Abstract
The packaging of DNA into proper chromatin structure contributes to transcriptional regulation. This packaging is environment sensitive, yet its role in adaptation to novel environmental conditions is completely unknown. We set out to identify candidate chromatin-remodeling loci that are differentiated between tropical and temperate populations in Drosophila melanogaster, an ancestrally equatorial African species that has recently colonized temperate environments around the world. Here we describe sequence variation at seven such chromatin-remodeling loci, four of which (chd1, ssrp, chm, and glu) exhibit strong differentiation between tropical and temperate populations. An in-depth analysis of chm revealed sequence differentiation restricted to a small portion of the gene, as well as evidence of clinal variation along the east coasts of both the United States and Australia. The functions of chd1, chm, ssrp, and glu point to several novel hypotheses for the role of chromatin-based transcriptional regulation in adaptation to a novel environment. Specifically, both stress-induced transcription and developmental homeostasis emerge as potential functional targets of environment-dependent selection.
THE level, timing, and spatial distribution of gene expression vary both within and between species (Oleksiak et al. 2002; Meiklejohn et al. 2003; Rifkin et al. 2003; Nuzhdin et al. 2004; Gilad et al. 2006). Recent work on the evolution of gene expression has focused largely on local regulatory elements (cis-regulation; Gompel et al. 2005; Wittkopp 2006) or on the expression/activity of proteins that interact with such sequences (trans-regulation; Wittkopp et al. 2004; Wang et al. 2007). These DNA-transcription factor interactions, however, compose only one component of gene regulation. The packaging of DNA into proper chromatin structure provides an additional level of transcriptional regulation (Elgin and Workman 2001). For example, variation in chromatin state (i.e., permissive or restrictive) may affect access of cis-regulatory sequence by transcription factors. Recent chromatin research, buttressed by new tools and key insights, is rapidly revealing the complex and elegant mechanisms of chromatin remodeling and their transcriptional consequences (Allis et al. 2007). Evolutionary biologists now have the unprecedented opportunity to explore how this fundamental component of gene regulation contributes to adaptation.
The packaging of DNA into chromatin is environment sensitive. Early Drosophila work demonstrated the temperature sensitivity of two classic chromatin-related phenotypes—position-effect variegation (PEV) (Gowen 1933) and polytene chromosome “puff” induction (Ashburner 1967). More recent studies demonstrate that silencing maintained by the Polycomb group genes (PcGs), which remodel chromatin at developmentally important loci to maintain developmental trajectories (Lewis 1978; Schwartz and Pirrotta 2007), increases with increasing temperature (Fauvarque and Dura 1993). Moreover, stress induces global histone hypoacetylation that results in a coordinated downregulation of transcription (Berthiaume et al. 2006), particularly at components of the translation apparatus (Causton et al. 2001). The promoters of stress response genes, in contrast, experience chromatin remodeling that results in transcriptional activation (yeast: Zhao et al. 2005; Uffenbeck and Krebs 2006; Drosophila: Leibovitch et al. 2002). Finally, stress responders such as heat-shock proteins (which are generalized environmental-response molecules) co-immunoprecipitate with such known chromatin-remodeling complexes as TAC1 (Smith et al. 2004) and the PcG complex, PRC1 (Wang and Brock 2003). Given that chromatin dynamics are environment sensitive, the evolution of proteins that remodel chromatin may contribute to adaptation to novel habitats.
Tolerance of environmental stresses varies both within and between species. The low-latitude species Drosophila virilis, for example, maintains both elevated thermotolerance and an elevated heat-shock protein (hsp) induction threshold relative to the closely related, high-latitude species, D. lummei (Garbuz et al. 2003). Similar variation in environmental tolerance occurs within species. For example, D. melanogaster, an ancestrally equatorial African species, has recently adapted to temperate environments (Lachaise et al. 1988). Krebs and Feder (1997) document natural variation in thermotolerance and hsp70 induction thresholds in D. melanogaster populations found in the midwestern United States (Krebs and Feder 1997). The association of genetic differentiation and environmental tolerance is perhaps most obvious in the context of variation along latitudinal clines, one of the most thoroughly documented cases of natural variation associated with habitat heterogeneity at both the genotypic (allozyme: Knibb et al. 1981; DNA: Verrelli and Eanes 2001 and Sezgin et al. 2004; inversions: Knibb et al. 1981) and phenotypic levels. Phenotypic clinal variation includes variation in tolerance of several environmental conditions, such as temperature tolerance (Hoffmann et al. 2002), starvation resistance (Karan et al. 1998), and dehydration resistance (Karan et al. 1998; but see Hoffmann et al. 2001). This clinal variation provides an opportunity to identify chromatin-regulated biological processes and the associated chromatin machinery, which may underlie phenotypic differences between populations experiencing different environmental stresses.
To search for candidate, clinally varying loci that function in chromatin/histone remodeling (hereafter, “chromatin remodeling”), we took advantage of a whole-genome tiling array analysis that used DNA hybridization patterns to describe, on a genomic scale, candidate regions of differentiation among tropical and temperate populations of D. melanogaster from multiple continents (Turner et al. 2008, accompanying article in this issue). Here we describe DNA sequence variation in tropical and temperate population samples from Australia for a set of chromatin-remodeling genes that exhibited tropical–temperate differences in hybridization patterns on tiling arrays. We explore one candidate locus, the PcG chameau (chm), in greater depth to delineate the physical extent of tropical–temperate differentiation and to test for clinal variation in both Australia and the United States. Our results raise several new hypotheses about how chromatin dynamics may contribute to adaptation.
MATERIALS AND METHODS
Generating a list of candidate tropical–temperate differentiated chromatin genes:
Here we briefly describe the tiling array experiment, details for which can be found in Turner et al. (2008, accompanying article). Female flies derived from isofemale lines from two northern Australian populations (Queensland) were pooled. DNA was extracted, fragmented, and hybridized to four replicate Affymetrix tilling arrays. The procedure was repeated for population samples from southern Australia (Tasmania) and for U. S. population samples from Maine and Florida (latitudes of population samples can be found in supplemental Table S1). Following normalization and t-tests, each array feature was assigned a q-value (false discovery rate). To generate windows of significant differentiation, the average q-value was calculated along each chromosome arm in windows of 20, 50, and 100 probes (∼800, 2000, 4000 bp, respectively), moving 10% of the window size at each measurement. Significance thresholds were determined by permutation test. Finally, the union of significant windows among all window sizes was determined. The resulting “regions of differentiation” spanned, on average, one to two genes (Turner et al. 2008). Using the FlyBase gene summaries (http://www.flybase.org), a list of chromatin-associated genes located within significant windows (supplemental Table S2) was compiled. Only windows that overlapped two or fewer genes, and for which the focal chromatin gene was located near the midpoint of the window, were considered further (but see below for one exception). This reduced set of windows contained eight chromatin genes. To provide finer-scale analysis of differentiation on the basis of the array data, a 20-probe (∼800 bp) sliding-window analysis was conducted to determine if variation in putative differentiation varied across the window, and if so, whether the focal chromatin gene overlapped the most differentiated region (data not shown). The window containing the chromatin gene Bj1 and its neighbor, CG33993, exhibited hybridization differences restricted largely to gene CG33993 (data not shown). Therefore, Bj1 was not considered for further sequence analysis. Table 1 shows the final set of seven candidates. The U. S. window containing chm contained three genes but was retained because it partially overlapped the Australian window.
TABLE 1.
Culled list of significantly differentiated windows from hybridization experiment and associated chromatin-remodeling genes
| Chromosome | Window coordinates | Window size | Continent | Gene |
|---|---|---|---|---|
| X | 20,213,337–20,218,718 | 5381 | Australia | HERC2 |
| X | 1,738,978–1,742,532 | 3554 | Australia | trr |
| 2L | 16,731,317–16,737,494 | 6177 | Australia | glu |
| 3L | 19,816,835–19,818,616 | 1781 | Australia | Mi-2 |
| 2L | 2,982,496–2,986,379 | 3883 | Australia | chd1 |
| 2L | 7,410,897–7,416,684 | 5787 | Australia | chm |
| 2L | 7,408,650–7,416,684 | 8034 | United States | chm |
| 2R | 19,311,720–19,315,687 | 3967 | Australia | ssrp |
Coordinates were obtained from D. melanogaster reference genome release 4.3.
Sequencing-based validation of candidate windows:
The goal of the DNA sequencing analysis was to document the magnitude and physical extent of genetic differentiation between tropical and temperate populations for the seven candidate genes. The differentiated regions detected in the tiling array analysis could be due to a small, highly differentiated region of DNA or to a larger, less dramatically differentiated region. Moreover, some regions, especially those at the less conservative significance thresholds (i.e., P = 0.05), may be false discoveries. Strong tropical–temperate sequence differentiation over a small physical scale that includes a candidate gene, however, would support the hypothesis that the gene is influenced by spatially varying selection.
For the single-gene windows, three regions were amplified: a single 1-kb region inside the window, and two 1-kb regions outside (one 5′, one 3′ of the window). If the window contained two genes, four 1-kb regions were amplified—two within the window and two outside the window. Sequence data were collected from 16 tropical (∼17° latitude, two populations) and 16 temperate (∼45° latitude, two populations) Australian isofemale lines. For chm only, we also sequenced 16 (sub)tropical (∼25° latitude) and 16 temperate (∼45° latitude) U. S. isofemale lines (supplemental Table S1). Multi-fly DNA preps were PCR amplified, cloned (TOPO TA kit, Invitrogen, San Diego), and sequenced with M13 primers. Rare errors introduced by Taq polymerase will not affect conclusions regarding geographic differentiation. Primer pairs used to generate all sequence data can be found in supplemental Table S4. Sequences have been submitted to GenBank under accession nos. EU414997–EU416170.
In-depth investigation of the chm locus—extent/magnitude of (sub)tropical–temperate differentiation, clinal variation, and linkage to In(2L)t:
As we describe below (results), the chm locus was differentiated in both the United States and Australia. We further investigated patterns of allele-frequency variation at chm by additional sequencing both inside and outside the gene region to determine more precisely the extent and magnitude of tropical and temperate sequence differentiation. We sequenced the same 64 isofemale lines described above (32 from Australia, 32 from the United States). Within the chm gene region, we amplified a total of five regions that each spanned between 1 and 1.3 kb and covered all eight exons. Outside chm, we sequenced a single 1-kb region 8 kb downstream in both the United States and Australia. We also sequenced two 1-kb regions 8 and 12 kb upstream of chm in the U. S. samples. In Australia, where upstream differentiation was more variable, we sequenced four 1-kb regions 4, 8, 12, and 17 kb 5′ of chm (see Figure 1). Because we sequenced cloned products from isofemale lines, we were unable to assess linkage disequilibrium among sites on different amplicons. Finally, one of the two significantly differentiated replacement sites shared among Australia and the United States (arginine/praline; see results) was assayed in 10 D. melanogaster alleles from Zimbabwe (Begun and Aquadro 1993).
Figure 1.—
FST across a 35-kb region (D. melanogaster reference genome release 4.3) containing the chm locus. Each point corresponds to the midpoint of a single amplicon (see materials and methods) listed in supplemental Table S2, with the exception of peak seven, which corresponds to a mosaic of two amplicons. All genes located in this chromosomal region are indicated. The two significant replacement SNPs are in exon 1 (indicated with asterisks), although they occur in different amplicons. The serine/proline polymorphism occurs at peak five, while the arginine-proline amino acid polymorphism occurs at peak six.
We also assessed clinal variation at chm by measuring allele-frequency variation in additional populations at several intermediate latitudes (with respect to the tropical and temperate samples, see supplemental Table S1) in the United States and Australia. A single clinal SNP was assayed from nine Australia populations that span 15°–45° latitude and seven U. S. populations that span 25°–45° latitude (names, latitudes, and sample sizes are listed in supplemental Table S1). To genotype the target SNP from clinal samples, a single male from each isofemale line was crossed to flies from the y;cn,bw;sp stock corresponding to the reference genome. A single offspring from each cross was assayed for the state of the target variant inherited from its father by PCR–restriction fragment length polymorphism (RFLP). The most significantly differentiated site in Australia that could be assayed by RFLP analysis was not the most significantly differentiated site in the United States that could be assayed with this method; therefore a different SNP was assayed on each continent. The Australian SNP (2L: 7,413,926, v. 4.3; FST = 0.36), a silent site in the coding region of chm, was assayed using HgaI (New England Biolabs, Ipswich, MA). The U. S. SNP (2L: 7,413,468, v. 4.3; FST = 0.20), also a silent site in the coding region of chm, was assayed using BstUI (New England Biolabs).
chm is located 5.7 Mb from the breakpoint of In(2L)t (http://www.flybase.org), a known clinal inversion (Mettler et al. 1977). To determine whether allele-frequency variation is explained by linkage to In(2L)t, the same chromosomes assayed for the target SNP were also assayed by PCR for the presence/absence of In(2L)t (Andolfatto and Kreitman 2000).
Analysis:
Multi-site FST, π, linkage disequilibrium, and Tajima's D estimates were made in DNAsp (Rozas et al. 2003). Site-by-site FST estimates and Monte Carlo sampling to determine P-values (following Berry and Kreitman 1993) were run using Python scripts. All other statistical tests were run in JMP (v. 5.1, SAS Institute, Cary, NC). The standard error for the allele-frequency estimates was estimated by √(p(1 − p)/n).
RESULTS
Chromatin/histone-remodeling genes ssrp, glu, chd1, and chm exhibit tropical–temperate sequence differentiation:
Seventeen significant windows (P ≤ 0.05) identified by the whole-genome tiling array analysis (Turner et al. 2008, accompanying article), representing regions of differential hybridization patterns of tropical–temperate population samples (see materials and methods), overlapped chromatin-remodeling genes (supplemental Table S2). Ten of those windows overlapped only a small portion of the chromatin-remodeling gene or contained three or more genes and were not considered further. The remaining seven windows and associated chromatin-remodeling genes (Table 1) were retained for DNA sequence analysis.
We found low levels of polymorphism in three of the seven regions (Mi-2, HERC, trr; see Table 2), all three of which had low estimates of FST. These three regions are located in areas of low crossing over (Kliman and Hey 1993), which is consistent with the observed lack of polymorphism (Begun and Aquadro 1992). We observed no sequence differentiation in the genes, suggesting that these three loci were false discoveries in the array experiment. Note that our list of candidate genes was inferred from the least conservative FDR class.
TABLE 2.
Estimates of sequence differentiation (FST) and polymorphism (π) for amplified regions inside window, overlapping chromatin gene, and outside window
| Focal region | Amplicon (midpoint) | Amplicon size | FST | π (all) | π (tropical/temperate) |
|---|---|---|---|---|---|
| chd1 | 2L: 2980275 | 936 | 0.030 | 0.005 | 0.005/0.004 |
| 2L: 2985171 | 1405 | 0.170 | 0.008 | 0.008/0.007 | |
| 2L: 2989305 | 1426 | 0.001 | 0.007 | 0.008/0.005 | |
| chm (Australia) | 2L: 7407345 | 1044 | 0.180 | 0.015 | 0.014/0.031 |
| 2L: 7412208 | 1250 | 0.151 | 0.015 | 0.017/0.010 | |
| 2L: 7414710 | 1251 | 0.422 | 0.011 | 0.015/0.003 | |
| 2L: 7417734 | 1326 | 0.041 | 0.012 | 0.015/0.009 | |
| chm (United States) | 2L: 740735 | 1044 | 0.000 | 0.013 | 0.015/0.013 |
| 2L: 7412208 | 1250 | 0.007 | 0.015 | 0.015/0.017 | |
| 2L: 7413459 | 1251 | 0.100 | 0.012 | 0.012/0.015 | |
| 2L: 7417734 | 1326 | 0.045 | 0.004 | 0.015/0.012 | |
| glu | NA | NA | NA | NA | NA |
| 2L: 16732566 | 1358 | 0.120 | 0.010 | 0.010/0.008 | |
| 2L: 16735626 | 1113 | 0.090 | 0.009 | 0.008/0.010 | |
| 2L: 16738423 | 943 | 0.070 | 0.004 | 0.004/0.003 | |
| ssrp | 2R: 19308927 | 1099 | 0.001 | 0.008 | 0.009/0.008 |
| 2R: 19312582 | 1139 | 0.018 | 0.012 | 0.013/0.011 | |
| 2R: 19315342 | 1000 | 0.130 | 0.005 | 0.007/0.004 | |
| 2R: 19320319 | 1121 | 0.001 | 0.003 | 0.003/0.004 | |
| Mi-2 | 3L: 19813845 | 1072 | — | 0.000 | 0.003/0.007 |
| 3L: 19816984 | 1248 | 0.002 | 0.001 | 0.001/0.001 | |
| 3L: 19822410 | 932 | 0.000 | 0.001 | 0.001/0.001 | |
| HERC | X: 20211332 | 1014 | 0.015 | 0.005 | 0.004/0.006 |
| X: 20216217 | 1194 | — | 0.000 | 0.000/0.000 | |
| X: 20221822 | 1039 | 0.018 | 0.005 | 0.005/0.005 | |
| trr | X: 1737499 | 1089 | 0.012 | 0.002 | 0.002/0.002 |
| X: 1741745 | 929 | 0.001 | 0.002 | 0.002/0.002 | |
| X: 1745278 | 1083 | 0.028 | 0.002 | 0.002/0.002 |
Boldface type indicates amplified regions inside window and italics indicate the region overlapping the chromatin gene. Locations are from D. melanogaster reference genome release 4.3.
The four remaining regions, which contained genes ssrp, glu, chd1, and chm, showed average levels of polymorphism (Shapiro et al. 2007) and significant sequence differentiation between tropical and temperate populations (Table 2). The FST estimates at the chromatin-associated loci range from 0.10 to 0.15, which are consistently elevated relative to regions immediately outside the putatively significant windows and relative to random regions of the genome (United States: FST = 0.040; Australia: FST = 0.036; Turner et al. 2008, this issue). Importantly, high levels of sequence differentiation were restricted to the focal gene, even in the two cases where two gene regions overlapped the window (ssrp, glu) (Table 2—the 5′ region outside the window containing glu was recalcitrant to amplification). The chm locus exhibits the most dramatic differentiation in Australia (FST = 0.42). FST at chm differs substantially between the two continents, with the Australian populations exhibiting considerably higher levels of differentiation, in agreement with the genomewide pattern (Turner et al. 2008, accompanying article in this issue). The elevated FST in both Australia and the United States inspired a more in-depth analysis of this gene region.
Tropical–temperate differentiation is restricted to the chm gene region:
The U. S. and Australian multi-site FST estimates peak in the first part of chm (Figure 1). Moreover, the fraction of significantly differentiated SNPs (P ≤ 0.05 by permutation test) also peaks in this 5′ region of chm (Table 3). Tropical–temperate differentiation is restricted to the chm gene region and is largely overlapping on the two continents (Figure 1), strongly suggesting that geographic variation at this chromatin-remodeling gene is the result of selection rather than demographic history.
TABLE 3.
Polymorphism estimates and breakdown of SNP number and class across 35-kb region near chm, corresponding to Figure 1
| Peak no. | Continent | Amplicon (bp) | No. of SNPs (diff/total) | Noncoding/coding (diff) | π (tropical/temperate) |
|---|---|---|---|---|---|
| 1 | Australia | 1010 | 3/38 | 3/0 | 0.015/0.010 |
| 2 | Australia | 1010 | 13/33 | 13/0 | 0.016/0.006 |
| United States | 0/29 | 0/0 | 0.014/0.017 | ||
| 3 | Australia | 1044 | 7/25 | 5/2 | 0.009/0.008 |
| United States | 1/32 | 1/0 | 0.014/0.014 | ||
| 4 | Australia | 1020 | 7/29 | 7/0 | 0.014/0.010 |
| 5 | Australia | 1250 | 11/34 | 9/2a | 0.017/0.010 |
| United States | 2/33 | 1/1a | 0.015/0.017 | ||
| 6 | Australia | 1251 | 19/30 | 6/13a | 0.014/0.007 |
| United States | 5/37 | 0/5a | 0.011/0.015 | ||
| 7 | Australia | 1251 | 23/34 | 17/6 | 0.015/0.003 |
| United States | 2/39 | 2/0 | 0.016/0.014 | ||
| 8 | Australia | 1320 | 1/40 | 1/0 | 0.015/0.009 |
| United States | 2/34 | 2/0 | 0.015/0.012 | ||
| 9 | Australia | 1250 | 4/26 | 4/0 | 0.009/0.009 |
| United States | 0/26 | 0/0 | 0.009/0.009 | ||
| 10 | Australia | 1080 | 0/21 | 0/0 | 0.009/0.011 |
| United States | 0/18 | 0/0 | 0.010/0.011 |
For each amplicon (“peak”) from Figure 1, the table lists the fraction of SNPs significantly differentiated between tropical and temperate regions (P ≤ 0.05 by permutation test; see materials and methods), as well as the number of SNPs significantly differentiated in the noncoding and coding regions of the amplicon. The peaks that overlap the chm gene region are in boldface. diff, significantly differentiated at the P ≤ 0.05 level.
Single replacement SNPs.
The total number of SNPs detected across the chm gene region is similar for Australia and the U. S. (Table 3), while the magnitude of FST and fraction of significantly differentiated SNPs differs substantially across the two continents (see y-axis of Figure 1). More specifically, of the 61 SNPs significantly differentiated between tropical and temperate zones (the union of all Australian and U. S. SNPs across chm—Table 3, boldface rows), 50 occur in Australia only, 3 in the U. S. only, and 8 are shared. In many cases SNPs differentiated only in Australia also show allele-frequency differences between Maine and Florida in the expected direction, but of smaller magnitude (supplemental Table S3). One exception is peak two (Figure 1), which is entirely intergenic and shows strong differentiation in Australia but not in the U. S. The different magnitudes of FST and fraction of differentiated SNPs on the two continents are consistent with differences observed at the genomewide scale (Turner et al. 2008, this issue) and likely reflect the larger latitudinal span in Australia.
Two of the eight SNPs that are significantly differentiated on both continents occur in the 5′-end of the first large intron and the remaining six occur in the chm coding region. Two of these coding variants are replacement SNPs. Intriguingly, none of the other 59 significantly differentiated SNPs are replacement variants (supplemental Table S3).
The first replacement site is similarly differentiated in Australia and the United States (FST = 0.18 and 0.20, respectively), but is not among the most differentiated sites for either continent. Nevertheless, this proline/serine polymorphism may be functionally significant, as the derived proline variant is a radical amino acid change from the ancestral serine allele, which is conserved in D. simulans, D. sechelia, and D. erecta (all species for which sequence was available and alignable; supplemental Figure S1).
The second replacement site is one of the most significantly differentiated sites in the United States (FST = 0.23), although not in Australia (FST = 0.22). The proline variant is also a radical amino acid change from the ancestral arginine allele, which is conserved in D. sechellia, D. yakuba, D. erecta, D. ananassae, and D. persimilis (supplemental Figure S2). Unfortunately, these two replacement sites were assayed on different amplicons, which precluded rigorous assessment of linkage disequilibrium between them (see materials and methods). Interestingly, variants that are virtually fixed in the temperate zones for both replacement sites correspond to the ancestral state (see supplemental Figures S1 and S2).
The most significantly differentiated sites in Australia occur in the first, large intron of chm (supplemental Table S3, three sites with FST = 0.66). These sites, however, are not significantly differentiated in the United States (although site 772 exhibits variation in the same direction; supplemental Table S3). The dominant temperate variant at site 772, like the amino acid variants described above, represents the ancestral state. Such a variant, if functionally significant, may affect adaptive gene expression variation across this latitudinal gradient. Interestingly, one of the best-studied clinally varying genes, Adh, also appears to harbor both selected amino acid and gene expression variants in D. melanogaster (summarized in Berry and Kreitman 1993). Moreover, Ldh in Fundulus heteroclitus exhibits both amino acid and gene expression variants along a north–south gradient (Place and Powers 1979; Schulte et al. 2000).
Differentiated site in chm exhibits linear cline along both continents independent of In(2L)t:
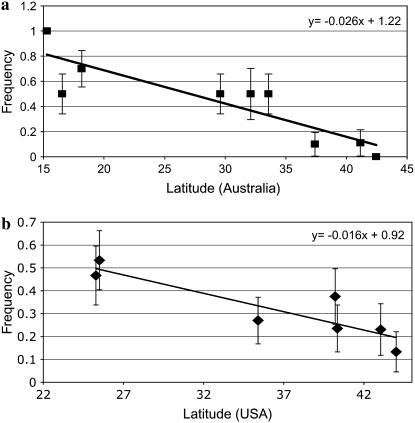
A single SNP was chosen for RFLP analysis in additional populations sampled along the east coasts of Australia and the United States (different SNPs were used on each continent; see materials and methods). We observed a significant correlation between allele frequency and latitude in Australia (R2 = 0.76, P < 0.001; Figure 2a) and in the U. S. (R2 = 0.79, P < 0.007; Figure 2b) for these SNPs. Analysis of standard [i.e., non-In(2L)t] chromosomes only also reveals a significant correlation with latitude, despite a reduced data set (Australia: R2 = 0.66, P = 0.007; United States: R2 = 0.47, P = 0.08). These data support previous findings that inversions explain clinal variation only at sites relatively close to the breakpoints (Kennington et al. 2006). Finally, both SNPs show strong linkage disequilibrium with the arginine/proline replacement SNP discussed above (D′ = 1.00 for both regions and continents; see supplemental Table S5 for haplotype frequencies), supporting the inference that the radical arginine/proline polymorphism is clinal on both continents.
Figure 2.—
Clinal variation at a differentiated SNP across latitudinal gradients in Australia (a) and the US (b), and equations for the regression lines.
Population genetic evidence of selection at chm in temperate populations:
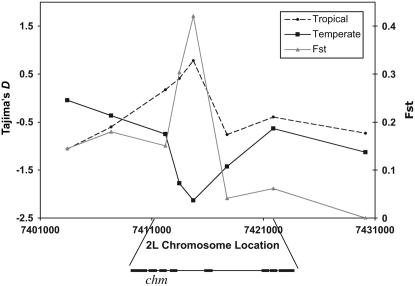
In both the tropical and temperate Australia data, Tajima's D (Tajima 1983) is near zero outside the region of significantly large FST values (Figure 3). Within the region associated with large FST values, however, Tajima's D is strongly negative in the temperate population and relatively positive (although not significantly so) in the tropical population in Australia. This pattern is not apparent in the United States, where differentiation is substantially smaller (data not shown). The skew toward rare variants in temperate Australia is consistent with a model in which the temperate population has a temperate-adapted, high-frequency haplotype and rare, less fit tropical haplotypes. These data create a puzzle when considered in combination with the observation of two, strongly differentiated, radical amino acid variants for which the common allele in temperate regions is the ancestral state. The classic model describes a derived, low-frequency sub-Saharan African variant rising to high frequency in a New World temperate zone (e.g., Schmidt et al. 2000; Sezgin et al. 2004). If the dominant temperate chm haplotype in southern Australian populations derives from a random sample of temperate haplotypes (defined by the presence of the putative selected ancestral variant) currently segregating in tropical populations, we would expect similar estimates of Tajima's D for both the temperate population and the “temperate haplotypes” segregating in tropical populations. Tajima's D for the latter population, however, is close to zero (D = 0.30). One possibility is the stochastic nature of selection associated with standing variation, such that a small subset of favored haplotypes may be disproportionately represented after selection (Orr and Betancourt 2001). Alternatively, there may be fitness differences among the haplotypes containing a putative favored, ancestral variant, such that only a small subset of such haplotypes are strongly favored in a novel environment. Epistatic fitness variation among ancestral variants during colonization of novel environments could, in principle, contribute to such dynamics. Several well-studied loci, such as Adh in D. melanogaster and Ldh in F. heterclitus, harbor multiple sites under selection along latitudinal clines (Place and Powers 1979; Berry and Kreitman 1993; Schulte et al. 2000).
Figure 3.—
Tajima's D across 35-kb region (D. melanogaster reference genome release 4.3) containing the chm locus in temperate and tropical Australia population samples. Each point corresponds to the midpoint of a single 1- to 1.3-kb amplicon.
A comparison of differentiation between Australian and U. S. temperate zones and between their tropical zones further supports the idea that selection has acted more strongly in the temperate zone in the recent evolutionary history of these D. melanogaster populations (Table 4). If temperate-adapted alleles were driven to high frequency independently on the two continents and the selected site(s) was segregating at a non-negligible frequency in the ancestral populations, different haplotypes could rise to high frequency in the Australian and U. S. temperate populations. Our observations are consistent with this hypothesis: FST calculated from the U. S. temperate region vs. the Australian temperate region is larger than the FST generated from the equivalent tropical comparison (Table 4). Moreover, we observed this difference only at the chm locus where the tropical–temperate FST is high.
TABLE 4.
Estimates of differentiation among similar climatic regions on different continents
| Peak no. | FST (temperate United States– temperate Australia) | FST (tropical United States– tropical Australia) |
|---|---|---|
| 2 | 0.120 | 0.063 |
| 3 | 0.052 | 0.046 |
| 5 | 0.040 | 0.000 |
| 6 | 0.160 | 0.021 |
| 7 | 0.260 | 0.000 |
| 8 | 0.036 | 0.041 |
| 9 | 0.005 | 0.000 |
| 10 | 0.010 | 0.000 |
Boldface type indicates peak U. S. differentiation; italics indicate peak Australian differentiation.
DISCUSSION
We have reported evidence of tropical–temperate differentiation at four chromatin-remodeling gene regions. A detailed analysis of one gene, chm, revealed that elevated geographic differentiation in the coding region was restricted to the 5′ end and that two replacement variants, both of which are radical changes, are significantly differentiated in both Australia and the United States. The amino acid variants at both replacement sites (see supplemental Figures S1 and S2) are virtually fixed in temperate zones of both continents, but occur at intermediate frequencies in the tropical zones. Interspecific comparisons (supplemental Figures S1 and S2) revealed that the high-frequency temperate variants are the ancestral rather than the derived states. Both variants at the arginine/proline replacement site were found in an African sample from Zimbabwe, with the ancestral variant occurring in 9 of the 10 alleles surveyed (data not shown).
Why might an ancestral variant (or variants), which is typically thought of as adapted for tropical environments, spread to high frequency under selection in temperate populations? One interesting possibility is selection associated with karyotypic variation. The ancestral karyotypic state (standard chromosomes) in D. melanogaster is found at high frequency in the temperate region (Mettler et al. 1977). Molecular data near inversion breakpoints suggest that the inversions are recent on the timescale of molecular evolution in D. melanogaster (Hasson and Eanes 1996; Andolfatto and Kreitman 2000; Matzkin et al. 2005) and that much of the polymorphism in the species is older than the inversions, despite the fact that the inversions are more abundant in equatorial Africa than in the recently established populations. This suggests the possibility that cases such as chm, in which temperate non-African populations have an ancestral variant(s) at high frequency while tropical non-African populations do not (Braverman et al. 2005), may be subject to selection pressures also associated with karyotypic variants.
The two continents do not share their respective, most significantly differentiated sites. This observation may be attributed to a combination of power differences (possibly related to the smaller latitudinal span in the United States) and difference in the organization of ancestral variation found among the chromosomes associated with the invasion of each continent. This lack of overlap, combined with evidence of elevated FST across the U. S. and Australian temperate zones (Table 4), support independent invasions of these two continents.
Previous work on evolution at chromatin-remodeling loci has focused largely on proteins that localize to the heterochromatin (Malik et al. 2002; Vermaak et al. 2005). These proteins are evolving adaptively perhaps in response to endogenous evolutionary forces such as centromeric drive. The group of genes identified in this study offers a less biased description (with respect to the diversity of chromatin functions) of how euchromatic chromatin remodeling may contribute to adaptation in response to exogenous forces, such as those found in a novel environment. However, these four loci by no means represent an exhaustive list of chromatin/histone-remodeling genes that may be influenced by spatially varying selection (see Turner et al. 2008 for limitations of hybridization technology).
CHD1, or chromo-ATPase/helicase-DNA-binding protein 1, is an ATP-utilizing chromatin assembly factor that acts on nucleosome spacing to confer a transcriptionally active chromatin conformation (Lusser et al. 2005; see Table 5). Stokes et al. (1996) document a nonrandom distribution of CHD1 on polytene chromosomes (Stokes et al. 1996), where CHD1 is restricted largely to a subset of interbands and puffed regions. CHD1 localizes specifically to developmental puffs (especially ecdysone-sensitive puffs) and heat-shock-induced puffs (Stokes et al. 1996).
TABLE 5.
Protein names, domain names, and functions encoded by the loci identified in this study
| Protein | Domain | Function |
|---|---|---|
| CHD1 | Chromodomain | Chromatin compaction, gene silencing (also found in HP1, PC) |
| DNA binding | Binds (A + T) minor grooves | |
| ATPase/helicase | Activates transcription via chromatin conformational changes | |
| SSRP1 | HMG box | DNA/chromatin binding |
| CHM | MYST | Acetyltransferase (transfers acetyl groups to histone tails) |
| SMC4 | ATPase | Chromatin condensation (in combination with other condensing components) |
| ATP binding | Chromatin condensation (in combination with other condensing components) |
Remarkably, CHD1 both colocalizes and interacts in vivo with SSRP1 (Kelley et al. 1999), another chromatin-remodeling protein identified in this study. SSRP1, or structure-specific recognition protein 1, is an HMG-box family protein (Table 5) that complexes with Spt16 to form the FACT complex (Orphanides et al. 1999). This complex localizes to genes actively transcribed by PolII and removes the histone dimer H2A-H2B as PolII passes (Belotserkovskaya et al. 2003). Like CHD1, the FACT complex is recruited to heat-shock loci following stress (Saunders et al. 2003) and to critical development loci during metamorphosis (Andrulis et al. 2000). Like CHM (discussed below), during embryogenesis the FACT complex modulates expression of several Hox genes (Shimojima et al. 2003).
The third gene identified in this study, chm, dominantly suppresses PEV and maintains Hox gene silencing by other Polycomb group proteins (Grienenberger et al. 2002). Specifically, chm dominantly enhances the aberrant sex-comb phenotype of Pc− mutants, designating chm as a Polycomb group gene. Like CHD1 and the FACT complex, Polycomb group proteins are also recruited to critical developmental loci during embryogenesis and metamorphosis (Ringrose and Paro 2004). Unlike CHD1 and FACT, however, Polycomb group proteins generally act as transcriptional repressors rather than activators. Nevertheless, the PcG CHM also activates transcription, specifically promoting histone 4 (H4) acetylation (Table 5) of AP-1 sites (dJun, cJun) associated with the JNK pathway (Miotto et al. 2006). This transcriptional activation enhances JNK pathway activity, which stimulates dorsal thorax closure during metamorphosis. In addition to this developmental role, the JNK pathway also contributes to stress response in both Drosophila (Wang et al. 2003) and mice (Tournier et al. 2000). More specifically, the interaction of chm with DFos and/or Djun is essential for JNK pathway response to both chemical and osmotic stress (Miotto and Struhl 2006). Finally, activation of JNK signaling in D. melanogaster inhibits the insulin/IGF signaling (IIS) pathway, resulting in increased life span (Wang et al. 2005). Intriguingly, longevity is one of several life-history traits known to vary along latitudinal clines (Schmidt et al. 2005).
Our final tropical–temperate differentiated locus, glu, codes for SMC4, an ATP-binding/ATPase that, as part of the condensin complex, contributes to chromosome condensation during the prometaphase of mitosis (Hirano and Mitchison 1994; Steffensen et al. 2001) and likely meiosis as well (Steffensen et al. 2001). By ensuring proper condensation, sister chromatids resolve appropriately prior to segregation, thereby minimizing nondisjunction. We have long understood that chromosome segregation in Drosophila is temperature sensitive, with low temperatures associated with elevated rates of meiotic nondisjunction in females (Tokunaga 1970a,b). Tropical–temperate sequence differentiation at the glu locus may contribute to faithful sister-chromatid segregation in the face of novel environmental conditions in recently established populations. Moreover, increased rates of nondisjunction are also associated with autosomal inversions (Roberts 1962). The likelihood of nondisjunction, potentially generated by clinal inversions [In(2L)t, In(3R)P, In(3L)P, In(2R)NS (Stalker 1976)] or due to temperature may be modulated by evolution at the glu locus. In addition to its role in prometaphase chromosome condensation, the condensin complex colocalizes with PcG proteins to their binding sites (Polycomb response elements) during interphase (Lupo et al. 2001). Localization with Polycomb group proteins suggests that, like CHD1, SSRP1, and CHM, SMC4 may carry out a regulatory role in development.
The regulation of developmental trajectories emerges as an unexpected biological process potentially targeted by spatially varying selection. All four chromatin-remodeling candidates investigated in this study either localize to developmental puffs and/or interact with the Polycomb group, which maintains long-term epigenetic silencing (through mitosis) of essential developmental regulators (Ringrose and Paro 2004). Long-term silencing, at least at Polycomb protein targets, is positively correlated with temperature (Fauvarque and Dura 1993). Given that cold temperatures (and possibly other stressors) disrupt the integrity of this chromatin-based silencing, it is tempting to speculate that evolution at chromatin proteins buffers these developmental trajectories from perturbations by novel environmental conditions. Moreover, low temperatures may also perturb transcriptional regulation at development puffs independently of the Polycomb group (Ashburner 1970), suggesting yet another potential source of selection driving the evolution at chromatin-remodeling proteins. At the phenotypic level, environment-induced developmental mistakes are common and are observed across diverse taxa, such as phenocopying in flies (Gloor 1947; Santamaria 1979; Gibson and Hogness 1996; Roberts and Feder 1999), temperature-induced neural tube defects in developing human fetuses (Chambers et al. 1998), and developmental instability as measured by fluctuating asymmetry across many taxa (e.g., Badyaev et al. 2000; Milton et al. 2003; Chang et al. 2007). Future research may determine that chromatin-associated aberrant transcription may underlie these environment-induced developmental mistakes found in naive populations.
In addition to developmental homeostasis, the four identified chromatin-remodeling loci reveal that stress response machinery, particularly those proteins associated with heat-shock gene transcription, may be targeted by spatially varying selection. Heat-shock genes, found at heat-shock puffs, are generalized stress responders to varied stimuli, such as osmotic stress, UV exposure, and cold stress (in addition to high temperature; Lindquist 1986). These proteins are remarkably well conserved across distantly related taxa (Voellmy 1984), which is consistent with the idea that selection on heat-shock protein function would likely occur at the level of transcriptional regulation rather than (or at least in addition to) the modification of chaperone function (Frydenberg et al. 2003). Promoter regions of many stress-induced and heat-shock loci have common cis-regulatory elements (Uffenbeck and Krebs 2006), further underscoring the potential for evolution in trans-activation. Such trans-activation at the level of chromatin organization provides coordinated, global gene regulation among disparate loci. Recent evolution at CHM, CHD1, and SSRP1 suggest that coordinately modulating disparate stress-response loci may be essential for maintaining a nonstressed biological state despite novel environmental conditions.
Adaptation to a novel habitat requires at least in part the buffering of environment-sensitive processes in the face of novel conditions. Given that chromatin packaging is environment sensitive, we expect many loci regulated by chromatin proteins to be aberrantly expressed in organisms exposed to novel environments. A small number of evolutionary changes in a few key chromatin-remodeling complexes may modulate the genomewide adverse effects of an (initially) suboptimal environment, thereby buffering critical biological processes and facilitating the invasion of novel habitat.
Acknowledgments
The authors thank P. Schmidt and A. Hoffmann for generously providing the clinal population samples from the United States and Australia, respectively. The inbred Raleigh, North Carolina, lines were generously provided by T. Mackay. The authors also thank U. Arshad, E. Nordman, and M. Eckert for technical assistance; T. Turner for insightful discussion; and A. Holloway and L. McBride for comments on previous versions of the manuscript. In addition, comments of two anonymous reviewers substantially improved the quality of the data presentation and discussion. This work was supported by National Science Foundation Graduate Research Fellowships to M.T.L. and National Institutes of Health grant GM071926 to D.J.B.
References
- Allis, C. D., T. Jenuwein, D. Reinberg and M. Caparros, 2007. Epigenetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Andolfatto, P., and M. Kreitman, 2000. Molecular variation at the In(2L)t proximal breakpoint site in natural populations of Drosophila melanogaster and D. simulans. Genetics 154 1681–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrulis, E. D., E. Guzman, P. Doring, J. Werner and J. T. Lis, 2000. High-resolution localization of Drosophila Spt5 and Spt6 at heat shock genes in vivo: roles in promoter proximal pausing and transcription elongation. Genes Dev. 14 2635–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner, M., 1967. Patterns of puffing activity in the salivary gland chromosomes of Drosophila. I. Autosomal puffing patterns in a laboratory stock of Drosophila melanogaster. Chromosoma 21 398–428. [DOI] [PubMed] [Google Scholar]
- Ashburner, M., 1970. Patterns of puffing activity in the salivary gland chromosomes of Drosophila. V. Responses to environmental treatments. Chromosoma 31 356–376. [DOI] [PubMed] [Google Scholar]
- Badyaev, A. V., K. R. Foresman and M. V. Fernandes, 2000. Stress and developmental stability: vegetation remodel causes increased fluctuating asymmetry in shrews. Ecology 81 336–345. [Google Scholar]
- Begun, D. J., and C. F. Aquadro, 1992. Levels of naturally occurring DNA polymorphism correlate with recombination rates in D. melanogaster. Nature 356 519–520. [DOI] [PubMed] [Google Scholar]
- Begun, D. J., and C. F. Aquadro, 1993. African and North American populations of Drosophila melanogaster are very different at the DNA level. Nature 365 548–550. [DOI] [PubMed] [Google Scholar]
- Belotserkovskaya, R., S. Oh, V. A. Bondarenko, G. Orphanides, V. M. Studitsky et al., 2003. FACT facilitates transcription-dependent nucleosome alteration. Science 301 1090–1093. [DOI] [PubMed] [Google Scholar]
- Berry, A., and M. Kreitman, 1993. Molecular analysis of an allozyme cline: alcohol dehydrogenase in Drosophila melanogaster on the east coast of North America. Genetics 134 869–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthiaume, M., N. Boufaied, A. Moisan and L. Gaudreau, 2006. High levels of oxidative stress globally inhibit gene transcription and histone acetylation. DNA Cell Biol. 25 124–134. [DOI] [PubMed] [Google Scholar]
- Braverman, J. M., B. P. Lazzaro, M. Aguade and C. H. Langley, 2005. DNA sequence polymorphism and divergence at the erect wing and suppressor of sable loci of Drosophila melanogaster and D. simulans. Genetics 170 1153–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causton, H. C., B. Ren, S. S. Koh, C. T. Harbison, E. Kanin et al., 2001. Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell 12 323–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers, C. D., K. A. Johnson, L. M. Dick, R. J. Felix and K. L. Jones, 1998. Maternal fever and birth outcome: a prospective study. Teratology 58 251–257. [DOI] [PubMed] [Google Scholar]
- Chang, X., B. Zhai, X. Liu and M. Wang, 2007. Effects of temperature stress and pesticide exposure on fluctuating asymmetry and mortality of Copera annulata (Selys) (Odonata: Zygoptera) larvae. Ecotoxicol. Environ. Saf. 67 120–127. [DOI] [PubMed] [Google Scholar]
- Elgin, S. C., and J. L. Workman, 2001. Chromatin Structure and Gene Expression. Oxford University Press, Oxford.
- Fauvarque, M. O., and J. M. Dura, 1993. Polyhomeotic regulatory sequences induce developmental regulator-dependent variegation and targeted P-element insertions in Drosophila. Genes Dev. 7 1508–1520. [DOI] [PubMed] [Google Scholar]
- Frydenberg, J., A. A. Hoffmann and V. Loeschcke, 2003. DNA sequence variation and latitudinal associations in hsp23, hsp26 and hsp27 from natural populations of Drosophila melanogaster. Mol. Ecol. 12 2025–2032. [DOI] [PubMed] [Google Scholar]
- Garbuz, D., M. B. Evgenev, M. E. Feder and O. G. Zatsepina, 2003. Evolution of thermotolerance and the heat-shock response: evidence from inter/intraspecific comparison and interspecific hybridization in the virilis species group of Drosophila. I. Thermal phenotype. J. Exp. Biol. 206 2399–2408. [DOI] [PubMed] [Google Scholar]
- Gibson, G., and D. S. Hogness, 1996. Effect of polymorphism in the Drosophila regulatory gene Ultrabithorax on homeotic stability. Science 271 200–203. [DOI] [PubMed] [Google Scholar]
- Gilad, Y., A. Oshlack, G. K. Smyth, T. P. Speed and K. P. White, 2006. Expression profiling in primates reveals a rapid evolution of human transcription factors. Nature 440 242–245. [DOI] [PubMed] [Google Scholar]
- Gloor, H., 1947. Phanokopie-versuche mit aether an Drosophila. Rev. Suisse Zool. 54 637. [Google Scholar]
- Gompel, N., B. Prud'homme, P. J. Wittkopp, V. A. Kassner and S. B. Carroll, 2005. Chance caught on the wing: cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature 433 481–487. [DOI] [PubMed] [Google Scholar]
- Gowen, J. W., and E. H. Gay, 1933. Effect of temperature on eversporting eye color in Drosophila melanogaster. Science 77 312–314. [DOI] [PubMed] [Google Scholar]
- Grienenberger, A., B. Miotto, T. Sagnier, G. Cavalli, V. Schramke et al., 2002. The MYST domain acetyltransferase Chameau functions in epigenetic mechanisms of transcriptional repression. Curr. Biol. 12 762–766. [DOI] [PubMed] [Google Scholar]
- Hasson, E., and W. F. Eanes, 1996. Contrasting histories of three gene regions associated with In(3L)Payne of Drosophila melanogaster. Genetics 144 1565–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano, T., and T. J. Mitchison, 1994. A heterodimeric coiled-coil protein required for mitotic chromosome condensation in vitro. Cell 79 449–458. [DOI] [PubMed] [Google Scholar]
- Hoffmann, A. A., R. Hallas, C. Sinclair and P. Mitrovski, 2001. Levels of variation in stress resistance in Drosophila among strains, local populations, and geographic regions: patterns for desiccation, starvation, cold resistance, and associated traits. Evolution 55 1621–1630. [DOI] [PubMed] [Google Scholar]
- Hoffmann, A. A., A. Anderson and R. Hallas, 2002. Opposing clines for high and low temperature resistance in Drosophila melanogaster. Ecol. Lett. 5 614–618. [Google Scholar]
- Karan, D., N. Dahiya, A. K. Munjal, P. Gibert, B. Moreteau et al., 1998. Desiccation and starvation tolerance of adult Drosophila: opposite latitudinal clines in natural populations of three different species. Evolution 52 825–831. [DOI] [PubMed] [Google Scholar]
- Kelley, D. E., D. G. Stokes and R. P. Perry, 1999. CHD1 interacts with SSRP1 and depends on both its chromodomain and its ATPase/helicase-like domain for proper association with chromatin. Chromosoma 108 10–25. [DOI] [PubMed] [Google Scholar]
- Kennington, W. J., L. Partridge and A. A. Hoffmann, 2006. Patterns of diversity and linkage disequilibrium within the cosmopolitan inversion In(3R)Payne in Drosophila melanogaster are indicative of coadaptation. Genetics 172 1655–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliman, R. M., and J. Hey, 1993. Reduced natural selection associated with low recombination in Drosophila melanogaster. Mol. Biol. Evol. 10 1239–1258. [DOI] [PubMed] [Google Scholar]
- Knibb, W. R., J. G. Oakeshott and J. B. Gibson, 1981. Chromosome inversion polymorphisms in Drosophila melanogaster. I. Latitudinal clines and associations between inversions in Australasian populations. Genetics 98 833–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs, R. A., and M. E. Feder, 1997. Natural variation in the expression of the heat-shock protein Hsp70 in a population of Drosophila melanogaster and its correlation with tolerance of ecologically relevant thermal stress. Evolution 51 173–179. [DOI] [PubMed] [Google Scholar]
- Lachaise, D., M. Cariou, J. R. David, F. Lemeunier, L. Tsacas et al., 1988. Historical Biogeography of the Drosophila melanogaster Species Subgroup. Plenum Press, New York.
- Leibovitch, B. A., Q. Lu, L. R. Benjamin, Y. Liu, D. S. Gilmour et al., 2002. GAGA factor and the TFIID complex collaborate in generating an open chromatin structure at the Drosophila melanogaster hsp26 promoter. Mol. Cell. Biol. 22 6148–6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, E. B., 1978. A gene complex controlling segmentation in Drosophila. Nature 276 565–570. [DOI] [PubMed] [Google Scholar]
- Lindquist, S., 1986. The heat-shock response. Annu. Rev. Biochem. 55 1151–1191. [DOI] [PubMed] [Google Scholar]
- Lupo, R., A. Breiling, M. E. Bianchi and V. Orlando, 2001. Drosophila chromosome condensation proteins Topoisomerase II and Barren colocalize with Polycomb and maintain Fab-7 PRE silencing. Mol. Cell 7 127–136. [DOI] [PubMed] [Google Scholar]
- Lusser, A., D. L. Urwin and J. T. Kadonaga, 2005. Distinct activities of CHD1 and ACF in ATP-dependent chromatin assembly. Nat. Struct. Mol. Biol. 12 160–166. [DOI] [PubMed] [Google Scholar]
- Malik, H. S., D. Vermaak and S. Henikoff, 2002. Recurrent evolution of DNA-binding motifs in the Drosophila centromeric histone. Proc. Natl. Acad. Sci. USA 99 1449–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzkin, L. M., T. J. Merritt, C. T. Zhu and W. F. Eanes, 2005. The structure and population genetics of the breakpoints associated with the cosmopolitan chromosomal inversion In(3R)Payne in Drosophila melanogaster. Genetics 170 1143–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiklejohn, C. D., J. Parsch, J. M. Ranz and D. L. Hartl, 2003. Rapid evolution of male-biased gene expression in Drosophila. Proc. Natl. Acad. Sci. USA 100 9894–9899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettler, L. E., R. A. Voelker and T. Mukai, 1977. Inversion clines in populations of Drosophila melanogaster. Genetics 87 169–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milton, C. C., B. Huynh, P. Batterham, S. L. Rutherford and A. A. Hoffmann, 2003. Quantitative trait symmetry independent of Hsp90 buffering: distinct modes of genetic canalization and developmental stability. Proc. Natl. Acad. Sci. USA 100 13396–13401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miotto, B., and K. Struhl, 2006. Differential gene regulation by selective association of transcriptional coactivators and bZIP DNA-binding domains. Mol. Cell. Biol. 26 5969–5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miotto, B., T. Sagnier, H. Berenger, D. Bohmann, J. Pradel et al., 2006. Chameau HAT and DRpd3 HDAC function as antagonistic cofactors of JNK/AP-1-dependent transcription during Drosophila metamorphosis. Genes Dev. 20 101–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuzhdin, S. V., M. L. Wayne, K. L. Harmon and L. M. McIntyre, 2004. Common pattern of evolution of gene expression level and protein sequence in Drosophila. Mol. Biol. Evol. 21 1308–1317. [DOI] [PubMed] [Google Scholar]
- Oleksiak, M. F., G. A. Churchill and D. L. Crawford, 2002. Variation in gene expression within and among natural populations. Nat. Genet. 32 261–266. [DOI] [PubMed] [Google Scholar]
- Orphanides, G., W. H. Wu, W. S. Lane, M. Hampsey and D. Reinberg, 1999. The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature 400 284–288. [DOI] [PubMed] [Google Scholar]
- Orr, H. A., and A. J. Betancourt, 2001. Haldane's sieve and adaptation from the standing genetic variation. Genetics 157 875–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Place, A. R., and D. A. Powers, 1979. Genetic variation and relative catalytic efficiencies: lactate dehydrogenase B allozymes of Fundulus heteroclitus. Proc. Natl. Acad. Sci. USA 76 2354–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rifkin, S. A., J. Kim and K. P. White, 2003. Evolution of gene expression in the Drosophila melanogaster subgroup. Nat. Genet. 33 138–144. [DOI] [PubMed] [Google Scholar]
- Ringrose, L., and R. Paro, 2004. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu. Rev. Genet. 38 413–443. [DOI] [PubMed] [Google Scholar]
- Roberts, P., 1962. Interchromosomal effects and the relation between crossing-over and nondisjunction. Genetics 47 1691–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, S. P., and M. E. Feder, 1999. Natural hyperthermia and expression of the heat shock protein Hsp70 affect developmental abnormalities in Drosophila melanogaster. Oecologia 21 323–329. [DOI] [PubMed] [Google Scholar]
- Rozas, J., J. C. Sanchez-DelBarrio, X. Messeguer and R. Rozas, 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19 2496–2497. [DOI] [PubMed] [Google Scholar]
- Santamaria, P., 1979. Heat shock induced phenocopies of dominant mutants of the bithorax complex in Drosophila melanogaster. Mol. Gen. Genet. 172 161–163. [DOI] [PubMed] [Google Scholar]
- Saunders, A., J. Werner, E. D. Andrulis, T. Nakayama, S. Hirose et al., 2003. Tracking FACT and the RNA polymerase II elongation complex through chromatin in vivo. Science 301 1094–1096. [DOI] [PubMed] [Google Scholar]
- Schmidt, P. S., D. D. Duvernell and W. F. Eanes, 2000. Adaptive evolution of a candidate gene for aging in Drosophila. Proc. Natl. Acad. Sci. USA 97 10861–10865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, P. S., L. Matzkin, M. Ippolito and W. F. Eanes, 2005. Geographic variation in diapause incidence, life-history traits, and climatic adaptation in Drosophila melanogaster. Evolution 59 1721–1732. [PubMed] [Google Scholar]
- Schulte, P. M., H. C. Glemet, A. A. Fiebig and D. A. Powers, 2000. Adaptive variation in lactate dehydrogenase-B gene expression: role of a stress-responsive regulatory element. Proc. Natl. Acad. Sci. USA 97 6597–6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, Y. B., and V. Pirrotta, 2007. Polycomb silencing mechanisms and the management of genomic programmes. Nat. Rev. Genet. 8 9–22. [DOI] [PubMed] [Google Scholar]
- Sezgin, E., D. D. Duvernell, L. M. Matzkin, Y. Duan, C. T. Zhu et al., 2004. Single-locus latitudinal clines and their relationship to temperate adaptation in metabolic genes and derived alleles in Drosophila melanogaster. Genetics 168 923–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro, J. A., W. Huang, C. Zhang, M. J. Hubisz, J. Lu et al., 2007. Adaptive genic evolution in the Drosophila genomes. Proc. Natl. Acad. Sci. USA 104 2271–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimojima, T., M. Okada, T. Nakayama, H. Ueda, K. Okawa et al., 2003. Drosophila FACT contributes to Hox gene expression through physical and functional interactions with GAGA factor. Genes Dev. 17 1605–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, S. T., S. Petruk, Y. Sedkov, E. Cho, S. Tillib et al., 2004. Modulation of heat shock gene expression by the TAC1 chromatin-modifying complex. Nat. Cell Biol. 6 162–167. [DOI] [PubMed] [Google Scholar]
- Stalker, H. D., 1976. Chromosome studies in wild populations of D. melanogaster. Genetics 82 323–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffensen, S., P. A. Coelho, N. Cobbe, S. Vass, M. Costa et al., 2001. A role for Drosophila SMC4 in the resolution of sister chromatids in mitosis. Curr. Biol. 11 295–307. [DOI] [PubMed] [Google Scholar]
- Stokes, D. G., K. D. Tartof and R. P. Perry, 1996. CHD1 is concentrated in interbands and puffed regions of Drosophila polytene chromosomes. Proc. Natl. Acad. Sci. USA 93 7137–7142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima, F., 1983. Evolutionary relationship of DNA sequences in finite populations. Genetics 105 437–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga, C., 1970. a Aspects of low-temperature-induced meiotic nondisjunction in Drosophila females. Genetics 66 653–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga, C., 1970. b The effects of low temperature and aging on nondisjunction in Drosophila. Genetics 65 75–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournier, C., P. Hess, D. D. Yang, J. Xu, T. K. Turner et al., 2000. Requirement of JNK for stress-induced activation of the cytochrome c-mediated death pathway. Science 288 870–874. [DOI] [PubMed] [Google Scholar]
- Turner, T. L., M. T. Levine and D. J. Begun, 2008. Genomic analysis of adaptive differentiation in Drosophila melanogaster. Genetics 179 455–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uffenbeck, S. R., and J. E. Krebs, 2006. The role of chromatin structure in regulating stress-induced transcription in Saccharomyces cerevisiae. Biochem. Cell Biol. 84 477–489. [DOI] [PubMed] [Google Scholar]
- Vermaak, D., S. Henikoff and H. S. Malik, 2005. Positive selection drives the evolution of rhino, a member of the heterochromatin protein 1 family in Drosophila. PLoS Genet. 1 96–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrelli, B. C., and W. F. Eanes, 2001. Clinal variation for amino acid polymorphisms at the Pgm locus in Drosophila melanogaster. Genetics 157 1649–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voellmy, R., 1984. The heat shock genes: a family of highly conserved genes with a superbly complex expression pattern. BioEssays 1 213–217. [Google Scholar]
- Wang, D., H. M. Sung, T. Y. Wang, C. J. Huang, P. Yang et al., 2007. Expression evolution in yeast genes of single-input modules is mainly due to changes in trans-acting factors. Genome Res. 17 1161–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, M. C., D. Bohmann and H. Jasper, 2003. JNK signaling confers tolerance to oxidative stress and extends lifespan in Drosophila. Dev. Cell 5 811–816. [DOI] [PubMed] [Google Scholar]
- Wang, M. C., D. Bohmann and H. Jasper, 2005. JNK extends life span and limits growth by antagonizing cellular and organism-wide responses to insulin signaling. Cell 121 115–125. [DOI] [PubMed] [Google Scholar]
- Wang, Y. J., and H. W. Brock, 2003. Polyhomeotic stably associates with molecular chaperones Hsc4 and Droj2 in Drosophila Kc1 cells. Dev. Biol. 262 350–360. [DOI] [PubMed] [Google Scholar]
- Wittkopp, P. J., 2006. Evolution of cis-regulatory sequence and function in Diptera. Heredity 97 139–147. [DOI] [PubMed] [Google Scholar]
- Wittkopp, P. J., B. K. Haerum and A. G. Clark, 2004. Evolutionary changes in cis and trans gene regulation. Nature 430 85–88. [DOI] [PubMed] [Google Scholar]
- Zhao, J., J. Herrera-Diaz and D. S. Gross, 2005. Domain-wide displacement of histones by activated heat shock factor occurs independently of Swi/Snf and is not correlated with RNA polymerase II density. Mol. Cell. Biol. 25 8985–8999. [DOI] [PMC free article] [PubMed] [Google Scholar]