Abstract
Aluminum toxicity is a major problem in agriculture worldwide. Among the cultivated Triticeae, rye (Secale cereale L.) is one of the most Al tolerant and represents an important potential source of Al tolerance for improvement of wheat. The Alt4 Al-tolerance locus of rye contains a cluster of genes homologous to the single-copy Al-activated malate transporter (TaALMT1) Al-tolerance gene of wheat. Tolerant (M39A-1-6) and intolerant (M77A-1) rye haplotypes contain five and two genes, respectively, of which two (ScALMT1-M39.1 and ScALMT1-M39.2) and one (ScALMT1-M77.1) are highly expressed in the root tip, typically the main site of plant Al tolerance/susceptibility. All three transcripts are upregulated by exposure to Al. High-resolution genetic mapping identified two resistant lines resulting from recombination within the gene cluster. These recombinants exclude all genes flanking the gene cluster as candidates for controlling Alt4 tolerance, including a homolog of the barley HvMATE Al-tolerance gene. In the recombinants, one hybrid gene containing a chimeric open reading frame and the ScALMT1-M39.1 gene each appeared to be sufficient to provide full tolerance. mRNA splice variation was observed for two of the rye ALMT1 genes and in one case, was correlated with a ∼400-bp insertion in an intron.
SOIL acidity and associated Al toxicity represents a major limitation to crop production. Acidity causes Al-containing compounds to release the phytotoxic Al3+ cation into the soil solution where it can cause root stunting, restricting the ability of crops to acquire water and nutrients. Over 50% of the world's arable land is acidic (pH < 5.0) (Von Uexküll and Mutert 1995; Bot et al. 2000). In addition to liming, the use of Al-tolerant germplasm offers a strategy for minimizing yield losses due to soil acidity.
The main documented mechanisms of Al tolerance in plants involve detoxification of Al3+ cations by chelation to organic acids (carboxylate compounds) such as malate and citrate, either within the symplast or after exudation of these compounds from root tips (Kochian et al. 2005). A role for organic acid exudation as a mechanism of Al tolerance has been highlighted by the recent cloning of four Al-tolerance genes that all encode organic acid transporters. The TaALMT1 gene controlling tolerance at the AltBH (Alt2) locus on wheat (Triticum aestivum L.) chromosome arm 4DL encodes the aluminum-activated malate transporter-1 (TaALMT1) protein representing the founding member of a family of plant-specific membrane-integral proteins (Sasaki et al. 2004; Delhaize et al. 2007). In Arabidopsis, a homolog of TaALMT1 (AtALMT1) makes a major contribution to the Al tolerance of wild-type plants (Hoekenga et al. 2006). In sorghum (Sorghum bicolor L.), the SbMATE Al-tolerance gene from the AltSB locus encodes a member of the large multidrug and toxic compound extrusion (MATE) family of membrane transporters (Magalhaes et al. 2007) whose members in prokaryotes and eukaryotes transport a variety of small molecules (Omote et al. 2006). The HvMATE (HvAACT1) Al-tolerance gene from the barley (Hordeum vulgare L.) Alp locus on chromosome 4H also encodes a MATE transporter (Furukawa et al. 2007; Wang et al. 2007). TaALMT1 and AtALMT1 are malate transporters and HvMATE and SbMATE are citrate transporters, which provide Al tolerance in tolerant genotypes by facilitating Al-triggered excretion of the respective Al-chelating organic acids from the root tips (Sasaki et al. 2004; Hoekenga et al. 2006; Furukawa et al. 2007; Magalhaes et al. 2007).
Within the cultivated Triticeae, tolerant genotypes of rye (Secale cereale L.) are equally or more Al tolerant than the most tolerant wheats and triticales (× Triticosecale Wittmack), while tolerant barleys are the least Al tolerant (Mugwira et al. 1976; Aniol and Gustafson 1984; Kim et al. 2001). Sets of wheat–rye addition lines, wheat chromosome translocation lines, and triticale substitution lines have demonstrated the existence of rye genes that can contribute Al tolerance in the presence of a wheat genetic background (Aniol and Gustafson 1984; Ma et al. 2000). Therefore, rye represents an important potential source of Al tolerance that could be used to improve wheat and perhaps other cereals through the use of chromosome engineering and/or genetic transformation approaches.
In rye, Al-tolerance loci have been identified by linkage mapping on chromosome arms 6RS and 7RS (Alt1 and Alt4 loci, respectively) (Gallego et al. 1998; Matos et al. 2005). An Al-tolerance locus originally referred to as Alt3 and reported to be on chromosome 4R was mapped using M39A-1-6 (tolerant) × M77A-1 (intolerant) F2 and RIL mapping populations (Miftahudin et al. 2002, 2004a, 2005). However, Benito et al. (2005) claimed that this locus was actually on 7RS and therefore the same as the Alt4 locus Matos et al. (2005) had mapped on 7RS using Ailés × Riodeva crosses. A rye homolog of TaALMT1 (ScALMT1-1) has been proposed as a candidate for the rye Alt4 tolerance gene, on the basis that it was closely linked to Alt4 and because its expression was found to be highest in the root tips, upregulated by Al, and higher in a tolerant line than an intolerant line (Fontecha et al. 2007). However, the Alt4 chromosome segment may also be related to the barley 4HL region containing the HvMATE (Alp locus) gene (Tang et al. 2000), raising the possibility that Alt4 is a MATE gene rather than an ALMT1 gene. The current study confirms that the locus segregating in the M39A-1-6 × M77A-1 cross (hereafter referred to as Alt4) is located on 7RS and reveals that the parents contain clusters of ALMT1 homologs at the Alt4 locus. The structure and function of these ALMT1 homologs are explored, together with the colinearity of ALMT1 and MATE genes, in the Triticeae and rice (Oryza sativa L.).
MATERIALS AND METHODS
Plant lines and DNA extraction:
Recombinants used for fine mapping had been selected from an F2 population of a cross between the rye inbred lines M39A-1-6 (Al tolerant) and M77A-1 (Al sensitive) (Miftahudin et al. 2005). This cross shows monogenic segregation for Al tolerance determined by the Alt4 locus and does not segregate for tolerance at any of the other Al-tolerance loci known in rye. The recombinants for the Alt4 interval were kindly provided by Miftahudin (Bogor Agricultural University). F2 recombinants had been grown in a glasshouse containing no other rye plants. F3 progeny of F2 recombinants were grown in the University of Adelaide quarantine glasshouse without isolating the heads to prevent outcrossing. However, F4 progeny of these plants used in tolerance assays were scored for PCR markers closely flanking the respective recombination points. Of the 186 F4 individuals assayed, 16 (8.6%) were found to have been derived from outcrossing and were removed from the analysis. F5 seed, which were also used in tolerance assays, were obtained exclusively from isolated heads. An F4 family that was derived from the M39A-1-6 × M77A-1 cross and segregating for Alt4 was analyzed with markers and individuals that were homozygous tolerant or intolerant were used to make tolerant and intolerant DNA bulks, respectively (10 individuals per bulk).
Genomic DNA for PCR and Southern analyses was extracted from leaf tissue using the method of Rogowsky et al. (1991). The method was also carried out in racks of 96 × 1.2 ml Collection Microtubes (Qiagen, Australia). Ball bearings (3.0 mm) were used to crush frozen samples by shaking in a MM300 Mixer Mill (Retsch, Germany) prior to removal of the balls with a magnet. Pellets were resuspended in RNaseA (40 ng/μl) (Sigma-Aldrich, St. Louis) and A260 measured to calculate total nucleic acid content.
Lines of Chinese Spring wheat containing individual chromosomes or chromosome arms of Imperial rye or Betzes barley used to assign markers to chromosomes were kindly provided by Ian Dundas and Rafiq Islam of the University of Adelaide, respectively. The presence of rye chromatin in individual wheat–rye addition line plants was checked by DNA dot-blot analysis using a probe for the R173 rye-specific repeat sequence (Rogowsky et al. 1992; nucleotides 2–442 of accession no. X64103). A rye line that was supplied to us as cv. Imperial often showed marker patterns different from those observed in the wheat–rye addition lines and therefore was not the same as the Imperial used to make the addition lines. Marker lanes representing this line, where included, are labeled “rye.” Seed of the barley Galleon × Haruna Nijo doubled-haploid population (Karakousis et al. 2003) were kindly provided by Peter Langridge (University of Adelaide).
Southern analysis:
Southern blots, hybridizations, and autoradiography were performed using standard methods. The rye ALMT1 cDNA probe fragment was obtained by reverse transcription (RT)–PCR from rye RNA using primers Alm-3 and Alm-5 (supplemental Table 7) and corresponds to nt 218–1357 of the ScALMT1-M39.1 ORF. RNA and cDNA were prepared as described in the ScALMT1 sequencing section. The fragment was cloned into pCR8/GW/TOPO TA vector (Invitrogen, Carlsbad, CA), reamplified from bacterial cells of a single colony, purified using the QIAquick PCR purification kit (Quiagen), sequenced using the BigDye version 3.1 kit (Applied Biosystems, Foster City, CA) to confirm identity, and the purified fragment used as template to make [α-32P]dCTP-labeled probes by random priming.
Markers:
For generation of new PCR markers, genes from the Alt4-related region of rice chromosome 3 were used in BLASTn searches against ESTs at NCBI (http://www.ncbi.nlm.nih.gov/) or the TIGR plant transcript assemblies (http://plantta.tigr.org/) to identify putative orthologs from rye or wheat. Primers matching rye/wheat exons and spanning a 0.5- to 1.5-kb interval in rice comprising mostly intron sequence were used to amplify the corresponding gene fragments from the rye mapping parents M39A-1-6 and M77A-1. PCRs were performed with hot-start immolase DNA polymerase (Bioline, Australia) in reactions of 20 μl containing ∼3.0 μg of total nucleic acid, using primers and conditions indicated in supplemental Table 1. Fragments were purified and sequenced in both directions. Sequences were aligned using Pregap4 and Gap4 programs (Bonfield et al. 1995) and restriction enzymes recognizing polymorphisms identified using the NEBcutter V2.0 program (http://tools.neb.com/NEBcutter2/index.php) or by analyzing ClustalW alignments (http://www.ebi.ac.uk/Tools/clustalw/) with CapsID (http://bbc.botany.utoronto.ca/capsid/input.spy). For scoring of cleaved amplified polymorphic sequence (CAPS) markers, 2–5 μl of unpurified PCR product were digested for 3–5 hr in 12-μl reactions containing 0.5 mg/ml acetylated BSA, 2 units of restriction enzyme, and 1× reaction buffer and the fragments visualized by agarose gel electrophoresis. The only polymorphism found in the STP fragment did not alter a restriction site and was therefore scored by direct sequencing of fragments amplified from recombinant plants. For markers B1, RPE, B11, B26, B6, and B4, new primers internal to the initial sequencing primers were made to make amplification more reliable by reducing amplicon size. All PCR markers used in linkage analysis are described in supplemental Table 2.
Primers and conditions used for PCR marker analysis of wheat–rye addition lines are shown in supplemental Table 6. Primers were based on rye sequences obtained in this study, the SCIM819-1434 marker sequence (AY587501), and rye ESTs and extended wheat EST contigs corresponding to RFLP probe sequences. PCR products were separated on 2–3% agarose gels. When PCR products of equal size were amplified from rye and wheat, the products were digested separately with a range of restriction enzymes (as for CAPS markers) to identify ones that would give rye-specific fragments.
Al-tolerance assays:
Tolerance assays were performed using F4 and F5 seed of families homozygous or segregating for recombinant chromosomes. All individuals in tolerance assays were also scored for PCR markers closely flanking the respective recombination points to assist in data interpretation. Al-tolerance assays were carried out using the Eriochrome Cyanine R method (Aniol 1983). Seeds were pregerminated on moist paper for 2–3 days and transferred to an aerated hydroponics system in a greenhouse. Growth solution was as described by Aniol (1983), except that 0.01 mm instead of 0.1 mm (NH4)2SO4 was used. The 1× solution was adjusted to pH 4.0 with HCl. Seedlings were grown without added aluminum for 4 days, then AlCl3 was added to a concentration of 150 μm (4 ppm). After 24 hr in Al, roots were thoroughly rinsed in water, stained for 10 min in a 0.1% solution of Eriochrome Cyanine R (Sigma-Aldrich), rinsed again, and the plants returned to Al-free growth solution. The dye mainly stains the root tips. After 48 hr in Al-free solution, each seedling was assessed for regrowth in the longest root following relief from Al stress by measuring the distance from the middle of the stained section to the end of the root.
Mapping:
The order of markers on the high-resolution map of the Alt4 region was determined from the graphical genotypes. The side of the Alt4 interval where the MATE/GAB marker pair was located was determined by three-point analysis in the M39A-1-6 × M77A-1 derived recombinants. The frequency of recombination between MATE/GAB and B1/B4 in the Blanco × M39A-1-6 rye F2 mapping population was converted to cM distance using the Kosambi mapping function.
ScALMT1 sequencing:
The primers Alm-1, -2, -3, -4, -6, and -7 (supplemental Table 7), based on exons 1 and 3 of the wheat TaALMT1 gene (Figure 2A), were used to amplify genomic fragments from rye ALMT1 genes. PCRs were performed with immolase DNA polymerase (Bioline) in 20 μl reactions containing 3.0 μg of total nucleic acid, using a program comprising (after an initial step of 95° for 7 min to activate the hot-start enzyme) 20–40 cycles of 95° for 10 sec, 60° for 30 sec, and 68° for 2 min, followed by a final extension step of 68° for 10 min. Amplification of multiple homologous DNA fragments with a single primer pair is known to produce a high proportion of recombinant fragments during the latter cycles of PCR (Judo et al. 1998). Therefore, PCR amplifications were performed in 5-cycle increments and the reactions that were found to give the first visible products on an agarose gel were the ones used for cloning. Products were cloned into the pCR8/GW/TOPO TA vector (Invitrogen) and clone insert fragments amplified from bacterial cells of individual colonies using the original primer pairs. Clone insert fragments were purified using the QIAquick PCR purification kit (Qiagen) and sequenced. Alternatively, some bands were gel-purified using a QIAquick gel extraction kit (Qiagen) and direct sequenced.
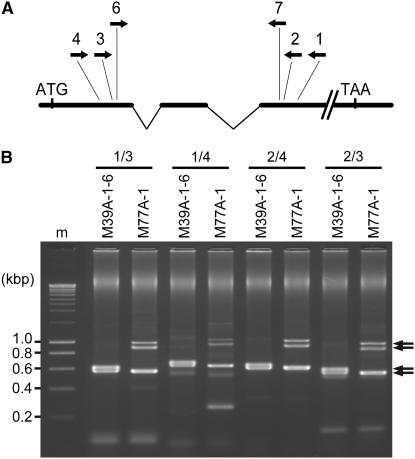
Figure 2.—
PCR amplification of rye ALMT1 gene partial genomic fragments. (A) Positions of primers used for PCR amplification of rye ALMT1 gene fragments in the wheat TaALMT1 gene sequence, relative to the first two introns and translation start/stop codons. (B) PCR amplifications from DNA of Al-tolerant parent M39A-1-6 and Al-intolerant parent M77A-1 using different primer pairs. Arrows indicate bands shown by cloning and sequencing to represent ALMT1 homologs. The two uppermost bands represent alternative secondary structure variants of the M77.1 gene fragment caused by the presence of the inverted-repeat MITE in this fragment (not shown). Sizes of molecular weight marker (m) bands are indicated.
Examples of the genomic PCRs are illustrated in Figure 2B. Product banding patterns obtained from recombinants 859 and 1135 resembled those obtained from M77A-1 and M39A-1-6, respectively (not shown). A total of 48, 40, 20, and 20 clones of the ∼0.5-kb size were sequenced from each of the M39A-1-6, M77A-1, 859, and 1135 lines, respectively. Half of the ∼0.5-kb clones sequenced from each line were derived from the Alm-6/Alm-7 primer combination and the other half from the Alm-2/Alm-3 combination. Six clones corresponding to the ∼0.9-kb doublet were sequenced from each of the M77A-1 and 859 lines. Additionally, the ∼0.9-kb doublets amplified from M77A-1 and 859, and the ∼0.27-kb band amplified from M77A-1 using the Alm-1/Alm-4 primers (Figure 2B), were gel purified and direct sequenced. Sequences obtained more than once from a given rye line were deemed genuine, while single-base differences unique to single clones were regarded as PCR errors. As a result of the minimum number of PCR cycles employed, PCR errors were detected in only a few clones.
Each total RNA sample for RACE–PCR and amplification of complete ScALMT1 ORFs was prepared from the end 3.0 mm of the root tips of 10 seedlings grown as for the tolerance assays, harvested 24 hr after the addition of 150 μm Al to the hydroponics solution. Root tips were collected in 2-ml Eppendorf tubes containing two 3.0-mm steel ball bearings, snap frozen in liquid nitrogen, and reduced to powder by shaking in a MM300 mixer mill. The RNA was extracted from plant tissues using Trizol reagent (Invitrogen). 5′- and 3′-RACE were performed using the SMART RACE cDNA amplification kit (Clontech, Mountain View, CA) with PowerScript reverse transcriptase (Clontech), according to the manufacturer's instructions. 5′-RACE was performed as a seminested PCR using the Alm-15 then the Alm-16 gene-specific primer while 3′-RACE was performed using specific primer Alm-17 (supplemental Table 7). To obtain entire ScALMT1 ORFs, first-strand cDNA was synthesized using an oligo-dT (17-mer) primer and SuperScript III (Invitrogen) reverse transcriptase according to the manufacturer's recommendations, except that a 50°-incubation temperature and DMSO at 6.0% was used to obtain efficient synthesis across the GC-rich 5′ ends of ScALMT1 transcripts. Full-length ScALMT1 ORFs were amplified with primers Alm-23 and Alm-27 (supplemental Table 7) in the presence of 5% DMSO, with (after 7 min at 95°) 25–40 cycles of 96° for 10 sec, 60° for 30 sec, and 72° for 2 min 30 sec, followed by a final extension step of 68° for 10 min. Products obtained with the minimum number of PCR cycles necessary to produce visible products on a gel were used for cloning. RACE and other RT–PCR products were gel purified using a QIAquick gel extraction kit (QIAGEN) and cloned and sequenced as for the genomic fragments, except that 5.0% DMSO was included in reactions to sequence 5′-RACE and full-length ORF products.
Expression analyses:
Plants were grown as for tolerance assays. Seven days after sowing, Al was added to the growth solution to 150 μm. Prior to the addition of Al (“0 hr”) and at 6 and 24 hr after adding Al, tissues were taken from 10 seedlings for each RNA sample. These tissues included root tip, root section, and leaf, as defined in the Figure 4 legend. Total RNA was extracted using Trizol reagent and treated with DNAse (DNA-free, Ambion, Austin, TX). First-strand cDNA was made in 20-μl reactions from 1.5 μg of total RNA, using an oligo-dT (17-mer) primer and SuperScript III reverse transcriptase (Invitrogen), according to the manufacturer's suggestions. cDNAs were diluted to 100 μl total volume. The reported expression levels refer to transcript copies per microliter of this cDNA dilution.
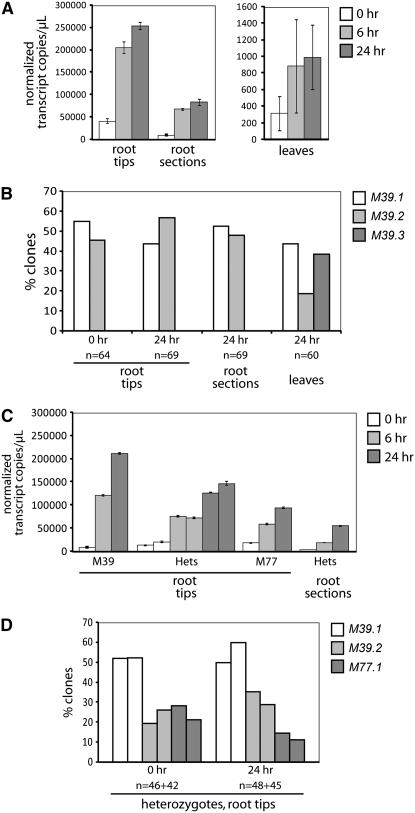
Figure 4.—
Expression analyses of rye ALMT1 homologs. A and B represent one experiment performed with M39A-1-6 plants only, while C and D represent another experiment performed using sib lines heterozygous for Alt4 (two replicates) or homozygous for the M39A-1-6 or M77A-1 Alt4 haplotypes (one replicate). The end 3 mm of the root (“root tip”), 7-mm root sections taken from just below the seed (“root sections”), and the 7-mm section of coleoptiles plus rolled up leaf within taken from just above the seed (leaves) were sampled from seedlings at 0, 6, or 24 hr after exposure to toxic (150 μm) Al. Total ScALMT1 gene expression was measured by Q-PCR with the primer pair Alm-6/Alm-10 that can potentially amplify from all ScALMT1 copies (A and C). Alm-10 spans the intron-2 splice site and would not be expected to amplify M77.1 splice variants lacking exon 2. Shown are means and standard deviations of normalized transcript copy numbers per microliter cDNA obtained from three replicate Q-PCR amplifications from the same cDNA. Q-PCR data from the different tissues were normalized independently (see materials and methods). The proportions of different ScALMT1 gene transcripts present in selected RNA samples were determined by cloning RT–PCR product mixtures and subjecting the indicated numbers of clone inserts to restriction analysis to identify from which gene each insert was derived (B and D).
Marker selection of plants for the second expression experiment was performed on Alt4 segregating families derived from the M39A-1-6 × M77A-1 cross at 6 days after sowing, using Alt4-flanking CAPS markers. After taking samples for RNA extraction, a second round of DNA extraction and marker analysis was performed to verify genotypes of the sampled plants.
ScALMT1 gene expression was measured by quantitative RT–PCR (Q-PCR) using primer pair Alm-6/Alm-10 (supplemental Table 7). Sequences corresponding to the 9 bases at the 5′ end of primer Alm-6 were not obtained from genes ScALMT1-M39.3, -M39.4 and -M39.5, and -M77.2. Otherwise, the genomic and full-length ORF sequence information that became available verified that all the ScALMT1 genes in M39A-1-6 and M77A-1 were identical at the primer binding sites. Primer Alm-10 spanning the splice sites of intron 2 (supplemental Figure 4) provided specificity for cDNA by being unable to prime from genomic DNA. Primers for Q-PCR of rye “housekeeping” genes, encoding glyceraldehyde-3-phosphate dehydrogenase, elongation factor 1α, cyclophilin, and actin were developed to provide controls for normalization (supplemental Table 9). Q-PCR from cDNA was carried out essentially as described by Burton et al. (2004), except that an RG 6000 real-time thermal cycler (Corbett Research, Australia) was used with half-scale reactions, employing a program comprising 3 min at 95°, followed by 45 cycles of 95° for 1 sec, 55° for 1 sec, and 72° for 30 sec, followed by a single step for 15 sec at the optimal acquisition temperature (supplemental Table 9). Normalization was performed as described by Vandesompele et al. (2002). Due to the substantial control gene expression pattern differences observed between the different tissues, data from the different tissues were normalized separately, using the three control genes showing the lowest internal control gene instability measure (M) for that particular tissue.
For selected cDNA samples used in Q-PCR, the relative expression levels of individual ScALMT1 genes in these samples were determined. PCR reactions of 20 μl were performed using 1 μl of cDNA template and primers Alm-6 and Alm-10. The PCR program comprised (after 7 min at 95°) 20–50 cycles of 95° for 10 sec, 60° for 30 sec, and 68° for 1 min 30 sec, followed by a single step of 68° for 10 min. The minimum number of PCR cycles required to give a visible product were identified and those reactions used in cloning with the pCR8/GW/TOPO TA vector (Invitrogen). Individual clone inserts were PCR amplified from cells of single bacterial colonies and digested with a set of restriction enzymes to determine which ScALMT1 transcript each fragment was derived from (supplemental Table 8). Digestion conditions were the same as those used for CAPS markers. One clone derived from each of the M39A-1-6 leaf-24-hr and root-tip-0-hr samples from the first experiment showed a novel combination of restriction patterns. Upon sequencing, these clones were found to be hybrids of multiple gene sequences. These likely represented artifacts of PCR that occur during amplification of mixtures of homologous sequences (Judo et al. 1998) and were removed from the data set. Each of the remaining clones gave restriction fragment profiles consistent with one of the known ScALMT1 gene sequences.
RESULTS
Refining the map of the Alt4 region:
Of the rye Alt4 genetic markers previously identified by colinearity to the corresponding interval on the rice (Oryza sativa L.) chromosome-3 sequence, PCR (CAPS) markers for the B1 and B4 genes (Miftahudin et al. 2004a) were modified, and the RPE, B11, B26, and B6 genes assayed by RFLP (Miftahudin et al. 2005) were converted to easy-to-use codominant PCR markers. PCR markers were also developed using 11 additional genes from the rice B1-B4 interval, the rye homolog of HvMATE, and a rye homolog of a neutral α-glucosidase AB precursor (GAB) gene, which is located close to the MATE gene in rice and barley (Wang et al. 2007). The details of the polymorphism screen undertaken here for marker development are summarized in supplemental Table 1. More than 17,500 bp of sequence was compared between the mapping parents M39A-1-6 and M77A-1, revealing 321 single nucleotide polymorphisms (SNPs) and 42 insertion/deletions (InDels), corresponding to an average frequency of one polymorphism every 48 bp. Details of all the developed PCR marker assays are provided in supplemental Table 2.
From a population of 1123 M39A-1-6 × M77A-1 F2 plants, Miftahudin et al. (2005) had reported 15 recombinants for the marker interval B1-B4 containing Alt4. In the current study, we recovered 13 of the reported recombinants (recombinants 257 and 965 were not recovered), plus an additional unreported recombinant (1135) isolated from the same population. All available PCR markers were scored in several F3 progeny of each F2 recombinant, enabling the genotypes for each recombinant chromosome to be determined as shown in supplemental Figure 1. Except for the KEL gene, which was found to be unlinked to the Alt4 region, all new markers from the rice B1-B4 rice interval mapped in a colinear fashion in the vicinity of Alt4, allowing construction of the revised molecular marker map shown in Figure 1. The rye MATE and GAB genes were found to be outside of the B1-B4 interval, consistent with their more distant location in rice. To more accurately determine their positions, the B1, B4, MATE, and GAB markers were mapped in a cv. Blanco × M39A-1-6 rye F2 mapping population of 96 individuals. In this population, no recombination was observed between B1 and B4 or between MATE and GAB, while the two pairs of markers mapped 27.5 cM (48 recombinants) from one another (Figure 1). Close linkage between the MATE and GAB gene, as observed in barley (Wang et al. 2007), confirmed that the MATE gene mapped in rye was the direct ortholog of the barley HvMATE (HvAACT1) Al-tolerance gene (Furukawa et al. 2007; Wang et al. 2007).
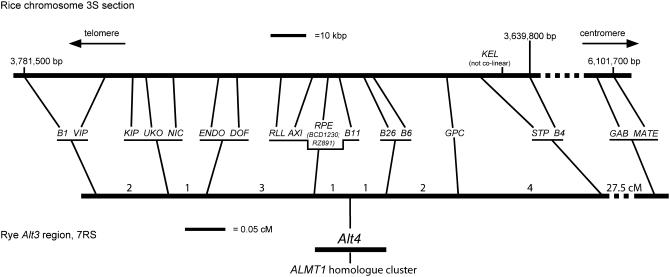
Figure 1.—
Genetic map of the Alt4 region of rye and colinearity with rice. Positions of genes on rye 7RS and their corresponding orthologs in a section of the short arm of rice chromosome 3 are shown by the lines connecting the maps. Numbers on the rye map indicate recombinants observed in each interval in the mapping population of 2246 gametes, except for the B4 to GAB/MATE interval, which was measured using a different population. Numbers above the rice sequence refer to nucleotide positions in chromosome 3 sequence assembly AP008209.1. The B1-B4 interval is contained entirely within the rice BAC clone OSJNBa0091P11 (AC073556). The ALMT1 gene cluster was not assigned a precise genetic position due to recombination observed within the cluster. The orientation of the mapped section with respect to the centromere of rye chromosome 7R was not determined.
The Alt4 genotypes of all the recombinants were checked using the Eriochrome Cyanine R method (Aniol 1983), which measures tolerance on the basis of the degree of root regrowth following relief from Al stress (Aniol 1983; supplemental Figure 2). Supplemental Tables 3–5 summarize the results from the tolerance assays, performed using F4 or F5 families either homozygous or segregating for the recombinant chromosomes. Alt4 tolerance behaved as a dominant trait. Tolerant and intolerant individuals showed regrowth after 2 days of 10–24 mm and 1–5 mm, respectively. Individual seedlings assayed for tolerance were also scored for PCR markers closely flanking the recombination sites to confirm when variation in tolerance was controlled by inheritance of the recombinant chromosome and to verify that individuals of informative genotype had been sampled. Alt4 genotypes agreed with those determined previously (Miftahudin et al. 2005), except that recombinant 862, previously reported to be homozygous tolerant, was found to be Alt4 heterozygous (supplemental Table 3). All families used in tolerance assays were also scored for the MATE marker. For the majority of the families the Alt4 tolerance genotype contrasted with the MATE marker genotype (supplemental Tables 3–5), demonstrating that the rye MATE gene does not determine rye Alt4 tolerance/intolerance, at least in this cross. The genotype data (supplemental Figure 1) combined to place Alt4 in the 0.09-cM interval between marker groups RLL/AXI/RPE/B11 and B26/B6, with one recombination event separating Alt4 from these groups of markers on each side (Figure 1). The corresponding 4.3-kb B11-B26 interval in rice contains a predicted gene for an unknown protein (TIGR locus LOC_Os03g07270; RiceGAAS predgene 21 of BAC clone OSJNBa0091P11), plus a sequence identified as a potential gene by Genescan only (RiceGAAS Predgene 20 of OSJNBa0091P11), which is repetitive in the rice genome and which matched no rice EST sequences in the database.
Reports disagree as to whether the rye Al-tolerance locus mapped using the M39A-1-6 × M77A-1 cross is located on rye 4R or 7RS (Miftahudin et al. 2002; Benito et al. 2005). To resolve this issue, PCR markers for RPE and B4 flanking this locus (Figure 1), PSR163 on 7RS, PSR129 on 7RL, and PSR167 on 4RL (Devos et al. 1993), and SCIM819-1434 located 7.4 cM from the Al-tolerance locus on 7RS described by Matos et al. (2005), were tested on lines of wheat cv. Chinese Spring containing individual chromosomes or chromosome arms from rye cv. Imperial. Details of the marker assays are provided in supplemental Table 6 and the results illustrated in supplemental Figure 3. PSR163, PSR129, and PSR167 markers identified sequences unique to the 7RS, 7RL, and 4RL addition lines, respectively, confirming that the identities of those lines were correct. RPE, B4, and SCIM819-1434 identified products unique to the 7RS addition line. Therefore, at least according to the chromosome arm definitions of Devos et al. (1993), the Al-tolerance locus segregating in the M39A-1-6 × M77A-1 cross is located on 7RS, and this is the same chromosome arm that contains the locus of Matos et al. (2005).
The RFLP probe BCD1230, homologous to the RPE marker gene (Figure 1), had been used to detect a locus close to the Alt4 locus in rye as well as a locus close to the AltBH Al-tolerance locus in wheat, leading to the proposal that these two tolerance loci may reside on related chromosome segments (Miftahudin et al. 2002). To test this possibility further, we located additional Alt4-linked markers in wheat, using deletion mapping data (Miftahudin et al. 2004b; http://wheat.pw.usda.gov/cgi-bin/westsql/map_locus.cgi). Three genes mapped close to Alt4 in rye (KIP, GPC, and B4; Figure 1) were found by BLASTn searches to be highly homologous to wheat ESTs BF474257, BM134287, and BE499019, respectively, which had all been located to deletion 4DL12-0.71-1.00. BCD1230/RPE was also found to be homologous to ESTs (BE497160/BE405275) mapped to this same deletion bin. This deletion bin, representing the distal third of 4DL, also contains the AltBH locus (Raman et al. 2005). Hence, it was confirmed that Alt4 and AltBH are located on related chromosome regions in the rye and wheat genomes.
A cluster of ALMT1 genes at the rye Alt4 locus:
Although the corresponding interval in rice lacks an ALMT1 homolog, the existence of an ALMT1 gene in the vicinity of Alt4 was expected from the relationship between the Alt4 and AltBH chromosome regions and the fact that rye and wheat are close relatives. When primers from exons 1 and 3 of TaALMT1 were used in PCRs from M39A-1-6 and M77A-1 DNA, similar patterns of multiple products were obtained using the different primer combinations (Figure 2, A and B). Gel purification and direct sequencing of the ∼0.27-kb band amplified from M77A-1 using the Alm-1/Alm-4 primers (Figure 2B) revealed it to be a sequence unrelated to TaALMT1. Cloning and sequencing of the ∼0.5-kb products revealed a number of TaALMT1-related sequences. Direct sequencing and sequencing of clones showed the doublet at ∼0.9 kb, amplified from M77A-1 (Figure 2B), to be comprised of a single ALMT1 sequence. This fragment appeared as a doublet because of secondary structure (data not shown). The number of ALMT1 genes identified in M39A-1-6 and M77A-1 was five (named ScALMT1-M39.1 to -M39.5) and two (ScALMT1-M77.1 and -M77.2), respectively. An alignment of the partial genomic sequences is shown in supplemental Figure 4. The gene represented by the ∼0.9-kb doublet, designated ScALMT1-M77.1, contained a ∼400-bp insertion in intron 2, which was unique among the ScALMT1 sequences. This insertion contained a 227-bp miniature inverted repeat transposable element (MITE).
In PCRs on DNA bulks of homozygous tolerant or intolerant individuals from the mapping population, the ∼0.9-kb doublet was amplified from the intolerant bulk only (not shown), indicating that the ScALMT1-M77.1 gene was linked to the Alt4 locus. Likewise, use of a PCR primer pair specific for ScALMT1-M39.4 (Alm-28 and Alm-29; supplemental Table 7; supplemental Figure 4), and digestion of Alm-6/Alm-7 genomic PCR product mixtures with a set of restriction enzymes that gave diagnostic restriction fragments for particular gene copies (supplemental Table 8), gave banding patterns from the tolerant and intolerant DNA bulks that were consistent with all seven ScALMT1 sequences being closely linked to Alt4 (not shown).
In Southern blots with a rye ALMT1 cDNA probe and the restriction enzyme EcoRV, all restriction fragments detected in M39A-1-6 were distinguishable in size from those detected in M77A-1 (Figure 3). In bulks of homozygous tolerant or intolerant individuals from the mapping population, and in individuals homozygous for the recombinant chromosomes, one weakly hybridizing fragment derived from M39A-1-6 (arrow in Figure 3) appeared unlinked to the Alt4 locus. Recombinants 859 and 1135, representing the closest recombination events to each side of Alt4 (supplemental Figure 1), showed novel banding patterns as a result of recombination within the ALMT1 gene cluster (see below). Otherwise, hybridization patterns in the homozygous recombinants were the same as those in M39A-1-6 or M77A-1 (Figure 3) and corresponded with the Alt4 tolerance allele present on each of the recombinant chromosomes. In this way, the ALMT1 gene cluster was located to the same 0.9-cM region as the Alt4 locus (Figure 1). PCR amplifications and Southern analysis with the wheat–rye addition lines (supplemental Figure 5, A and B) confirmed the presence of an ALMT1 gene cluster on 7RS of rye cv. Imperial and revealed no homologs on any other rye chromosomes. Hence, both Al-tolerant and Al-intolerant rye lines contain clusters of ALMT1 gene homologs at the Alt4 Al-tolerance locus. This contrasts with previous reports in which solitary ALMT1 gene copies were described for the tolerance and intolerance alleles of the rye Alt4 locus and wheat AltBH locus (Sasaki et al. 2004; Fontecha et al. 2007).
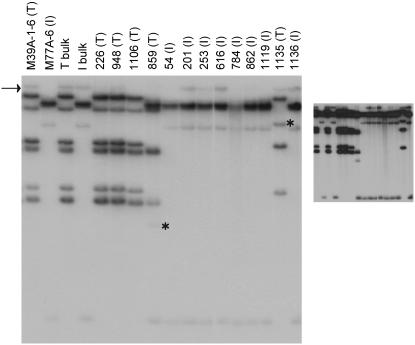
Figure 3.—
Southern blot analysis of rye ALMT1 homologs. Rye genomic DNAs cut with EcoRV were probed with a rye ALMT1 cDNA. Included are the inbred mapping parents M39A-1-6 (tolerant) and M77A-1 (intolerant), bulks of homozygous tolerant or intolerant segregants, and lines homozygous for Alt4-region recombinant chromosomes. I, intolerant; T, tolerant. The arrow indicates a fragment not linked to Alt4. Asterisks indicate novel-sized restriction fragments in 859 and 1135. A longer exposure of the same blot (inset) more clearly illustrates weakly hybridizing fragments.
ScALMT1 expression patterns, full-length ORFs, and splice variants:
ScALMT1 gene expression was monitored by Q-PCR. We also developed Q-PCR primers for rye genes encoding glyceraldehyde-3-phosphate dehydrogenase, elongation factor 1α, cyclophilin, and actin to provide controls for normalization (supplemental Table 9). Q-PCR from tissues of M39A-1-6 (tolerant) seedlings, 0, 6, and 24 hr after exposure to toxic levels (150 μm = 4 ppm) of Al revealed low, intermediate, and high expression in leaves, root midsections, and root tips, respectively, with expression in root tips and root midsections being upregulated upon exposure to Al (Figure 4A). Using a set of restriction enzymes that could distinguish potential RT–PCR products derived from any of the ScALMT1 genes (supplemental Table 8), individual RT–PCR fragments cloned from leaf-24-hr, root-tip-0-hr, and root-tip-24-hr amplification product mixtures were assigned to particular M39A-1-6 gene copies to estimate the relative expression levels of each gene in each of the samples. ScALMT1-M39.1 and -M39.2 were the only transcripts detected in the root-tip-0-hr and root-tip-24-hr samples and were present in roughly equal proportions in both samples (Figure 4B), indicating that these genes were equally upregulated by Al exposure. RT–PCR product mixtures amplified from M77A-1 root tips were also subjected to cloning and restriction analysis. All 96 clones analyzed were derived from ScALMT1-M77.1 transcripts, indicating that M77.1 was the only gene expressed in root tips from the M77A-1 haplotype.
In another experiment (Figure 4, C and D), seedlings that were homozygous for the M39A-1-6 or M77A-1 Alt4 locus haplotypes, or heterozygous, were selected with PCR markers from segregating families, and plants of the three genotypes were analyzed separately for ScALMT1 gene expression in roots at 0, 6, and 24 hr after exposure to 150 μm Al. In M77A-1 root tips, total ScALMT1 gene expression (representing the M77.1 gene) was upregulated by Al exposure (Figure 4C). Samples from heterozygotes provided the opportunity to test the relative expression of the ScALMT1 genes in a common (tolerant) background. In heterozygotes, total ScALMT1 expression increased approximately sixfold after Al exposure (Figure 4C). By 24 hr, the proportions of ScALMT1 transcripts that were derived from the M39.1 and M39.2 genes remained relatively unchanged, while the proportion derived from the M77.1 gene appeared to decrease slightly (by 50%; Figure 4D), suggesting that the M77.1 gene may undergo a lower fold increase in expression level following Al stress than the other two genes. In heterozygotes, transcript expression levels of M39.2 appeared to be reduced relative to those of M39.1 (to ∼50% of M39.1 levels; Figure 4D). In both homozygotes and heterozygotes, transcript levels of M77.1 appeared to be a half to a quarter of those of M39.1 (Figure 4, C and D).
RACE–RT–PCR performed on M39A-1-6 root-tip-24-hr RNA using primers based on published ALMT1 sequences from wheat (Sasaki et al. 2004) and rye (Fontecha et al. 2007) yielded complete 3′-UTR sequence and partial 5′-UTR sequence for ScALMT1-M39.1 and -M39.2. Primers Alm-23 and Alm-27 based on the UTR sequences were then used in RT–PCR amplifications of full open reading frames of the ScALMT1-M39.1 and -M39.2 and -M77.1 genes. Comparison of the cereal ALMT1 cDNA sequences (supplemental Figure 6) revealed identities of 98–99% between the rye sequences. All the rye sequences have the same 21-bp in-frame deletion relative to the wheat TaALMT1 ORF and are otherwise ∼91% identical with the wheat sequence. The transcripts from each rye gene, including M77.1, all encode predicted full-length proteins with high levels of similarity to the published rye and wheat sequences (supplemental Figure 7). Hydrophobicity plots for the rye proteins were essentially identical to those of the TaALMT1 protein (not shown), which has seven hydrophobic domains, only six of which are membrane spanning (Motoda et al. 2007). In the TaALMT1 topology model described by Motoda et al. (2007), the seven-amino-acid region of TaALMT1 that is deleted in the rye proteins occurs entirely within the N-terminal extracellular tail, at the junction between the first hydrophobic (extracellular) domain and the second hydrophilic domain.
Cloning and sequencing of the RT–PCR products revealed mRNA splice variants for the ScALMT1-M39.2 and -M77.1 genes. A quarter (2 of 8) of the clones obtained for M39.2 were precisely missing exon 4 (113 bp), resulting in a predicted frameshift and truncated product (EU146238), which is the same as the full-length ScALMT1-M39.2 protein up until the 232nd amino acid (in boldface type and underlined in supplemental Figure 7) and ends in a novel 68-amino-acid C-terminal extension. Approximately half (15 of 24) of the clones obtained for M77.1 were precisely missing exon 2 (150 bp) and would encode a predicted protein (EU146240) with an in-frame deletion of 50 amino acid residues (underlined in supplemental Figure 7). The deleted region begins in the second transmembrane domain and ends in the fourth transmembrane domain (third to fifth hydrophobic domains, respectively) of the full-length protein, as defined by the membrane topology model of Motoda et al. (2007). The M77.1 splice variant was visible as a smaller product in RT–PCR amplifications from M77A-1 root tip RNA (supplemental Figure 8). The proportions of correctly and incorrectly spliced M77.1 transcripts in root tips of M77A-1 plants did not appear to change after Al exposure, judging from the results of RT–PCRs that detected both splice variants at once (not shown).
The 859 and 1135 recombinant lines:
Lines containing the homozygous 859 and 1135 recombinant chromosomes showed Southern blot ALMT1 banding patterns unlike either of the parents (Figure 3). To explore the possibility that these lines were derived by pollen or seed contamination, all the marker fragments listed in supplemental Table 1, except for DOF, GPC, STP, KEL, GAB, and MATE, were amplified from each of the two homozygous recombinants and sequenced. The results were entirely consistent with these lines having arisen from single recombination events between M39A-1-6 and M77A-1 chromosomes. In line 859, the sequences of all markers to the B1 and B4 sides of Alt4 were identical to M39A-1-6 and M77A-1, respectively, while in the 1135 line the reverse was true. The sequenced fragments totaled over 10.9 kb and covered 219 sites of polymorphism between M39A-1-6 and M77A-1, making it very unlikely that the 859 and 1135 recombinant chromosomes were derived from any rye haplotypes other than M39A-1-6 and M77A-1.
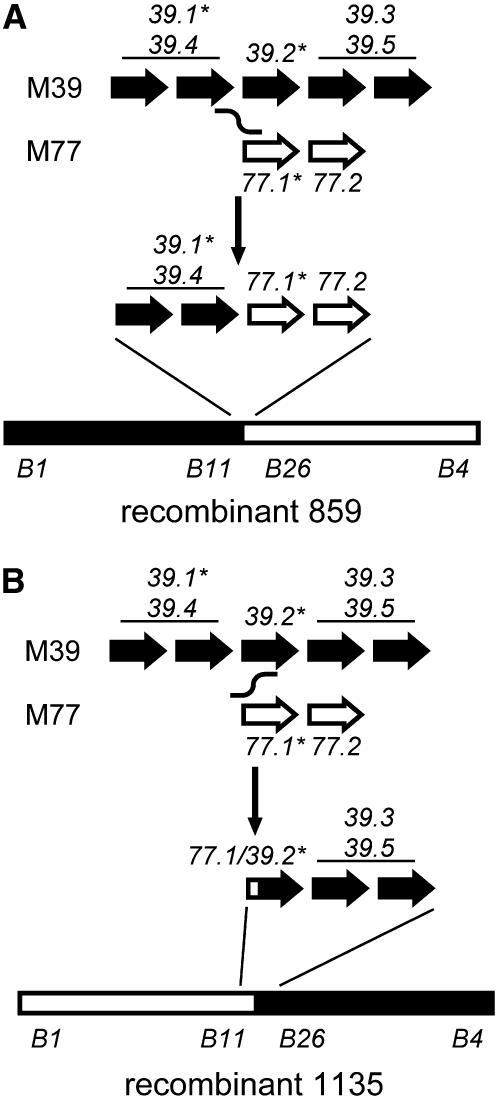
The location of the 859 and 1135 recombination events within the smallest (B11-B26) Alt4 marker interval (supplemental Figure 1) suggested that these lines may have arisen from recombination between the ALMT1 gene clusters. Therefore, the nature of these lines was investigated further by using the same procedure as was applied to the parents to determine the number and sequence of ALMT1 gene copies present in the lines and to identify the ALMT1 full-length ORFs expressed in their root tips after exposure to Al. The results of these analyses were all completely consistent with the 859 and 1135 recombinants having arisen from recombination between the M39A-1-6 and M77A-1 ALMT1 gene clusters, as represented in Figure 5. In this model, a similar pairing of the M39A-1-6 and M77A-1 ALMT1 clusters at meiosis was followed by recombination at similar locations to give rise to the 859 and 1135 recombinant chromosomes representing almost perfectly reciprocal products (Figure 5). For line 1135, recombination occurred between the 5′ ends of the M77.1 and M39.2 ORFs to produce an expressed gene (ScALMT1-1135.1) with a hybrid ORF comprised of the 5′ end of M77.1 and 3′ end of M39.2. The hybrid ORF differed from the M39.2 sequence at only two base positions near the 5′ end, only one of which encoded an amino acid difference (supplemental Figures 6 and 7). Among the Alm-6/Alm-7 genomic PCR fragment sequences obtained from the 1135 line were ones identical to the M39.2 sequence shown in supplemental Figure 4, which were presumably derived from the hybrid gene. The combined cDNA and genomic sequence information enabled the recombination point in the 1135.1 hybrid gene to be located between the 14th base position of the ORF and the 11th base position of intron 1. For line 859, recombination appeared to have occurred 5′ of the M77.1 and M39.2 ORFs. In addition to the M39.1 transcript, this line expressed a transcript identical to the M77.1 sequence shown in supplemental Figure 6, locating the 859 recombination point 5′ of the first known position that is polymorphic between M77.1 and M39.2 (base position 18 in the 5′-UTRs as defined by supplemental Figure 6). The single novel-sized ScALMT1-hybridizing restriction fragment identified in the 1135 line (asterisk in Figure 3) most likely represents the hybrid 1135.1 gene, while the presence of another novel fragment in the 859 line (asterisk in Figure 3) suggests that the 859 recombination event also occurred close to or within an ALMT1 ORF.
Figure 5.—
Model for the origin of the (A) 859 and (B) 1135 recombinants. Positions and orientations of ScALMT1 genes in the parent and recombinant haplotypes are represented by arrows. The relative orders of genes within the pairs M39.1/M39.4 and M39.3/M39.5 could not be inferred. The locations of the gene clusters within the overall structures of the recombinant chromosomes are shown. Black and white fill indicates genes and chromosome regions originating from the M39A-1-6 and M77A-1 parents, respectively. Asterisks mark genes transcribed in root tips. M39.3 was also expressed in leaves.
The same aberrant M77.1 transcript lacking exon 2 observed in the M77A-1 parent line was also observed for the M77.1 gene inherited by the 859 recombinant chromosome and was also visible as a smaller product in RT–PCR amplifications from this line (supplemental Figure 8). Likewise, the aberrant M39.2 splicing event leading to omission of exon 4, seen in the M39A-1-6 line, was also seen for the 1135.1 hybrid gene comprised mostly of M39.2 sequences. Evidently, the amount of correctly spliced transcript from this hybrid gene was sufficient to provide tolerance to the tolerant 1135 line.
Location of an ALMT1 gene in barley:
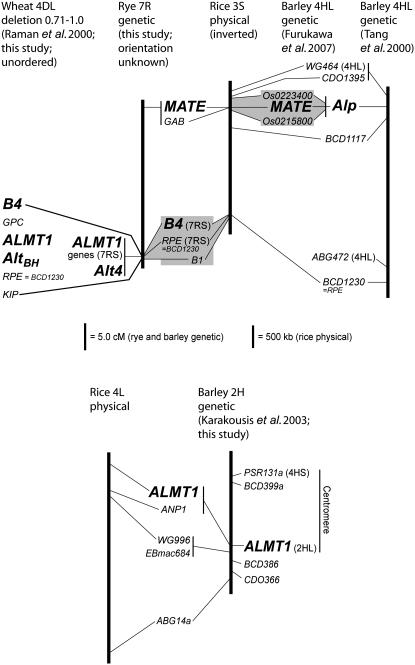
Fontecha et al. (2007) reported that barley contains a homolog of the wheat TaALMT1 gene on chromosome 2H, whereas Wang et al. (2007) reported such a barley homolog only on chromosome 4H. To explore this possible discrepancy, we used the rye ALMT1 cDNA probe in Southern blot analysis of Chinese Spring wheat lines containing individual chromosomes and chromosome arms of barley cv. Betzes. With four restriction enzymes, ≥1 hybridizing fragments were observed in Betzes barley, and the barley-specific fragments only appeared in the addition lines containing 2H or 2HL (not shown). A BamHI RFLP was mapped in a Galleon × Haruna Nijo population of 112 doubled-haploids and cosegregated with the ANP1 marker, near the centromere on 2HL (Figure 6). BLAST searches with nearby marker sequences established colinearity with the corresponding region on rice chromosome 4, showing that the ALMT1 gene in barley was in a position corresponding to the closest homolog of TaALMT1 in rice (TIGR locus LOC_Os04g34010; Figure 6), which has been named OsALMT5 (Delhaize et al. 2007).
Figure 6.—
Locations of genes and markers related to Al tolerance in rice, wheat, and barley. Lines connect markers and genes common to the illustrated chromosomes or chromosome arm segments. The orientation of the mapped section on rye chromosome 7R and the orders of genes and loci located to wheat 4DL deletion bin 0.71–1.0 relative to the centromeres are unknown. The rice 3S map is inverted to align with barley. Otherwise, all maps are oriented with the short arm toward the top. Shaded areas connect areas shown to have extensive colinearity (Furukawa et al. 2007; Wang et al. 2007; this study). The chromosome arm locations of markers are shown, where this has been determined using chromosome addition lines.
DISCUSSION
ALMT1 homologs as Al-tolerance genes in rye:
Solitary ALMT1 genes were reported at the AltBH locus in wheat Al-tolerant or Al-intolerant lines (Sasaki et al. 2004; Raman et al. 2005). Similarly, single ALMT1 genes at the rye Alt4 locus of Al-intolerant (cv. Riodeva) and Al-intolerant (cv. Riodeva) genotypes were described (Fontecha et al. 2007). In contrast, we identified a cluster of ALMT1 homologs at the Alt4 locus of rye, in both the tolerant M39A-1-6 line (five genes) and the intolerant M77A-1 line (two genes). Therefore, different rye genotypes can contain different numbers of ALMT1 homologs at the Alt4 locus, ranging from perhaps one copy, up to at least five copies. The gene copy numbers in this study were determined using multiple primers on genomic and cDNA (including RACE–PCR) and sequencing of many clones, from both the parent lines and the 859 and 1135 recombinants. However, it is possible that these lines contained highly related (recently duplicated) copies that were identical in the amplified region. The latter possibility aside, we think it very unlikely that these lines contained additional ALMT1 gene copies that went undetected by sequencing.
Fontecha et al. (2007) observed perfect cosegregation between a codominant marker for a rye ALMT1 homolog and the dominant Al-tolerance trait encoded by the Alt4 locus in a population of 381 F2 plants. The current study tested this association to a higher level of genetic resolution. In the recombinants derived from a population of 1123 F2 individuals, the ALMT1 homologs mapped to the same 0.09-cM region as the Alt4 locus. Furthermore, the 859 and 1135 recombinant chromosomes resulting from recombination within the ALMT1 cluster carry reciprocal allele types for molecular markers flanking the Alt4 locus yet both confer tolerance (Figure 5; supplemental Figure 1), essentially ruling out all genes that flank the cluster as candidates for encoding the Alt4 tolerance. Consequently, these recombinants very strongly indicate that the ALMT1 genes are responsible for controlling Al tolerance at the Alt4 locus. Two of the genes from the tolerant M39A-1-6 haplotype (ScALMT1-M39.1 and -M39.2) showed mRNA expression in root tips (Figure 4, B and D) and encode full-length predicted proteins with high similarity to TaALMT1 (supplemental Figure 7). M39.1 was inherited by the 859 recombinant chromosome, and a gene comprising most of the M39.2 ORF and the active M77.1 promoter was present in the 1135 recombinant chromosome (Figure 5). M39.1 and M39.2 therefore each appeared to be potentially active and sufficient to proved tolerance, offering an explanation as to why the two recombinants, carrying reciprocal flanking marker alleles and derived from recombination at nearly the same position within the ALMT1 cluster (Figure 5), could both be tolerant.
In a group of 13 wheat accessions, TaALMT1 mRNA expression levels showed a strong positive correlation with tolerance, suggesting that ALMT1 gene expression levels may be an important determinant of tolerance/susceptibility (Raman et al. 2005). Wheat TaALMT1 mRNA expression is not upregulated by Al, but the organic acid transporting activity of the TaALMT1 protein appears to be activated post-translationally by the presence of toxic levels of Al (Sasaki et al. 2004). In rye, mRNA expression levels of the M39.1 and M39.2 genes increased in root tips in the first 6 hr after exposure to toxic levels of Al and increased again in the next 18 hr (Figure 4). Expression concentrated in the root tip, and Al-triggered transcriptional upregulation, were also reported for the previously described rye ScALMT1-1 gene (Fontecha et al. 2007). The contrasting Al inducibility of the wheat and rye genes may be related to the different patterns of excretion of organic acids observed in wheat and rye. The Al-tolerant wheat cv. Atlas 66 exuded malate at high levels 2–10 hr after Al exposure, whereas malate and citrate exudation by rye cv. King increased at a steady state throughout this period, suggesting a different mode of regulation (Li et al. 2000). Therefore, rye may be slower to reach maximum rates of organic acid exudation following Al exposure because ScALMT1 proteins are present at low levels in the absence of Al and then increase upon exposure due to transcriptional upregulation, whereas tolerant wheat may be able to respond more quickly to Al because the Al-activated TaALMT1 transporter is expressed at a maximum level before exposure to Al.
In tolerant wheat and rye, Al-triggered excretion of organic acids occurs mainly in the first few millimeters of the root apex (Delhaize et al. 1993; Li et al. 2002), which is also the part of the roots that exhibit the most conspicuous Al-toxicity damage, as manifested by root-tip malformation and swelling, in intolerant genotypes (Delhaize et al. 2004; supplemental Figure 2). In wheat, TaALMT1 expression was not detected in leaves and in roots was detected at severalfold higher levels in the apex than in the other parts (Sasaki et al. 2004). Rye ALMT1 gene expression was found to be approximately threefold higher in root tips than in other parts of the roots, before and after exposure to Al, and was up to 130–250 times lower in leaves than in root tips (Figure 4, A and C). Therefore, the expression patterns of the rye ALMT1 genes were consistent with a potential involvement in an Al-tolerance mechanism involving organic acid excretion from the root tips.
ScALMT1 splice variants:
Around half of the transcripts produced by the ScALMT1-M77.1 gene lacked exon 2 due to aberrant splicing, resulting in an in-frame 50-amino-acid deletion within the predicted product, which may or may not destroy the protein's activity. Results of the expression study using Alt4 heterozygotes (Figure 4D) also suggested that the M77.1 promoter may be less active and/or less Al inducible than the M39.1 and M39.2 promoters. If the interpretation of the 1135 recombinant (Figure 5) is correct, the only gene conferring Al tolerance in the 1135 line expresses a functional ORF comprising mostly M39.2 coding sequence, driven by the M77.1 promoter. No splice variants lacking exon 2 were detected for this hybrid gene, probably because the sequences causing this splice defect in M77.1 were located 3′ of the recombination site and therefore were not inherited by the hybrid gene. Of the 18 F4 individuals from the segregating 1135-1 family assessed for tolerance (supplemental Table 4) 9 contained the 1135 recombinant chromosome paired with the M77A-1 nonrecombinant chromosome and were fully tolerant, indicating that a single hemizygous copy of the hybrid gene driven by the M77.1 promoter was sufficient for tolerance. Therefore, if a halving of the transcript abundance in the 1135 hemizygote did not affect tolerance, an effective halving of the amount of functional M77.1 transcript as a result of the aberrant splicing would not appear to fully account for the failure of the M77.1 gene to provide tolerance in homozygotes. Consequently, the M77.1 full-length protein encoded by the correctly spliced transcript may have lower activity than the M39.1, M39.2, and 1135.1 proteins. The full-length M77.1 protein has three amino acid substitutions relative to the M39.1, M39.2, and 1135.1 functional rye proteins (Figure 7; Cys125 to Tyr, Arg178 to Ile, Ala324 to Thr), which may potentially impair activity.
The aberrant M77.1 transcript lacked exon 2, due to joining of the 5′ splice (donor) site of intron 1 to the 3′ splice (acceptor) site of intron 2. The sequence at the several bases surrounding splice sites is either essential or influential for splice-site choice in plants (Reddy 2007). However, ScALMT1 genes showed no sequence variation around the splice sites of either intron 1 or 2 (supplemental Figure 4). The missplicing in M77.1 could have been caused by the unique ∼400-bp insertion, including the 227-bp MITE in intron 2 of M77.1 (supplemental Figure 4). This insertion may have interfered with the nucleophilic attack of the intron-2 5′ splice site by the branch point in intron 2, leading instead to occasional attack of the 5′ splice site of intron 1 by the intron-2 branch point. Various stresses are known to affect the relative frequencies of splice variants of particular genes (Reddy 2007). However, in RT–PCRs that detected both M77.1 splice variants at once, the proportions of the two products appeared unchanged in root tips following exposure to Al stress (not shown). At the same point of the ∼400-bp insertion in ScALMT1-M77.1, the -M39.5 and -1 genes have 5-bp and 39-bp insertions unrelated in sequence to the insertion in M77.1 (supplemental Figure 4), indicating that this may be a hot spot for sequence insertion and/or deletion. However, the functional significance of these other insertions is unknown, since no transcripts of any type were detected from the M39.5 gene, and no alternative splicing was reported for ScALMT1-1.
Transcripts missing exon 4 were sometimes produced by the M39.2 gene and by the hybrid 1135.1 gene comprised of M39.2 sequence from the 5′ end of intron 1 onwards. The aberrant transcripts with the 113-bp deletion occurred at a lower frequency than the M77.1 transcripts with the 150-bp deletion, based on the poor visibility of the smaller transcripts in RT–PCR amplifications from the M39A-1-6 and 1135 lines (supplemental Figure 8), and the smaller proportion of cDNA clones obtained for these variants (2/8 and 1/23 cDNA clones from M39.2 and 1135.1, respectively). Skipping of exon 4 causes a frameshift and severe truncation of the predicted product, most likely abolishing protein function. However, there was no evidence that this occasional splicing defect had an impact on Al tolerance, since the 1135.1 gene was shown to be sufficient to provide Al tolerance.
The rye ALMT1 gene cluster:
A characteristic of some clusters of tandemly repeated genes is the ability to align in a variable way during meiotic chromosome pairing (analogous to a slide rule), resulting in recombination products containing different numbers of repeats. This can result in spontaneous loss of the phenotypic trait when recombination generates a new haplotype containing only inactive gene copies. The maize rp1-D gene-cluster haplotype pairs with itself in a wide variety of ways (Sun et al. 2001), whereas evidence for only one type of pairing between the Cf-4 and Cf-9 haplotypes of the complex tomato MW locus was obtained (Wulff et al. 2004). The extent and distribution of homologies in intergenic regions are thought to be determinants of the recombination behaviors of different complex loci (Wulff et al. 2004). The 859 and 1135 recombinants resulted from the same alignment of the M39A-1-6 and M77A-1 haplotypes and recombination at a similar position, at or near the 5′ ends of the paired M39.2 and M77.1 genes (Figure 5), suggesting a bias in how M39A-1-6 and M77A-1 haplotypes recombine. However, further recombinants will need to be analyzed to determine if these haplotypes can recombine in other ways.
Recombination at Alt4 can generate novel coding sequences, as demonstrated by the chimeric ScALMT1-1135.1 gene. The patchwork pattern of sequence polymorphisms between ScALMT1 genes also indicates a history of recombination between the ORFs. For example, the ScALMT1-2 cDNA sequence is identical to ScALMT1-M39.2 at four polymorphic base positions between the end of exon 1 and the start of exon 2, but then diverges from ScALMT1-M39.2 and matches ScALMT1-1 sequence for two polymorphic base positions before becoming different from ScALMT1-1 again in exon 4 (supplemental Figure 6). Rye is an outcrossing, wind-pollinated species. Therefore, in heterogeneous rye populations, one would expect there to be frequent opportunities for recombination between different Alt4 locus haplotypes with the potential to generate new variation in ALMT1 copy number and coding sequence.
Differences in gene copy number and accompanying changes in expression levels are regarded as important determinants of trait variation, both within and between species (Dumas et al. 2007). The overall amount of ScALMT1 mRNA produced by the two active ScALMT1 copies from the M39A-1-6 haplotype was threefold or more greater than that produced by the only gene transcribed from the M77A-1 haplotype (Figure 4D). However, a reduced level of correctly spliced transcript produced by the M77A-1 haplotype does not appear to account fully for its failure to provide Al tolerance, and the encoded M77.1 protein would appear to be at least partially defective. We also saw no evidence that the 859 and 1135 haplotypes, each carrying a single active ScALMT1 gene copy, conferred any less Al tolerance than the parental M39A-1-6 haplotype containing two active tolerance genes, even when the former were paired with the intolerant haplotype in Alt4 heterozygotes. Further tests with higher levels of Al stress will need to be carried out to fully evaluate the effect of ScALMT1 gene copy number on Al tolerance in rye.
Expression of gene M39.3 was observed in the samples comprising part of the coleoptile and rolled-up leaf base taken from just above the seed (“leaves,” Figure 4B), but was not detected in roots. It is possible that M39.3 was expressed at similar low levels in the roots but was not detected there because high expression of M39.1 and M39.2 in roots prevented its detection. Expression of M39.1 and M39.2 was also detected in leaves. Metal homeostasis, Al tolerance in the apoplast, and many important biochemical processes in plants involve transport of organic acids such as malate, or their metal complexes, between subcellular compartments (Holtum et al. 2005; Kochian et al. 2005; De Angeli et al. 2007; Delhaize et al. 2007; Haydon and Cobbett 2007). Potentially, rye ALMT1 proteins, or other members of the ALMT protein family (Delhaize et al. 2007), could be involved in these processes in root tips and other tissues.
Relationships between cereal Al-tolerance genes:
We showed that the Alt4 Al-tolerance locus identified with the M39A-1-6 × M77A-1 cross is located on the short arm of rye chromosome 7R as it is defined on the RFLP map of Devos et al. (1993) (supplemental Figure 3). This agrees with the claim by Benito et al. (2005) that this locus was on 7RS, but disagrees with the chromosome 4R location originally determined by Miftahudin et al. (2002). The 4R location was determined by RFLP analysis using a probe cloned from a single-linked AFLP band. However, the AFLP band was reported to be comprised of a mixture of at least two different sequences, only one of which was low copy and could be used for mapping (Miftahudin et al. 2002). Hence, an incorrect chromosome assignment may have been obtained by mapping with the wrong cloned fragment. An Al-tolerance locus mapped using rye Ailés × Riodeva crosses was also located to 7RS (Matos et al. 2005). The common 7RS location of the loci detected in the different crosses led Benito et al. (2005) to propose that these rye loci were the same. Their equivalence is now confirmed by the finding that ALMT1 genes reside at both loci. The locus identified using the M39A-1-6 × M77A-1 cross was originally named Alt3. However, in light of the current findings, we suggest that the 7RS locus be referred to as Alt4, and that the name Alt3 be reserved for the potential Al-tolerance gene Aniol and Gustafson (1984) identified on 4R by chromosome addition line analysis.
Both the rye Alt4 locus and the AltBH Al-tolerance locus on wheat 4DL had been mapped close to RFLP loci detected with the probe BCD1230 (RPE, Figure 1; Miftahudin et al. 2002). In the current study, analysis of additional markers confirmed that these loci were on related chromosome segments (Figure 6). The relationship between wheat 4L and rye 7RS is consistent with the known relationships between the wheat and rye genomes, including discontinuities in colinearity caused by chromosome translocations that occurred since the divergence of wheat and rye from a common ancestor. Relationships between rye 4R and wheat group 4 chromosomes are limited to the short arms, whereas relationships to wheat 4L are found only on rye 7RS and 5RL (Devos et al. 1993). Consistent with the occurrence of the wheat AltBH and rye Alt4 loci on related chromosome segments, we demonstrated that tolerance at Alt4 is controlled by genes homologous to the AltBH tolerance gene TaALMT1.
The barley chromosome 4H region containing the Alp Al-tolerance locus is also related to the rye Alt4 and wheat AltBH regions (Tang et al. 2000; Figure 6). However, the HvMATE gene, which is completely unrelated to TaALMT1, controls Al tolerance at Alp (Furukawa et al. 2007; Wang et al. 2007). The rye ortholog of HvMATE was mapped 27.5 cM from the Alt4 locus and recombined with Alt4 tolerance in most of the rye families characterized for Al tolerance (Figures 1 and 6; supplemental Tables 3–5), clearly ruling out the rye MATE gene as a candidate for controlling Al tolerance at Alt4. Therefore, while rye contains an ortholog of the barley HvMATE gene close to the Alt4 locus, there is no evidence that it contributes to Al tolerance in rye, at least in the lines investigated.
We identified a barley homolog of TaALMT1 only on chromosome 2H, as did Wang et al. (2007). In contrast, Fontecha et al. (2007) reported identifying a barley ALMT1 homolog only on chromosome 4H using the same Chinese Spring–Betzes barley chromosome addition lines that we used initially to locate a copy to 2H. While a 4H location is expected from the overall colinearity between group 4 chromosome regions carrying Al-tolerance loci in wheat, rye, and barley, the locus position we determined on 2H is the one expected from colinearity with rice (Figure 6). The chromosome 4H position was determined by PCR (Fontecha et al. 2007) and may require verification. The locations of ALMT1 genes on rye 7RS and wheat 4DL do not fit with the overall colinearity with rice (Figure 6). Within the Triticeae, wheat and rye diverged from a common ancestor more recently than wheat/rye diverged from barley (Huang et al. 2002). Therefore, one interpretation would be that ALMT1 on barley 2H represents the original location and that ALMT1 moved to the chromosome group 4 position in the wheat/rye lineage after the divergence from the barley lineage. No Al-tolerance loci have been reported on barley chromosome 2H. Furthermore, Al-tolerance QTL have been reported on rice chromosome 4 (Nguyen et al. 2002; Xue et al. 2006), but not in the vicinity of the TaALMT1 homolog (data not shown). Therefore, of the two ALMT1 positions detected in cereals, the one represented by the wheat/rye copies appears to be the only one associated with an ability to confer Al tolerance.
In contrast to the ALMT1 genes, the MATE genes were located in colinear positions in barley, rye, and rice (Figure 6). Al-tolerance QTL have been identified on rice 3S (Wu et al. 2000; Nguyen et al. 2003) in a region containing the closest rice homolog of the barley HvMATE citrate transporter gene. Al tolerance from the wild rice species Oryza rufipogon was most closely associated with markers CDO1395 and RG391 located only 0.5 cM apart, and the nearest flanking marker on the upper side (lower side in Figure 6) was located 11.0 cM away (Nguyen et al. 2003). The genetic distance between the MATE gene and the position corresponding to Alt4 in rice (Figure 6) is 12.3–13.2 cM (http://rgp.dna.affrc.go.jp/cgi-bin/statusdb/irgsp-status.cgi). Hence, the position of this rice QTL corresponds well with the MATE gene. Rice roots excrete citrate in response to Al stress (Chen et al. 2006), which also fits with the known citrate transporting capability of the homolog in barley. Therefore, the MATE gene could be considered as a good candidate for the gene underlying the rice 3S QTL effect.
Acknowledgments
We thank Miftahudin, Peter Langridge, Ian Dundas, and Rafiq Islam for provision of rye lines, and Stephen Fletcher, Andrew Jacobs, Irandokht Fathi, Zahra Shoaei, and Ursula Langridge for technical assistance. The Australian Centre for Plant Functional Genomics is supported by the Australian Research Council, the Grains Research and Development Council, and the South Australian State Government.
References
- Aniol, A., 1983. Aluminum uptake by roots of two winter wheat varieties of different tolerance to aluminum. Biochem. Physiol. Pflanz. 178 11–20. [Google Scholar]
- Aniol, A., and J. P. Gustafson, 1984. Chromosome location of genes controlling aluminum tolerance in wheat, rye, and triticale. Can. J. Genet. Cytol. 26 701–705. [Google Scholar]
- Benito, C., G. Fontecha, J. Silva-Navas, V. Hernández-Riquer, M. Eguren et al., 2005. Chromosomal location of molecular markers linked to aluminum tolerance genes in rye. Czech J.Genet. Plant Breed. 41 288. [Google Scholar]
- Bot, A. J., F. O. Nachtergaele and A. Young, 2000. Land Resource Potential and Constraints at Regional and Country Levels. Food and Agricultural Organization of the United Nations, Rome.
- Bonfield, J. K., K. F. Smith and R. Staden, 1995. A new DNA sequence assembly program. Nucleic Acids Res. 23 4992–4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton, R. A., N. J. Shirley, B. J. King, A. J. Harvey and G. B. Fincher, 2004. The CesA gene family of barley. Quantitative analysis of transcripts reveals two groups of co-expressed genes. Plant Physiol. 134 224–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, R. F., R. F. Shen, P. Gu, X. Y. Dong, C. W. Du et al., 2006. Response of rice (Oryza sativa) with root surface iron plaque under aluminium stress. Ann. Bot. 98 389–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Angeli, A., S. Thomine, J. M. Frachisse, G. Ephritikhine, F. Gambale et al., 2007. Anion channels and transporters in plant cell membranes. FEBS Lett. 581 2367–2374. [DOI] [PubMed] [Google Scholar]
- Delhaize, E., P. R. Ryan and P. J. Randall, 1993. Aluminum tolerance in wheat (Triticum-aestivum L). II. Aluminum-stimulated excretion of malic acid from root apices. Plant Physiol. 103 695–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaize, E., P. R. Ryan, D. M. Hebb, Y. Yamamoto, T. Sasaki et al., 2004. Engineering high-level aluminum tolerance in barley with the ALMT1 gene. Proc. Natl. Acad. Sci. USA 101 15249–15254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaize, E., B. D. Gruber and P. R. Ryan, 2007. The roles of organic anion permeases in aluminium resistance and mineral nutrition. FEBS Lett. 581 2255–2262. [DOI] [PubMed] [Google Scholar]
- Devos, K. M., M. D. Atkinson, C. N. Chinoy, H. A. Francis, R. L. Harcourt et al., 1993. Chromosomal rearrangements in the rye genome relative to that of wheat. Theor. Appl. Genet. 85 673–680. [DOI] [PubMed] [Google Scholar]
- Dumas, L., Y. H. Kim, A. Karimpour-Fard, M. Cox, J. Hopkins et al., 2007. Gene copy number variation spanning 60 million years of human and primate evolution. Genome Res. 17 1266–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontecha, G., J. Silva-Navas, C. Benito, M. A. Mestres, F. J. Espino et al., 2007. Candidate gene identification of an aluminum-activated organic acid transporter gene at the Alt4 locus for aluminum tolerance in rye (Secale cereale L.). Theor. Appl. Genet. 114 249–260. [DOI] [PubMed] [Google Scholar]
- Furukawa, J., N. Yamaji, H. Wang, N. Mitani, Y. Murata et al., 2007. An aluminum-activated citrate transporter in barley. Plant Cell Physiol. 48 1081–1091. [DOI] [PubMed] [Google Scholar]
- Gallego, F. J., B. Calles and C. Benito, 1998. Molecular markers linked to the aluminium tolerance gene Alt1 in rye (Secale cereale L.). Theor. Appl. Genet. 97 1104–1109. [Google Scholar]
- Haydon, M. J., and C. S. Cobbett, 2007. Transporters of ligands for essential metal ions in plants. New Phytol. 174 499–506. [DOI] [PubMed] [Google Scholar]
- Hoekenga, O. A., L. G. Maron, M. A. Pineros, G. M. A. Cancado, J. Shaff et al., 2006. AtALMT1, which encodes a malate transporter, is identified as one of several genes critical for aluminum tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 103 9738–9743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtum, J. A. M., J. A. C. Smith and H. E. Neuhaus, 2005. Intracellular transport and pathways of carbon flow in plants with crassulacean acid metabolism. Funct. Plant Biol. 32 429–449. [DOI] [PubMed] [Google Scholar]
- Huang, S., A. Sirikhachornkit, X. J. Su, J. Faris, B. Gill et al., 2002. Genes encoding plastid acetyl-CoA carboxylase and 3-phosphoglycerate kinase of the Triticum/Aegilops complex and the evolutionary history of polyploid wheat. Proc. Natl. Acad. Sci. USA 99 8133–8138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judo, M. S. B., A. B. Wedel and C. Wilson, 1998. Stimulation and suppression of PCR-mediated recombination. Nucleic Acids Res. 26 1819–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakousis, A., A. R. Barr, J. M. Kretschmer, S. Manning, S. J. Logue et al., 2003. Mapping and QTL analysis of the barley population Galleon x Haruna Nijo. Aust. J. Agr. Res. 54 1131–1135. [Google Scholar]
- Kim, B. Y., A. C. Baier, D. J. Somers and J. P. Gustafson, 2001. Aluminum tolerance in triticale, wheat, and rye. Euphytica 120 329–337. [Google Scholar]
- Kochian, L. V., M. A. Pineros and O. A. Hoekenga, 2005. The physiology, genetics and molecular biology of plant aluminum resistance and toxicity. Plant Soil 274 175–195. [Google Scholar]
- Li, X. F., J. F. Ma and H. Matsumoto, 2000. Pattern of aluminum-induced secretion of organic acids differs between rye and wheat. Plant Physiol. 123 1537–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X. F., J. F. Ma and H. Matsumoto, 2002. Aluminum-induced secretion of both citrate and malate in rye. Plant Soil 242 235–243. [Google Scholar]
- Ma, J. F., S. Taketa and Z. M. Yang, 2000. Aluminum tolerance genes on the short arm of chromosome 3R are linked to organic acid release in triticale. Plant Physiol. 122 687–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magalhaes, J. V., J. Liu, C. T. Guimaraes, U. G. P. Lana, V. M. C. Alves et al., 2007. A gene in the multidrug and toxic compound extrusion (MATE) family confers aluminum tolerance in sorghum. Nat. Genet. 39 1156–1161. [DOI] [PubMed] [Google Scholar]
- Matos, M., M. V. Camacho, V. Perez-Flores, B. Pernaute, O. Pinto-Carnide et al., 2005. A new aluminum tolerance gene located on rye chromosome arm 7RS. Theor. Appl. Genet. 111 360–369. [DOI] [PubMed] [Google Scholar]
- Miftahudin, G. J. Scoles and J. P. Gustafson, 2002. AFLP markers tightly linked to the aluminum-tolerance gene Alt3 in rye (Secale cereale L.). Theor. Appl. Genet. 104 626–631. [Google Scholar]
- Miftahudin, G. J. Scoles and J. P. Gustafson, 2004. a Development of PCR-based codominant markers flanking the Alt3 gene in rye. Genome 47 231–238. [DOI] [PubMed] [Google Scholar]
- Miftahudin, K. Ross, X. F. Ma, A. A. Mahmoud, J. Layton et al., 2004. b Analysis of expressed sequence tag loci on wheat chromosome group 4. Genetics 168 651–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miftahudin, T. Chikmawati, K. Ross, G. J. Scoles and J. P. Gustafson, 2005. Targeting the aluminum tolerance gene Alt3 region in rye, using rice/rye micro-colinearity. Theor. Appl. Genet. 110 906–913. [DOI] [PubMed] [Google Scholar]
- Motoda, H., T. Sasaki, Y. Kano, P. R. Ryan, E. Delhaize et al., 2007. The membrane topology of ALMT1, an aluminum-activated malate transport protein in wheat (Triticum aestivum). Plant Signal. Behav. 2 467–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugwira, L. M., S. M. Elgawhary and K. I. Patel, 1976. Differential tolerances of triticale, wheat, rye, and barley to aluminum in nutrient solution. Agron. J. 68 782–787. [Google Scholar]
- Nguyen, B. D., D. S. Brar, B. C. Bui, T. V. Nguyen, L. N. Pham et al., 2003. Identification and mapping of the QTL for aluminum tolerance introgressed from the new source, Oryza rufipogon Griff., into indica rice (Oryza sativa L.). Theor. Appl. Genet. 106 583–593. [DOI] [PubMed] [Google Scholar]
- Nguyen, V. T., B. D. Nguyen, S. Sarkarung, C. Martinez, A. H. Paterson et al., 2002. Mapping of genes controlling aluminum tolerance in rice: comparison of different genetic backgrounds. Mol. Genet. Genomics 267 772–780. [DOI] [PubMed] [Google Scholar]
- Omote, H., M. Hiasa, T. Matsumoto, M. Otsuka and Y. Moriyama, 2006. The MATE proteins as fundamental transporters of metabolic and xenobiotic organic cations. Trends Pharmacol. Sci. 27 587–593. [DOI] [PubMed] [Google Scholar]
- Raman, H., K. R. Zhang, M. Cakir, R. Appels, D. F. Garvin et al., 2005. Molecular characterization and mapping of ALMT1, the aluminium-tolerance gene of bread wheat (Triticum aestivum L.). Genome 48 781–791. [DOI] [PubMed] [Google Scholar]
- Reddy, A. S. N., 2007. Alternative splicing of pre-messenger RNAs in plants in the genomic era. Annu. Rev. Plant Biol. 58 267–294. [DOI] [PubMed] [Google Scholar]
- Rogowsky, P. M., F. L. Y. Guidet, P. Langridge, K. W. Shepherd and R. M. D. Koebner, 1991. Isolation and characterization of wheat-rye recombinants involving chromosome arm 1DS of wheat. Theor. Appl. Genet. 82 537–544. [DOI] [PubMed] [Google Scholar]
- Rogowsky, P. M., J. Y. Liu, S. Manning, C. Taylor and P. Langridge, 1992. Structural heterogeneity in the R173 family of rye-specific repetitive DNA sequences. Plant Mol. Biol. 20 95–102. [DOI] [PubMed] [Google Scholar]
- Sasaki, T., Y. Yamamoto, B. Ezaki, M. Katsuhara, S. J. Ahn et al., 2004. A wheat gene encoding an aluminum-activated malate transporter. Plant J. 37 645–653. [DOI] [PubMed] [Google Scholar]
- Sun, Q., N. C. Collins, M. Ayliffe, S. M. Smith, J. Drake et al., 2001. Recombination between paralogues at the rp1 rust resistance locus in maize. Genetics 158 423–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, Y., M. E. Sorrells, L. V. Kochian and D. F. Garvin, 2000. Identification of RFLP markers linked to the barley aluminum tolerance gene Alp. Crop Sci. 40 778–782. [Google Scholar]
- Vandesompele, J., K. De Preter, F. Pattyn, B. Poppe, N. Van Roy et al., 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Uexküll, H. R., and E. Mutert, 1995. Global extent, development and economic impact of acid soils. Plant Soil 171 1–15. [Google Scholar]
- Wang, J. P., H. Raman, M. X. Zhou, P. R. Ryan, E. Delhaize et al., 2007. High-resolution mapping of the Alp locus and identification of a candidate gene HvMATE controlling aluminium tolerance in barley (Hordeum vulgare L.). Theor. Appl. Genet. 115 265–276. [DOI] [PubMed] [Google Scholar]
- Wu, P., C. Y. Liao, B. Hu, K. K. Yi, W. Z. Jin et al., 2000. QTLs and epistasis for aluminum tolerance in rice (Oryza sativa L.) at different seedling stages. Theor. Appl. Genet. 100 1295–1303. [Google Scholar]
- Wulff, B. B. H., C. M. Thomas, M. Parniske and J. D. G. Jones, 2004. Genetic variation at the tomato Cf-4/Cf-9 locus induced by EMS mutagenesis and intralocus recombination. Genetics 167 459–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, Y., J. M. Wan, L. Jiang, C. M. Wang, L. L. Liu et al., 2006. Identification of quantitative trait loci associated with aluminum tolerance in rice (Oryza sativa L.). Euphytica 150 37–45. [Google Scholar]