Abstract
Myosin VI is an actin-based motor that has been implicated in many cellular processes. Studies in vertebrates have demonstrated that animals lacking this ubiquitously expressed myosin are viable. However in Drosophila, myosin VI loss of function has been thought to be lethal. We show here that complete loss of myosin VI is not lethal in flies and that the previously reported lethality of the null mutation (jar322) is most likely due to deletion of a neighboring gene. Maternally provided myosin VI does not account for the survival of myosin VI null animals. Mutant animals are recovered at a lower than expected Mendelian frequency, suggesting that myosin VI participates in processes which contribute to normal development, but its participation is not essential.
MYOSIN VI is an actin motor that is conserved from flies and worms to humans. This motor is unique among myosins in that it can move along actin filaments toward the pointed or minus end, a direction opposite other characterized myosins. In addition, myosin VI can both move processively and bind tightly to an actin filament, stalling under backward load. These properties may allow it to act as a transporter in some processes and an anchor in other processes (Buss et al. 2004; Frank et al. 2004; Sweeney and Houdusse 2007).
Myosin VI has been implicated in a variety of cellular functions in Drosophila using loss-of-function techniques. Microinjection of myosin VI antibodies into syncytial blastoderm embryos caused defects in actin pseudocleavage furrow formation. This phenotype was hypothesized to be the result of defects in transport of furrow components (Mermall and Miller 1995). Promoter mutations (jar1 and jarmmw14), which prevent myosin VI expression in the testes, cause male sterility due to a failure of spermatid individualization. Defects in the actin structures that mediate the separation of the syncytial spermatids are observed (Hicks et al. 1999; Noguchi et al. 2006). Using a Gal4-UAS targeted expression system combined with antisense RNA, which allows for disruption of function in specific groups of cells at different developmental stages, myosin VI has been implicated in border cell migration in oogenesis, tissue integrity during late embryogenesis, cuticle development in third star larvae, and imaginal disc morphogenesis during metamorphosis (Deng et al. 1999; Geisbrecht and Montell 2002). Myosin VI promoter mutations (jarR39 and jarR235), which cause a loss of myosin VI expression in late embryogenesis and expression of a myosin VI dominant negative protein that lacks the ATP binding site cause lethality in late embryos due to defects in the integrity of the epithelial cell layers during dorsal closure (Millo et al. 2004). A null allele of myosin VI (jar322) causes abnormalities in asymmetric localization of determinants and, therefore, neural fate determination (Petritsch et al. 2003). Animals homozygous for this mutation die as first or second instar larvae.
In contrast to Drosophila, studies in vertebrates show that myosin VI loss of function is not lethal. Myosin VI mutations in mice, fish, and humans result in deafness due to degeneration of hair cell stereocilia (Avraham et al. 1995; Self et al. 1999; Ahmed et al. 2003; Kappler et al. 2004). In addition, vertebrate myosin VI has been implicated in processes similar to those affected in Drosophila, such as endocytosis, cell adhesion, and basolateral sorting (Dance et al. 2004; Ameen and Apodaca 2007; Au et al. 2007; Maddugoda et al. 2007; Morriswood et al. 2007). Since myosin VI is involved in similar processes in vertebrates as Drosophila, the lethality of myosin VI in flies but not in vertebrates seems surprising.
Because the jar322 mutation deletes all of the amino acid coding sequences in the myosin VI gene, it offers the best means to understand myosin VI's role in vivo. However, this small deletion affects both myosin VI and a neighboring gene, CG5706. Thus, it is unclear which gene is responsible for the lethal phenotype. To determine if this lethality is due to loss of myosin VI, we have examined the jar322 allele in transheterozygous combination with two large deletions in this region. Our experiments demonstrate that myosin VI null animals can survive, but do so at less than the expected Mendelian frequency. This phenotype is consistent with myosin VI playing a role in a variety of processes during development but not being essential for them. The ability to examine myosin VI null animals at all stages of the Drosophila life cycle provides an opportunity to investigate its contribution to a wide variety of cellular processes and in many developmental contexts.
MATERIALS AND METHODS
Fly stocks and fertility tests:
All Drosophila melanogaster fly stocks and crosses were maintained on standard Drosophila cornmeal food at 25° unless otherwise noted. For crosses and fertility tests, parents were removed after 5 days and progeny were counted after 18 days. For fertility tests, three males or females of the test genotype were placed with three Oregon-R males or females in a vial supplemented with instant Drosophila medium (Carolina Biologicals, Burlington, NC). A total of five vials were scored for each test genotype. The previously described jaguar322 (jar322) mutant (Petritsch et al. 2003) was obtained from C. Petritsch and Y. N. Jan. Deletion chromosomes Df(3R)S87-5 and Df(3R)S87-4 were obtained from the Bloomington Drosophila Stock Center (Bloomington, IN). Zygotic null myosin VI animals were generated by crossing jar322/TM3 Sb ubx lacZ e pp females to Df(3R)S87-5/TM3 Sb e pp males. To generate flies lacking maternal myosin, jar322/Df(3R) S87-5 females from the above cross were back-crossed to jar322/TM3 Sb ubx lacZ e pp males. For rescue of jar322 in the Df(3R)S87-5 background, w; P[C95w+]; Df(3R)S87-5 st e/TM3 Sb e pp females were crossed to jar322/TM3 Sb ubx lacZ e pp males. P[C95 w+] is a transgene containing a myosin VI cDNA under the control of a hsp83 promoter, which is ubiquitously expressed without heat shock. Animals were not heat-shocked in rescue experiments, since basal expression resulted in myosin VI amounts similar to endogenous protein levels. P[C95 w+] flies were generated and first reported by (Hicks et al. 1999). Flies lacking both myosin VI and a neighboring gene, CG5706, were generated by crossing jar322/TM3 Sb ubx lacZ e pp males to Df(3R)S87-4 st e/TM3 Ser e females. To test the ability of myosin VI expression to rescue jar322 in the Df(3R)S87-4 background, w; P[C95w+]; Df(3R)S87-4 st e/TM3 Sb e pp females were crossed to jar322/TM3 Sb ubx lacZ e pp males. A total of two vials were scored for each test genotype.
Genomic PCR:
PCR was performed on DNA extracted from a single fly and products were analyzed on a 1.8% agarose gel. Primers used were as follows: set 1, CAGGCGAGTGAAAGTGGT CGGGGCC/CTTGGTCTCTATGGGACCGGCACTG; set 2, TACATGCTCCTCGCCGGAG CTC/TGCATCACTCGGGATACCAGGG; set 3, GCGAATCACTATCGCCTGGGTC/ACGC GATGGATAACCGTGCTCC; set 4, CTGGGAGCCCTCTGCGTGATCAAGC/GATTTCCT CGCGCTGGCGCTGCTCC; set 5, GGAGCAGCGCCAGCGCGAGGAAATC/CACCTGGC CATTGGACTCGTTGGCC; set 6, GGCCAACGAGTCCAATGGCCAGGTG/GGGAGACGC GTCATTGACACCACTG; set 7, GCCCAACAACAGGCCCTGGGCAAGC/CCAGGCGTG GTACACCTTCAAGCGG; and set 8, AACCGCAAGCGCACCACCATGGATG/GTTTCTG CATTGCTGCAGCCGGCCC.
RT–PCR:
RNA was extracted from OreR and jar322/Df(3R)S87-5 adult flies using RNAeasy mini kit (QIAGEN, Valencia, CA). RT–PCR was performed with the above primers using the One-Step RT–PCR kit (QIAGEN).
Western analysis:
Ovaries (20), testes (20), or adults (5) were homogenized in 75 μl 1× testes IP buffer (100 mm NaCl, 20 mm Hepes, 1% NP4O, pH 7.5), protease inhibitor cocktail (Sigma, St. Louis) and 1mm PMSF supplemented with 4× SDS–PAGE sample buffer. Samples with whole adults were homogenized and then briefly centrifuged to remove insoluble debris before 4× sample buffer was added. Proteins were separated on a 7.5% SDS polyacrylamide gel and blotted onto nitrocellulose using conventional methods. Detection was performed using Supersignal West Pico chemiluminescence substrate (Pierce, Rockford, IL). Antibodies used were mouse monoclonal anti-myosin VI (3C7) (Kellerman and Miller 1992) at 1:20 dilution and a monoclonal anti-β-tubulin antibody, DM1-A (Sigma) at 1:1000 dilution. Flies were fed yeast for several days before collecting ovaries.
Fluorescence microscopy:
jar322/TM3 Sb Green males were crossed to jar322/Df(3R)S87-5 females and progeny were collected. Embryos were scored as either expressing GFP [jar322/TM3 Sb Green, Df(3R)S87-5/TM3 Sb Green] or not expressing [jar322/Df(3R)S87-5, jar322/jar322] at approximately stage 16 using fluorescence of the Bolvic's organ as a marker (Casso et al. 2000). Third instar mutant larvae of the genotype jar322/Df(3R)S87-5 and GFP-expressing larvae [jar322/TM3 Sb Green, Df(3R)S87-5/TM3 Sb Green] were selected, put into vials with instant food (which increases survival), and scored for the number of adults obtained. GFP expression was visualized using a Nikon Eclipse TE 2000 microscope (Nikon, Japan) equipped with a GFP band pass filter.
RESULTS AND DISCUSSION
jar322/Df(3R)S87-5 flies lack myosin VI protein expression:
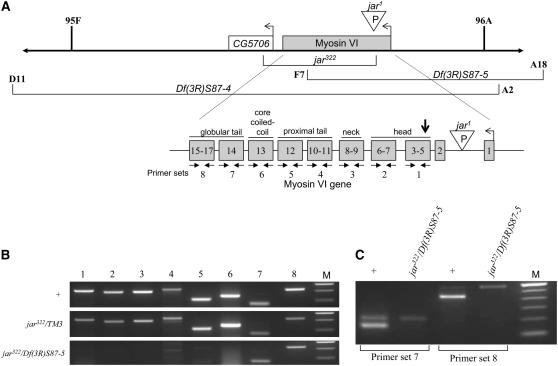
jar322/Df(3R)S87-4 flies are not viable while jar322/Df(3R)S87-5 flies are viable, but male sterile (Petritsch et al. 2003; J. K. Morrison, unpublished observations). Previous studies showed that jar1/Df(3R)S87-5 were male sterile due to the loss of myosin VI expression in the testis but not other tissues (Hicks et al. 1999). The complementation observed among the myosin VI alleles and deficiencies used in this study is shown in Table 1. To determine if jar322/Df(3R)S87-5 animals are sterile due to loss of myosin VI function only in the testis [similar to jar1/Df(3R)S87-5], we examined expression in adult testis and other tissues using Western blot analysis with monoclonal anti-myosin VI antibody (3C7). This antibody recognizes a determinant in the predicted core coiled-coil region of the tail (Figure 2A, exon 13), which immediately precedes the predicted globular region at the C terminus of the molecule (exons 14–17). Carcass, ovaries, and testes of jar322/Df(3R)S87-5 adults and jar322/Df(3R)S87-5 adults expressing a myosin VI cDNA transgene were selected. No myosin VI was detected in the jar322/Df(3R)S87-5 adults (Figure 1A). Additionally, myosin VI expression was restored in these flies when the myosin VI cDNA transgene was present (Figure 1C).
TABLE 1.
Complementation among the myosin VI alleles and deficiencies used in this study
| jar322 | jar1 | jarmmw14 | |
|---|---|---|---|
| jar322 | Lethala | — | — |
| jar1 | Viable, male sterileb | Viable, male sterilec | — |
| jarmmw14 | Viable, male sterileb | Viable, male sterilec | Lethalce |
| Df(3R)S87-5 | Viable, male steriled | Viable, male sterilec | Viable, male sterilec |
| Df(3R)S87-4 | Lethald | Viable, male sterilec | Viable, male sterilec |
J. K. Morrison and K. G. Miller (unpublished results).
This study.
The lethality in this genotype is unlinked to myosin VI.
Figure 2.—
Characterization of the genomic region of myosin VI in the Df(3R)S87-5 deletion chromosome. (A) Schematic depicts a portion of the right arm of the third chromosome in Drosophila, showing the position of the myosin VI gene, the deficiencies that remove portions of this region, and the mutations used. The unfilled arrows indicate the transcription start site and direction. Below, an expanded schematic illustrates the exons encoding different parts of the myosin VI protein with eight primer sets (solid arrows) designed to detect different regions (not drawn to scale). The vertical arrowhead indicates the translation start, which is encoded in exon 3. (B) PCR products generated in amplification reactions of genomic DNA using the primers indicated in A, resolved on a 1.8% gel. Size markers (M) are in increments of 100 bp starting with 600 bp at the top. Note that in jar332/Df(3R)S87-5, no PCR products are obtained from exons 3–13, but products are obtained from exons 14–17. (C) RT–PCR products obtained from amplification of total RNA using the primers indicated, resolved on a 1.8% gel. In wild-type animals the higher band is derived from genomic DNA and the lower band from cDNA generated in the RT reaction. Note the lack of the lower band in the mutant genotype. The bands corresponding to genomic DNA amplification were verified by reactions in which the reverse transcription step was not performed (not shown). Size markers (M) are in increments of 100 bp starting with 600 bp at the top.
Figure 1.—
jar322/Df(3R)S87-5 adults do not express the myosin VI protein in any tissue. Western blot analysis of carcasses, ovaries, and testes in a (A) zygotic null myosin VI mutant, (B) maternal and zygotic null myosin VI mutant, and (C) zygotic null myosin VI mutant that expresses a myosin VI cDNA transgene. One-half of a whole fly, two ovaries, and two testes were loaded except for the mutant expressing the myosin VI transgene (one-quarter of an ovary).
Df(3R)S87-5 endpoint maps in the region of exon 13 of myosin VI:
Since we could not detect intact myosin VI protein in jar322/Df(3R)S87-5 adults and they were viable, we wondered whether a low level of expression of myosin VI earlier in development from sequences still present in Df(3R)S87-5 allowed these animals to bypass a larval lethal phase and account for their survival. We therefore wished to examine the myosin VI genomic region in the Df(3R) S87-5 chromosome to determine if a functional myosin VI protein was encoded. The myosin VI gene lies on the right arm of the third chromosome at cytological map position 95F6-95F8 (Figure 2A). Downstream (centromere proximal) of myosin VI lies a predicted gene, CG5706, which encodes a phenylalanine tRNA ligase. The jar322 mutation was generated by imprecise excision of the P element inserted in myosin VI's first intron in the male sterile allele jar1. In the jar322 mutation, both myosin VI (exons 3–17) and at least the first exon of CG5706 are absent (Petritsch et al. 2003). There are two deficiencies that uncover this region. Df(3R)S87-4 removes the region 95D11-96A2, thus uncovering both myosin VI and CG5706. Df(3R)S87-5 removes the region 95F7-96A18, but its exact endpoint was unknown. Eight primer sets were designed to span sequences across the entire myosin VI amino acid coding region to determine if any part of the myosin VI gene was retained in the Df(3R)S87-5 chromosome (Figure 2A). PCR amplification of genomic DNA using these primers showed that jar322/Df(3R)S87-5 animals only contain sequences corresponding to the globular tail (exons 14–17) of the myosin VI gene (Figure 2B). Molecularly, the left endpoint of Df(3R)S87-5 thus lies in the region of exon 13 of myosin VI.
jar322/Df(3R)S87-5 flies lack myosin VI transcripts:
It was possible that the globular tail sequences (exons 14–17) retained in jar322/Df(3R)S87-5 might encode a fragment that provided some myosin VI function and accounted for our ability to recover these flies (although this seemed unlikely). Neither of the antibodies (monoclonal 3C7 and polyclonal) that recognize myosin VI can detect the C-terminal globular tail fragment (M. Isaji, unpublished observations). To determine if a globular tail fragment was expressed, RT–PCR was performed using the primers that amplified this region. While we were able to detect transcripts for the CG5706 gene in jar322/Df(3R)S87-5 flies (data not shown), we were unable to detect transcripts corresponding to the myosin VI tail (Figure 2C). We conclude that no portion of myosin VI is expressed in jar322/Df(3R)S87-5 flies and, therefore, expression of a truncated myosin VI protein does not account for their viability.
Together, these data resolve three important issues. First, jar322/Df(3R)S87-5 adults completely lack myosin VI protein and are viable. Second, Df(3R)S87-5 removes most of the myosin VI coding region, through exon 13, and no myosin VI sequences are transcribed. Third, Df(3R)S87-5 does not remove the sequences encoding CG5706. These data indicate that a complete myosin VI loss of function is not lethal in D. melanogaster.
Maternally contributed myosin VI is not essential for viability:
It was possible that maternally contributed myosin VI enabled null animals to survive. To test this idea, we compared viability of animals with and without maternally contributed myosin VI. jar322/Df(3R)S87-5 adults with no maternal myosin VI eclosed at the same frequency as jar322/Df(3R)S87-5 adults that had maternally contributed myosin VI [18% (n = 415) and 16% (n = 443) of total progeny, respectively]. In addition, jar322/Df(3R)S87-5 females derived from embryos with or without maternal myosin VI were fertile. When compared to control genotypes, both types of mutant females produced progeny in numbers equal to nonmutant animals (data not shown). Males were sterile, as expected based on the jar1 phenotype. These adult flies expressed no intact myosin VI protein (Figure 1B), as expected. Thus, maternally provided myosin VI is not responsible for our ability to obtain jar322/Df(3R)S87-5 adults, further supporting the idea that complete myosin VI loss of function is not lethal.
jar322/Df(3R)S87-5 mutants display partially penetrant heterogeneous lethality:
Although we were able to obtain jar322/Df(3R)S87-5 flies, they were found at ∼40% of the expected Mendelian frequency (Table 2). No obvious abnormalities were observed in the null mutant adults that would suggest a developmental defect responsible for this partially penetrant lethality. The viability of these myosin VI null animals was completely rescued by basal expression (without heat shock) of the hsp83-myosin VI cDNA transgene (Table 2). To investigate the arrest point of myosin VI deficient animals, we used a GFP balancer to identify genotypes of progeny during development. jar322/Df(3R)S87-5 virgin myosin VI null females were crossed to jar322/TM3 Green males. If the number of non-GFP-expressing embryos [jar322/jar322, jar322/Df(3R)S87-5] are equivalent to the number of GFP-expressing embryos [jar322/TM3 Green, Df(3R)S87-5/TM3 Green], then no defect occurred during oogenesis due to lack of myosin VI in ovaries of adult females. Of 165 embryos, 37% expressed GFP (contained the balancer) and 63% did not express GFP (did not contain the balancer and thus were null for myosin VI). Since we see >50% myosin VI null embryos, developmental defects during oogenesis do not account for the 40% viability we observed. Recovery of only 37% GFP-expressing embryos (i.e., nonmutant), rather than the expected 50%, may be explained by the observation that animals carrying both the TM3 balancer and Df(3R)S87-5 appear somewhat unhealthy. In this cross, it is impossible to distinguish jar322 homozygotes and jar322/Df(3R)S87-5 transheterozygotes, so examining phenotypes during embryogenesis and early larval development specifically attributable to loss of myosin VI function was not possible. Since jar322 homozygotes die during the first to second instar larval stage (Petritsch et al. 2003; J. K. Morrison, unpublished observations), we were able to unambiguously identify non-GFP-expressing third instar larvae as jar322/Df(3R)S87-5 and compare their viability to GFP- expressing control animals (jar322/TM3 Green). Third instar jar322/Df(3R)S87-5 larvae eclosed in reduced numbers compared with controls [77% (n = 239) and 87% (n = 499), respectively]. Examination of jar322/Df(3R)S87-5 pupae revealed that some were normal appearing but did not eclose, some died trying to eclose, and some did not complete metamorphosis. Because myosin VI null animals are present at the expected frequency at egg laying and only 10% of the animals die during metamorphosis, null animals must die at other developmental stages to account for our inability to recover 60% of the expected number of progeny.
TABLE 2.
Viability of jar322/Df(3R)S87-5 adult flies
| Cross | na | jar322/Df(3R)S87-5 viability (%)b |
|---|---|---|
| Df(3R)S87-5/TM3 × jar322/TM3 | 443 | 37 |
| jar322/Df(3R)S87-5 × jar322/TM3 | 415 | 44 |
| P[C95]; jar322/Df(3R)S87-5 × P[C95]; jar322/TM3c | 398 | 105 |
Total number of progeny from three crosses.
Calculated as observed/expected × 100.
Expressing myosin VI cDNA transgene.
Conclusion:
Previous experiments suggested that in Drosophila, myosin VI loss of function was lethal. However, the studies reported here using genetic null animals demonstrate conclusively that myosin VI loss of function is not lethal in flies and that the lethality attributed previously to loss of myosin VI function in jar322 deletion must be caused by loss of the neighboring gene. Interestingly, our results are consistent with results in other species that loss of myosin VI function is not lethal. Despite the fact that animals completely lacking myosin VI can survive, they are not obtained at the expected frequency. This partially penetrant lethality is attributable to myosin VI loss of function, because we can rescue this defect by ubiquitous expression of a myosin VI cDNA transgene. The same transgene fails to rescue the lethality of the jar322 homozygotes and jar322/Df(3R)S87-4 transheterozygotes. Thus, the lethality observed in these animals is not associated with myosin VI loss of function.
In addition to observations reported here, an ENU mutagenesis screen for new myosin VI mutants in which we screened both for lethal and male sterile alleles lends additional support to the idea that myosin VI loss of function is not lethal. We did not obtain any myosin VI lethal mutations, even though this screen was successful in identifying three new male sterile myosin VI alleles (J. K. Morrison, unpublished observations).
Because of the phenotypes previously observed using other loss-of-function techniques and other myosin VI alleles, we were surprised that the null mutant animals were viable. The phenotypes attributed to myosin VI loss of function in other mutant alleles, including jar322 homozygous mutants, and defects generated by other loss of function techniques, such as antibody injection and antisense expression, must be reassessed in the jar322/Df(3R)S87-5 background. In neuroblasts, both expression of dominant negative fragments and lack of myosin VI (jar322 homozygotes) resulted in a partially penetrant defect in basal determinant localization (Petritsch et al. 2003), but perhaps such a defect is not severe enough to cause lethality. During early embryonic development, function-blocking antibody injection prevented metaphase pseudocleavage furrow formation (Mermall and Miller 1995), leading to massive defects in nuclear division. This severe phenotype would be expected to cause embryonic lethality. However, loss of myosin VI does not cause embryonic lethality. Perhaps the furrow defects caused by antibody injection can be explained by the sequestration of myosin VI-associated proteins, rather than as a direct effect of inhibiting myosin VI function. When myosin VI is absent (in null embryos), the myosin VI-associated proteins may still be able to function in furrow formation. Transgenic antisense expression caused severe defects in epithelial integrity and morphogenesis at a number of times in development and these animals were not viable (Deng et al. 1999; Millo et al. 2004). It is unclear why antisense expression causes more severe defects than the null mutation, but one explanation is off-target effects of the antisense. Possibly, impairment of epithelial integrity and defects in morphogenesis due to lack of myosin VI could contribute to the partially penetrant lethality we observe. However, the processes in question cannot be completely blocked by loss of myosin VI function, since 40% of the null animals survive and these survivors have no obvious defects. The significant differences in phenotype of this null mutation compared to those observed using other functional manipulations reinforces the importance of careful genetic analysis of loss-of-function alleles in understanding in vivo functions.
Acknowledgments
We thank current and former members of the Miller lab for their valuable advice and comments. Deborah Frank was especially helpful in discussions and in her commentary on the manuscript. We also thank Claudia Petritsch and the Bloomington Drosophila Stock Center for providing us with fly lines. This work was supported by National Institutes of Health grant GM-060494.
References
- Ahmed, Z., R. J. Morell, S. Riazuddin, A. Gropman, S. Shaukat et al., 2003. Mutations of MYO6 are associated with recessive deafness, DFNB37. Am. J. Hum. Genet. 72 1315–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameen, N., and G. Apodaca, 2007. Defective CFTR apical endocytosis and enterocyte brush border in myosin VI-deficient mice. Traffic 8 998–1006. [DOI] [PubMed] [Google Scholar]
- Au, J. S., C. Puri, G. Ihrke, J. Kendrick-Jones and F. Buss, 2007. Myosin VI is required for sorting of AP-1B-dependent cargo to the basolateral domain in polarized MDCK cells. J. Cell Biol. 177 103–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avraham, K. B., T. Hasson, K. P. Steel, D. M. Kingsley, L. B. Russell et al., 1995. The mouse Snell's waltzer deafness gene encodes an unconventional myosin required for structural integrity of inner ear cells. Nat. Genet. 11 369–375. [DOI] [PubMed] [Google Scholar]
- Buss, F., G. Spudich and J. Kendrick-Jones, 2004. Myosin VI: cellular functions and motor properties. Annu. Rev. Cell. Dev. Biol. 20 649–676. [DOI] [PubMed] [Google Scholar]
- Casso, D., F. Ramirez-Weber and T.B. Kornberg, 2000. GFP-tagged balancer chromosomes for Drosophila melanogaster. Mech. Dev. 91 451–454. [DOI] [PubMed] [Google Scholar]
- Dance, A. L., M. Miller, S. Seragaki, P. Aryal, B. White et al., 2004. Regulation of myosin-VI targeting to endocytic compartments. Traffic 5 798–813. [DOI] [PubMed] [Google Scholar]
- Deng, W., K. Leaper and M. Bownes, 1999. A targeted gene silencing technique shows that Drosophila myosin VI is required for egg chamber and imaginal disc morphogenesis. J. Cell Sci. 112 3677–3690. [DOI] [PubMed] [Google Scholar]
- Frank, D. J., T. Noguchi and K. G. Miller, 2004. Myosin VI: a structural role in actin organization important for protein and organelle localization and trafficking. Curr. Opin. Cell Biol. 16 189–194. [DOI] [PubMed] [Google Scholar]
- Geisbrecht, E. R., and D. J. Montell, 2002. Myosin VI is required for E-cadherin-mediated border cell migration. Nat. Cell Biol. 4 616–620. [DOI] [PubMed] [Google Scholar]
- Hicks, J. L., W. M. Deng, A. D. Rogat, K. G. Miller and M. Bownes, 1999. Class VI unconventional myosin is required for spermatogenesis in Drosophila. Mol. Biol. Cell 10 4341–4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappler, J. A., C. J. Starr, D. K. Chan, R. Kollmar, A. J. Kollmar et al., 2004. A nonsense mutation in the gene encoding a zebrafish myosin VI isoform causes defects in hair-cell mechanotransduction. Proc. Natl. Acad. Sci. USA 101 13056–13061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellerman, K. A., and K. G. Miller, 1992. An unconventional myosin heavy chain gene from Drosophila melanogaster. J. Cell Biol. 119 823–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddugoda, M. P., M. S. Crampton, A. M. Shewan and A. S. Yap, 2007. Myosin VI and vinculin cooperate during the morphogenesis of cadherin cell-cell contacts in mammalian epithelial cells. J. Cell Biol. 178 529–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermall, V., and K. G. Miller, 1995. The 95F unconventional myosin is required for proper organization of the Drosophila syncytial blastoderm. J. Cell Biol. 129 1575–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millo, H., K. Leaper, V. Lazou and M. Bownes, 2004. Myosin VI plays a role in cell-cell adhesion during epithelial morphogenesis. Mech. Dev. 121 1335–1351. [DOI] [PubMed] [Google Scholar]
- Morriswood, B., G. Ryzhakov, C. Puri, S. D. Arden, R. Roberts et al., 2007. T6BP and NDP52 are myosin VI binding partners with potential roles in cytokine signaling and cell adhesion. J. Cell Sci. 120 2574–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi, T., M. Lenartowska and K. G. Miller, 2006. Myosin VI stabilizes an actin network during Drosophila spermatid individualization. Mol. Biol. Cell 17 2559–2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petritsch, C., G. Tavosanis, C. W. Turck, L. Y. Jan and Y. N. Jan, 2003. The Drosophila myosin VI Jaguar is required for basal protein targeting and correct spindle orientation in mitotic neuroblasts. Dev. Cell 4 273–281. [DOI] [PubMed] [Google Scholar]
- Self, T., T. Sobe, N. G. Copeland, N. A. Jenkins, K. B. Avraham et al., 1999. Role of myosin VI in the differentiation of cochlear hair cells. Dev. Biol. 214 331–341. [DOI] [PubMed] [Google Scholar]
- Sweeney, H. L., and A. Houdusse, 2007. What can myosin VI do in cells? Curr. Opin. Cell Biol. 19 57–66. [DOI] [PubMed] [Google Scholar]