ability of either lin-24(n432)/lin-24(+) or lin-33(n1043)/lin-33(+) animals to lay eggs (97% suppression for both; n = 20 and 50, respectively). When lin-24(n432) or lin-33(n1043) was heterozygous, alleles of additional cell-death genes were able to suppress the vulvaless phenotype. For example, the partial loss-of-function allele ced-10(n1993) strongly suppressed both lin-24(n432)/lin-24(+) and lin-33(n1043)/lin-33(+), supporting the view that ced-10 might act with ced-2, ced-5, and ced-12 in promoting the cytotoxicity.
Unexpectedly, these experiments also revealed roles for other genes that function in programmed cell death. For example, loss of the BH3-only killer gene egl-1, the function of which is required for essentially all somatic programmed cell deaths in C. elegans (Conradt and Horvitz 1998), strongly suppressed the vulvaless phenotype of animals heterozygous for lin-24(n432) or for lin-33(n1043) (88 and 40%, respectively; n = 67 and 63, respectively). Additionally, the lin-24(n432)/lin-24(+) strain was suppressed by loss-of-function mutations of ced-4, ced-1, and ced-6 (58, 50, and 77%, respectively; n = 68, 70, and 67, respectively). The penetrance of the vulvaless phenotype of lin-24(n432) mutants is less than that of lin-33(n1043) mutants, especially when heterozygous, and presumably for this reason is more easily modified by the loss of function of genes that act in programmed cell death.
To determine the effect of simultaneously eliminating the two partially redundant engulfment pathways on the cytotoxicity, we constructed strains mutant in both ced-5 and ced-6 in combination with homozygous lin-24(n432) or lin-33(n1043). The elimination of both engulfment pathways suppressed the vulvaless phenotype to a greater extent than did the elimination of either single engulfment pathway (Figure 5, C and D).
To determine if the mutations that most strongly suppressed the vulvaless phenotype of lin-24(n432) and lin-33(n1043) animals [i.e., ced-2(e1752) and the combination of ced-5(n1812) and ced-6(n1813)] affected Pn.p cell survival, we counted the number of Pn.p cell nuclei present in these strains. Consistent with the observed suppression of the vulvaless phenotype, Pn.p cell death was suppressed in each of the strains examined (Table 6). Unlike the lin-24(n432) and lin-33(n1043) strains, when Pn.p cells were missing in the suppressed strains, a cell corpse was usually present at the location of the missing Pn.p cell. This observation suggests that when a Pn.p cell dies, its removal requires the genes that mediate the removal of apoptotic corpses. Consistent with this genetically established role for the process of engulfment in Pn.p cell death, our electron microscopic studies of Pn.p cells in lin-24 and lin-33 mutants indicate that dying Pn.p cells can be engulfed by neighboring cells (supplemental Figure 2).
TABLE 6.
lin-33(n1043)- and lin-24(n432)-induced Pn.p cell death is partially dependent on genes that mediate the removal of apoptotic corpses
| Genotype | Average no. of Pn.p nuclei ± SD |
|---|---|
| Wild type | 11 |
| lin-33(n1043) | 6.5 ± 1.4 |
| ced-2(e1752) lin-33(n1043) | 9.0 ± 1.1 |
| ced-6(n1813); lin-33(n1043) ced-5(n1812) | 10.1 ± 0.8 |
| lin-24(n432) | 7.6 ± 1.6 |
| ced-2(e1752) lin-24(n432) | 10.6 ± 0.6 |
| ced-6(n1813); ced-5(n1812) lin-24(n432) | 9.8 ± 0.9 |
The average number of Pn.p nuclei per animal was determined by counting the number of Pn.p nuclei present ∼24 hr after hatching using Nomarski optics. Twenty animals were examined for each strain.
Together the observed suppression both of the vulvaless phenotype and of Pn.p cell death indicates that the engulfment process, and most importantly the engulfment pathway involving ced-2, ced-5, and ced-12, plays an important role in promoting the cytotoxicity that affects both Pn.p cell fate and Pn.p cell survival caused by gain-of-function mutations in lin-24 and lin-33.
DISCUSSION
Mutations in lin-24 and lin-33 dominantly cause the Pn.p hypodermal blast cells, a subset of which gives rise to the vulva, to die or to fail to undergo cell division, thereby resulting in a vulvaless phenotype. We have characterized lin-24 and lin-33 genetically and molecularly. We found that the mutations that result in Pn.p cell cytotoxicity are gain-of-function mutations and that lin-24 and lin-33 act together to cause the abnormalities of the Pn.p cells. Although the Pn.p cell cytotoxicity shares some genetic and morphological characteristics with both apoptotic and necrotic cell deaths that occur in C. elegans, this cytotoxicity is genetically and morphologically distinct from these other cell death processes. It is also distinct from the recently described linker cell death, which is controlled by an apparently cell-autonomous program and does not involve such engulfment cell-death genes as ced-2, ced-5, and ced-12 (Abraham et al. 2007), which have substantial roles in lin-24- and lin-33-induced cell death. The death process caused by mutations in the uncloned gene pvl-5, which affects Pn.p cells in a way that, on the basis of observations using Nomarski optics, seems similar to the effects of lin-24 and lin-33 mutations (Joshi and Eisenmann 2004).
lin-24- and lin-33-induced cytotoxicity does not require most of the genes that act in the execution of programmed cell death, since the cytotoxicity, as measured by the suppression of the vulvaless phenotype, persists in animals homozygous for mutations in the Bcl-2 family member ced-9, the Apaf-1 homolog ced-4, and the caspase gene ced-3. The partial suppression of the weaker vulvaless phenotype seen in animals heterozygous for either lin-24(n432) or lin-33(n1043) by the BH3-only killer gene egl-1 is surprising, as it suggests that egl-1, which is the most upstream gene that functions in all somatic programmed cell deaths (Conradt and Horvitz 1998), might, in cell deaths other than those occurring through the process of apoptotic programmed cell death, function independently from ced-3, ced-4, and ced-9. The lin-24- and lin-33-induced cytotoxicity is most strongly dependent on the functions of ced-2, ced-5, and ced-12, which act in one of the two partially redundant pathways that mediate the engulfment of cell corpses generated by both programmed and necrotic cell deaths (Ellis et al. 1991; Chung et al. 2000; Gumienny et al. 2001; Wu et al. 2001; Zhou et al. 2001). Simultaneous elimination of the function of genes from both engulfment pathways even more strongly prevents the cytotoxicity, suggesting a more general role for engulfment that extends beyond ced-2, ced-5, and ced-12.
What are the molecular activities of mutant and wild-type LIN-24 and LIN-33?
The toxin-like domain found in LIN-24 suggests a possible mechanism for the lin-24- and lin-33-mediated cytotoxicity. The toxins with similarity to LIN-24 are thought to form membrane complexes that alter eukaryotic membrane permeability and cause osmotic lysis (Gilbert 2002). Perhaps the lin-24 and lin-33 gain-of-function mutations inappropriately activate a toxin-like activity of the toxin-like domain of LIN-24, causing sickness and cell death. The results of our dosage studies, which indicate a novel, abnormal activity for the allele lin-24(n432), are consistent with such a model.
Given the similarity of LIN-24 to bacterial toxins, what might be the wild-type function of the lin-24 gene product in C. elegans? Neither loss of lin-24 function alone nor loss of lin-24 function in combination with the loss of its only homolog lntl-1 results in any obvious abnormal phenotype. Almost all genes similar to lin-24 are found in bacteria, which C. elegans eats. These bacterial toxins are capable of killing animals, such as mosquitoes and cattle, by forming oligomers that increase membrane permeability and cause osmotic lysis and cellular destruction (De Maagd et al. 2003). Perhaps wild-type LIN-24 normally functions to interact with bacterial toxins and inactivate them, possibly by a mechanism that requires LIN-33. The ability of wild-type LIN-24 to antagonize mutant LIN-24 supports this possibility. Such an activity might enable C. elegans to safely consume or survive exposure to bacteria that produce these deadly toxins. Consistent with this possibility is the observation that lin-24 is expressed in both the pharynx and intestine, the main tissues that are exposed to bacteria (Hunt-Newbury et al. 2007).
The role of engulfment in cell death:
The observation that mutations in ced-2, ced-5, and ced-12 reduce lin-24- and lin-33-induced cytotoxicity is similar to the observation that engulfment genes are involved not only in engulfing corpses generated by programmed cell death but also in promoting the death process itself (Hoeppner et al. 2001; Reddien et al. 2001). The programmed cell death of at least one cell type, one of the pair of cells B.alapaav and B.arapaav in the male tail, appears to be entirely dependent on engulfment (Hedgecock et al. 1983). Additional studies of lin-24- and lin-33-induced cytotoxicity could provide valuable insight not only into how engulfment contributes to cytotoxicity but also into how engulfment contributes to cell killing in programmed cell death.
In contrast to programmed cell death and to lin-24- and lin-33-induced cytotoxicity, necrotic cell death is not dependent on the function of the engulfment genes. Specifically, engulfment genes act in the engulfment and removal of the necrotic corpses seen in animals with gain-of-function mutations in mec-4, deg-3, and deg-1, but mutations that eliminate the function of the engulfment genes appear to affect only the timing of the disappearance of these corpses and not cell survival (Chung et al. 2000). This difference demonstrates that although common mechanisms contribute to the removal of cell corpses generated by different cell-death processes, there are critical differences among these processes.
ced-2, ced-5, ced-10, and ced-12 function in parallel to ced-1, ced-6, and ced-7 in the rapid removal of corpses generated by programmed and necrotic cell death (Ellis et al. 1991; Chung et al. 2000; Gumienny et al. 2001; Wu et al. 2001; Zhou et al. 2001). Whereas the function of the ced-1, -6, -7 pathway is unclear, the ced-2, -5, -10, -12 pathway is likely a signal transduction pathway that drives a cytoskeletal rearrangement (Reddien and Horvitz 2004; Mangahas and Zhou 2005). CED-2 is an Src-homology 2 (SH2) and Src-homology 3 (SH3) domain-containing adapter protein (Reddien and Horvitz 2000), CED-10 is a Rac-like GTPase (Reddien and Horvitz 2000), and CED-5 and CED-12 likely activate CED-10/Rac1 by exchanging GDP for GTP (Wu and Horvitz 1998; Gumienny et al. 2001; Wu et al. 2001; Zhou et al. 2001; Brugnera et al. 2002). The mammalian counterparts of CED-2, CED-5, and CED-12 (Crk, DOCK180, and ELMO1, respectively) regulate the CED-10 homolog Rac and function in cytoskeletal reorganization (Brugnera et al. 2002; Akakura et al. 2005; Lu and Ravichandran 2006), which is important for cell migration, axonal outgrowth, and for phagocytosis in programmed cell death.
Whereas the ced-1, -6, -7 pathway appears to be largely specific for the engulfment of cell corpses, the ced-2, -5, -10, -12 pathway is involved in several other processes, including gonadal migration and axonal outgrowth (Reddien and Horvitz 2004; Mangahas and Zhou 2005), as well as the promotion of lin-24- and lin-33-induced cytotoxicity. We suggest that the ced-2, -5, -10, -12 pathway responds to a signal presented on the surface of a cell that is sick or dying (Figure 6). It is possible that the signal presented by the cells affected by lin-24- and lin-33-induced cytotoxicity is the same as that recognized by the ced-2, -5, -10, -12 pathway in programmed cell death. The ced-1, -6, -7 pathway, by contrast, might recognize a different signal that is more specific to corpses generated by programmed cell death.
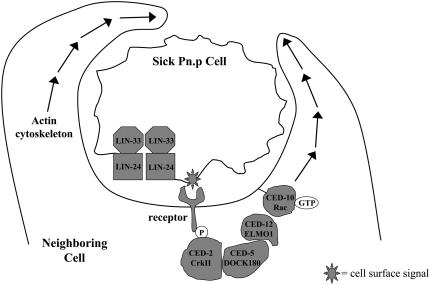
Figure 6.—
A model for the effects of engulfment on lin-24- and lin-33-induced cytotoxicity. Gain-of-function mutations in lin-24 and lin-33 make the Pn.p cells sick, possibly by forming membrane complexes that alter membrane permeability in a process that is similar to that of the bacterial toxins that share similarity with LIN-24. A consequence of this sickness is that a signal is presented on the cell membrane and recognized by a neighboring cell. The neighboring cell then promotes the cytotoxicity in a process that requires the engulfment genes ced-2, ced-5, ced-10, and ced-12, which likely act by reorganizing the actin cytoskeleton (adapted from Reddien and Horvitz 2004). Additional genes that function in programmed cell death, such as ced-6 and egl-1, might also play a role in lin-24- and lin-33-induced cytotoxicity.
The role of the engulfment genes in promoting the cytotoxicity that affects the fate and survival of the Pn.p cells in lin-24 and lin-33 mutant animals expands the established role for engulfment in cell killing. If the promotion of death by engulfing cells is a general phenomenon for many sick, injured, or apoptotic cells, pharmacological inhibition of engulfment might broadly promote the survival of dying cells. The ability to simultaneously promote cell survival of both apoptotic and injured cells could greatly limit tissue damage in critical clinical situations, such as after traumatic brain injury or a myocardial infarction, during which cells die by both apoptotic and nonapoptotic mechanisms (Krijnen et al. 2002; Mehta et al. 2007).
Acknowledgments
We thank Erika Hartwieg and Victoria Hatch for generating the EM images; Beth Castor, Na An, Shannon McGonagle, and Andrew Hellman for technical assistance; Yuji Kohara for providing a cDNA clone; and Hillel Schwartz, Niels Ringstad, and Craig Ceol for critical reading of this manuscript. B.D.G. was supported in part by a Howard Hughes Medical Institute Predoctoral Fellowship. H.R.H. is the David H. Koch Professor of Biology at MIT and is an Investigator of the Howard Hughes Medical Institute. This work was supported by grants GM24663 and GM24943 and by the Howard Hughes Medical Institute.
References
- Abraham, M. C., Y. Lu and S. Shaham, 2007. A morphologically conserved nonapoptotic program promotes linker cell death in Caenorhabditis elegans. Dev. Cell. 12 73–86. [DOI] [PubMed] [Google Scholar]
- Akakura, S., B. Kar, S. Singh, L. Cho, N. Tibrewal et al., 2005. C-terminal SH3 domain of CrkII regulates the assembly and function of the DOCK180/ELMO Rac-GEF. J. Cell. Physiol. 204 344–351. [DOI] [PubMed] [Google Scholar]
- Bargmann, C. I., E. Hartwieg and H. R. Horvitz, 1993. Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell 74 515–527. [DOI] [PubMed] [Google Scholar]
- Barstead, R. J., and R. H. Waterston, 1989. The basal component of the nematode dense-body is vinculin. J. Biol. Chem. 264 10177–10185. [PubMed] [Google Scholar]
- Berger, A. J., A. C. Hart and J. M. Kaplan, 1998. G alphas-induced neurodegeneration in Caenorhabditis elegans. J. Neurosci. 18 2871–2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom, L., and H. R. Horvitz, 1997. The Caenorhabditis elegans gene unc-76 and its human homologs define a new gene family involved in axonal outgrowth and fasciculation. Proc. Natl. Acad. Sci. USA 94 3414–3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner, S., 1974. The genetics of Caenorhabditis elegans. Genetics 77 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugnera, E., L. Haney, C. Grimsley, M. Lu, S. F. Walk et al., 2002. Unconventional Rac-GEF activity is mediated through the Dock180-ELMO complex. Nat. Cell Biol. 4 574–582. [DOI] [PubMed] [Google Scholar]
- Chalfie, M., and J. Sulston, 1981. Developmental genetics of the mechanosensory neurons of Caenorhabditis elegans. Dev. Biol. 82 358–370. [DOI] [PubMed] [Google Scholar]
- Chalfie, M., and E. Wolinsky, 1990. The identification and suppression of inherited neurodegeneration in Caenorhabditis elegans. Nature 345 410–416. [DOI] [PubMed] [Google Scholar]
- Chung, S., T. L. Gumienny, M. O. Hengartner and M. Driscoll, 2000. A common set of engulfment genes mediates removal of both apoptotic and necrotic cell corpses in C. elegans. Nat. Cell. Biol. 2 931–937. [DOI] [PubMed] [Google Scholar]
- Clark, D. V., and D. L. Baillie, 1992. Genetic analysis and complementation by germ-line transformation of lethal mutations in the unc-22 IV region of Caenorhabditis elegans. Mol. Gen. Genet. 232 97–105. [DOI] [PubMed] [Google Scholar]
- Conradt, B., and H. R. Horvitz, 1998. The C. elegans protein EGL-1 is required for programmed cell death and interacts with the Bcl-2-like protein CED-9. Cell 93 519–529. [DOI] [PubMed] [Google Scholar]
- de Maagd, R. A., A. Bravo, C. Berry, N. Crickmore and H. E. Schnepf, 2003. Structure, diversity, and evolution of protein toxins from spore-forming entomopathogenic bacteria. Annu. Rev. Genet. 37 409–433. [DOI] [PubMed] [Google Scholar]
- Degterev, A., M. Boyce and J. Yuan, 2003. A decade of caspases. Oncogene 22 8543–8567. [DOI] [PubMed] [Google Scholar]
- DeLong, L., L. P. Casson and B. J. Meyer, 1987. Assessment of X chromosome dosage compensation in Caenorhabditis elegans by phenotypic analysis of lin-14. Genetics 117 657–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll, M., and M. Chalfie, 1991. The mec-4 gene is a member of a family of Caenorhabditis elegans genes that can mutate to induce neuronal degeneration. Nature 349 588–593. [DOI] [PubMed] [Google Scholar]
- Driscoll, M., and B. Gerstbrein, 2003. Dying for a cause: invertebrate genetics takes on human neurodegeneration. Nat. Rev. Genet. 4 181–194. [DOI] [PubMed] [Google Scholar]
- Edinger, A. L., and C. B. Thompson, 2004. Death by design: apoptosis, necrosis and autophagy. Curr. Opin. Cell Biol. 16 663–669. [DOI] [PubMed] [Google Scholar]
- Ellis, R. E., D. M. Jacobson and H. R. Horvitz, 1991. Genes required for the engulfment of cell corpses during programmed cell death in Caenorhabditis elegans. Genetics 129 79–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson, E. L., and H. R. Horvitz, 1985. Identification and characterization of 22 genes that affect the vulval cell lineages of the nematode Caenorhabditis elegans. Genetics 110 17–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson, E. L., P. W. Sternberg and H. R. Horvitz, 1987. A genetic pathway for the specification of the vulval cell lineages of Caenorhabditis elegans. Nature 326 259–267. [DOI] [PubMed] [Google Scholar]
- Gilbert, R. J., 2002. Pore-forming toxins. Cell. Mol. Life Sci. 59 832–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumienny, T. L., E. Lambie, E. Hartwieg, H. R. Horvitz and M. O. Hengartner, 1999. Genetic control of programmed cell death in the Caenorhabditis elegans hermaphrodite germline. Development 126 1011–1022. [DOI] [PubMed] [Google Scholar]
- Gumienny, T. L., E. Brugnera, A. C. Tosello-Trampont, J. M. Kinchen, L. B. Haney et al., 2001. CED-12/ELMO, a novel member of the CrkII/Dock180/Rac pathway, is required for phagocytosis and cell migration. Cell 107 27–41. [DOI] [PubMed] [Google Scholar]
- Hall, D. H., G. Gu, J. Garcia-Anoveros, L. Gong, M. Chalfie et al., 1997. Neuropathology of degenerative cell death in Caenorhabditis elegans. J. Neurosci. 17 1033–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedgecock, E. M., J. E. Sulston and J. N. Thomson, 1983. Mutations affecting programmed cell deaths in the nematode Caenorhabditis elegans. Science 220 1277–1279. [DOI] [PubMed] [Google Scholar]
- Hengartner, M. O., and H. R. Horvitz, 1994. C. elegans cell survival gene ced-9 encodes a functional homolog of the mammalian proto-oncogene bcl-2. Cell 76 665–676. [DOI] [PubMed] [Google Scholar]
- Hengartner, M. O., R. E. Ellis and H. R. Horvitz, 1992. Caenorhabditis elegans gene ced-9 protects cells from programmed cell death. Nature 356 494–499. [DOI] [PubMed] [Google Scholar]
- Hockenbery, D., G. Nunez, C. Milliman, R. D. Schreiber and S. J. Korsmeyer, 1990. Bcl-2 is an inner mitochondrial membrane protein that blocks programmed cell death. Nature 348 334–336. [DOI] [PubMed] [Google Scholar]
- Hoeppner, D. J., M. O. Hengartner and R. Schnabel, 2001. Engulfment genes cooperate with ced-3 to promote cell death in Caenorhabditis elegans. Nature 412 202–206. [DOI] [PubMed] [Google Scholar]
- Horvitz, H. R., 1999. Genetic control of programmed cell death in the nematode Caenorhabditis elegans. Cancer Res. 59 1701s–1706s. [PubMed] [Google Scholar]
- Hunt-Newbury, R., R. Viveiros, R. Johnsen, A. Mah, D. Anastas et al., 2007. High-throughput in vivo analysis of gene expression in Caenorhabditis elegans. PLoS Biol. 5 e237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen, G., E. Hazendonk, K. L. Thijssen and R. H. Plasterk, 1997. Reverse genetics by chemical mutagenesis in Caenorhabditis elegans. Nat. Genet. 17 119–121. [DOI] [PubMed] [Google Scholar]
- Joshi, P., and D. M. Eisenmann, 2004. The Caenorhabditis elegans pvl-5 gene protects hypodermal cells from ced-3-dependent, ced-4-independent cell death. Genetics 167 673–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble, J., 1981. Alterations in cell lineage following laser ablation of cells in the somatic gonad of Caenorhabditis elegans. Dev. Biol. 87 286–300. [DOI] [PubMed] [Google Scholar]
- Kimble, J., and D. Hirsh, 1979. The postembryonic cell lineages of the hermaphrodite and male gonads in Caenorhabditis elegans. Dev. Biol. 70 396–417. [DOI] [PubMed] [Google Scholar]
- Korswagen, H. C., J. H. Park, Y. Ohshima and R. H. Plasterk, 1997. An activating mutation in a Caenorhabditis elegans Gs protein induces neural degeneration. Genes Dev. 11 1493–1503. [DOI] [PubMed] [Google Scholar]
- Krijnen, P. A., R. Nijmeijer, C. J. Meijer, C. A. Visser, C. E. Hack et al., 2002. Apoptosis in myocardial ischaemia and infarction. J. Clin. Pathol. 55 801–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, C. C., K. Hong, M. Kinnell, M. Chalfie and M. Driscoll, 1996. Sequence and transmembrane topology of MEC-4, an ion channel subunit required for mechanotransduction in Caenorhabditis elegans. J. Cell Biol. 133 1071–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettre, G., and M. O. Hengartner, 2006. Developmental apoptosis in C. elegans: a complex CEDnario. Nat. Rev. Mol. Cell Biol. 7 97–108. [DOI] [PubMed] [Google Scholar]
- Liu, J. W., A. G. Porter, B. Y. Wee and T. Thanabalu, 1996. New gene from nine Bacillus sphaericus strains encoding highly conserved 35.8-kilodalton mosquitocidal toxins. Appl. Environ. Microbiol. 62 2174–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, L. X., J. M. Spoerke, E. L. Mulligan, J. Chen, B. Reardon et al., 1999. High-throughput isolation of Caenorhabditis elegans deletion mutants. Genome Res. 9 859–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, M., and K. S. Ravichandran, 2006. Dock180-ELMO cooperation in Rac activation. Methods Enzymol. 406 388–402. [DOI] [PubMed] [Google Scholar]
- Mangahas, P. M., and Z. Zhou, 2005. Clearance of apoptotic cells in Caenorhabditis elegans. Semin. Cell Dev. Biol. 16 295–306. [DOI] [PubMed] [Google Scholar]
- Mehta, S. L., N. Manhas and R. Raghubir, 2007. Molecular targets in cerebral ischemia for developing novel therapeutics. Brain Res. Rev. 54 34–66. [DOI] [PubMed] [Google Scholar]
- Mello, C. C., J. M. Kramer, D. Stinchcomb and V. Ambros, 1991. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien, P. W., and H. R. Horvitz, 2000. CED-2/CrkII and CED-10/Rac control phagocytosis and cell migration in Caenorhabditis elegans. Nat. Cell Biol. 2 131–136. [DOI] [PubMed] [Google Scholar]
- Reddien, P. W., and H. R. Horvitz, 2004. The engulfment process of programmed cell death in Caenorhabditis elegans. Annu. Rev. Cell Dev. Biol. 20 193–221. [DOI] [PubMed] [Google Scholar]
- Reddien, P. W., S. Cameron and H. R. Horvitz, 2001. Phagocytosis promotes programmed cell death in C. elegans. Nature 412 198–202. [DOI] [PubMed] [Google Scholar]
- Riddle, D. L., T. Blumenthal, B. I. Meyer and J. R. Priess, 1997. C. elegans II. Cold Spring Harbor Laboratory Press, Plainview, N.Y. [PubMed]
- Robertson, A. M. G., and J. N. Thomson, 1982. Morphology of programmed cell death in the ventral nerve cord of C. elegans larvae. J. Embryol. Exp. Morphol. 67 89–100. [Google Scholar]
- Shaham, S., and H. R. Horvitz, 1996. Developing Caenorhabditis elegans neurons may contain both cell-death protective and killer activities. Genes Dev. 10 578–591. [DOI] [PubMed] [Google Scholar]
- Sternberg, P. W., and H. R. Horvitz, 1986. Pattern formation during vulval development in C. elegans. Cell 44 761–772. [DOI] [PubMed] [Google Scholar]
- Sulston, J. E., and H. R. Horvitz, 1977. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 56 110–156. [DOI] [PubMed] [Google Scholar]
- Sulston, J. E., and J. G. White, 1980. Regulation and cell autonomy during postembryonic development of Caenorhabditis elegans. Dev. Biol. 78 577–597. [DOI] [PubMed] [Google Scholar]
- Sulston, J. E., D. G. Albertson and J. N. Thomson, 1980. The Caenorhabditis elegans male: postembryonic development of nongonadal structures. Dev. Biol. 78 542–576. [DOI] [PubMed] [Google Scholar]
- Sulston, J. E., E. Schierenberg, J. G. White and J. N. Thomson, 1983. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100 64–119. [DOI] [PubMed] [Google Scholar]
- Syntichaki, P., K. Xu, M. Driscoll and N. Tavernarakis, 2002. Specific aspartyl and calpain proteases are required for neurodegeneration in C. elegans. Nature 419 939–944. [DOI] [PubMed] [Google Scholar]
- Treinin, M., and M. Chalfie, 1995. A mutated acetylcholine receptor subunit causes neuronal degeneration in C. elegans. Neuron 14 871–877. [DOI] [PubMed] [Google Scholar]
- Trent, C., N. Tsuing and H. R. Horvitz, 1983. Egg-laying defective mutants of the nematode Caenorhabditis elegans. Genetics 104 619–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaux, D. L., S. Cory and J. M. Adams, 1988. Bcl-2 gene promotes haemopoietic cell survival and cooperates with c-myc to immortalize pre-B cells. Nature 335 440–442. [DOI] [PubMed] [Google Scholar]
- Vaux, D. L., I. L. Weissman and S. K. Kim, 1992. Prevention of programmed cell death in Caenorhabditis elegans by human bcl-2. Science 258 1955–1957. [DOI] [PubMed] [Google Scholar]
- Wicks, S. R., R. T. Yeh, W. R. Gish, R. H. Waterston and R. H. Plasterk, 2001. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat. Genet. 28 160–164. [DOI] [PubMed] [Google Scholar]
- Williams, B. D., B. Schrank, C. Huynh, R. Shownkeen and R. H. Waterston, 1992. A genetic mapping system in Caenorhabditis elegans based on polymorphic sequence-tagged sites. Genetics 131 609–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Y. C., and H. R. Horvitz, 1998. C. elegans phagocytosis and cell-migration protein CED-5 is similar to human DOCK180. Nature 392 501–504. [DOI] [PubMed] [Google Scholar]
- Wu, Y. C., M. C. Tsai, L. C. Cheng, C. J. Chou and N. Y. Weng, 2001. C. elegans CED-12 acts in the conserved crkII/DOCK180/Rac pathway to control cell migration and cell corpse engulfment. Dev. Cell 1 491–502. [DOI] [PubMed] [Google Scholar]
- Wyllie, A. H., 1981. Cell death: a new classification separating apoptosis from necrosis, pp. 9–34 in Cell Death in Biology and Pathology, edited by I. D. Bowen and R. A. Lockshin. Chapman & Hall, London.
- Xu, K., N. Tavernarakis and M. Driscoll, 2001. Necrotic cell death in C. elegans requires the function of calreticulin and regulators of Ca(2+) release from the endoplasmic reticulum. Neuron 31 957–971. [DOI] [PubMed] [Google Scholar]
- Yuan, J., S. Shaham, S. Ledoux, H. M. Ellis and H. R. Horvitz, 1993. The C. elegans cell death gene ced-3 encodes a protein similar to mammalian interleukin-1 beta-converting enzyme. Cell 75 641–652. [DOI] [PubMed] [Google Scholar]
- Zhou, Z., E. Caron, E. Hartwieg, A. Hall and H. R. Horvitz, 2001. The C. elegans PH domain protein CED-12 regulates cytoskeletal reorganization via a Rho/Rac GTPase signaling pathway. Dev. Cell 1 477–489. [DOI] [PubMed] [Google Scholar]
- Zong, W. X., and C. B. Thompson, 2006. Necrotic death as a cell fate. Genes Dev. 20 1–15. [DOI] [PubMed] [Google Scholar]