Abstract
An olfactory receptor (OR) multigene family is responsible for the well-developed sense of smell possessed by terrestrial tetrapods. Mammalian OR genes had diverged greatly in the terrestrial environment after the fish–tetrapod split, indicating their importance to land habitation. In this study, we analysed OR genes of marine tetrapods (minke whale Balaenoptera acutorostrata, dwarf sperm whale Kogia sima, Dall's porpoise Phocoenoides dalli, Steller's sea lion Eumetopias jubatus and loggerhead sea turtle Caretta caretta) and revealed that the pseudogene proportions of OR gene repertoires in whales were significantly higher than those in their terrestrial relative cattle and also in sea lion and sea turtle. On the other hand, the pseudogene proportion of OR sequences in sea lion was not significantly higher compared with that in their terrestrial relative (dog). It indicates that secondary perfectly adapted marine vertebrates (cetaceans) have lost large amount of their OR genes, whereas secondary-semi-adapted marine vertebrates (sea lions and sea turtles) still have maintained their OR genes, reflecting the importance of terrestrial environment for these animals.
Keywords: olfactory receptor, secondary-adapted marine vertebrates, Cetacea, sea lion, sea turtle, marine and terrestrial environment
1. Introduction
Terrestrial mammals can distinguish millions of different odours (Beets 1970). The molecular basis of this ability relies on the diversity of olfactory receptor (OR) genes. The intronless OR genes, the largest multigene (approx. 1000 genes) family in the terrestrial mammalian genome (Firestein 2001), belong to the superfamily of seven transmembrane G-protein-coupled receptors (Buck & Axel 1991).
Rouquier et al. (2000) found that the number of OR pseudogenes has increased in the primate lineage leading to humans, which is suggested to reflect a reduced dependence on olfaction by the acquisition of trichromatic colour vision (Gilad et al. 2004). These results suggest that the proportion of OR pseudogenes dependt1s on the importance of olfactory capabilities in the life history of the species (Ache & Young 2005).
OR repertoires in terrestrial tetrapods have been reported to differ greatly from those in marine fishes (Freitag et al. 1998; Mezler et al. 1999) and have diverged widely since the fish–tetrapod split (Niimura & Nei 2005). These findings suggest that animals living in marine and terrestrial environments require different types of OR genes: ORs of marine fishes have evolved to detect water-soluble molecules and that of terrestrial tetrapods have evolved to detect volatile compounds (Freitag et al. 1998; Ache & Young 2005).
In contrast to terrestrial tetrapods, only limited data of OR genes in marine tetrapods had ever been reported previously. In this study, we analysed OR genes of secondary-adapted marine vertebrates, including Cetacea, Pinnipedia and Cheloniidae, and compared the genomic differences with their terrestrial relatives to understand the importance of ORs possessed by terrestrial tetrapods in marine environment.
2. Material and methods
(a) Tissue preparation, DNA extraction, amplification, cloning and sequencing
Muscle tissue of adult minke whale Balaenoptera acutorostrata was purchased from a fish market in Japan. On 26 August 2004, a dead female dwarf sperm whale Kogia sima was stranded on the Bansho-zaki beach, Shirahama, Wakayama (Kishida et al. 2004) and the muscle tissue of the animal was collected. Muscle tissue of adult Dall's porpoise Phocoenoides dalli was kindly provided by Dr Azusa Hayano. Liver tissue of a male adult Steller's sea lion Eumetopias jubatus was kindly provided by Dr Kaoru Hattori. Muscle tissue of a male adult loggerhead sea turtle Caretta caretta was collected immediately after the death of an animal stranded on the Kitahama beach, Shirahama, Wakayama. Genomic DNA of each sample was extracted using the protocol modified from Blin & Stafford (1976).
Two pairs of OR-specific degenerate primers, OR5B–OR3B (Ben-Arie et al. 1994), and ORI-F (5′-CT(inosine)CAYSARCCCATGTWCYW(inosine) TTYCT-3′)—ORI-R (5′-TA(inosine)AYRATRGGRTTSAK(inosine)R(inosine)DGG(inosine)GG-3′) were used to amplify open reading frame (ORF) part of OR genes. These primers were employed in polymerase chain reaction (PCR) amplification under the following conditions: 94°C for 1 min, 43°C for 2 min, 72°C for 5 min (5 cycles) and 94°C for 30 s, 47°C for 1 min, 72°C for 2 min (30 cycles). PCR products were subcloned into the pGEM-T vector (Promega, WI, USA) and amplified with DH5α competent cells (Takara, Otsu, Japan). Sequencing reactions were performed in both directions using BigDye terminator v. 1.1 cycle sequencing kit (Applied Biosystems, CA, USA), and DNA sequences were determined on an ABI310 automated sequencer (Applied Biosystems).
(b) Collecting cattle and dog OR sequences
Whole genome sequence data of an even-toed ungulate cattle Bos taurus, which is closely related to cetaceans (Graur & Higgins 1994), were downloaded from Ensembl FTP server (ftp://ftp.ensembl.org/pub/) on 27 February 2006. Each cetacean OR sequences cloned in this study was searched against the cattle genomes using the FastA3 program (nucleotide–nucleotide, Pearson & Lipman 1988) to obtain the most similar, potentially orthologous cattle OR sequence. As a result, we obtained as many cattle OR sequences as cetacean ORs we cloned, containing several sets of sequences coding the same OR genes. Finally, we merged the sequences coding same OR genes.
An almost complete OR repertoire of a terrestrial carnivore, dog Canis canis has been reported previously (Olender et al. 2004) and we quote their data.
(c) Genetic analysis
An OR sequence was classified as a pseudogene when termination codons and/or frame shifts were observed in its ORF. Based on the distance matrix calculated using the FastA3 program, a phylogenetic tree of cattle–Cetacea OR sequences was inferred by the neighbour-joining method (Saitou & Nei 1987), and two lamprey OR sequences LFor1 and LFor2 (GenBank accession nos. AJ012708 and AJ012709, respectively) were used as outgroups.
3. Results and discussion
Table 1 shows the number of OR sequences obtained in this study. The estimated proportions of pseudogenes were 77% in dwarf sperm whale, 78% in Dall's porpoise, 58% in minke whale, 37% in Steller's sea lion and 31% in loggerhead sea turtle, although these proportions may be underestimated because these sequences are partial. Twenty-nine cattle OR sequences were obtained by the procedure mentioned above (§2b).
Table 1.
Pseudogene proportions in the OR gene repertoire.
| order/species (abbreviation) | no. of pseudogenes | no. of sequences analysed | pseudogene proportion (%) |
|---|---|---|---|
| Cetacea | |||
| dwarf sperm whale (KS) | 10 | 13 | 77 |
| Dall's porpoise (PD) | 7 | 9 | 78 |
| minke whale (BA) | 11 | 19 | 58 |
| Artiodactyla | |||
| cattle (BT) | 5 | 29 | 17 |
| marine carnivora | |||
| Steller's sea lion | 10 | 27 | 37 |
| terrestrial carnivora | |||
| dog | 258 | 971 | 27 |
| testudines | |||
| loggerhead sea turtle | 4 | 13 | 31 |
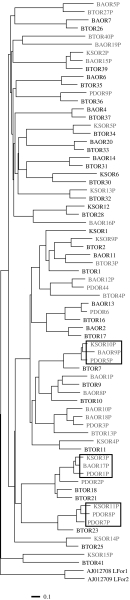
Pseudogene proportions of OR sequences were analysed statistically using Fisher's exact test for the significant differences between cattle–cetaceans, sea lion–dog, cetaceans–sea lion and cetaceans–sea turtle (table 2). We found that the pseudogene proportion of OR sequences in marine cetaceans, might be underestimated, but was significantly higher than that of their most similar, potentially orthologous cattle OR sequences (p<0.01), indicating that a large-scale pseudogenization of OR genes had occurred in cetacean lineage after cattle–Cetacea split. Three sets of cetacean OR sequences were observed to have evolved into pseudogenes at the same positions, while no cattle OR sequences were observed to have evolved into pseudogenes at the same positions as cetacean OR sequences (figure 1). It indicates that a number of cetacean ORs had evolved into pseudogenes between cattle–Cetacea split and mysticetes–odontocetes split, when cetaceans had been beginning to adapt to the marine habitat. In turn, in sea lion, the pseudogene proportion of OR sequences was not significantly higher compared with that in their terrestrial relatives. Furthermore, the pseudogene proportions of sea lion and sea turtle ORs were significantly (p<0.05) lower than those of marine cetaceans.
Table 2.
Comparison of the proportion of OR pseudogenes in terrestrial and aquatic tetrapods. (Note. *p-value is calculated by Fisher's exact test.)
| intact genes | pseudogenes | pseudogene proportion (%) | p-value* | |
|---|---|---|---|---|
| cattle | 24 | 5 | 17.2 | |
| cetaceans | 13 | 28 | 68.3 | >0.01 |
| dog | 713 | 258 | 26.6 | |
| sea lion | 17 | 10 | 37.0 | 0.27 |
| cetaceans | 13 | 28 | 68.3 | |
| sea lion | 17 | 10 | 37.0 | >0.05 |
| cetaceans | 13 | 28 | 68.3 | |
| sea turtle | 9 | 4 | 30.8 | >0.05 |
Figure 1.
A phylogenetic tree of cattle and cetacean OR genes identified in this study. The tree was inferred by the NJ method, based on the distance matrix calculated using FastA3 program and two lamprey ORs were used as outgroups. Black text indicates intact genes; grey text indicates pseudogenes. The first two letters of each gene indicate the species, i.e. BT, B. taurus (cattle); BA, B. acutorostrata (minke whale); KS, K. sima (dwarf sperm whale); PD, P. dalli (Dall's porpoise). The boxed sets of genes have the same mutations which turned these genes into pseudogenes.
These results suggest that cetaceans, which do not rely on terrestrial environment at all in their life history, lost large number of OR genes when they adapted completely to the marine habitat. On the other hand, sea lion and sea turtle seem to have maintained their OR repertoires and it could be due to their reliance on land habitat for breeding and/or other several important roles. Our results coincide with the anatomical and ecological facts that the olfactory bulbs and nerves, which function in terrestrial mammals for sensing airborne odours, are reduced or absent in whales (Breathnach 1960) and that the sense of olfaction has been either lost or greatly reduced among modern cetaceans (Tyack 2000). We could conclude that most of the OR genes which have diverged widely in terrestrial environment have little function in the marine environment, and that the pressure of natural selection to maintain OR genes relaxed as the cetaceans entered and adapted completely in the marine habitat, supporting the idea that ORs in terrestrial tetrapods have evolved to detect airborne chemical substances. In contrast to olfaction, there have been some suggestions of use of pheromones among cetaceans (Norris 1991; Tyack 2000) in spite of the fact that not only the OR gene repertoires, but also the vomeronasal receptor gene repertoires in terrestrial tetrapods differ greatly from that in marine fish (Shi & Zhang 2007). Further extended studies, including vomeronasal receptors and taste receptors, will lead us to an understanding of the molecular basis of underwater chemical recognition among tetrapods.
The nucleotide sequence data cloned in this study are available in the DDBJ/EMBL/GenBank databases under the following accession numbers: AB301617–AB301697.
Acknowledgments
We thank Dr Azusa Hayano, Kyoto University for providing the muscle tissue of Dall's porpoise; Dr Kaoru Hattori, Hokkaido National Fisheries Research Institute for providing the liver tissue of Steller's sea lion; Dr P. Robin Rigby, Kyoto University for checking our English text; all colleagues in Seto Marine Biological Laboratory, Kyoto University for their kindly help.
Supplementary Material
Twenty-nine cattle olfactory receptor genes analyzed in this study
References
- Ache B.W, Young J.M. Olfaction: diverse species, conserved principles. Neuron. 2005;48:417–430. doi: 10.1016/j.neuron.2005.10.022. doi:10.1016/j.neuron.2005.10.022 [DOI] [PubMed] [Google Scholar]
- Beets M. The molecular parameters of olfactory response. Pharm. Rev. 1970;22:23–34. [PubMed] [Google Scholar]
- Ben-Arie N, et al. Olfactory receptor gene cluster on human chromosome 17: possible duplication of an ancestral receptor repertoire. Hum. Mol. Genet. 1994;3:229–235. doi: 10.1093/hmg/3.2.229. doi:10.1093/hmg/3.2.229 [DOI] [PubMed] [Google Scholar]
- Blin N, Stafford D.W. A general method for isolation of high molecular weight DNA from eukaryotes. Nucleic Acids Res. 1976;3:2303–2308. doi: 10.1093/nar/3.9.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breathnach A.S. The cetacean central nervous system. Biol. Rev. 1960;35:187–230. [Google Scholar]
- Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. doi:10.1016/0092-8674(91)90418-X [DOI] [PubMed] [Google Scholar]
- Firestein S. How the olfactory system makes sense of scents. Nature. 2001;413:211–218. doi: 10.1038/35093026. doi:10.1038/35093026 [DOI] [PubMed] [Google Scholar]
- Freitag J, Ludwig G, Andrini I, Rossler P, Breer H. Olfactory receptors in aquatic and terrestrial vertebrates. J. Comp. Physiol. A. 1998;183:635–650. doi: 10.1007/s003590050287. doi:10.1007/s003590050287 [DOI] [PubMed] [Google Scholar]
- Gilad Y, Wiebe V, Przeworski M, Lancet D, Paabo S. Loss of olfactory receptor genes coincides with the acquisition of full trichromatic vision in primates. PLoS Biol. 2004;2:120–125. doi: 10.1371/journal.pbio.0020005. doi:10.1371/journal.pbio.0020005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graur D, Higgins D.G. Molecular evidence for the inclusion of cetaceans within the order Artiodactyla. Mol. Biol. Evol. 1994;11:357–364. doi: 10.1093/oxfordjournals.molbev.a040118. [DOI] [PubMed] [Google Scholar]
- Kishida T, Kubota S, Kobayashi A, Tanase T. A kogiid whale stranded on an open beach of Bansho-zaki, Shirahama, Wakayama, Japan. J. Jpn Driftol. Soc. 2004;2:33–34. [Google Scholar]
- Mezler M, Konzelmann S, Freitag J, Rossler P, Breer H. Expression of olfactory receptors during development in Xenopus laevis. J. Exp. Biol. 1999;202:365–376. doi: 10.1242/jeb.202.4.365. [DOI] [PubMed] [Google Scholar]
- Niimura Y, Nei M. Evolutionary dynamics of olfactory receptor genes in fishes and tetrapods. Proc. Natl Acad. Sci. USA. 2005;102:6039–6044. doi: 10.1073/pnas.0501922102. doi:10.1073/pnas.0501922102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris K.S. W. W. Norton; New York, NY: 1991. Dolphin days: the life and times of the spinner dolphin. [Google Scholar]
- Olender T, Fuchs T, Linhart C, Shamir R, Adams M, Kalush F, Khen M, Lancet D. The canin olfactory subgenome. Genomics. 2004;83:361–372. doi: 10.1016/j.ygeno.2003.08.009. doi:10.1016/j.ygeno.2003.08.009 [DOI] [PubMed] [Google Scholar]
- Pearson W.R, Lipman D.J. Improved tools for biological sequence comparison. Proc. Natl Acad. Sci. USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. doi:10.1073/pnas.85.8.2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouquier S, Blancher A, Giorgi D. The olfactory receptor gene repertoire in primates and mouse: evidence for reduction of the functional fraction in primates. Proc. Natl Acad. Sci. USA. 2000;97:2870–2874. doi: 10.1073/pnas.040580197. doi:10.1073/pnas.040580197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Shi P, Zhang J. Comparative genomic analysis identifies an evolutionary shift of vomeronasal receptor gene repertoires in the vertebrate transition from water to land. Genome Res. 2007;17:166–174. doi: 10.1101/gr.6040007. doi:10.1101/gr.6040007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyack P.L. Functional aspects of cetacean communication. In: Mann J, Conner R.C, Tyack P.L, Whitehead H, editors. Cetacean societies: field studies of dolphins and whales. The University of Chicago Press; Chicago, IL: 2000. pp. 270–307. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Twenty-nine cattle olfactory receptor genes analyzed in this study