Abstract
C57BL/6J (B6) mice containing the Mus domesticus poschiavinus Y chromosome, YPOS, develop ovarian tissue, whereas testicular tissue develops in DBA/2J or 129S1/SvImJ (129) mice containing the YPOS chromosome. To identify genes involved in sex determination, we used a congenic strain approach to determine which chromosomal regions from 129Sl/SvImJ provide protection against sex reversal in XYPOS mice of the C57BL/6J.129-YPOS strain. Genome scans using microsatellite and SNP markers identified a chromosome 11 region of 129 origin in C57BL/6J.129-YPOS mice. To determine if this region influenced testis development in XYPOS mice, two strains of C57BL/6J-YPOS mice were produced and used in genetic experiments. XYPOS adults homozygous for the 129 region had a lower incidence of sex reversal than XYPOS adults homozygous for the B6 region. In addition, many homozygous 129 XYPOS fetuses developed normal-appearing testes, an occurrence never observed in XYPOS mice of the C57BL/6J-YPOS strain. Finally, the amount of testicular tissue observed in ovotestes of heterozygous 129/B6 XYPOS fetuses was greater than the amount observed in ovotestes of homozygous B6 XYPOS fetuses. We conclude that a chromosome 11 locus derived from 129Sl/SvImJ essentially protects against sex reversal in XYPOS mice. A number of genes located in this chromosome 11 region are discussed as potential candidates.
INITIATING the male-determining cascade in mammals begins with the expression of the Y chromosome gene Sry (Sinclair et al. 1990; Ross and Capel 2005). In both humans and mice, mutations in Sry cause XY sex reversal (Jäger et al. 1990; Lovell-Badge and Robertson 1990), and presence of exogenous Sry (Fechner et al. 1993; Koopman et al. 1991) causes XX sex reversal. Because cases of human sex reversal are relatively rare and the heterogeneity found within our species makes genotype-to-phenotype correlations difficult, mouse models of inherited sex reversal are particularly useful. All identified nonsynonymous SRY polymorphisms in humans are associated with varying degrees of XY gonadal dysgenesis or disrupted gonadal development (Hawkins et al. 1992). Although there is selection bias when analyzing such affected individuals, to date the human genome project has not identified any benign nonsynonymous polymorphisms. By contrast, at least 15 distinct Sry alleles are present among different species of Mus (Albrecht and Eicher 1997), and most Sry alleles of Mus domesticus and M. musculus origin direct normal male sex determination when present in C57BL/6J mice. This is quite striking, considering that the corresponding predicted SRY proteins vary in size due to a difference in the number of polyglutamine repeats within mouse Sry genes (Bowles et al. 1999), but not their human ortholog, and M. domesticus Sry genes encode a protein that is significantly shorter than the protein encoded for by M. musculus Sry genes (Albrecht and Eicher 1997).
Some M. domesticus Y chromosomes cause XY sex reversal when present on C57BL/6J background, an inbred laboratory strain that normally carries a M. musculus Y chromosome, i.e., M. musculus Sry gene (Albrecht and Eicher 1997). This inherited phenomenon, referred to as C57BL/6J-YDOM sex reversal, has been extensively studied for the M. domesticus poschiavinus Y chromosome (YPOS) (Eicher et al. 1982), as well as for the related M. domesticus tirano Y chromosome (YTIR) (Lee and Taketo 1994, 2001). Since the current genetic investigation is based on C57BL/6J-YPOS, and previous genetic experiments have been exclusively performed using that strain, we have concentrated only on it and not on other M. domesticus models. The failure to develop testes in these mice stems from an incompatibility of the SryPOS gene to initiate normal testicular development when C57BL/6J autosomal and/or X-linked factors are present. All XYPOS C57BL/6J-YPOS individuals develop some ovarian tissue (Eicher et al. 1982); half develop exclusively ovarian tissue, thus are completely sex reversed, and the remainder develop ovarian and testicular tissue, thus are partially sex reversed (Eicher et al. 1982; Eicher and Washburn 2001). An investigation using linkage analysis concluded that C57BL/6J-YPOS sex reversal is inherited as a complex trait influenced by at least three different autosomal loci and the SryPOS gene (Eicher et al. 1996).
Although protein isoform differences among M. domesticus Sry alleles have been demonstrated to play a role in the precise gonadal phenotype each causes when transferred to C57BL/6J (Albrecht et al. 2003), the molecular mechanisms through which those differences arise are unknown. In addition, the SryPOS gene, when introduced as a transgene into XX C57BL/6J mice, is capable of directing testicular development (Albrecht et al. 2003). This latter result suggests that overexpression of the SryPOS gene is capable of initiating testis development in C57BL/6J mice and it is the improper expression of the single copy of SryPOS carried on the YPOS chromosome that is the major factor underlying sex reversal, with the protein isoform difference contributing to a lesser degree.
In mice, Sry is normally expressed between E10.5 and E12.5 (where E is the embryonic developmental age) in the XY genital ridge (Koopman et al. 1990; Jeske et al. 1995; Bullejos and Koopman 2001), with peak Sry expression occurring in normal XY C57BL/6J genital ridges at ∼E11.5, i.e., at the 16–18 tail somite stage of development (Bullejos and Koopman 2005). In contrast, expression of the SryPOS gene peaks at ∼10–14 hr later in the genital ridges of XYPOS C57BL/6J-YPOS fetuses (Albrecht et al. 2003; Bullejos and Koopman 2005). However, SryPOS is expressed at the normal time and normal levels in the genital ridges of XYPOS (C57BL/6J-YPOS × DBA/2J)F1 mice (Albrecht et al. 2003), which supports the notion that upstream factors derived from the C57BL/6J genome, when homozygous, delay and reduce Sry expression levels and therefore contribute to C57BL/6J-YPOS sex reversal.
We used a congenic approach to identify loci that modify the C57BL/6J-YPOS sex-reversal phenotype with the goal of gaining further insight into male sex determination and regulation of Sry expression. Genomic elements from the 129 mouse strain were transferred onto C57BL/6J-YPOS background with selection for fully masculinized males at each backcross generation. The XYPOS mice of the resulting congenic strain, C57BL/6J.129-YPOS, were “protected” from sex reversal even though the majority of the genome was of C57BL/6J origin (Whitney et al. 2000). The focus of the presented study was to characterize the genetic make up of this congenic strain, with the aims of identifying the location of the protective 129-derived congenic region(s) and understanding how those regions affect testicular development.
MATERIALS AND METHODS
Genome scans:
The microsatellite genome scan was performed on DNA isolated from C57BL/6J.129-YPOS mice received at the University of California to detect 129 vs. B6 microsatellite markers. PCR amplification was performed using fluorescently labeled primer pairs consisting of labeled and unlabeled oligonucleotides. Individual PCR conditions were optimized for each primer pair. PCR products were analyzed using an ABI 3700 capillary sequencer (Applied Biosystems, Foster City, CA) at the University of California, Los Angeles (UCLA) Human Genetics Sequencing and Genotyping Core Facility. Data analysis was performed using the ABI PRISM Genotyper software (Applied Biosystems). One hundred forty-nine markers were used in the microsatellite scan (supplemental Table 1).
Two genome scans were performed using SNP markers. Approximately 183 SNP markers using DNA obtained from C57BL/6J.129-YPOS strain mice received at UCLA, with 37 markers located on chromosome 11 (supplemental Table 2). A genome scan also was performed on DNA obtained from C57BL/6J.129-YPOS strain maintained at The Jackson Laboratory. In this case, 404 SNP markers were used, 11 of which are located on chromosome 11 (supplemental Table 3). SNP genotyping was performed by The Jackson Laboratory JAX Services SNP-based genome scanning commercially available service (The Jackson Laboratory, Bar Harbor, ME). The results from the University of California SNP scan were reported by JAX services as SNP genotypes only for those SNPs that differed from C57BL/6J and for all SNPs on chromosome 11. For all other SNPs the data were reported as matching C57BL/6J. The full combined data from all three scans for chromosome 11 are depicted in supplemental Table 4.
The SNP scans also identified a single marker, rs13480100 on chromosome 9 as being of non-C57BL/6J origin. Interestingly, C57BL/6J is the only strain reported to have a T base pair at this locus, differing even from C57BL/6N, a closely related strain (data not shown). During the reconstruction of the C57BL/6J.129-YPOS strain the chromosome 9 rs13480100 region became of C57BL/6J origin.
Mouse strains:
Animals were housed according to the guidelines of UCLA's Division of Laboratory Animal Medicine. All experiments were approved by the Institutional Animal Care and Use Committees of The Jackson Laboratory and UCLA. The Jackson Laboratory is fully accredited by the American Association for Accreditation of Laboratory Animal Care.
Construction of the C57BL/6J.129-YPOS strain:
The original C57BL/6J.129-YPOS strain (Whitney et al. 2000) was produced by mating a C57BL/6J female to a 129-YPOS male. The nomenclature used to define the 129 strain used in this study, “129S1/Sv” is not in use today, making the actual 129 strain used unknown. On the basis of current nomenclature it is most likely that the strain used was 129Sl/SvImJ. For this reason, 129Sl/SvImJ was used in this study as a control, but the possibility exists that this is not identical to the 129 strain that was used in making the congenic C57BL/6J.129-YPOS strain. In the original experiment an F1 male was backcrossed to a C57BL/6J female, and an N2 XY normal-appearing male (on the basis of external and internal genitalia and testis size) was backcrossed to a C57BL/6J female. This mating scheme, normal-appearing male mated to C57BL/6J female was continued until the N13 backcross generation was reached. Thereafter, sister by brother matings were conducted to produce the C57BL/6J.129-YPOS strain. XY mice of the C57BL/6J.129-YPOS strain differ from XY mice of the C57BL/6J-YPOS strain in the following ways: (1) C57BL/6J.129-YPOS mice contain one or more chromosomal regions derived from the 129S1/SvImJ strain, and (2) XY C57BL/6J.129-YPOS adult mice are not sex reversed.
The reported ratio of phenotypic females to males in C57BL/6J.129-YPOS mice was 58:42, which does not differ significantly from 1:1, but is different from the sex ratio reported in C57BL/6J-YPOS mice (Eicher et al. 1982; Eicher and Washburn 2001). Thus, the selection process used selected for protection against XYPOS sex reversal. However, the location of the congenic region(s) derived from the 129 strain was not known, and fetal gonad analysis was not performed to investigate whether ovarian tissue was present during fetal development.
Construction of B6.129-Chr11-YPOS and B6-YPOS lines:
Mice from the C57BL/6J.129-YPOS congenic strain (Whitney et al. 2000) were a gift from Tom Abney and Barry Whitney, Medical College of Georgia. Embryos from the C57BL/6J.129-YPOS strain are preserved in The Jackson Laboratory frozen embryo bank.
Unfortunately, the C57BL/6J.129-YPOS females sent to UCLA failed to breed. Because presence of the 129-derived chromosome 11 region was detected before the breeding problem was evident, it was possible to essentially reestablish the C57BL/6J.129-YPOS genome. To accomplish this, a male offspring, obtained from mating a C57BL/6J.129-YPOS male to C57BL/6J female, was crossed to a C57BL/6J female. The N2 offspring were typed for the chromosomal 11 region to distinguish 129/B6 and B6/B6 individuals with microsatellite markers D11Mit71 and D11Mit4 or D11Mit320 and D11Mit284. In addition the Sry gene was sequenced to confirm that it was of YPOS origin (data not shown). A 129/B6 F1 female was mated to a 129/B6 F1 male. From this mating, a 129/129 female and male were identified and used to establish the B6.129-Chr11-YPOS line.
To establish a control B6-YPOS line, an N2 B6/B6 male (see above) was mated to a C57BL/6J female and thereafter XYPOS individuals were mated to C57BL/6J females. As is the case for XYPOS C57BL/6J-YPOS mice, XYPOS B6-YPOS mice are females or hermaphrodites. Some of these hermaphrodites develop sufficient testicular tissue to masculinize their internal and external genitalia, and these mice breed as males.
Sex chromosome genotyping:
DNA was extracted using the standard phenol/chloroform method. Chromosomal sex was determined with a PCR-based assay using a single primer pair, 5′-CCGCTGCCAAATTCTTTGG-3′ and 5′-TGAAGCTTTTGGCTTTGAG-3′ that detects the X-linked Smcx gene (330 bp) and the Y-linked Smcy gene (301 bp) (Bullejos and Koopman 2005). XX DNA produces a single PCR band, while XY DNA produces 2 bands when resolved on 2% agarose gel by electrophoresis. PCR amplification was performed using REDTaq ReadyMix PCR Reaction Mix (Sigma-Aldrich, St. Louis). An initial denaturation step at 94° for 3 min was followed by 35 cycles of 94° for 30 sec, 57° for 30 sec, 72° for 30 sec, and the process completed by a final extension at 72° for 7 min.
Confirming presence of the SryPOS gene in C57BL/6J.129-YPOS mice:
The Sry open reading frame was sequenced using DNA isolated from C57BL/6J.129-YPOS males. Sry was amplified using primers 5′-TTGATTTTTAGTGTTCAGCCCTACAGCC-3′ and 5′-AGCTGTTGCTGTCTTTGTGCTAGCC-3′ resulting in a band of ∼1.6 kb, which was sequenced using those same primers and primers 5′-GGAGTAGAGCTGCACACCTGTACTCC-3′ and 5′-CCAGTGTCATGAGACTGCCAACC-3′ (Albrecht and Eicher 1997). PCR amplification was performed using REDTaq ReadyMix PCR Reaction Mix (Sigma-Aldrich). An initial denaturation step at 94° for 3 min was followed by 35 cycles of 94° for 30 sec, 57° for 30–90 sec depending on product size, 72° for 30 sec, and the process completed by a final extension at 72° for 7 min. PCR products were sequenced on an ABI 3700 capillary sequencer (Applied Biosystems, Foster City, CA) at UCLA's Human Genetics Sequencing and Genotyping Core Facility.
Classification at 3–6 weeks of age:
At 3–6 weeks of age, individual XYPOS B6.129-Chr11-YPOS, B6-YPOS, and F1 mice were classified as female, hermphrodite, or male on the basis of the following: A mouse was classified as a female if female external genitalia, yellow mammary-associated hair pigmentation, bilateral uterine horns, and normal-appearing ovaries or dysgenetic gonads (located in the abdominal region normally occupied by ovaries) were present; as a hermaphrodite if ambiguous genitalia, some yellow mammary-associated hair pigmentation, and an ovary or dysgenetic gonad and a contralateral ovotestis or testis were present; and as a male if normal-appearing male genitalia, no yellow mammary-associated hair pigmentation, and two testes were present. The gonads of some mice classified as male differed in size, or one or both were smaller than the testes of normal controls; these males likely would have been classified as hermaphrodites if analyzed as fetuses.
Classification of fetuses:
Midday of the day a vaginal plug was detected was considered E0.5 (where E is the embryonic day of development). The exact fetal stage of development was determined by limb morphology (Theiler 1989). Individual gonads from E14.5–16 fetuses were classified as an ovary (O), an ovotestis (OT), or a testis (T) (Eicher et al. 1982). Fetuses were further classified into one of five phenotypic categories on the basis of the gonadal pair (O-O, O-OT, OT-OT, OT-T, and T-T). The approximate amount of testicular tissue in each ovotestis was estimated at the time of dissection and confirmed using a captured image. If ≥50% testicular tissue was present, the ovotestis was designated as oT. If <50% testicular tissue was present, the ovotestis was designated as Ot.
Database use:
The NCBI http://www.ncbi.nlm.nih.gov/, UCSC Genome Bioinformatics http://genome.ucsc.edu/ and the Mouse Genome Informatics http://www.informatics.jax.org/ sites were used in retrieving gene, marker, and sequence information. The genomic location information used is based on Mouse Build 36, February 2006/dbSNP build 126 as well as updated information from Mouse Build 37, December 2007/dbSNP build 128.
Statistical analysis:
Categorical and ordinal data analyses for the genotype–phenotype correlations were performed with the statistical software Stata's tabulate and ologit options (http://www.stata.com/, StataCorp LP, College Station, TX). The significance of the associations was assessed using Fisher's exact tests for rxc tables or likelihood ratio tests for the ordinal treatment of the data. The dose-effect hypothesis was tested using Stata's ologit and mlogit options.
RESULTS
Genome scans:
Microsatellite and SNP markers were used to identify the chromosomal regions derived from 129 that protect against sex reversal in XYPOS C57BL/6J.129-YPOS mice (Whitney et al. 2000). DNA from the original 129 strain used in making the congenic was not available as a control. Results obtained from other 129 strains, including 129S1/SvImJ, which is likely to be the strain most closely related to the 129 strain used in the original study, were used in this analysis. A region on chromosome 11 was identified as being of non-C57BL/6J (B6) origin (supplemental Tables 1–4, and Figure 1). This region appeared fragmented, comprised of non-B6 segments interspersed within B6 segments. This result is compatible with multiple recombination events occurring during construction of the congenic strain, given that construction occurred over a span of several years. However, we cannot rule out the possibility that markers appearing to match B6 are not from a different strain of origin. For that reason markers that appear of B6 origin, but are interspersed between non-B6 markers were considered part of the congenic region when delineating its boundaries in Figure 1. In addition, some markers within the congenic region matched neither 129 nor B6 in origin. It was not feasible to determine if this indicated that the 129 or B6 markers have mutated over time and/or a strain other than 129S1/SvImJ was the original donor strain. Irrespective of the origin of the congenic region, once this congenic region was identified, we proceeded to investigate its association with sex reversal.
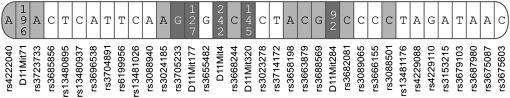
Figure 1.—
Chromosome 11 schematic. Lightly shaded regions represent regions on chromosome 11 that are of 129S1/SvImJ background and darkly shaded regions represent regions that match neither 129S1/SvImJ nor C57BL/6J. Representative markers from the three combined genome scans are depicted; see supplemental materials for more marker information. Markers are in the most likely order on the basis of current location information. The congenic regions appear to be between the start of chromosome 11 and marker rs3685856 (position 14.267381 Mbp) and again between markers rs3088940 (position 56.329569 Mbp) and rs13481176 (position 97.413258 Mbp). Markers that match B6, but that are interspersed between non-B6 markers, were considered part of the congenic region. Image is not to scale.
Association of the congenic region and sex reversal in XYPOS adults:
The initial description of the C57BL/6J.129-YPOS congenic strain stated that genetic elements derived from 129 protected against XYPOS sex reversal: The ratio of phenotypic males to females in the colony was indistinguishable from 1:1 and that the size and appearance of the testes of the adult XYPOS males were normal (Whitney et al. 2000). To determine if the 129-derived region of chromosome 11 protected against sex reversal, two lines of mice were constructed: B6.129-Chr11-YPOS (homozygous 129 for the chromosome 11 region) and B6-YPOS (homozygous B6 for the chromosome 11 region). The phenotype distributions by overall genotype (B6/B6, 129/B6, and 129/129) are shown in Figure 2A, and the distributions by region (a dose-effect or multiplicative model, B6 vs. 129) are shown in Figure 2B. Adult XYPOS animals fell into three distinct phenotypes: Female (fully sex reversed), hermaphrodite (partially sex reversed), and male. Only 2 of 20 129/129 adult XYPOS individuals were females, and no hermaphrodites were observed within this genotype category. This result is in contrast with the B6/B6 group, where the majority of XYPOS individuals were females and hermaphrodites. Thus, the B6/B6 genotype category is similar to what is observed in the original C57BL/6J-YPOS strain, for which similar ratios of sex reversal have been reported (Eicher et al. 1982). The present data also are in accord with the initial congenic description (Whitney et al. 2000). Within the B6.129-Chr11-YPOS colony, the phenotypic ratio of males to females is indistinguishable from 1:1 (data not shown), which can be attributed to the low occurrence of XYPOS females and the difficulty of recognizing hermaphrodites on the basis of external and internal phenotypes.
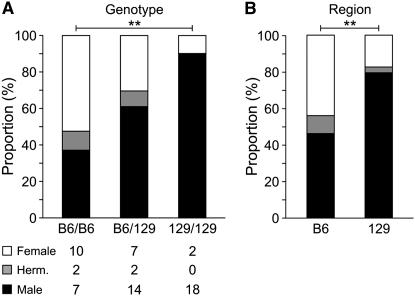
Figure 2.—
Adult phenotypic analysis. (A) Adult distribution by genotype of chromosome 11. The overall comparison of gender distribution among the three genotypes using Fisher's exact test resulted in a P-value = 0.007. The largest difference was observed between the B6/B6 and 129/129 homozygotes (**) (P-value = 0.002). (B) Adult distribution by region of chromosome 11. Using a multiplicative model, the B6 and 129 regions were also found to be different with respect to adult gender distribution (**) (P-value = 0.0005). The same results were obtained when the phenotype categories were treated as ordinal.
Having identified the 129-derived chromosome 11 region allowed us to further evaluate the effect of B6 vs. 129 dosage. Heterozygous 129/B6 XYPOS adults had a phenotype distribution intermediate between B6/B6 and 129/129 (Figure 2A), which suggested that this region derived from 129 exerts a dose effect by reducing the incidence of sex reversal and that two doses of 129 reduce the incidence further than a single dose. We formally tested whether a codominant-genotype model provided a significantly better fit to the data than the dose-effect (multiplicative) model using polychotomous logistic regression. The dose-effect model could not be rejected (P = 0.552). Similar results were obtained when the phenotype categories were treated as ordinal rather than categorical. In summary, the phenotype distributions between inheritance of the 129 vs. B6 region confirmed that there is a difference in gonad development depending on which region is present (Figure 2B) such that presence of the 129 region decreased the frequency of sex reversal relative to the B6 region.
Association of the congenic region and sex reversal in XYPOS fetuses:
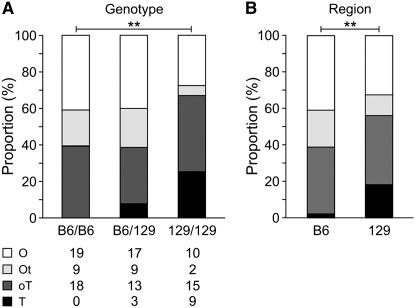
Determination of whether one or both types of gonadal tissue are present in an individual gonad is observed between E14.5–16, when testicular cords have unequivocally formed, and development of ovarian tissue is clearly visible (Eicher et al. 1982). For this reason, experiments also were conducted at this developmental age on B6/B6, 129/B6, and 129/129 XYPOS mice. XYPOS fetuses were classified into one of five categories: O-O, O-OT, OT-OT, OT-T and T-T. The results are depicted in Figure 3, A and B, respectively. The strongest difference was observed between the B6/B6 and 129/129 groups in the overall genotype comparison (Figure 3A), whereas the dose-effect model results reinforced the previous findings that a protective dosage effect occurs if the 129 region of chromosome 11 is present (Figure 3B). Taken together, the results from the two developmental stages (adult and fetus) demonstrate that the strain origin of the chromosome 11 region affects the gonad phenotype distribution in XYPOS mice. The 129 congenic contribution is dosage sensitive, protecting more in the homozygous 129/129 state than in the heterozygous 129/B6 state. Furthermore, inheritance of the single 129 region in the 129/B6 group is still protective compared to the homozygous B6/B6 group.
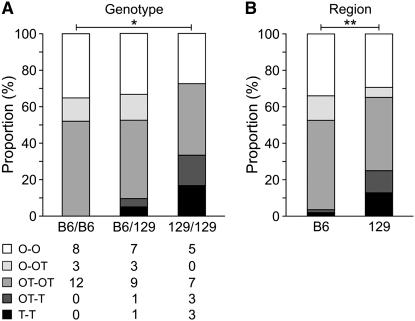
Figure 3.—
Embryo phenotypic analysis. (A) Embryonic distribution by genotype of chromosome 11. The overall comparison of gonad constitution distribution among the three genotypes using Fisher's exact test resulted in a P-value = 0.174. The largest difference was observed between the B6/B6 and 129/129 homozygotes (*) (P-value = 0.031). (B) Embryonic distribution by region of chromosome 11. Using a multiplicative model, the B6 and 129 regions were also found to be different with respect to embryonic gonad constitution distribution (**) (P-value = 0.007). Similar results were obtained by treating the embryo classes as ordinal. O, ovary; OT, ovotestis; T, testis.
Association of the congenic region on an individual gonad level:
In addition to observing less sex reversal in XYPOS 129/129 fetuses, we noted that ovotestes in these individuals contained more testicular tissue than the ovotestes of XYPOS B6/B6 and 129/B6 fetuses. To further explore this observation, we divided ovotestes into those with <50% testicular tissue, (Ot), and those with >50% testicular tissue, (oT). Representative images and schematics of each gonad type, as well as the numbers in each genotype (B6/B6, 129/B6, and 129/129) are shown in Figure 4. The same gonad type distribution by genotype is graphically represented in Figure 5A and the comparison between the B6 and 129 regions is depicted in Figure 5B. Most striking was the finding that 9 of 36 129/129 gonads and 3 of 42 129/B6 gonads developed as testes, whereas no testes were observed in the B6/B6 gonads (Figure 4). The lack of testes in the B6/B6 group was expected, given that XYPOS C57BL/6J-YPOS fetuses develop ovarian tissue in each gonad (Eicher et al. 1982), and our unprotected B6/B6 group is the genetic equivalent of that strain.
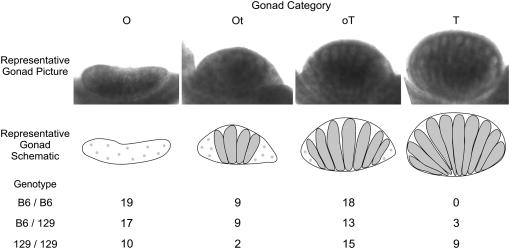
Figure 4.—
Embryo gonad numbers. Individual gonads were classified as an ovary (O), ovotestis having <50% testicular tissue (Ot), ovotestis having >50% testicular tissue (oT), or testis (T). Representative images and schematics for each category are shown.
Figure 5.—
Embryo gonad analysis. (A) Gonad distribution by genotype of chromosome 11. The overall comparison of gonad type distribution among the three genotypes using Fisher's exact test resulted in a P-value = 0.004. The largest difference was observed between the B6/B6 and 129/129 homozygotes (**) (P-value = 0.001). (B) Gonad distribution by region of chromosome 11. Using a multiplicative model, the B6 and 129 regions were also found to be different with respect to gonad type distribution (**) (P-value = 0.001). Similar results were obtained when the gonad categories were treated as ordinal rather than categorical.
The presence of fully developed testes in XYPOS mice that inherit the 129-derived chromosome 11 region has strong biological significance, attesting to the ability of this region to attenuate the sex-reversal phenotype. Analysis of the distribution of gonad types across the three genotypes showed greatest difference between B6/B6 and 129/129, demonstrating that more testicular tissue is formed in 129/129 fetal gonads. Heterozygous 129/B6 fetuses formed less testicular tissue than homozygous 129 fetuses and more testicular tissue than homozygous B6/B6 fetuses (Figure 5A). Again, using either ordinal or polychotomous logistic regression, the null hypothesis of a dose effect could not be rejected. The dose-effect comparison also confirmed that the 129 chromosome 11 region, in a dosage-sensitive manner, influences the amount of testicular tissue formed at the individual gonad level (Figure 5B).
Candidate genes from the congenic region:
The chromosome 11 congenic region delineated by the genome scan was evaluated for genes that could play a role in sex determination. The strongest candidates, listed in Table 1, were selected on the basis of gonadal expression and/or relevant protein function. Where available, mouse models and their phenotypes are noted.
TABLE 1.
Candidate genes within the congenic region on chromosome 11
| Gene symbol | Gene name | Location (bp) | Protein function | Expression | Mouse model |
|---|---|---|---|---|---|
| Srebf1 | Sterol regulatory element binding factor 1 | 60015284 | Transcriptional activator, cholesterol synthesis | Testis, germ cells | Knockout viable, disrupted cholesterol synthesis |
| Ncor1 | Nuclear receptor corepressor 1 | 62132885 | Negative regulation of transcription | Ovary, vagina | Knockout embryonic/perinatal lethal |
| Trp53 | Transformation related protein 53 | 69396176 | Cell cycle and apoptosis | Germ cells, ovary, testis, uterus | Knockout displays early-onset tumor incidence, disrupted vasculature |
| Shbg | Sex hormone binding globulin | 69431011 | Lipid binding, transport | Sertoli cells | |
| Sox15 | SRY-box containing gene 15 | 69471232 | Regulation of transcription | Testicular cords, ovary, urogenital system | Knockout viable, muscle abnormalities; no sex determination phenotype observed |
| Dvl2 | Dishevelled 2, dsh homolog (Drosophila) | 69816821 | Wnt signaling | Ovary, testis | Knockout incompletely perinatal lethal, surviving animals predominantly female |
| Foxn1 | Forkhead box N1 | 78174483 | Transcriptional regulator involved in development | Testis | Targeted and natural mutants show keratinocyte and immune abnormalities |
| Nle1 | Notchless homolog 1 (Drosophila) | 82716966 | Nuclear localization | Testis, ovary | Knockout lethal prior to somite formation |
| Lhx1 | LIM homeobox protein 1 | 84335666 | Regulation of transcription | Urogenital sytem | Knockout embryonic lethal lacking kidneys and gonads |
| Tbx2 | T-box 2 | 85649273 | Regulation of transcription | Urogenital ridge | Knockout embryonic lethal with cardiac and vascular abnormalities |
| Tbx4 | T-box 4 | 85713027 | Regulation of transcription | Oocyte | Knockout embryonic lethal, impaired allantois growth, lack of hindlimb buds |
Genes were selected on the basis of a combination of location within the congenic region on chromosome 11, gonadal expression, and function during development. Knockout mouse models and their phenotypes are listed when available.
DISCUSSION
The congenic approach we used successfully identified a locus on chromosome 11 that is associated with sex determination and modifies the C57BL/6J-YPOS sex-reversal phenotype. Interestingly, this chromosome 11 region was not noted in the earlier experiment aimed at locating the chromosomal regions involved in C57BL/6J-YPOS sex reversal. In this case, a backcross mapping approach was used in combination with the DBA/2J strain and loci were mapped to chromosomes 2, 4, and 5 (Eicher et al. 1996). One possibility for this difference is that the interaction between the M. domesticus poschiavinus Sry and the 129 vs. DBA/2 backgrounds is different. Alternatively, the process of making a congenic also is a fundamentally different approach than is a linkage backcross approach: The linkage approach identified genomic regions implicated in contributing to sex reversal in XYPOS mice of the C57BL/6J-YPOS strain, whereas the congenic approach identified regions that can rescue or correct the sex-reversal phenotype. The chromosome 11 region we identified could contain one or more genes that, when of 129 origin, can rescue the C57BL/6J-YPOS sex-reversal phenotype, not because they are the genes involved in C57BL/6J-YPOS sex reversal, but because they may be in similar pathways or have redundant functions that compensate for the actual genes causing the sex reversal. Still, on the basis of the limitations of the experiments performed, it cannot be determined whether a congenic approach using DBA/2 would have yielded the same results as with 129 or whether the effect we observed is specific to 129.
Complete sex reversal was rare in XYPOS B6.129-Chr11-YPOS adults, a finding in contrast to what is observed in XYPOS C57BL/6J-YPOS adults where ∼50% present as females (Eicher et al. 1982). In addition, most XYPOS B6.129-Chr11-YPOS adults presented as males containing two normal-appearing testes, a finding not noted in XYPOS C57BL/6J-YPOS adults. In contrast to what was observed in adults, we did observe that ovotestes were present in some of the XYPOS B6.129-Chr11-YPOS fetuses but again the number of individuals containing ovotestes was less that what is observed in XYPOS C57BL/6J-YPOS fetuses (Eicher et al. 1982). We conclude that inheritance of the 129-derived congenic region in the homozygous state increases the amount of testicular tissue that develops in an individual XYPOS gonad thus reducing the probability that an XYPOS individual will be sex reversed. Why is only partial protection against sex reversal observed? One possibility would be that, since multiple C57BL/6J regions have been shown to contribute to C57BL/6J-YPOS sex reversal using linkage analysis (Eicher et al. 1996), any single region rescuing this sex-reversal condition is likely to correct only partially, as we noted here for the XYPOS B6.129-Chr11-YPOS mice in which a few adults were female and some fetuses were hermaphrodites. Another, more likely, possibility is that the congenic approach selected only for phenotypically male adults. As the selection process continued with each backcross generation, the 129 region required for this phenotype was narrowed by recombinational events. However, it is likely that other 129-derived regions were lost that contained additional genes contributing to the protection. The fact that some of the congenic fetal gonads contain ovarian tissue and some congenic males are not fully protected is in support of this idea.
Our experimental approach also allowed us to determine that protection from XYPOS sex reversal was dosage dependent on the 129-derived chromosome 11 region. For example, when the amount of testicular tissue was considered in ovotestes of 129/129 vs. 129/B6 fetuses, more testicular tissue was present in 129/129 ovotestes. Similarly, when the amount of testicular tissue was considered in ovotestes of 129/B6 vs. B6/B6 fetuses, more testicular tissue was present in 129/B6 ovotestes. The multiplicative model could not be rejected when gonad categories were treated as categorical or ordinal, and thus this model supports the hypothesis of a dosage effect.
We used three adult- and five fetal-phenotype groups to best represent the gonadal phenotypes observed, which precluded analysis between developmental stages. It was clear during the course of the research, however, that assigning a gonad phenotype to an adult was different than assigning a gonad phenotype to genetically identical fetuses. The reason is that a small amount of ovarian tissue present in a fetal ovotestis is easily missed in an adult gonad unless histological analysis is performed, and thus an ovotestis could be classified as a testis. It is possible that fetal gonads classified as oT or even Ot in a fetus could “catch up” developmentally, so that in the adult animal they may present as normal-appearing testes. Because the original claim of protection in the congenic strain was demonstrated in adult animals, we conducted both the adult analysis to compare our result to the earlier study and the more detailed fetal analysis for better understanding of the protection phenomenon. Despite the difference between the developmental stages analyzed in this study, there is still protection at both stages when compared to analysis of the original unprotected C57BL/6J-YPOS strain (Eicher et al. 1982).
Analysis of candidate genes is outside the scope of the study; however, we compiled a set of strong candidates, most notably genes involved in transcription and signaling, for further investigation (Table 1). The chromosome 11 congenic region contains representatives from numerous developmental genes that are active during testicular formation. Ncor1 interacts with Dax1 (Crawford et al. 1998) to repress transcription of Sf1, a factor well established to be upstream of Sry (De Santa Barbara et al. 2001). Sox15 expression is not necessary for normal male sex determination (Lee et al. 2004), but it is expressed in the developing testis (Sertoli cells), and not in the developing ovary (Sarraj et al. 2003). Sox15 is a transcriptional repressor (Béranger et al. 2000) and it could have redundant function in testicular development and possibly even affect Sry transcription. Lhx1 is involved in gonadal development prior to sex determination and differentiation (Shawlot and Behringer 1995). Recently, Tbx2, a transcriptional repressor (Nef et al. 2005), was reported to have sexually dimorphic expression, with higher levels in E12.5 XY gonads (Carreira et al. 1998). However, mice homozygous for a null allele of Tbx2 are not reported to have an abnormal gonadal phenotype (Harrelson et al. 2004). Members of the forkhead family of proteins (Schmidt et al. 2004) and the Wnt signaling pathway (Vainio et al. 1999) participate in gonadal development, more specifically in ovarian development. Foxn1 and Dvl2, members of those two groups respectively, are located in the chromosome 11 region discussed here. Transcription regulators and signaling factors are key players during patterning of the embryonic testis, and some or several of these genes are likely to play a role in the C57BL/6J.129-YPOS protection phenotype.
Considering the previous findings that proper Sry expression is disrupted in XYPOS C57BL/6J-YPOS mice, and the current investigation showing that the congenic contribution derived from 129 protects against sex reversal in XYPOS B6.129-Chr11-YPOS mice, it is likely that the gene or genes underlying the protection affect Sry expression. Some of the candidate genes discussed have sexually dimorphic expression during sex determination. Factors involved in initiating Sry expression are present at equal levels in males and females prior to initiation of testicular development, but may or may not be subject to feedback regulation following Sry expression. Analyzing Sry expression in XYPOS B6.129-Chr11-YPOS fetal gonads by qPCR or in situ hybridization in future studies could determine if the timing and/or levels of Sry expression are altered compared to those in the unprotected C57BL/6J-YPOS strain, which would help elucidate the mechanism of protection.
Better understanding of the gonadal phenotype present in XYPOS C57BL/6J.129-YPOS mice helps to define the processes and gene networks that the chromosome 11 congenic region is likely to affect, but identifying the factors causing the protection phenotype is still a daunting task. The precise mechanism of Sry action itself is still under investigation, which confounds the process of identifying additional factors potentially involved in the sex-determining cascade. Upstream factors, interacting partners, and downstream targets of Sry could all play a role. We focused on transcriptional regulators or other factors with a possible role upstream of Sry, given that sex reversal in XYPOS C57BL/6J-YPOS mice is in large part due to delayed and reduced Sry expression. Future analyses, including evaluation of the sequence and expression pattern of the candidate genes present in congenic present in C57BL/6J.129-YPOS mice using in vitro and in vivo models, analysis of Sry expression, and further restricting the congenic region, will help focus in on the gene or combination of genes responsible for the protection sex-reversal phenotype in XYPOS C57BL/6J.129-YPOS mice.
Acknowledgments
We thank Linda L. Washburn for her expertise in embryonic gonad classification and Alice Fleming and Emmanuele Délot for the helpful conversations and for reviewing this manuscript. This work was supported by National Institutes of Health grants HD-44513 (E.V.), RR-01183 (E.M.E.), GM-20919 (E.M.E.), and CA-34296 (The Jackson Laboratory). Ganka Nikolova was supported by a predoctoral fellowship from the Howard Hughes Medical Institute.
References
- Albrecht, K. H., and E. M. Eicher, 1997. DNA sequence analysis of Sry alleles (subgenus Mus) implicates misregulation as the cause of C57BL/6J-Y(POS) sex reversal and defines the SRY functional unit. Genetics 147 1267–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht, K. H., M. Young, L. L. Washburn and E. M. Eicher, 2003. Sry expression level and protein isoform differences play a role in abnormal testis development in C57BL/6J mice carrying certain Sry alleles. Genetics 164 277–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Béranger, F., C. Méjean, B. Moniot, P. Berta and M. Vandromme, 2000. Muscle differentiation is antagonized by SOX15, a new member of the SOX protein family. J. Biol. Chem. 275 16103–16109. [DOI] [PubMed] [Google Scholar]
- Bowles, J., L. Cooper, J. Berkman and P. Koopman, 1999. Sry requires a CAG repeat domain for male sex determination in Mus musculus. Nat. Genet. 22 405–408. [DOI] [PubMed] [Google Scholar]
- Bullejos, M., and P. Koopman, 2001. Spatially dynamic expression of Sry in mouse genital ridges. Dev. Dyn. 221 201–205. [DOI] [PubMed] [Google Scholar]
- Bullejos, M., and P. Koopman, 2005. Delayed Sry and Sox9 expression in developing mouse gonads underlies B6-Y(DOM) sex reversal. Dev. Biol. 278 473–481. [DOI] [PubMed] [Google Scholar]
- Carreira, S., T. J. Dexter, U. Yavuzer, D. J. Easty and C. R. Goding, 1998. Brachyury-related transcription factor Tbx2 and repression of the melanocyte-specific TRP-1 promoter. Mol. Cell. Biol. 18 5099–5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford, P. A., C. Dorn, Y. Sadovsky and J. Milbrandt, 1998. Nuclear receptor DAX-1 recruits nuclear receptor corepressor N-CoR to steroidogenic factor 1. Mol. Cell. Biol. 18 2949–2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santa Barbara, P., C. Méjean, B. Moniot, M. H. Malclès, P. Berta et al., 2001. Steroidogenic factor-1 contributes to the cyclic-adenosine monophosphate down-regulation of human SRY gene expression. Biol. Reprod. 64 775–783. [DOI] [PubMed] [Google Scholar]
- Eicher, E. M., and L. L. Washburn, 2001. Does one gene determine whether a C57BL/6J-Y(POS) mouse will develop as a female or as an hermaphrodite? J. Exp. Zool. 290 322–326. [DOI] [PubMed] [Google Scholar]
- Eicher, E. M., L. L. Washburn, J. B. Whitney, III, and K. E. Morrow, 1982. Mus poschiavinus Y chromosome in the C57BL/6J murine genome causes sex reversal. Science 217 535–537. [DOI] [PubMed] [Google Scholar]
- Eicher, E. M., L. L. Washburn, N. J. Schork, B. K. Lee, E. P. Shown et al., 1996. Sex-determining genes on mouse autosomes identified by linkage analysis of C57BL/6J-YPOS sex reversal. Nat. Genet. 14 206–209. [DOI] [PubMed] [Google Scholar]
- Fechner, P. Y., S. M. Marcantonio, V. Jaswaney, G. Stetten, P. N. Goodfellow et al., 1993. The role of the sex-determining region Y gene in the etiology of 46,XX maleness. J. Clin. Endocrinol. Metab. 76 690–695. [DOI] [PubMed] [Google Scholar]
- Harrelson, Z., R. J. Kelly, S. N. Goldin, J. J. Gibson-Brown, R. J. Bollag et al., 2004. Tbx2 is essential for patterning the atrioventricular canal and for morphogenesis of the outflow tract during heart development. Development 131 5041–5052. [DOI] [PubMed] [Google Scholar]
- Hawkins, J. R., A. Taylor, P. Berta, J. Levilliers, B. Vanderauwera et al., 1992. Mutational analysis of Sry—nonsense and missense mutations in XY sex reversal. Hum. Genet. 88 471–474. [DOI] [PubMed] [Google Scholar]
- Jäger, R. J., M. Anvret, K. Hall and G. Scherer, 1990. A human XY female with a frame shift mutation in the candidate testis-determining gene Sry. Nature 348 452–454. [DOI] [PubMed] [Google Scholar]
- Jeske, Y. W., J. Bowles, A. Greenfield and P. Koopman, 1995. Expression of a linear Sry transcript in the mouse genital ridge. Nat. Genet. 10 480–482. [DOI] [PubMed] [Google Scholar]
- Koopman, P., A. Munsterberg, B. Capel, N. Vivian and R. Lovell-Badge, 1990. Expression of a candidate sex-determining gene during mouse testis differentiation. Nature 348 450–452. [DOI] [PubMed] [Google Scholar]
- Koopman, P., J. Gubbay, N. Vivian, P. Goodfellow and R. Lovell-Badge, 1991. Male development of chromosomally female mice transgenic for Sry. Nature 351 117–121. [DOI] [PubMed] [Google Scholar]
- Lee, C.H., and T. Taketo, 1994. Normal onset, but prolonged expression, of Sry gene in the B6.YDOM sex-reversed mouse gonad. Dev. Biol. 165 442–452. [DOI] [PubMed] [Google Scholar]
- Lee, C.H., and T. Taketo, 2001. Low levels of Sry transcripts cannot be the sole cause of B6-Y(TIR) sex reversal. Genesis 30 7–11. [DOI] [PubMed] [Google Scholar]
- Lee, H. J., W. Goring, M. Ochs, C. Muhlfeld, G. Steding et al., 2004. Sox15 is required for skeletal muscle regeneration. Mol. Cell. Biol. 24 8428–8436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell-Badge, R., and E. Robertson, 1990. XY female mice resulting from a heritable mutation in the primary testis-determining gene, Tdy. Development 109 635–646. [DOI] [PubMed] [Google Scholar]
- Nef, S., O. Schaad, N. R. Stallings, C. R. Cederroth, J. L. Pitetti et al., 2005. Gene expression during sex determination reveals a robust female genetic program at the onset of ovarian development. Dev. Biol. 87 361–377. [DOI] [PubMed] [Google Scholar]
- Ross, A. J., and B. Capel, 2005. Signaling at the crossroads of gonad development. Trends Endocrinol. Metab. 16 19–25. [DOI] [PubMed] [Google Scholar]
- Sarraj, M. A., H. P. Wilmore, P. J. McClive and A. H. Sinclair, 2003. Sox15 is up regulated in the embryonic mouse testis. Gene Expr. Patterns 3 413–417. [DOI] [PubMed] [Google Scholar]
- Schmidt, D., C. E. Ovitt, K. Anlag, S. Fehsenfeld, L. Gredsted et al., 2004. The murine winged-helix transcription factor Foxl2 is required for granulosa cell differentiation and ovary maintenance. Development 131 933–942. [DOI] [PubMed] [Google Scholar]
- Shawlot, W., and R. R. Behringer, 1995. Requirement for Lim1 in head-organizer function. Nature 374 425–430. [DOI] [PubMed] [Google Scholar]
- Sinclair, A. H., P. Berta, M. S. Palmer, J. R. Hawkins, B. L. Griffiths et al., 1990. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature 346 240–244. [DOI] [PubMed] [Google Scholar]
- Theiler, K., 1989. The House Mouse: Atlas Of Embryonic Development. Springer-Verlag, New York.
- Vainio, S., M. Heikkila, A. Kispert, A., N. Chin and A. P. McMahon, 1999. Female development in mammals is regulated by Wnt-4 signaling. Nature 397 405–409. [DOI] [PubMed] [Google Scholar]
- Whitney, J. B., T. M. Mills, R. W. Lewis, R. Wartell and T. O. Abney, 2000. A single genetic determinant that prevents sex reversal in C57BL-YPOS congenic mice. Biochem. Genet. 38 119–137. [DOI] [PubMed] [Google Scholar]