Abstract
Hedgehog (Hh) signaling is critical for many developmental processes and for the genesis of diverse cancers. Hh signaling comprises a series of negative regulatory steps, from Hh reception to gene transcription output. We previously showed that stability of antagonistic regulatory proteins, including the coreceptor Smoothened (Smo), the kinesin-like Costal-2 (Cos2), and the kinase Fused (Fu), is affected by Hh signaling activation. Here, we show that the level of these three proteins is also regulated by a microRNA cluster. Indeed, the overexpression of this cluster and resulting microRNA regulation of the 3′-UTRs of smo, cos2, and fu mRNA decreases the levels of the three proteins and activates the pathway. Further, the loss of the microRNA cluster or of Dicer function modifies the 3′-UTR regulation of smo and cos2 mRNA, confirming that the mRNAs encoding the different Hh components are physiological targets of microRNAs. Nevertheless, an absence of neither the microRNA cluster nor of Dicer activity creates an hh-like phenotype, possibly due to dose compensation between the different antagonistic targets. This study reveals that a single signaling pathway can be targeted at multiple levels by the same microRNAs.
THE Hedgehog (Hh) pathway controls multiple developmental processes such as pattern formation, differentiation, and proliferation in diverse animal phyla ranging from Drosophila to humans (McMahon et al. 2003). Aberrant Hh pathway activation in humans has also been implicated in the growth and maintenance of a variety of cancers (Ruiz i Altaba 1999; Watkins et al. 2003; Fan et al. 2004; Kayed et al. 2004). A better knowledge of Hh signaling would thus provide a greater understanding of the genesis of these cancers.
Hh signaling controls the activity of the Gli/Ci transcription factors. In Drosophila, the activity of Ci, originally called Cubitus Interruptus, is controlled by a large intracellular multiprotein complex that includes the kinesin-like protein Costal-2 (Cos2), the serine-threonine protein kinase Fused (Fu), and the transmembrane protein Smoothened (Smo). Nuclear translocation of Ci is also controled by the Su(fu) protein, another cytoplasmic anchor. In the absence of Hh, Ci associates with this complex (Ruel et al. 2003). When secreted Hh binds to its receptor, the transmembrane protein Patched (Ptc), Ci dissociates from the protein complex, enters the nucleus, and induces the expression of target genes such as decapentaplegic (dpp), the homolog of the mitogenic factor TGFβ.
To identify novel regulators of the Hh pathway, a misregulation screen was previously carried out in our laboratory using the Drosophila wing imaginal disc as a model (Rorth et al. 1998). In this tissue, Hh is produced and released by posterior cells and then diffuses into the anterior compartment, where it induces various target genes. The correct expression of these target genes is necessary for the wild-type patterning of the adult wing. The study described here of one particular line, named PUAS-29B3, showed that the hh target gene misregulation resulted from the overexpression of a single cluster of three microRNAs.
MicroRNAs are small, endogenous, noncoding RNAs, typically 21–23 nucleotides in length, that direct negative posttranscriptional regulation of gene expression in plants and animals. MicroRNAs are transcribed as long, primary, mono- or polycistronic precursor transcripts that are processed in a two-step process involving the nuclear and cytosolic RNase III-type endonucleases Drosha and Dicer (Dcr), respectively. MicroRNAs bind to specific complementary sequences in the 3′-untranslated regions (3′-UTRs) of messenger RNAs (mRNAs), inducing either mRNA degradation or the inhibition of protein synthesis, depending on the degree of miRNA:mRNA complementarity (Bartel 2004). MicroRNAs have been implicated in a variety of developmental and physiological processes, including the control of developmental timing, cell proliferation, cell fate specification, apoptosis, morphogenesis, fat metabolism, and insulin secretion (reviewed in Di Leva et al. 2006). In addition, links between cancer development and microRNA expression have been described (reviewed in Zhang et al. 2007).
In this study, we provide evidence that the overexpression of a microRNA cluster induces a specific modification of the Hh pathway in the wing imaginal disc. Indeed two microRNAs, miR-12 and miR-283, are able to regulate the 3′-UTRs of the mRNAs encoding Cos2, Fu, and Smo and decrease the levels of the encoded proteins, resulting in ectopic expression of dpp. Flies lacking the microRNA cluster were produced by homologous recombination. Suprisingly, an absence of neither the microRNA cluster nor of Dcr induced any modifications in the Hh signaling pathway. Nevertheless, analysis of cells mutant for either the microRNA cluster or for Dicer function shows that both the cos2 and smo 3′-UTR sequences are regulated by microRNAs, strongly suggesting that antagonistic components of the Hh signaling pathway are regulated by microRNAs in the wing disc. One possible role for miR-12 and miR-283 might be to dampen down the levels of Hh pathway components, particularly Cos2 and Smo, to prevent the accidental activation or downregulation of the pathway.
MATERIALS AND METHODS
Drosophila stocks and genetic experiments:
PUAS-29B3 was isolated using Gal4-directed overexpression of EPg elements as described (Teleman et al. 2005). Scalloped-GAL4 (Sd-GAL4), MS1096-GAL4, engrailed-GAL4 (en-GAL4), and patched-GAL4 (ptc-GAL4) lines are described in FlyBase (http://flystocks.bio.indiana.edu/). Overexpression of clones in the wing imaginal discs was produced using the “flip-out” technique (Basler and Struhl 1994) on heat-shocked early third instar larvae hatched from dpp-lacZ; act<CD2<Gal4, UAS-GFPnls males crossed to yw hs-flp122; PUAS-miR12 or yw hs-flp122, PUAS-miR283 or yw hs-flp122; PUAS-miR304 or PUAS-29B3; hs-flpSb/TM6b females. To generate Δ3miR and dcr-1 mutant clones, 24- to 36-hr-old larvae with the following genotype were heat-shocked at 37° for 1 hr: [hs-πM] FRT19A/Δ3miR FRT19A; hs-flpSb/+ and flp122; FRT82B arm-lacZ/FRT82B dcr-1Q1147X (Lee et al. 2004).
Transgenic lines:
PUAS-microRNA lines:
EcoRI–XhoI PCR fragments were obtained from genomic DNA using oligos 5′-CGGAATTCTCCGTTGTCGTATGCCCTTGTTCTCTCT and 5′-CCGCTCGAGTGCTTTGCTGCTTATTGAGTCCCACCTTAT, 5′-CGGAATTCCCAACCACGTGTAGCTCCCCAAAACTGTAT and 5′-GCTCTAGATGAGGTGAGGTGAGCAGTTAACAGCCAATA, or 5′-CGGAATTCACCCCTTACGCTGCCCCACACTTCTATT and 5′-GCTCTAGAGGGCATACGACAACGGAAACAGATCAGA, and the fragments were then cloned into the pUAS vector to create the PUAS-miR12, PUAS-miR283, and PUAS-miR304 vectors, respectively. Two transgenic lines were tested for each microRNA and gave similar results.
Sensor lines:
Two copies of the 30 nt containing perfect miR-12, miR-283, or miR-304 target sites were cloned into the 3′-UTR of the tub-EGFP vector (Brennecke et al. 2003) to obtain the miR12-sensor, miR283-sensor, and miR304-sensor lines, respectively. Two to three transgenic lines were tested for each sensor and gave similar results.
The cos2, smo, and fu 3′-UTRs were amplified from genomic DNA with the following primers: 5′-tgtgaagcaatagctcagatcctg and 5′-tcacacgctgatattgagggaac, 5′-CGCCTAGGtagcaagactaaataagcaattgatg and 5′-CCGCTCGAGcgaggatttaaaatcgtttattagtt, or 5′-CGCCTAGGccggcactttcttttattgcgctcag and 5′-CCGCTCGAGaggtacccaacattatatcagacgctag, respectively. The 3′-UTRs were cloned into the tub-EGFP plasmid (Brennecke et al. 2003). Two independent transgenic strains were assayed for each construct.
Targeted homologous recombination:
Ends-out homologous recombination was performed essentially as described (Gong and Golic 2003). The left and right arms of the targeting pw25 vector were prepared using PCR fragments generated from genomic DNA with the following oligos: 5′-AGCAGGCGCGCCTCGCACTAACCGCATGACCAACAACcaacg and 5′-TACCCGTACGgcggggttttttggggtcggattatttcgg for the downstream flank, and 5′-AGCAGCGGCCGCGCTGGTCAGGTGGCGAGTCTCAAACAATTA and 5′-AGCAGCGGCCGCAACAGTTGGTCAGCGAGCACGAGAAGAAGA for the upstream flank of the microRNA cluster, respectively. Flies carrying the targeting construct were crossed to flies with heat-inducible FLP recombinase and I-SceI endonuclease (70FLP and 70I-SceI, Bloomington Stock Center), and the resulting progeny were heat-shocked for 90 min at 37° on day 3 of development. Mosaic-eyed females were crossed to FM7i males, and targeting events were mapped to the sex chromosome in the next generation. The absence of the microRNA locus was verified by genomic PCR.
In situ hybridization and immunostaining:
Embryo immunostaining and in situ hybridization were performed as described previously (Friggi-Grelin et al. 2006). Disc immunolabeling was performed as described previously (Gallet et al. 2003). Antibodies were used at the following dilutions: rabbit anti-dGMAP (homemade) 1:1000; mouse anti-Smo 1:100; purified rabbit anti-Cos2 (homemade) 1:1000; purified rabbit anti-Fu (homemade) 1:1000; monoclonal 2A1 rat anti-Ci (gift from R. Holmgren) 1:20; monoclonal 4D9 mouse anti-En (DSHB) 1:1000; monoclonal 5E10 mouse anti-Ptc (gift from P. Ingham) 1:400; monoclonal mouse anti-Myc (Santa Cruz) 1:200; monoclonal mouse anti-β-Gal (Promega, Madison, WI) 1:1000; rabbit anti-β-Gal (Capel) 1:1000. Secondary antibodies labeled with fluorescent Cy5 or Cy3 (Jackson, West Grove, PA) or alexa488 (Molecular Probes, Eugene, OR) were used at a dilution of 1:200. Images were obtained by confocal microscopy (Leica DMR TCS_NT).
RESULTS
Identification of a new locus, overexpression of which induces a modification of the Hh pathway:
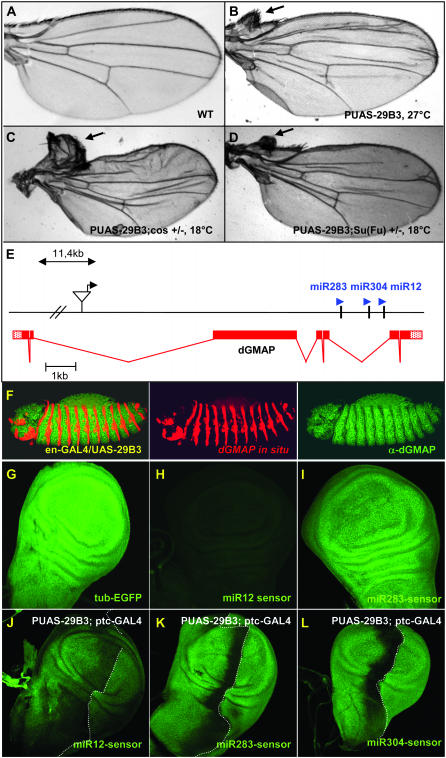
During the course of a misregulation screen to search for new regulators of the Hedgehog pathway in Drosophila we identified a new EP line, named PUAS-29B3, that induces the outgrowth of anterior structures in the wing costa region when induced in the wing imaginal disc (Figure 1B). Several lines of evidence indicated that the defects observed in this line involved the misregulation of the Hh pathway. First, a similar outgrowth phenotype was obtained upon overexpression of Hh (data not shown). Second, the PUAS-29B3 line led to ectopic expression of the Hh target gene, dpp (Figure 2, A and B) (Capdevila and Guerrero 1994; Felsenfeld and Kennison 1995). Finally, the PUAS-29B3-induced outgrowth is sensitive to the Hh signaling level. Specifically, the outgrowth phenotype was enhanced when PUAS-29B3 was induced in a background heterozygous for mutations in cos2 and suppressor of fused [Su(fu)], two negative components of the Hh pathway (Figure 1, C and D, respectively). Moreover, the outgrowth was absent in animals in which both PUAS-29B3 and Cos2 expressions were induced (data not shown). These data encouraged us to further examine the relationship between PUAS-29B3 and the Hh pathway.
Figure 1.—
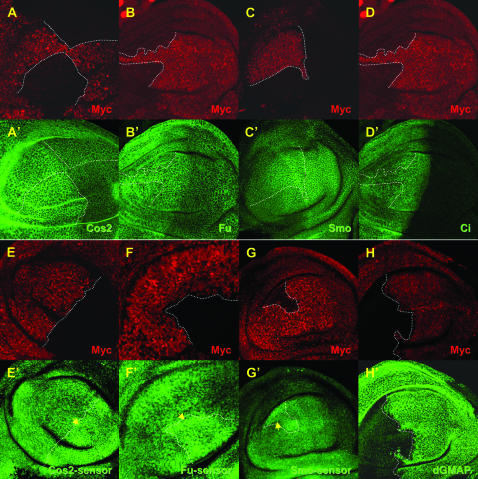
The PUAS-29B3 line controls the expression of a microRNA cluster and induces anterior wing outgrowth. (A) Wild-type adult wing and (B) PUAS-29B3/Sd-GAL4 wing obtained at 27°. Note that when flies were grown at 18°, a temperature at which GAL4 activity is much lower, this phenotype is not observed (data not shown). (C) PUAS-29B3/Sd-GAL4; cos2w1/+ and (D) PUAS-29B3/Sd-GAL4; Su(Fu)LP/+ wings obtained at 18°. Arrows indicate outgrowth of anterior wing tissue in the costa region. (E) Localization of the PUAS-29B3 element in gene CG33206, which encodes dGMAP. The microRNA cluster, composed of miR-283, miR-304, and miR-12, is present in the fifth intronic sequence of dGMAP. (F) In situ hybridization for dGMAP mRNA (red), followed by dGMAP immunostaining (green) in PUAS-29B3; en-GAL4 embryos. No increase in dGMAP protein was observed in the en domain. Note that endogenous dGMAP mRNAs are present in all cells but levels are low compared to ectopic RNA expression (middle). Note also that dGMAP proteins are present in all ectodermal cells but their level is artifactually decreased in the en cells where the dGMAP in situ detection is very high (right). (G–L) GFP expression in tub-EGFP (G), miR12-sensor (H and J), miR283-sensor (I and K), miR304-sensor (L), wt discs (G–I), or in PUAS-29B3; ptc-GAL4 discs (J–L). The pictures shown in G, H, and I were obtained with a similar laser setting on the confocal microscope. Note that the laser setting is different for H and J (in which laser intensity has been increased) and thus sensor level is not comparable for these two panels. G–I are illustrative of three independent lines for each sensor. Dotted lines indicate the A/P border. (G–L) Magnification, 200×.
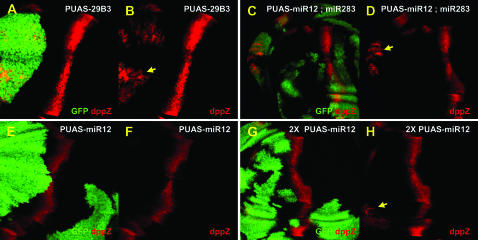
Figure 2.—
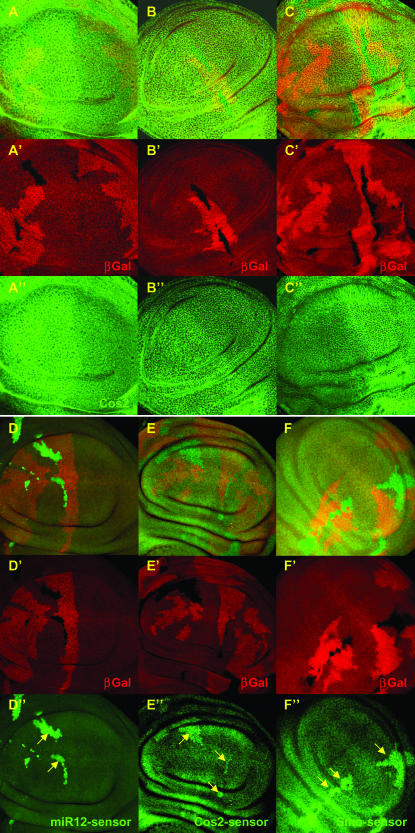
Ectopic dpp expression is due to overexpression of two microRNAs, miR-12 and miR-283. dpp expression is visualized with the dpp-lacZ reporter gene (red) in wing imaginal discs. Clones of cells driving expression of PUAS-29B3 (A and B), PUAS-miR12 alone (E and F), or PUAS-miR12 together with PUAS-miR283 (C and D), are labeled with GFP (green). Ectopic dppZ expression (arrows) is observed in more anterior clones following overexpression of PUAS-29B3, two copies of miR-12, or one copy each of miR-12 and miR-283. Note that ectopic dpp expression is mostly observed at the anterior edge of the wing pouch, as if this region is more sensitive to the activation of the Hh pathway. We also note that the expression of Dpp in the clones is variable, likely due to tissue deformation in the outgrowth.
PUAS-29B3 induces overexpression of a microRNA cluster:
To identify the gene responsible for the outgrowth phenotype, we first localized the PUAS element of the 29B3 line. This element was present in an intron of the dGMAP gene (Friggi-Grelin et al. 2006) (Figure 1E). When the PUAS-29B3 insertion was expressed in the engrailed (en) expression domain, dGMAP transcripts were observed in the characteristic en embryonic stripes, but no dGMAP protein was produced likely due to the absence of the normal initiation codon in the induced transcripts (Figure 1F). Further, the overexpression of the corresponding cDNA did not affect the Hh pathway (data not shown). Interestingly, a microRNA cluster (Lagos-Quintana et al. 2001; Aravin et al. 2003; Lai et al. 2003) was identified in the intronic sequence of the gene (Figure 1E). The PUAS-29B3 locus contains a cluster of three previously uncharacterized microRNAs, miR-12, miR-283, and miR-304 (Lagos-Quintana et al. 2001; Aravin et al. 2003; Lai et al. 2003). Because this cluster is embedded within the dGMAP gene and is oriented in the same direction as the gene, we reasoned that it would be coexpressed with the dGMAP transcript. To verify this and to follow the expression of the microRNAs, sensor lines (Brennecke et al. 2003) for the three microRNAs were generated. Two complementary sites with perfect matches for each microRNA were placed downstream of an EGFP gene controlled by the ubiquitously expressed tubulin (tub) promoter. The binding of the microRNAs to their complementary 3′-UTR sequences was expected to block the translation of EGFP mRNA and thereby decrease the level of EGFP. In the three sensor lines, we observed a uniform decrease in the EGFP level (Figure 1, H and I, and data not shown) compared to the tub-EGFP control line (Figure 1G), the control line lacked microRNA sites downstream of the EGFP gene and was analyzed using a similar laser setting. This indicated that, like dGMAP, the intronic microRNAs are uniformly expressed in the wing imaginal disc, suggesting that they are subjected to the same regulation as the host gene, as previously proposed (Friggi-Grelin et al. 2006). The miR12-sensor line, however, showed a stronger decrease in the level of EGFP (Figure 1H) than did the miR-283 and miR-304 lines (Figure 1I and data not shown), suggesting that it was more highly expressed in the disc. These data are in agreement with the finding that miR-12 is one of the most abundant Drosophila microRNAs (Lagos-Quintana et al. 2001) and are also consistent with a Northern analysis of the three microRNAs reported in another study (Leaman et al. 2005). It is therefore possible that miR-12, which is processed from the same transcript as miR-283 and miR-304, is more stable than the two other microRNAs or might just be more efficient.
To verify that PUAS-29B3 induces the expression of this microRNA cluster, we tested the sensitivity of each sensor line to PUAS-29B3 activation. When PUAS-29B3 was induced in the patched (ptc) expression domain along the A/P border of the wing imaginal disc (using the ptc-GAL4 driver), we observed a decrease in the level of GFP at the A/P border in the three sensor lines (Figures 1, J–L).
miR-12 and miR-283 overexpression affects the Hh pathway:
In view of the above results, it seemed possible that the induced PUAS-29B3 phenotype resulted from the combinatorial expression of one or more of the three microRNAs. To test this possibility, we constructed new PUAS transgenic lines to be able to independently express miR-12, miR-283, or miR-304. We induced clones of cells overexpressing 29B3, miR-12, miR-283, or miR-304 in the wing imaginal discs. In PUAS-29B3 clones located in the A compartment, ectopic dpp expression was observed (100% of the clones, Figure 2, A and B), consistent with the outgrowth observed in adult wings (Figure 1B). When miR-12 or miR-283 were induced separately, however, no ectopic dpp expression was observed (Figure 2, E and F, and data not shown). But ectopic dpp expression was observed when the miR-12 level was increased by expressing two copies of the transgene at the anterior edge of the wing pouch (100% of the clones, Figure 2, G and H) or when miR-12 and miR-283 were overexpressed together (100% of the clones located at the edge, Figure 2, C and D). No modifications in Hh target gene expression were observed in clones overexpressing miR-304 (data not shown). Together, these results suggest that the PUAS-29B3 phenotype is caused by the overexpression of both miR-12 and miR-283. The limited activation of dpp by individual miR expression might be due to a decreased stability of miR transcripts compared to the precursor mRNAs induced by PUAS-29B3.
Several components of the Hh pathway are potential targets of the microRNA cluster:
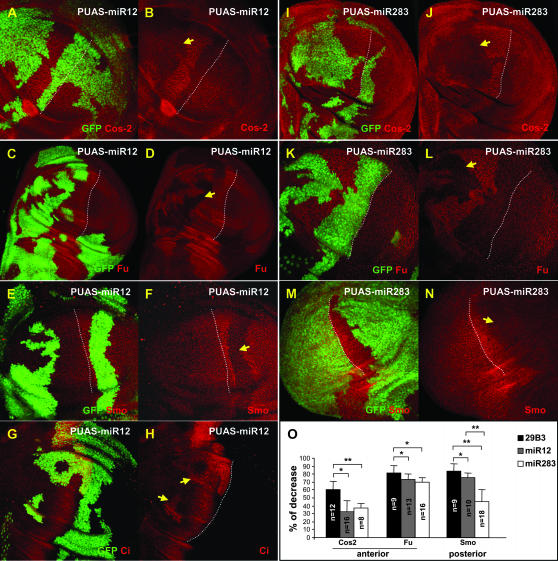
To determine which components of the Hh pathway are targeted by miR-12 and miR-283, we analyzed the levels of several proteins in the pathway in cells overexpressing either of the two microRNAs. In the Hh pathway, a multicomponent cytoplasmic complex including the Smo, Fu, and Cos2 proteins controls the stability and activity of the transcriptional factor Ci, which in turn regulates the expression of Hh target genes. In cells overexpressing miR-304, no modification was observed, however, in cells overexpressing either miR-12 (Figure 3, A–F) or miR-283 (Figure 3, I–N), all three proteins—Cos2 (Figure 3, A, B, I, and J), Fu (Figure 3, C, D, K, and L), and Smo (Figure 3, E, F, M, and N)—were destabilized. In addition, Ci was stabilized (Figure 3, G and H, and data not shown), likely due to the decreased level of Cos2 (Wang and Holmgren 1999). The decrease in the levels of these three proteins in 29B3 clones was greater than in miR-12 or miR-283 clones (Figure 3O). Thus, ectopic dpp expression was probably observed in the PUAS-29B3 clones (Figure 2, A and B), but not in the miR-12 clones, because of the higher destabilization of Cos2 protein in the former context.
Figure 3.—
Overexpression of miR-12 or miR-283 modifies the levels of Cos2, Fu, Smo, and Ci proteins. Clones of cells driving expression of PUAS-miR12 (A–H) or PUAS-miR283 (I–L) are labeled with GFP. Cos2 (A, B, I, and J), Fu (C, D, K, and L), Smo (E, F, M, and N), and Ci (G and H) proteins are visualized by immunofluorescence (red). Induction of microRNA expression decreases the level of Fu, Cos2, and Smo proteins in both anterior and posterior clones, whereas Ci is stabilized in anterior clones. Dotted lines indicate the A/P border. Arrows indicate the most significant clones. Note that the antibody against Cos2 gives a higher background than the one against Fu, which likely accounts for the seemingly stronger affects on Fu than on Cos2. (O) Percentage of the decrease in the levels of Cos2, Fu, and Smo proteins in PUAS-29B3 (solid bars), PUAS-miR12 (shaded bars), or PUAS-miR283 (open bars) clones compared to surrounding wild-type cells. In PUAS-29B3 clones, the decrease in Cos2 and Fu levels is more pronounced than in PUAS-miR12 or PUAS-miR283 clones. **P < 0.001; *P < 0.04.
smo, cos2, and fu are regulated by the microRNAs cluster:
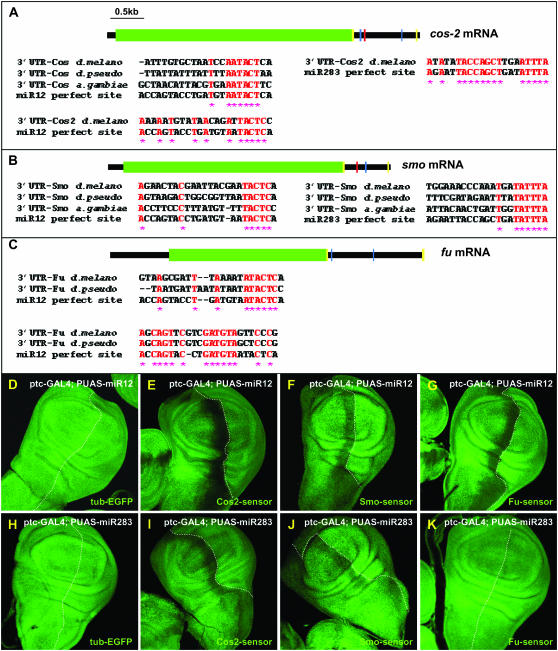
In view of this data we hypothesized that these genes might represent direct targets of the microRNAs. Binding sites of microRNAs are possible to identify because a 6- to 8-base “seed” of contiguous pairing is present between the 3′ end of the binding site and the 5′ end of the microRNA (Brennecke et al. 2005; Rajewsky 2006) and is conserved among the 3′-UTRs of homologous genes in related species. Using these criteria, we identified potential miR-12 and miR-283 binding sites on both the cos2 and smo 3′-UTRs (Figure 4, A and B, respectively), as well as two potential miR-12 sites on the fu 3′-UTR (Figure 4C).
Figure 4.—
smo, cos2, and fu mRNAs are regulated by the 29B3-microRNA cluster. Schematics of cos2 (A), smo (B), and fu (C) mRNAs. Green squares indicate the open reading frames of the transcripts. Blue and red bars represent potential 3′-UTR binding sites for miR-12 and miR-283, respectively. Each site is presented below, showing the conservation between Drosophila pseudoobscura and Anopheles gambiae and theoretical miR-12 or miR-283 binding sites. Conserved residues are shaded in red, with stars below. Yellow bars represent the primers used to amplify the 3′-UTR sequences to establish the Cos2-, Smo-, and Fu-sensor lines. (D–K) GFP expression in tub-EGFP (D and H), Cos2- (E and I), Smo- (F and J), and Fu- (G and K) sensor lines in ptc-GAL4/UAS-miR12 imaginal discs (D–G) or in ptc-GAL4/UAS-miR283 (H–K) wing imaginal discs. Note that no modifications in EGFP expression were observed when miR-12 or miR-283 were overexpressed in the tub-EGFP control line (D and H).
To determine whether the microRNAs can regulate the 3′-UTRs of cos2, smo, and fu, we established new sensor lines in which the EGFP gene was fused to the 3′-UTRs of these genes. In the absence of microRNA induction, these sensor lines displayed uniform EGFP expression in wing imaginal discs (data not shown). However, when miR-12 was overexpressed in the ptc domain, EGFP expression decreased in all three sensor lines at the A/P boundary (Figure 4, E–G), strongly suggesting that miR-12 can regulate, likely directly, the 3′-UTRs of cos2, smo, and fu and thereby diminish the expression of the encoded proteins. When the same experiments were performed with miR-283, we obtained similar results in the Cos2 and Smo-sensor lines (Figure 4, I and J, respectively), but not in the Fu-sensor lines (Figure 4K). We suspect that the decreased levels of Fu observed upon miR-283 overexpression (Figure 3, I and J) were likely a consequence of the decreased level of Cos2, consistent with results from a previous study (Ruel et al. 2003) and not due to the direct regulation of fu mRNA by the microRNA. No modifications in the levels of the three sensor lines were observed upon miR-304 overexpression.
Deletion of the microRNA cluster does not lead to Hh pathway deregulation:
To assess the importance of these microRNAs for fly development, flies lacking the cluster were produced by homologous recombination (Gong and Golic 2003) and were named Δ3miR. The mutant line was verified by PCR and sequencing (supplemental Figure 1 and data not shown). Mutants lacking the three microRNAs presented strong lethality before the L1 stage (88.3%). Because most of the mutant embryos died well before the end of embryogenesis, we were unable to analyze the cuticular pattern of the dead embryos, although we did stain these embryos with phalloidin, DAPI, and a Golgi marker. No phenotypes were observed that were reminiscent of Hh lack of function (supplemental Figure 2). In Δ3miR dead embryos, nuclei are observed (supplemental Figure 2A) but cell membranes are lacking (supplemental Figure 2B), suggesting that lethality in these animals takes place before cellularization, whereas Δ3miR escaper embryos presented wild-type organization (supplemental Figure 2G). We also analyzed Wg expression in the escaper embryos that survived until later stages (supplemental Figure 2I). Because Wg expression is dependent upon Hh signaling activity between 3 and 6–7 hr after egg laying, any mutations affecting the Hh pathway should affect the Wg pattern. However, no differences in the Wg pattern were identified between the wt and the Δ3miR mutant embryos. Finally, in view of the role of Hh in the egg chamber, homeostasis of germ line and follicular cells, we also stained the egg chamber of Δ3miR adult escaper mutants with DAPI and phalloidin. No defects in the egg chamber organization were observed (supplemental Figure 2, K–K″). Adult structures of the Δ3miR escaper mutants, e.g., wings, eyes, legs, and abdomen, were also analyzed. No differences from wild-type animals were observed (data not shown). We also sensitized the background for Hh signaling and analyzed the Δ3miR escaper mutants in heterozygous mutant backgrounds for Cos2, Smo, and Su(fu). We did not observe any Hh-like phenotypes in Δ3miR; cos2w1/+, in Δ3miR; smoIIX43/+, or in Δ3miR; SufuLP/+ mutant animals (data not shown).
A likely explanation for the observed lethal phenotype was that these three microRNAs are involved in essential processes unrelated to Hh signaling regulation (Lim et al. 2005). For example, miR-12, miR-283, and miR-304 could have targets other than smo, cos2, and fu, and the regulation of these other targets could be critical for animal viability. Alternatively, it also seemed possible that the lethality was due to the reduced levels of dGMAP observed in the Δ3miR line (Figure 5, H–H′, and supplemental Figure 2, H and K″), possibly due to modified splicing of the microRNA-containing intron. To discriminate between these hypotheses, we asked whether expressing dGMAP or different microRNAs could rescue the lethality of the Δ3miR mutant. We found that ubiquitous expression of individual microRNAs driven by arm-Gal4 rescues the viability of the Δ3miR mutant by 17 to 25%, whereas dGMAP did not significantly increase the viability of mutant animals (supplemental Figure 3). These results demonstrated that the lethality of the Δ3miR mutants is mainly the consequence of the absence of the microRNA cluster and not of GMAP activity but it is possible that the absence of dGMAP might still contribute to the lethality observed in the Δ3miR mutants. Moreover the microRNA-rescue animals look comparable to wild type. Indeed, we did not observe any phenotypes reminiscent of the Hh phenotype. Taken together, these results show that the lethality of the Δ3miR mutant is not due to Hh signaling misregulation.
Figure 5.—
An absence of the microRNA cluster does not lead to Hh phenotype but changes the level of the sensor constructs. (A–H) Clones of cells homozygous mutant for Δ3miR are marked by the absence of Myc (red). Cos2 (A–A′), Fu (B–B′), Smo (C–C′), Ci (D–D′) and dGMAP (H–H′) proteins are visualized by immunofluorescence (green). GFP expression of Cos2- (E–E′), Fu- (F–F′) and Smo- (G–G′) sensor constructs is in green. No modifications in the levels of the proteins (A–D) were observed except for dGMAP (H). In contrast GFP level of the sensor constructs modestly increased (E–G, arrows) in Δ3miR mutant clones.
Smo, cos2, and fu 3′-UTRs are physiological targets of mir-12 and mir-283:
To confirm that the Hh pathway is not modified in Δ3miR mutant animals, we induced clones of cells mutant for the microRNA cluster in the wing discs at the L1 stage (Figure 5). We observed normal levels of Hh pathway components such as the Cos2, Fu, Smo, and Ci proteins (Figure 5, A–A′, B–B′, C–C′, and D–D′, respectively), as well as normal expression of Hh target genes (data not shown). Because the absence of the microRNA cluster had no consequences for the Hh pathway, it seemed possible that, although expressed in this tissue, the cluster has no functional role in the wing imaginal disc. To test this hypothesis, we analyzed GFP expression from the Cos2- (Figure 5, E–E′), Fu- (Figure 5, F–F′), and Smo- (Figure 5, G–G′) sensor constructs in the Δ3miR mutant clones. Clones were induced at the L1 stage and discs dissected 4 days later to minimize the possibility that perdurance of the microRNAs might mask a loss-of-function phenotype. As shown in Figure 5, E–G, a small increase in the GFP level was observed in the mutant cells, suggesting that miR-12 and miR-283 do indeed regulate the 3′-UTRs of the three genes in wing imaginal discs.
Absence of Dicer activity does not lead to Hh pathway deregulation:
We were surprised to only observe a modest increase in GFP sensor levels in the Δ3miR mutant cells. Redundancy with other microRNAs might explain why no strong increase in the GFP sensor level was seen. This seemed to be a reasonable hypothesis, since the Drosophila genome is predicted to encode as many as 100 miRNAs, and it is common to find individual mRNAs targeted by multiple miRNAs (Brennecke et al. 2005). It was thus possible that potential phenotypes resulting from a lack of the microRNA cluster were masked by functional redundancy with other microRNAs that can also bind smo, cos2, and fu mRNAs.
One way to circumvent this potential problem was to block all microRNA synthesis. In Drosophila, Dcr-1 is essential for microRNA-directed repression as it participates in the digestion of pre-microRNAs to form microRNAs (Lee et al. 2004). In dcr-1 mutants, therefore, microRNA synthesis and function are inhibited. To test our hypothesis, we created dcr-1 mutant clones in the wing imaginal discs (Figure 6). We also induced mutant clones in a Minute background but we did not obtain significantly bigger clones (data not shown). A strong increase in the GFP level of the miR12-sensor was observed in the dcr-1 mutant cells (Figure 6, D–D″), confirming that the microRNAs are absent in these clones. Nevertheless these mutant clones contained normal levels of Hh pathway components such as Cos2, Fu, and Smo (Figure 6, A–A″, B–B″, and C–C″, respectively), as well as normal expression of Hh target genes (data not shown). In contrast, GFP levels in the Cos2- (Figure 6, E–E″) and Smo- (Figure 6, F–F″) sensor lines were strongly increased (by 40%) in the dcr-1 mutant clones. Thus, although the sensor lines for the cos2 and smo 3′-UTRs showed increased expression in the dcr-1 mutant clones, confirming that a loss-of-function phenotype was not masked by perdurance of the miRNAs, the absence of Dcr activity affected neither the Cos2 and Smo protein levels nor the Hh pathway.
Figure 6.—
Absence of Dcr-1 function changes Cos2- and Smo-sensor level without affecting their protein levels. (A–E) Clones of cells homozygous mutant for dcr-1 are marked by the absence of β-Gal (red). Cos2 (A–A″), Fu (B–B″), and Smo (C–C″) proteins are visualized by immunofluorescence (green). No modifications in the levels of the proteins were observed. GFP expression of miR12 (D–D″), Cos2- (E–E″), and Smo- (F–F″) sensor lines is in green. Note that GFP level strongly increases in dcr-1 mutant clones. Note also that, although dcr-1 mutant clones have been induced in L1, as attested by the big size of the wt twin spots (bright red), they reach a small size only likely due to cell lethality. Due to technical difficulties we could not analyze Fu-sensor constructs in dcr-1 mutant clones.
DISCUSSION
We found that cos2, fu, and smo mRNA can be regulated by a cluster of microRNAs, including miR-12 and miR-283, in Drosophila wing disc. The overexpression of this cluster decreases the levels of Smo, Cos2, and Fu proteins and activates the Hh pathway, as evidenced by the induction of dpp expression in the wing imaginal discs and by the adult wing outgrowth. The experiments presented here with the 3′-UTR sensors of smo, fu, or cos2 are in favor of a direct binding. To constitute a real proof of a direct effect, further experiments as direct biochemical binding assay or compensatory mutation between the 3′-UTR and the miRNAs will be necessary to perform.
The three programs (Stark et al. 2003; Grun et al. 2005; Griffiths-Jones et al. 2006) that have been created to genomewide predictions of Drosophila miRNA targets provide lists of presumptive miR-12, and miR-283 regulated genes. In addition to our in vivo validations, miR-12 binding sites are predicted on the 3′-UTR of ci and no sites were found on the 3′-UTR of the Su(fu) gene (Griffiths-Jones et al. 2006). We did not observe a decrease in either of these two proteins in the microRNA cluster overexpressing clones (data not shown). It is interesting to note that Flynt et al. (2007) recently showed that Su(fu) mRNA, encoding another negative regulator of Hedgehog signaling, is targeted by miR-214 in zebrafish. Absence of miR-214 results in the reduction of muscle cell types, the specification of which is dependent on Hh pathway activity. Nevertheless, our study shows that in Drosophila wing discs an absence of microRNA does not modify the Hh pathway, raising the question of what the role of microRNAs in Drosophila Hh pathway regulation is.
Could the microRNAs overexpression phenotype that we identified be artifactual and simply the result of forced overexpression of the microRNA cluster in a tissue in which it should be silent? We believe the answer is no, because Northern blot analysis (Leaman et al. 2005) and the increase of our miR-sensor in the dcr-1 mutant clones (Figure 6) showed that the microRNA cluster is indeed expressed in this tissue. This, to us, suggests that the cluster likely has a role in this tissue in which it is normally present. Is the microRNA cluster regulation of the cos2 and smo 3′-UTRs physiological? We think so, because an absence of either the microRNA cluster or of Dicer in the wing imaginal disc induces an increase in the Cos2- and Smo-sensor lines. This, to us, signifies that the microRNAs expressed from the cluster regulate the cos2 and smo 3′-UTRs and thus display some functionality in the disc during larval development. Altogether, these data clearly show that we have not created an artifactual situation in which the microRNA cluster is expressed in a tissue in which it should not be present. The miRs overexpression was also tested on embryonic patterning but it did not lead to any phenotype (data not shown), suggesting that the miR cluster regulation on the Hh pathway is specific to larval tissues.
As miR-12 and miR-283, and likely redundant miRs, are present in every cell of the wing disc, one possibility is that their normal roles are to dampen down the levels of Hh pathway components, particularly Cos2 and Smo, to prevent the accidental activation or downregulation of the pathway. Indeed, expressing both the microRNA cluster and its targets in the same tissue could provide a means of “buffering stochastic fluctuations” in mRNA levels or in protein translation rates within the Hh signaling pathway, as has been proposed for other processes (Hornstein and Shomron 2006).
Absence of the microRNA cluster does not lead to Hh pathway deregulation:
Our data possibly indicate that miRNAs are able to regulate two antagonistic components of the pathway, Cos2 and Smo. We have previously shown that the stability of these two proteins is “interdependent”: an increased level of Cos2 in the wing imaginal disc lowers the level of Smo, and, in the opposite direction, increased Smo decreases the level of Cos2 (Ruel et al. 2003). We propose that the interregulation of Cos2/Smo levels is independent of their relative activities because Cos2 effect on Smo levels is observed in posterior cells in which Cos2 activity is strongly inhibited by the constitutive activation of the pathway. Therefore, eliminating the miRNA-mediated inhibition of Cos2 and Smo in Δ3miR or dcr-1 mutant cells likely initially increased the levels of both proteins, but then the resulting higher levels of each protein presumably downregulated the other; the net variation of Cos2 and Smo levels would therefore be null. We favor this hypothesis because the independent Smo- and Cos2-sensor lines, which are unaffected by this Cos2/Smo interregulation, showed increased levels of GFP staining in Δ3miR and dcr-1 mutant animals (Figures 5, E–G, and 6, E and F, respectively). This suggests that the levels of both Cos2 and Smo are increased in the mutant animals but, because of the downregulation of each protein by the other, no ultimate alterations in the levels of the proteins are observed. If so, we would not necessarily expect to see an Hh phenotype in the miR mutant.
MicroRNAs misexpression and cancers:
Our screen created a situation in which the expression of the microRNA cluster is deregulated, ultimately destabilizing Cos2 protein levels and thereby activating Ci and Hh target gene expression. Importantly, a similar situation might be encountered during tumoral development. Aberrant Hh signaling activity is known to trigger the development of diverse cancers (Ruiz i Altaba 1999; Watkins et al. 2003; Fan et al. 2004; Kayed et al. 2004). While several of these tumors have been linked to mutations in Hh signaling components, not all of them have, leaving open the possibility that they are caused by other factors such as microRNA misexpression. Interestingly, more than half of the known human microRNA genes are located near chromosomal breakpoints associated with cancer (Calin et al. 2004a,b), and in some documented cases the microRNAs are amplified, leading to overexpression (Lu et al. 2005). Some upregulated microRNAs are possibly able to bind mRNAs encoding negative regulators of Hh signaling, such as Su(fu) or Ptc, and could thus induce the misactivation of the Hh pathway, as is observed in some cancers. Therefore, a fine analysis of microRNA expression levels and the levels of known Hh components should be considered in studies of Hh pathway-related cancers.
What does our manuscript add to the current knowledge about miRNA regulation? Our study shows for the first time that a cluster of three microRNAs can target several antagonistic components of the same pathway in vivo. This is novel and unexpected. This raises the question of how to interpret the miRNA expression signatures observed in human tumors. Indeed, as stated above, it has been proposed that miRNAs are differentially expressed in human cancers and contribute to cancer development. The working hypothesis in the cancer/miRNAs field is that key cancer genes are regulated by aberrant expression of miRNAs. The identification of a specific miRNA:mRNA interactor pair is generally accepted as being of biological importance when the mRNA encodes a tumor suppressor or an oncogene whose expression is modified in the tumor. Our study shows indirectly this is an oversimplified view, because identifying an oncogene or tumor suppressor as a target of a miRNA may not provide a full explanation for tumor development if the same miRNA hits other antagonistic components of the same pathway that nullify the effect of the identified miRNA:mRNA interactor pair.
Acknowledgments
We thank S. Cohen for providing the tub-EGFP vector and for transgenic lines, R. Carthew for the dicer-1 mutant line, and all members of P. Thérond's lab for discussions. F.F.G. was supported by a postdoctoral fellowship from Fondation de France. This work was supported by grants from Fondation de France, Centre National de la Recherche Scientifique, and Ligue Nationale Contre le Cancer “équipe labellisée 2008” to P.P.T.
References
- Aravin, A. A., M. Lagos-Quintana, A. Yalcin, M. Zavolan, D. Marks et al., 2003. The small RNA profile during Drosophila melanogaster development. Dev. Cell 5 337–350. [DOI] [PubMed] [Google Scholar]
- Bartel, D. P., 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116 281–297. [DOI] [PubMed] [Google Scholar]
- Basler, K., and G. Struhl, 1994. Compartment boundaries and the control of Drosophila limb pattern by hedgehog protein. Nature 368 208–214. [DOI] [PubMed] [Google Scholar]
- Brennecke, J., D. R. Hipfner, A. Stark, R. B. Russell and S. M. Cohen, 2003. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113 25–36. [DOI] [PubMed] [Google Scholar]
- Brennecke, J., A. Stark, R. B. Russell and S. M. Cohen, 2005. Principles of microRNA-target recognition. PLoS Biol. 3 e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin, G. A., C. G. Liu, C. Sevignani, M. Ferracin, N. Felli et al., 2004. a MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc. Natl. Acad. Sci. USA 101 11755–11760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin, G. A., C. Sevignani, C. D. Dumitru, T. Hyslop, E. Noch et al., 2004. b Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. USA 101 2999–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capdevila, J., and I. Guerrero, 1994. Targeted expression of the signaling molecule decapentaplegic induces pattern duplications and growth alterations in Drosophila wings. EMBO J. 13 4459–4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Leva, G., G. A. Calin and C. M. Croce, 2006. MicroRNAs: fundamental facts and involvement in human diseases. Birth Defects Res. C Embryo Today 78 180–189. [DOI] [PubMed] [Google Scholar]
- Fan, L., C. V. Pepicelli, C. C. Dibble, W. Catbagan, J. L. Zarycki et al., 2004. Hedgehog signaling promotes prostate xenograft tumor growth. Endocrinology 7 7. [DOI] [PubMed] [Google Scholar]
- Felsenfeld, A. L., and J. A. Kennison, 1995. Positional signaling by hedgehog in Drosophila imaginal disc development. Development 121 1–10. [DOI] [PubMed] [Google Scholar]
- Flynt, A. S., N. Li, E. J. Thatcher, L. Solnica-Krezel and J. G. Patton, 2007. Zebrafish miR-214 modulates Hedgehog signaling to specify muscle cell fate. Nat. Genet. 39 259–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friggi-Grelin, F., C. Rabouille and P. Therond, 2006. The cis-Golgi Drosophila GMAP has a role in anterograde transport and Golgi organization in vivo, similar to its mammalian ortholog in tissue culture cells. Eur. J. Cell. Biol. 85 1155–1166. [DOI] [PubMed] [Google Scholar]
- Gallet, A., R. Rodriguez, L. Ruel and P. P. Therond, 2003. Cholesterol modification of hedgehog is required for trafficking and movement, revealing an asymmetric cellular response to hedgehog. Dev. Cell 4 191–204. [DOI] [PubMed] [Google Scholar]
- Gong, W. J., and K. G. Golic, 2003. Ends-out, or replacement, gene targeting in Drosophila. Proc. Natl. Acad. Sci. USA 100 2556–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones, S., R. J. Grocock, S. van Dongen, A. Bateman and A. J. Enright, 2006. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 34 D140–D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grun, D., Y. L. Wang, D. Langenberger, K. C. Gunsalus and N. Rajewsky, 2005. microRNA target predictions across seven Drosophila species and comparison to mammalian targets. PLoS Comput. Biol. 1 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornstein, E., and N. Shomron, 2006. Canalization of development by microRNAs. Nat. Genet. 38 S20–S24. [DOI] [PubMed] [Google Scholar]
- Kayed, H., J. Kleeff, S. Keleg, J. Guo, K. Ketterer et al., 2004. Indian hedgehog signaling pathway: expression and regulation in pancreatic cancer. Int. J. Cancer 110 668–676. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana, M., R. Rauhut, W. Lendeckel and T. Tuschl, 2001. Identification of novel genes coding for small expressed RNAs. Science 294 853–858. [DOI] [PubMed] [Google Scholar]
- Lai, E. C., P. Tomancak, R. W. Williams and G. M. Rubin, 2003. Computational identification of Drosophila microRNA genes. Genome Biol. 4 R42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaman, D., P. Y. Chen, J. Fak, A. Yalcin, M. Pearce et al., 2005. Antisense-mediated depletion reveals essential and specific functions of microRNAs in Drosophila development. Cell 121 1097–1108. [DOI] [PubMed] [Google Scholar]
- Lee, Y. S., K. Nakahara, J. W. Pham, K. Kim, Z. He et al., 2004. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 117 69–81. [DOI] [PubMed] [Google Scholar]
- Lim, L. P., N. C. Lau, P. Garrett-Engele, A. Grimson, J. M. Schelter et al., 2005. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433 769–773. [DOI] [PubMed] [Google Scholar]
- Lu, J., G. Getz, E. A. Miska, E. Alvarez-Saavedra, J. Lamb et al., 2005. MicroRNA expression profiles classify human cancers. Nature 435 834–838. [DOI] [PubMed] [Google Scholar]
- McMahon, A. P., P. W. Ingham and C. J. Tabin, 2003. Developmental roles and clinical significance of hedgehog signaling. Curr. Top. Dev. Biol. 53 1–114. [DOI] [PubMed] [Google Scholar]
- Rajewsky, N., 2006. microRNA target predictions in animals. Nat. Genet. 38 S8–S13. [DOI] [PubMed] [Google Scholar]
- Rorth, P., K. Szabo, A. Bailey, T. Laverty, J. Rehm et al., 1998. Systematic gain-of-function genetics in Drosophila. Development 125 1049–1057. [DOI] [PubMed] [Google Scholar]
- Ruel, L., R. Rodriguez, A. Gallet, L. Lavenant-Staccini and P. P. Therond, 2003. Stability and association of Smoothened, Costal2 and Fused with Cubitus interruptus are regulated by Hedgehog. Nat. Cell Biol. 5 907–913. [DOI] [PubMed] [Google Scholar]
- Ruiz i Altaba, A., 1999. Gli proteins and Hedgehog signaling: development and cancer. Trends Genet. 15 418–425. [DOI] [PubMed] [Google Scholar]
- Stark, A., J. Brennecke, R. B. Russell and S. M. Cohen, 2003. Identification of Drosophila microRNA targets. PLoS Biol. 1 E60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teleman, A. A., Y. W. Chen and S. M. Cohen, 2005. Drosophila Melted modulates FOXO and TOR activity. Dev. Cell 9 271–281. [DOI] [PubMed] [Google Scholar]
- Wang, Q. T., and R. A. Holmgren, 1999. The subcellular localization and activity of Drosophila cubitus interruptus are regulated at multiple levels. Development 126 5097–5106. [DOI] [PubMed] [Google Scholar]
- Watkins, D. N., D. M. Berman, S. G. Burkholder, B. Wang, P. A. Beachy et al., 2003. Hedgehog signalling within airway epithelial progenitors and in small-cell lung cancer. Nature 422 313–317. [DOI] [PubMed] [Google Scholar]
- Zhang, B., X. Pan, G. P. Cobb and T. A. Anderson, 2007. microRNAs as oncogenes and tumor suppressors. Dev. Biol. 302 1–12. [DOI] [PubMed] [Google Scholar]