Abstract
We investigated the function of chlorophyll a/b binding antenna proteins Chlorophyll Protein 26 (CP26) and CP24 in light harvesting and regulation of photosynthesis by isolating Arabidopsis thaliana knockout lines that completely lacked one or both of these proteins. All three mutant lines had a decreased efficiency of energy transfer from trimeric light-harvesting complex II (LHCII) to the reaction center of photosystem II (PSII) due to the physical disconnection of LHCII from PSII and formation of PSII reaction center depleted domains in grana partitions. Photosynthesis was affected in plants lacking CP24 but not in plants lacking CP26: the former mutant had decreased electron transport rates, a lower ΔpH gradient across the grana membranes, reduced capacity for nonphotochemical quenching, and limited growth. Furthermore, the PSII particles of these plants were organized in unusual two-dimensional arrays in the grana membranes. Surprisingly, overall electron transport, nonphotochemical quenching, and growth of the double mutant were restored to wild type. Fluorescence induction kinetics and electron transport measurements at selected steps of the photosynthetic chain suggested that limitation in electron transport was due to restricted electron transport between QA and QB, which retards plastoquinone diffusion. We conclude that CP24 absence alters PSII organization and consequently limits plastoquinone diffusion.
INTRODUCTION
In plants, photosynthetic reaction centers (RCs) exploit solar energy to drive electrons from water to NADP+. This transport is coupled to H+ transfer from the chloroplast stroma to the thylakoid lumen, which builds a proton gradient for ATP synthesis. The capacity of light absorption is increased by the pigment binding proteins composing the antenna system. In higher plants, the antenna system surrounding the plastid-encoded photosystem II (PSII) core is composed of the nuclear-encoded chlorophyll a/b binding light-harvesting complexes (Lhc). LHCII is the major component of the outer antenna and comprises different heterotrimers of LHCB1, LHCB2, and LHCB3 gene products, while minor antenna complexes (Chlorophyll Protein 29 [CP29], CP26, and CP24) are encoded by LHCB4, LHCB5, and LHCB6 genes, respectively, and are found as monomers (Bassi et al., 1996; Jansson, 1999). Structural analysis of PSII and Lhcb supercomplex organization within grana membranes has revealed that minor complexes CP26 and CP29 are located in between the core complex and the trimeric LHCII (Harrer et al., 1998; Boekema et al., 1999). Additional LHCII trimers, depending on growth light intensity (Dekker and Boekema, 2005; Morosinotto et al., 2006; Ballottari et al., 2007), complete the PSII structure and require CP24 for connection to PSII core by forming a complex with CP29 (Bassi and Dainese, 1992; Yakushevska et al., 2003; Dekker and Boekema, 2005). Similarly, PSI has four Lhca antenna proteins, yielding a total of 10 distinct Lhc isoforms in higher plants (Jansson, 1999). These gene products have been conserved during at least 350 million years of evolution, strongly indicating that each pigment-protein complex has a specific function in the highly variable conditions of the natural subaerial environment (Durnford, 2003; Ganeteg et al., 2004).
Rapid changes in light intensity, temperature, and water availability easily lead to overexcitation of photosystems when the absorbed light exceeds the capacity to use reducing equivalents. Incomplete photochemical quenching leads to an increased chlorophyll excited state (1Chl*) lifetime and increased probability of chlorophyll a triplet formation (3Chl*) by intersystem crossing. Chlorophyll triplets react with oxygen (3O2) and form harmful reactive oxygen species responsible for photoinhibition and oxidative stress (Barber and Andersson, 1992). These harmful events are counteracted by photoprotection mechanisms that either scavenge the reactive oxygen species produced (Asada, 1999) or prevent their production through deexcitation of excessive 1Chl* (Niyogi, 2000). This latter process is known as nonphotochemical quenching (NPQ) since it is observed as light-dependent quenching of Chl fluorescence. The largest NPQ component is rapidly reversible and dependent on the formation of a low thylakoid lumen pH and is thus defined as energy quenching (qE; Briantais et al., 1980; Niyogi, 1999).
The qE developing within the first minute after overexcitation is largely zeaxanthin (Zea) independent and is followed by a slower component that depends on Zea synthesis (Horton et al., 1996), which is promoted by acidic pH in the lumen through the activation of violaxanthin deepoxidase. Zea is rapidly produced under conditions of high light intensity and is bound to Lhc proteins, mainly to CP24 and CP26 (Morosinotto et al., 2002), where it displaces violaxanthin (Viola) and induces conformational changes that result in a quenched state (Crimi et al., 2001). This event is coupled to the rise of a slower component of the NPQ process and to a sustained dissipation of light energy known as qI (Dall'Osto et al., 2005). Besides violaxanthin deepoxidase activation, low lumenal pH exerts control over the thylakoid membrane by reversibly protonating exposed acidic residues, as suggested by the inhibition of NPQ by dicyclohexylcarbodiimide (DCCD), a reagent that modifies acidic residues that undergo reversible protonation (Ruban et al., 1992). Whereas 14C DCCD binding antenna proteins CP26 and CP29 are located between the inner antenna and the LHCII (Walters et al., 1996, Pesaresi et al., 1997), the site of DCCD inhibition of qE is located in PsbS (Li et al., 2004), an Lhc-like protein (Li et al., 2000) that likely does not bind pigments (Sundaresan et al., 1995; Dominici et al., 2002) but exerts its function by interacting with Lhc proteins (Bonente et al., 2007; Teardo et al., 2007).
Consistently, antenna proteins are needed for full expression of NPQ (Briantais, 1994). Functional dissection of individual Lhc isoforms has been undertaken using antisense and knockout approaches. Antisense inhibition of CP29, CP26 (Andersson et al., 2001), and Lhcb1+Lhcb2 (Andersson et al., 2003) expression did not disrupt NPQ, while deletion of the Lhcb6 gene encoding CP24 did reduce this function (Kovacs et al., 2006). We have isolated and characterized CP24 and CP26 knockout (koCP24 and koCP26) plants and confirmed the phenotype described in previous work for koCP24. Surprisingly, the limitation in NPQ and growth rate of koCP24 was reversed in the double koCP24/26 mutant, suggesting that the koCP24 phenotype was not due to specific properties of CP24 but rather to an effect on the organization of photosynthetic complexes within grana partitions, which affected electron transport rate (ETR) and proton pumping into the thylakoid lumen.
RESULTS
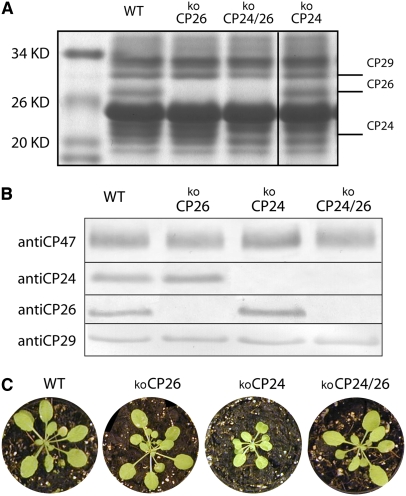
We identified kolhcb6 and kolhcb5 homozygous lines in seed pools obtained from the Nottingham Arabidopsis Stock Centre (NASC) by immunoblot analysis using specific antibodies raised against CP26 and CP24 antenna proteins (Di Paolo et al., 1990). Similarly, the kolhcb5 kolhcb6 double mutant was obtained by selection of the progeny of single mutant crossings. Thylakoid membranes from kolhcb5, kolhcb6, and kolhcb5 kolhcb6 were depleted in the corresponding gene products (Figures 1A and 1B). We will henceforth refer to these genotypes as koCP26 (kolhcb5), koCP24 (kolhcb6), and koCP24/26 (kolhcb5 kolhcb6). Single knockouts did not differ in chlorophyll content per leaf area compared with wild-type plants in regular lighting, but koCP24/26 showed a small decrease in chlorophyll content (Table 1). Pigment composition was similar in all dark-adapted plants. Nevertheless, when plants were exposed to high light intensity for 30 min to induce Zea synthesis, deepoxidation was significantly lower in koCP24 than in wild-type, koCP26, and koCP24/26 plants (Table 1). When grown in control conditions (100 μmol photons m−2 s−1, 24°C, 8/16 day/night) for 3 weeks, koCP26 plants did not show significant reduction in growth with respect to the wild type, while koCP24 plants were much smaller than wild-type plants (Figure 1C). Surprisingly, koCP24/26 plants were less affected in their growth than koCP24 plants and appeared more similar to the wild type.
Figure 1.
Polypeptide Composition of Thylakoid Membranes from Wild-Type and Knockout Mutants.
(A) SDS/PAGE analysis of wild-type and mutants thylakoid proteins. Selected apoprotein bands are marked. Fifteen micrograms of chlorophylls were loaded in each lane.
(B) Immunoblot analysis of thylakoid membranes with antibodies directed against minor antenna proteins CP29, CP26, and CP24 and against the PSII core subunit CP47.
(C) Phenotype of wild-type and mutant plants grown in control conditions for 3 weeks (100 μmol photons m−2 s−1, 25°C, 8/16 h day/night).
Table 1.
Photosynthetic Pigment Content of the Wild Type and Mutants
| Wild Type | koCP24 | koCP26 | koCP24/26 | ||
|---|---|---|---|---|---|
| Chlorophyll (mg cm−2) | 16.8 ± 1.0 | 16.4 ± 0.6 | 15.9 ± 1.7 | 14.3 ± 0.8* | |
| Chlorophyll a/b | 3.04 ± 0.06 | 3.20 ± 0.10 | 3.06 ± 0.10 | 2.96 ± 0.09 | |
| Chlorophyll/carotenoid | 3.29 ± 0.04 | 3.23 ± 0.05 | 3.19 ± 0.13 | 3.18 ± 0.16 | |
| Dark-Adapted Leaves | Neo | 4.7 ± 0.2 | 4.9 ± 0.1 | 4.8 ± 0.1 | 5.1 ± 0.2 |
| Viola | 3.9 ± 0.9 | 4.1 ± 0.2 | 3.6 ± 0.1 | 4.0 ± 0.1 | |
| Anthera | – | – | – | – | |
| Lute | 14.5 ± 0.5 | 14.4 ± 0.3 | 15.4 ± 0.4 | 15.7 ± 1.0 | |
| β-Carotenoid | 7.1 ± 0.1 | 7.3 ± 0.1 | 7.2 ± 0.1 | 6.3 ± 0.5 | |
| Light-Treated Leaves | Viola | 1.4 ± 0.1 | 2.1 ± 0.5 | 1.3 ± 0.1 | 1.4 ± 0.1 |
| Anthera | 1.0 ± 0.1 | 1.3 ± 0.1 | 0.8 ± 0.1 | 1.2 ± 0.1 | |
| Zea | 1.2 ± 0.1 | 0.7 ± 0.2* | 1.3 ± 0.1 | 0.9 ± 0.1 | |
| (Z + 0.5A)/(V + A + Z) | 0.47 ± 0.04 | 0.32 ± 0.08* | 0.51 ± 0.06 | 0.42 ± 0.05 |
Pigment content is expressed as mol/100 mol chlorophylls. Viola, antheraxanthin, and Zea content was determined after leaves were illuminated for 30 min at 1000 μmol m−2 s−1. The (Z+1/2A)/(Z+A+V) ratio quantifies the operation of the xanthophyll cycle. Data are mean values of four experiments. Significantly different values with respect to the wild type are marked with an asterisk (P > 0.05).
Chloroplast Organization
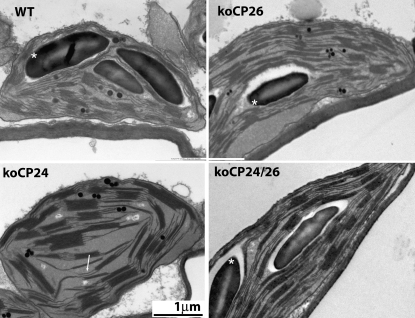
Chloroplast structure was analyzed by transmission electron microscopy on leaf samples harvested at the middle of the light period (Figure 2). Under these growth conditions, wild-type chloroplasts showed a characteristic organization of stroma membranes, interconnecting grana stacks, and large starch granules in most sections. koCP24 plants differed in that a large number of their stroma membranes had blunt ends not engaged in grana stacks and they completely lacked starch granules. koCP26 chloroplasts, on the other hand, had starch granules and a thylakoid organization similar to the wild type. Chloroplasts from the double mutant koCP24/26 accumulated starch granules normally but had a higher ratio of stroma membranes to grana stacks than wild-type chloroplasts and their grana membranes had fewer partitions.
Figure 2.
Transmission Electron Micrographs of Plastids from Mesophyll Cells of the Wild Type and Mutants.
Leaf samples were harvested at the midpoint of the light period from plants grown in short-day conditions (100 μmol photons m−2 s−1, 25°C, 8/16 h day/night). Starch granules (marked with asterisks) can be distinguished from plastoglobules (small black dots). Stroma membranes with blunt ends not engaged in grana stacks in koCP24 chloroplasts are indicated by an arrow.
Organization and Stoichiometry of Chlorophyll Proteins
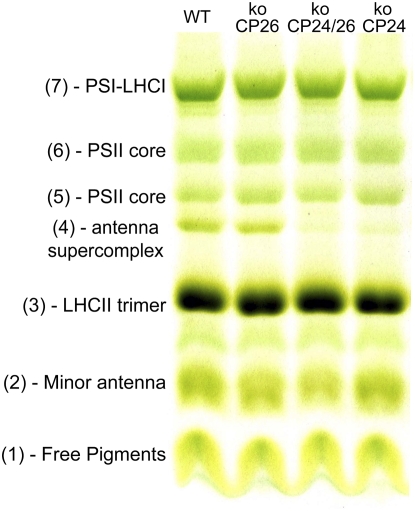
The organization of pigment-protein complexes was analyzed by nondenaturing Deriphat-PAGE. In agreement with a previous report (Havaux et al., 2004), seven major green bands were resolved upon solubilization of thylakoid membranes with 0.8% dodecyl-α-d-maltoside (α-DM) (Figure 3). The uppermost band (band 7) contained the supramolecular PSI-LHCI complex. PSII-LHCII dissociated into its components, namely, the PSII core dimer and monomer (bands 6 and 5, respectively) and antenna moieties, including the CP29-CP24-(LHCII)3 supercomplex (band 4; Bassi and Dainese, 1992), LHCII trimer (band 3), and monomeric Lhcbs (band 2). Band 1 was composed of free pigments that dissociated during solubilization. A faint band was also detected between bands 2 and 3, containing the PSII core subunit CP43. Chlorophyll distribution between pigment proteins was not strongly modified in mutant thylakoids compared with the wild type. The major difference was that koCP24 and koCP24/26 had reduced levels of band 4. Also, the relative amount of band 2 was affected, being lowest in the double mutant and highest in the wild type. The ratio between monomeric and dimeric PSII core complexes was constant in all plants tested. Densitometric analysis of the green gels allowed evaluation of the total chlorophyll associated with PSI-LHCI (band 7) versus PSII + Lhcb components (bands 2 to 6). koCP24 showed a slightly higher PSII-core/Lhcb ratio (0.26) and a lower PSI-LHCI/PSII-Lhcb ratio (koCP24 = 1.26) than the wild type (0.22 and 1.84, respectively). koCP26 did not show significant differences with respect to the wild type, while the koCP24/26 had a higher PSII/Lhcb ratio (0.28).
Figure 3.
Analysis of Pigment-Protein Complexes of the Wild Type and Mutant.
Thylakoid pigmented complexes were separated by nondenaturing Deriphat-PAGE.
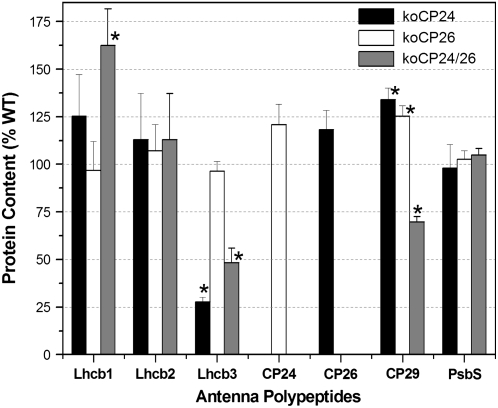
We then verified alterations in Lhc stoichiometry in thylakoids extracted from the different mutants (Figure 4) by quantitative immunoblot analysis using CP47 as an internal standard (Ballottari et al., 2007). In koCP24, besides the complete lack of CP24, the LHCII component Lhcb3 was also strongly decreased, while CP29 and CP26 were increased with respect to the wild type. We also detected a very small increase in Lhcb1 and Lhcb2 but below statistical significance. In koCP26, the only clear difference was the increase in CP29 and in CP24 content, while the remaining Lhcb proteins did not change within the error of the determination. The double mutant koCP24/26 showed a strong decrease in Lhcb3 and, to a lesser extent, of CP29 while Lhcb1 was increased by 60%. Lhcb2 was also increased, although to a lower extent. Remarkably, PsbS subunit content did not show significant differences between all genotypes (Figure 4).
Figure 4.
Immunological Quantification of Lhc Proteins in Thylakoid Membranes of Wild-Type and Mutant Plants.
Lhc proteins of purified thylakoid membranes were immunodetected with specific antibodies. The mean optical density of bands developed in four lanes (loaded with 1.0, 0.75, 0.50, and 0.25 μg of chlorophyll, respectively) was plotted against amount of chlorophyll loaded to assess the linearity of response and compared with the optical density of reaction bands of an antibody directed to the PSII core subunit CP47. Total amount of each subunit is expressed as a percentage of the corresponding wild-type content. Data are expressed as means ± sd (n = 4). Significantly different values from wild-type membranes are marked with an asterisk (according to Student's t test, P < 0.05).
Photosynthetic Functions: NPQ of Chlorophyll Fluorescence
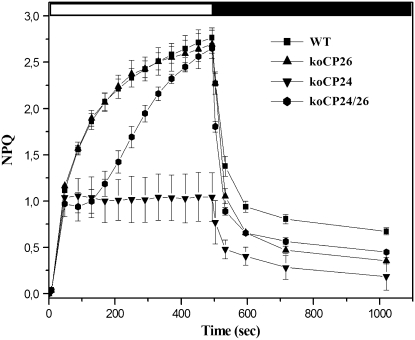
Since antenna polypeptides have been implicated in energy dissipation (Walters et al., 1996), we evaluated the capacity of different mutants to activate the NPQ (Figure 5). Wild-type and koCP26 plants grown in control conditions had a NPQ of 2.7 after 8 min of illumination at 1200 μmol photons m−2 s−1, consistent with literature data (Niyogi, 1999). In the same conditions, the NPQ of koCP24 was clearly different: similar to the wild type, it showed a rapid rise to a value of 1.0 in the first minute of illumination at 1200 μmol photons m−2 s−1 but then reached a plateau that lasted for the remaining 7 min of illumination. The double koCP24/26 mutant also showed a fast rise to a value of 1.0; however, a delay of 1.5 to 2 min was then evident before resuming rise and reaching, after 8 min of illumination, an NPQ value similar to the wild type. The dark recovery of fluorescence was clearly different, with the wild type retaining a quenching level (qI) of 0.65, while koCP26 and koCP24/26 further released quenching to 0.4 and koCP24 to 0.18. Thus, koCP24/26 showed the most complete relaxation of quenching.
Figure 5.
NPQ Analysis of Wild-Type and Mutant Genotypes.
Kinetics of NPQ induction and relaxation were recorded with a pulse-amplitude modulated fluorometer. Chlorophyll fluorescence was measured in intact, dark-adapted leaves, during 8 min of illumination at 1260 μmol m−2 s−1 followed by 9 min of dark relaxation. All NPQ values of mutant plants after 530 s (dark recovery) are significantly lower than the corresponding wild-type values (means ± sd, n = 4, Student's t test, P < 0.05).
Determination of Light-Induced Proton Gradient by 9-Aminoacridine Quenching
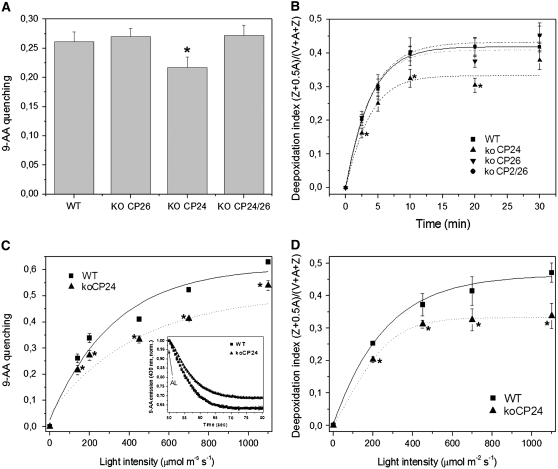
NPQ amplitude has been reported to be dependent on the concentration of PsbS (Li et al., 2002a) and on the lumenal pH (Horton et al., 1996). Since PsbS content was the same in all genotypes analyzed (Figure 4), we determined the capacity of intact chloroplasts to produce changes in thylakoid pH by following the light-induced quenching of 9-aminoacridine (9-AA) in the presence of methylviologen as the final electron acceptor (Johnson et al., 1994). The only genotype significantly affected in proton pumping into the chloroplast lumen in these conditions was koCP24 (Figure 6A), while the others performed similar to the wild type. Differences between the wild type and koCP24 were confirmed over a wide range of light intensities (Figure 6C). This is consistent with the hypothesis that the limitation in NPQ described above for koCP24 is (at least in part) associated with a reduced acidification of the lumen upon illumination.
Figure 6.
Measurement of Trans-Thylakoid ΔpH.
(A) The light-dependent quenching of 9-AA fluorescence in intact chloroplasts was quantified as a measure for trans-thylakoid ΔpH.
(B) Time course of violaxanthin deepoxidation in wild-type and mutant plants. Leaf discs from dark-adapted leaves were illuminated at 450 μmol m−2 s−1 (white actinic light). At different times, discs were frozen in liquid nitrogen and total pigment extracted.
(C) Amplitude of light-dependent quenching of 9-AA fluorescence measured at different light intensities on wild-type and koCP24 intact chloroplast. Inset: traces of 9-AA fluorescence emission (430 nm) during ΔpH buildup (induced by red actinic light, 450 μmol m−2 s−1) shows a slower lumen acidification in mutant chloroplasts. AL, red actinic light turned on.
(D) Amplitude of violaxanthin deepoxidation was measured on leaf discs from the wild type and koCP24 after illumination (10 min) at different light intensities. All data are expressed as mean ± sd (n = 4). Significantly different values according to Student's t test (P < 0.05) are marked with an asterisk.
Viola deepoxidation is also dependent on low lumenal pH, and it has been reported to be slower in the pgr1 mutant, which has a reduced proton gradient (Munekage et al., 2001). Deepoxidation rate was slower in koCP24 than in either wild-type or koCP24/26 plants (Figures 6B and 6D), thus providing an independent confirmation that pH generation is affected in the CP24-less genotype. Measurement of NPQ in the isolated chloroplast preparation used for 9-AA quenching yielded similar qE amplitudes in the wild type, koCP26, and koCP24/26 but was two times lower in koCP24, implying that the relation between NPQ and ΔpH was conserved in the conditions used for transmembrane gradient determination (see Supplemental Table 1 online).
ETR
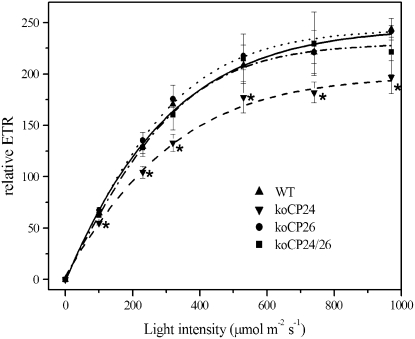
Differences in transmembrane gradient could be due to changes in electron transport (ET) capacity. To test this hypothesis, ET rate was evaluated in vivo on plants grown in control light conditions by fluorescence analysis at different light intensities under saturating CO2 conditions (1%) (Figure 7). In the wild type, the light-dependent increase in ET approached saturation at 650 μmol photons m−2 s−1, and after this value no further increase was observed. koCP24 showed significantly lower rates of ETR, also at very low light intensities. By contrast, koCP24/26 and koCP26 plants showed ETR behavior not significantly different with respect to the wild type.
Figure 7.
ETR Measurements.
Relative ETR as a function of quantum flux density of PAR was measured fluorometrically in light-adapted leaves under saturating CO2 (1%). Data represent an average of five to eight independent measurements and are expressed as mean ± sd. Significantly different values from the wild type (P < 0.05, Student's t test) are marked with an asterisk.
Fluorescence Transient Analysis
To determine if the mutations affected the capacity of the antenna system to transfer absorbed energy to reaction centers, we measured the functional antenna size of PSII by estimating the rise time of florescence in the presence of 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU). No significant differences were observed between the different genotypes considered in this study (Table 2), suggesting that the light-harvesting capacity is not affected despite the depletion in some antenna subunits.
Table 2.
Analysis of Room Temperature Chlorophyll Fluorescence
| Wild Type | koCP24 | koCP26 | koCP24/CP26 | |
|---|---|---|---|---|
| F0 | 451 ± 40 | 646 ± 64* | 508 ± 47 | 695 ± 53* |
| Fv/Fm | 0.83 ± 0.02 | 0.71 ± 0.03* | 0.80 ± 0.01 | 0.73 ± 0.03* |
| tm (ms) | 213.1 ± 25.8 | 1042.3 ± 113.7* | 192.0 ± 24.9 | 218.5 ± 30.7 |
| Sm/tm | 0.103 ± 0.008 | 0.0328 ± 0.0023* | 0.114 ± 0.015 | 0.112 ± 0.013 |
| T2/3 (ms) | 388 ± 68 | 423 ± 71* | 371 ± 42 | 356 ± 43 |
Photosynthetic parameters were provided by analysis of chlorophyll fluorescence measured with green light (7 μmol m−2 s−1 or 1100 μmol m−2 s−1; see Methods for details) on leaves of the wild type and mutants. The two-thirds time of the fluorescence rise (T2/3) was measured in 3.0 10−5 M DCMU infiltrated leaves using a flash of green light (7 μmol m−2 s−1, 8 s). The T2/3 parameter is inversely related to the incident photon flux and is an index of the functional antenna size of PSII. Significantly different values with respect to the wild type are marked with an asterisk (P > 0.05).
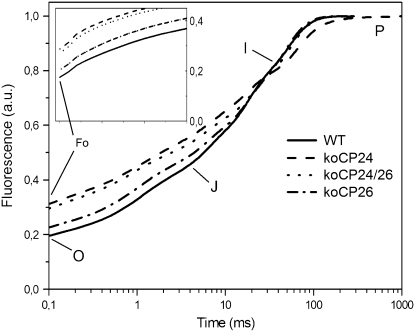
Further insights into the light-harvesting and ET activity were obtained by analyzing the fluorescence induction in dark-adapted leaves. We determined F0 (a parameter inversely related to the efficiency of energy transfer from antenna pigments to open PSII reaction centers), Fv/Fm (an estimate of the maximum quantum efficiency of PSII photochemistry [Butler and Strasser, 1978]), tm (the time to reach the maximal fluorescence), and area (the area above the fluorescence transient). The former two parameters (F0 and Fv/Fm) refer to the structure and function of PSII only, while the latter two yield information on ET activity after QA−, the first electron acceptor of PSII (Strasser et al., 1995). A first observation was that all knockout mutants have a higher F0 value than the wild type. The increase was rather small, although significant, in koCP26 mutants and larger in koCP24 and in the double mutant. This suggests that a larger fraction of absorbed energy is lost as fluorescence in the mutants, implying that the connection between the major LHCII complex and PSII RC is less efficient in the absence of minor antenna proteins (Table 2).
The maximum quantum efficiency of PSII photochemistry (Fv/Fm) was similar in the wild type and koCP26, while it was reduced in koCP24 and koCP24/26 plants (Table 2).
When examining later steps of fluorescence induction, it appeared that koCP24 was slower in reaching Fm: fluorescence rose till the end of the measurement window, while the other genotypes were already declining toward Fs. In koCP24, tm was significantly longer than in any other genotype (Table 2). Since Fm is reached when the plastoquinone (PQ) pool is fully reduced, these results suggest the existence of restrictions in electron transfer to the PSII acceptor PQ.
For a more detailed analysis of the ET contribution to the fluorescence induction curve, we calculated the integrated area between the measured fluorescence signal and the maximal measured fluorescence Fm, given by: 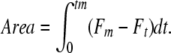
This area value has to be normalized by Fv to compare different samples, yielding a parameter called Sm. The Sm to tm ratio expresses the average redox state of QA in the time span from 0 to tm and, thus, the average fraction of open reaction centers during the time needed to complete their closure. This parameter therefore allows a quantification of the ET activity (Strasser et al., 1995). koCP24 had a lower Sm/tm value than the wild type, meaning that it had a higher average fraction of closed reaction centers. This result implies that ET activity was limited after QA−. The other genotypes, on the contrary, had a similar Sm/tm value to the wild type, thus suggesting a similar ET to QA (Table 2).
Fluorescence induction curves also yield information on ET downstream of QA. Curves are characterized by three rapid rises (0-J, J-I, and I-P) divided by plateau phases (Strasser et al., 1995). Differences were observed in koCP24 with respect to the wild type and the other genotypes (Figure 8): the second rise (J-I) was faster, while the third (I-P) was slower. koCP24/26 plants were affected similarly to koCP24 in their O-J phases of the induction curves, while they had similar kinetics to koCP26 and the wild type at longer times (I-P interval), thus rapidly reaching Fm without revealing restrictions in ET between QA and QB.
Figure 8.
PSII Fluorescence Induction Kinetics Normalized to the Fm Value.
Fluorescence rise was induced on dark-adapted leaf, using a saturating flash of green light (1200 μmol m−2 s−1, 1 s). Inset: the initial rise (sector O-J) of the induction curves. F0 values increase in the order wild type < koCP26 < koCP24 < koCP24/26. Data are expressed as mean values of at least 10 fluorescence curves. A.u., arbitrary units.
Fluorescence parameter analysis thus suggests impairment in PQ reduction rates specifically in koCP24. This phenotype is not retained in the koCP24/26 double mutant, which behaves similarly to wild-type and koCP26 plants.
Partial ET Reactions
ET activity using artificial donor/acceptors was performed to determine the efficiency of different steps of the transport chain and thus to elucidate the nature and location of ETR restriction in koCP24. Whole-chain ETR was measured in isolated thylakoids by following O2 evolution using NADP+ as electron acceptor and was expressed as μmol O2 mg Chl−1 h−1 (Table 3). Wild-type thylakoids exhibited an ETR consistent with previous results (Johnson et al., 1994). koCP26 and koCP24/26 exhibited the same rate of O2 evolution, while koCP24 activity was decreased by 40%, consistent with ETR estimation by fluorescence analysis (Figure 7). ETR from water to PQ was measured using p-benzoquinone (PBQ), which accepts electrons at the QB site of PSII. The ETR in this partial electron chain was consistent with results from the whole-chain assay: koCP24 had a lower O2 evolution than the wild type; while koCP26 and koCP24/26 showed a lower rate of ET to PBQ than the wild type, both mutants showed a significantly higher ETR with respect to koCP24. This result suggests that, even if the PBQ was added in excess, QA to QB e− transport was lower in the koCP24 mutant with respect to the wild type, possibly by limited diffusion of the electron acceptor to the QB site.
Table 3.
Effect of Electron Donors and Acceptors on the ETR
| Wild Type | koCP24 | koCP26 | koCP24/CP26 | |
|---|---|---|---|---|
| μmol O2 mg−1 Chlorophyll h−1 | ||||
| Whole-chain ET H2O → NADP+ | 15.30 ± 1.02 | 8.96 ± 0.71* | 13.74 ± 0.84 | 13.80 ± 0.93 |
| Partial ET H2O → PBQ (PSII) | 45.33 ± 0.38 | 19.33 ± 0.34* | 40.00 ± 1.50* | 33.51 ± 1.32* |
| Partial ET | μmol NADPH mg−1 Chlorophyll h−1 | |||
| DPIPH2 → NADP+ (PQH2 → PSI) | 82.19 ± 6.28 | 86.72 ± 2.86 | 101.93 ± 5.62* | 85.66 ± 1.64 |
| TMPDH2 → NADP+ (PC → PSI) | 143.69 ± 10.04 | 159.74 ± 11.79 | 151.87 ± 12.81 | 149.25 ± 14.41 |
Data are expressed as mean ± sd (n = 4). Significantly different values with respect to the wild type are marked with an asterisk (according to Student's t test, P < 0.05).
ETRs downstream from the plastoquinol (PQH2) and from plastocyanin (PC) to NADP+ (Table 3) have been analyzed spectrophotometrically by following NADP+ reduction. There were not major differences among genotypes, consistent with the hypothesis that the restriction in ET in koCP24 is localized between the QA site and the cytochrome b6f complex. The only significant difference we observed was that koCP26 had a higher rate of e− transport from PQH2 to PSI than the wild type.
Kinetics of QA Reoxidation
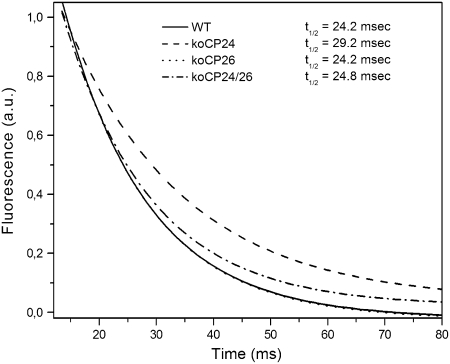
The above suggestion that ET is restricted from QA to QB in koCP24 plants was verified by a further independent measurement. The PQ diffusion step is accessible to analysis through the evaluation of QA reoxidation kinetics by measuring leaf chlorphyll fluorescence decay after a single turn-over flash. In short, when PSII is excited by a very short flash of saturating light, QA is fully reduced and fluorescence reaches its maximal value, after which it decreases with a rate dependent on reoxidation of QA by PQ diffusing from the surrounding membrane domains. Thus, the kinetics of fluorescence decay depend on the rate of PQ diffusion to the PSII QB site (Sane et al., 2003). Fluorescence recovery kinetics were clearly slower in koCP24 than in the wild type and koCP26, implying that the accessibility of the QB site to PQ was restricted (Figure 9). Also, koCP24/26 kinetics was somewhat slower than the wild type and koCP26, but the effect was much smaller than in koCP24. To verify that these results were not due to differences in PQ content in different genotypes, we evaluated the total amount of reducible PQ by comparing fluorescence induction in DCMU-infiltrated leaves with dibromothymoquinone-infiltrated leaves (Bennoun, 2001), which did not show significant differences.
Figure 9.
QA− Reoxidation Kinetics.
Chlorophyll fluorescence decay kinetics were measured after single-turnover flash illumination in dark-adapted leaves. Drawn lines are fits for the experimental data points. Experimental fluorescence curves were normalized to the corresponding Fm values and represent averages from 12 separate experiments. The experimental data set is shown in Supplemental Figure 6 online.
Structural and Functional Analysis of Isolated Grana Membranes
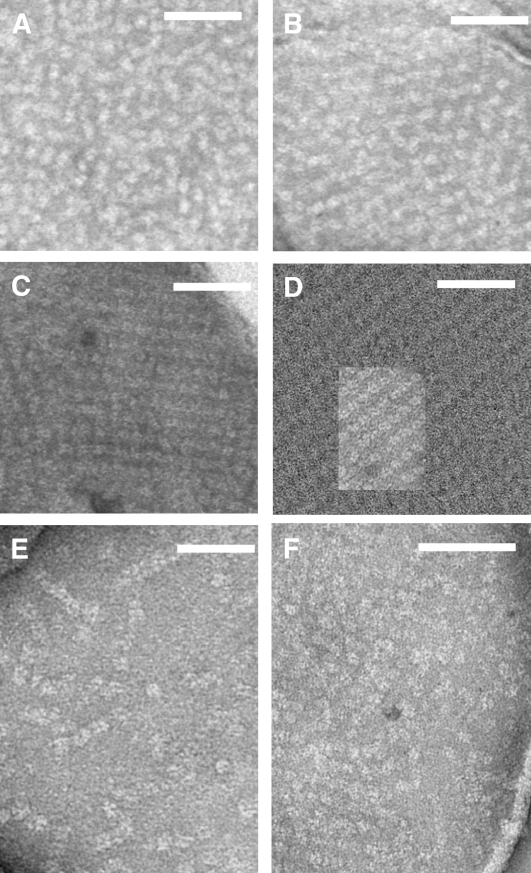
All results presented above support the idea of a restriction of PQ reduction rate in the koCP24 mutant with respect to the other genotypes under study. To find possible explanations for this phenotype, we analyzed the organization of PSII complexes in grana partitions by transmission electron microscopy. Grana membranes were isolated by α-DM fractionation of stacked thylakoid membranes and observed after negative staining as previously described (Morosinotto et al., 2006). The preparation consisted of circular patches of membranes with diameters between 0.7 and 1 μm (see Supplemental Figure 1 online), consistent with derivation from grana partitions (Simpson, 1983). SDS-PAGE analysis of the grana preparations from the different genotypes showed large depletion of ATPase and PSI components and enrichment in LHCII and PSII core polypeptides (see Supplemental Figure 2 online). When observed at high resolution (Figure 10A), grana membranes from the wild type are characterized by stain-excluding particles with a tetrameric structure randomly distributed in a negative stain background and identifiable as PSII cores (Simpson, 1979; Tremmel et al., 2003; Morosinotto et al., 2006). Tetrameric particles in wild-type grana occurred at an average density of 3.5 × 10−4 PSII tetramers nm−2, similar to the case of koCP26. The latter showed, however, that membrane patches where particles were loosely organized into rows (Figure 10B). Samples from koCP24 were clearly different, being characterized by highly ordered arrays of tetrameric particles that covered most of the membrane surface and had a repetition size of 170 × 221 Å (Figure 10C). At the periphery of the membrane circles, these tetrameric particles were less ordered and more widely distributed into a negatively stained background (Figure 10F). Grana partitions from the koCP24/26 double mutant were characterized by the presence of particle rows of four to seven tetrameric particles widely spaced, yielding a particle density of 9.9 × 10−4 nm−2 (Figure 10E). We superimposed the array lattice from koCP24 on a background of membranes isolated from the barley (Hordeum vulgare) mutant vir zb63, chosen as a reference because a high resolution density map is available of this mutant (Morosinotto et al., 2006). This showed that the unit cell of the arrays (16.5 × 25 nm) was the same in the two mutant membranes (Figure 10D). The array in koCP24 thus corresponds to C2S2 supercomplexes as determined at high resolution (Morosinotto et al., 2006).
Figure 10.
EM of Negatively Staining Grana Partition Membranes Obtained by Partial Solubilization with α-DM.
(A) to (C) and (E) High-resolution micrographs show the distribution of stain-excluding tetrameric particles: Wild type (A), koCP26 (B), koCP24 (C), and koCP24/26 (E).
(D) A two-dimensional array from koCP24 was superimposed on a larger array from the grana membranes of the barley mutant vir zb63, showing that the crystal lattice is identical in the two samples.
(F) koCP24 periphery membrane areas in which tetrameric particles were less ordered and more widely distributed into a negatively stained background. The bar is 100 nm long.
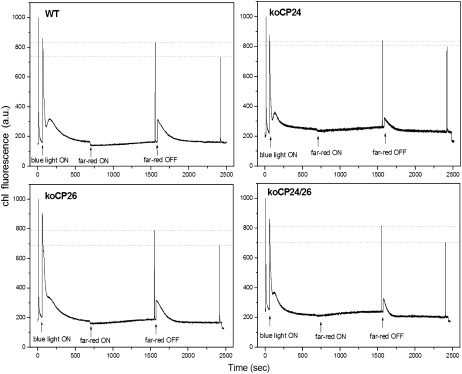
State I–State II Transitions
The above results show that both PQ diffusion within grana membranes and the connection between PSII core complexes and outer LHCII are affected in koCP26, koCP24, and koCP24/26 double mutants. The process of state transitions consists in the adjustment of PSI versus PSII antenna size based on the transfer of phosphorylated LHCII from PSII. Since LHCII phosphorylation is induced by overreduction of the PQ pool (Allen, 1992), state transition measurements are a good indicator of the modifications undergone by PQ redox state. According to a well-established procedure, State I to State II transitions were measured from the changes in chlorophyll fluorescence level of leaves when PSI light was overimposed to PSII light, and then PSI light was switched off to induce PQ reduction (Jensen et al., 2000). The amplitude of state transition of the wild type, koCP26, and koCP24/26, measured as decrease in Fm′ upon reduction of PQ, is essentially the same (Figure 11). A smaller decrease of F′m amplitude was instead observed in koCP24. Differences between koCP24 and others were also observed in the amplitude and rate of the stationary fluorescence (Fs), which reflects the redox state of PQ pools, observed upon switching on far-red light, which oxidizes PQ. While the fluorescence decrease was fast in wild-type and koCP26 plants (0.8 s), it was 20-fold slower in koCP24 (18 s) (see Supplemental Figure 3 online). A further difference was observed in the rate of the transition from State II to State I upon switching off far-red light (Figure 11): while the half-time of the transition was similar in the wild type and koCP26 (88 and 94 s, respectively), it was approximately twice as fast in koCP24 and koCP24/26.
Figure 11.
Measurement of State 1–State 2 Transitions.
Plants, upon dark adaptation for 1 h, were illuminated with blue light (40 μmol m−2 s−1, wavelength <500 nm) for 15 min to reach State II. Far-red light source was used to induce transition to State I. Values of Fm, Fm′, and Fm″ were determined by light saturation pulses (4500 μmol m−2 s−1, 0.6 s).
Functional Characterization of an Independent Allele of koCP24
It is worth noting that the correspondence between the limitation in ETR and the koCP24 mutation was further confirmed by the isolation and characterization of an independent allele of the koCP24 genotype in a different ecotype (Landsberg erecta versus Columbia). We show here that this different allele (and ecotype) has the same alteration of photosynthetic parameters described above for koCP24 and that the double koCP24/26 mutant recovers photosynthetic ETR similar to the wild type (see Supplemental Figure 4 online).
DISCUSSION
Deleting CP24, CP26, or both of these components of the PSII antenna system did not severely affect pigment composition and chloroplast structure. Only koCP24 plants showed a reduction in the rate of Zea synthesis upon exposure to strong light. Nevertheless, this genotype also showed alteration in several photosynthetic parameters and a reduced growth (Figure 1C). All these symptoms were suppressed in the case of the double koCP24/26 mutant, suggesting that phenotypes are not caused merely by the absence of CP24 but rather due to pleiotropic or compensatory effects. This is consistent with the higher reduction in fitness of plants lacking CP24 than of plants lacking CP26 (Ganeteg et al., 2004).
The Functional Phenotypes Are Caused by Pleiotropic Effects Rather Than by Lack of Function Specifically Associated with Individual Gene Products
The mechanistic reason for the above phenotypes is not obvious. Therefore, we have investigated changes in the composition/function of the antenna system by several methods, including the kinetics of fluorescence rise in the presence of DCMU, the pigment distribution among chlorophyll proteins, and the stoichiometry of Lhcb apoproteins. The kinetics of fluorescence in DCMU yields a functional evaluation of the antenna size (i.e., the flux of photons trapped per reaction center). The photon flux reaching the PSII RC is not significantly different between genotypes (Table 2). Clear differences were, however, detected in the Lhcb polypeptide composition of the different mutants (Figure 4). The effects of deleting a subunit within the PSII-LHCII supercomplex can be various: removal of CP29 was shown to decrease the stability of CP24 (Andersson et al., 2001). Alternatively, the loss of Lhcb1 and Lhcb2 was accompanied by the compensatory overaccumulation of CP26 and Lhcb3 (Ruban et al., 2003). We show that lack of CP26 was accompanied by the increase of CP29 and CP24 and, conversely, lack of CP24 increased CP29 and CP26 (Figure 4), suggesting functional compensation within the group of monomeric Lhcb proteins. An additional effect was observed in both koCP24 and the the koCP24/26 double mutant consisting of a change in the relative abundance of the components of the major LHCII antenna: while Lhcb3 is decreased by 55% (koCP24/26) and 70% (koCP24), Lhcb1 and Lhcb2 are overaccumulated in the double mutant (by 65 and 15%, respectively) and, to a lesser extent, in koCP24 plants. This is likely due to the participation of CP24 in a supramolecular antenna complex that also includes CP29, Lhcb1, Lhcb2, and Lhcb3 polypeptides (Bassi and Dainese, 1992). It is interesting to note that CP24-less plants do not lose CP29, suggesting that this complex is stabilized by direct interaction with the PSII core complex. The compensatory relationship observed within antenna polypeptides in the single mutants is broken in the double mutant (Figure 4): upon genetic deletion of both CP26 and CP24, CP29 is also decreased, yielding a PSII strongly depleted in minor antenna complexes. We suggest that the interaction between the PSII core and minor Lhcbs is cooperative, thus leading to decreased affinity when two of them are lacking.
The efficiency of excitation energy transfer to the PSII RC is affected by depletion of monomeric Lhc, as can be inferred by the analysis of the initial fluorescence level (F0): F0 level is inversely related to the efficiency of energy transfer from LHCII to PSII RC. We observed a steady increase in F0 in the order wild type< koCP26< koCP24 = koCP24/26 (Figure 8, Table 2). This is a clear indication that the connection between the PSII core and the bulk trimeric LHCII was partially impaired in the mutants. Cooperativity between PSII centers has been reported to be affected in koCP24, possibly as the result of the clustering of LHCII and/or PSII RC particles observed by electron microscopy (EM) analysis (Kovacs et al., 2006). This implies that exciton migration between many LHCII trimers decreases the probability that an exciton visiting a closed PSII center is then quenched by a neighboring open reaction center. While functional antenna size of different genotypes was essentially the same in wild-type and knockout plants, F0 increases steadily when monomeric Lhcb proteins are deleted and is maximal in koCP24 and koCP24/26 where organization of LHCII into clusters separated from PSII RC is maximal (Figure 10). This can be reconciled by the hypothesis that exciton transfer is slower in the absence of monomeric Lhcs, which decreases the probability of trapping by PSII RC and thus reduces PSII quantum yield.
State transitions, the mechanism by which photosystems balance their complement of light-harvesting antennas depending on the reduction state of the intermediate electron carrier PQ, are triggered by changes of the relative affinity of LHCII for either PSI or PSII, which is regulated by a reversible phosphorylation (Jensen et al., 2000). Neither CP24 nor CP26 are phosphorylated in higher plants (Bassi et al., 1988); thus, changes in state transitions are not expected. While this was verified for koCP26, koCP24 was affected in its capacity to activate state transitions (Figure 11). This effect has been attributed to a decreased presence of PSII-connected LHCII-type M trimers (Kovacs et al., 2006). This statement is not consistent with our finding that the LHCII trimer complement is not significantly affected in our genotypes (Figure 3) and appears to be rather efficient in transferring excitation energy to PSII RC (Table 2). Furthermore, the koCP24/26 mutant, although showing increased F0, is fully able to perform state transitions. Rather, we observe that in koCP24 and koCP24/26, the fluorescence changes induced by switching off the far-red light are faster than the wild type, implying that the transiently reduced state of the free PQ pool is more promptly relaxed in koCP24 and koCP24/26 than the wild type by migration of the LHCII to the RC of PSI. However, koCP24 and koCP24/26 differ in their capacity to undergo reduction of the PQ pool and to activate state transitions (Figure 11).
The Topology of Grana Membranes Is Affected by Mutations
Grana partitions are made up essentially of proteins with very little lipids, which are tightly bound to photosynthetic complexes (Tremolieres et al., 1994). Previous work on negatively stained PSII membranes and cryo-EM analysis of negatively and unstained membranes has shown that tetrameric PSII particles protrude from the membrane plane, while Lhc particles are located in the dark background (Simpson, 1979). In grana membranes from the wild type, the distribution of tetrameric PSII particles is homogeneous through the whole surface. This is not the case for koCP24, where most of the area is occupied by arrays of tetrameric particles and the remaining patches are formed by a stained background with rare stain-excluding particles. The PSII arrays in koCP24 are composed of C2S2 supercomplexes (Figure 10D) since they have the same basic unit as is found in vir zb63, a genotype with a strongly reduced Lhc antenna system lacking CP24 and a large fraction of the LHCII trimers (Morosinotto et al., 2006). koCP24, on the other hand, has a full complement of LHCII trimers (Figures 1 and 4). We conclude that grana membranes of koCP24, besides having arrays of C2S2 particles, contain discrete patches of LHCII trimers that are interspersed by a few PSII core complexes (Figure 10F). We conclude that in some discrete areas of koCP24 grana membranes, the LHCII/PSII core ratio is strongly increased: in these grana partitions, LHCII fluorescence is not efficiently quenched photochemically, thus yielding increased F0. When both CP24 and CP26 are missing, the PSII core appears to be randomly distributed within a network of trimeric LHCII, underlining lack of organized interactions between the PSII RC and its antenna. Depletion in CP24 leads to the formation of C2S2 arrays. We can speculate that the array formation is due to the lack of connection between the inner antenna system and the outer LHCII trimer population, which exposes interaction sites between CP26 of one supercomplex and CP26 of the neighboring complex (Morosinotto et al., 2006). This hypothesis is consistent with the report of CP26 forming trimers when overaccumulated in antisense LHCII plants (Ruban et al., 2003), showing that CP26–CP26 interactions might be strong. In the double mutant, lack of CP26 subunits is thus probably responsible for the disruption of arrays.
How Does the Lack of CP24 Affect Growth and ETR?
The only genotype with a drastically reduced growth rate is koCP24. However, lack of this antenna subunit in itself is unlikely to limit plant growth since PSII quantum yield is only marginally affected (Table 2), while it has been reported that acclimation of wild-type plants to high light yields into 80% decrease in CP24 content without affecting neither plant growth nor the amplitude NPQ (Ballottari et al., 2007). Decreased NPQ, moreover, cannot be considered as the cause for low growth rate in koCP24 since the npq4 mutant, lacking qE, is affected only in harsh stress conditions (Li et al., 2002b) and even grows better than the wild type in low light (Dall'Osto et al., 2005), a feature not found in koCP24 plants. Reduced growth of koCP24 has been attributed to increased F0 and decreased connectivity with respect to the wild type (Kovacs et al., 2006), but this is in contrast with the observation that F0 is even higher in the double mutant, which shows normal growth and ETR (Figures 1C and 7). An effect of the mutation is a strong depletion in Lhcb3. However, this polypeptide is also depleted in the koCP24/26 mutant, and koLhcb3 plants have a normal ETR phenotype (L. Dall'Osto, unpublished data).
EM analysis of chloroplasts shows that photosynthesis is affected in koCP24 since starch grains, accumulated within the chloroplasts of wild-type, koCP26, and koCP24/26 plants, cannot be detected in koCP24. This effect correlates with a reduced ETR both in leaves (Figure 7) and isolated chloroplasts of koCP24 (Table 3). Partial ET reactions localize the restricted step to between the QA site of PSII and the cytochrome b6f complex, since electron donors to cytochrome b6f are effective in sustaining NADP+ reduction at similar rates in all genotypes. We conclude that lack of CP24 leads to restriction of PQH2 diffusion from the PSII QB site to the cytochrome b6f complex, which is the limiting step for photosynthetic ET (Joliot and Joliot, 1977). We cannot formally exclude that ET restriction upon CP24 deletion is located between QA and QB within the PSII core. Nevertheless, this hypothesis seems highly unlikely since it would imply that lack of one Lhcb can have an impact on the PSII core, while lack of two Lhcbs restores full function. Moreover, CP24 subunit, together with all antenna proteins, is lacking in Chlorina f2 and yet the ETR is higher than in the wild type (Guo et al., 2007).
While koCP24 has most of the membrane partition surface occupied by tightly packed arrays of PSII supercomplexes, this feature is not evident in wild-type, koCP26, and koCP24/26 plants (Figure 10), which have normal rates of ET. We conclude that the restriction in ET is associated with the regular organization of PSII particles into arrays in the koCP24 mutant. Indeed, the restriction in ET was confirmed by fluorescence induction in the isolated grana membrane preparation used in the EM analysis (see Supplemental Figure 5 online).
ET from the PSII QB site to cytochrome b6f is mediated by the small diffusible transporter PQ, whose diffusion in the membrane bilayer strongly depends on the organization of intrinsic membrane proteins, which are extremely crowded in grana partitions (Tremmel et al., 2003). PSII organized into ordered arrays restricts protein dynamics and limits the PQ diffusion. While the surface occupancy of randomly organized PSII and LHCII particles is 0.72 to 0.77 (Tremmel et al., 2003), ordered C2S2 arrays leave very little space in between particles (Morosinotto et al., 2006), thus allowing PQ diffusion only in boundary lipids tightly bound to membrane complexes (Tremolieres et al., 1994). This is fully consistent with the analysis of fluorescence induction curves (Figure 8; see Supplemental Figure 5 online), where the last phase (J-P), reflecting the reduction of acceptors downstream of PSII, primarily PQ, is delayed by five times in koCP24 with respect to the wild type. Moreover, the Sm/tm value, expressing the average fraction of open reaction centers during the time needed to complete their closure (Strasser et al., 1995), is three times smaller in koCP24 than in the wild type, implying a higher average fraction of closed reaction centers in the mutants. Together with the observation that ET from cytochrome b6f to PC is equally efficient in all genotypes, this implies a restricted diffusion of PQH2 between site QB and cytochrome b6f that increases QA− reduction. A final confirmation of the above hypothesis was obtained by the measurement of QA reoxidation kinetic, which clearly showed a reduced rate of ET from QA to PQ pool (Figure 9).
Alternatively, a longer average diffusion distance between QA and cytochrome b6f could produce the same effect on ET. Such an effect would likely be present in membrane domains organized into C2S2 arrays that would confine cytochrome b6f complexes, which are normally present in both grana and stroma exposed membranes (Vallon et al., 1991), to grana margins, and/or to stroma membranes. However, our data suggest that neither changes in activity of cytochrome b6f (Table 3) nor the increased distance between QA and cytochrome b6f (see Supplemental Figure 5 online) are responsible for ET limitations. We observed a reduced ET activity from water to PBQ in koCP24 chloroplasts, while steps downstream were unaffected (Table 3). Since oxidized PBQ is present in excess, this effect cannot be due to a lower activity of cytochrome b6f but only to a limited accessibility of PQB to PSII. Furthermore, the slower QA reoxidation kinetics in koCP24 was not due to a higher average distance between QB sites and cytochrome b6f in this genotype, since fluorescence kinetic differences would be removed by dibromothymoquinone treatments (see Supplemental Figure 5 online).
Why Does Restriction in PQ Diffusion Affect qE?
qE is triggered by low lumenal pH (Briantais et al., 1980), while the major lumen acidification step is realized by proton pumping concomitant to PQH2 oxidation by the cytochrome b6f complex. Decreased PQH2 diffusion will thus result in decreased proton pumping. This was confirmed by our observation that the koCP24 mutant had a decreased capacity to generate a pH gradient and synthesized Zea at a slower rate than the wild-type, koCP26, and koCP24/26 plants (Figures 6B and 6D). It is worth noting that koCP24, besides a slower rate of Zea synthesis, also has a lower deepoxidation index at saturating light than the wild type. This can be explained by considering that the release of Viola from outer binding sites of LHCII, which is promoted by lumen acidification, is likely limited in this genotype (Caffarri et al., 2001). We conclude that koCP24 is a proton gradient regulation (pgr) mutant. Its NPQ phenotype, similarly to pgr mutants (Munekage et al., 2001, 2002), is mainly due to a decreased capacity for proton accumulation at the transition from dark to light. koCP24 generates a pH-dependent quenching similar to wild-type plants in the first minute of illumination (Figure 5), but quenching does not develop further beyond this point. In the first seconds of illumination, lumen pH decreases in the mutant and in the wild type. A further decrease in the mutant, however, is limited by its restricted proton transport, and ΔpH does not reach the same amplitude. In addition, reduced Zea synthesis and limited protonation of DCCD binding sites in CP29, CP26, and PsbS might contribute to limitation of the second part of NPQ development. Previous work with koCP24 has underlined the importance of membrane organization and protein–protein interaction between Lhc subunits for the proper operation and full expression of qE (Kovacs et al., 2006). This is consistent with our finding that the restriction in ET is to be ascribed to changes in the PQ diffusion rate caused by tight interaction between C2S2 modules in regular arrays. The alternative view that disconnection of a trimeric LHCII fraction hosting the quenching site is responsible for decreased qE (Kovacs et al., 2006) is inconsistent with our finding that disconnected LHCII domains are far more extended in koCP24/26 than koCP24 plants, while koCP24/26 have a wild-type level of qE (Figure 5).
The NPQ Rise Kinetics Are Affected by Lack of Zea-Exchanging Lhc Proteins
A different pattern of NPQ rise kinetics is observed in the double mutant than in the wild type: although reaching the same qE amplitude at 8 min light, there is a clear plateau between 1 and 3 min, after which the kinetics ascend again. This genotype lacks both CP26 and CP24, the two most effective Lhc proteins in Viola>Zea exchange (Morosinotto et al., 2003) and has reduced CP29. Since Zea has been shown to decrease the activation energy required for the transition from unquenched to quenched conformation (Wentworth et al., 2003), we interpret these results as the effect of a slower transduction of conformational change signal, upon protonation of PsbS, to Lhc proteins, where quenching is catalyzed even in the absence of Zea bound (Briantais, 1994; Bonente et al., 2007). The high levels of qE in koCP24/26 mutants, although with slower kinetics than the wild type, suggest that the major LHCII, which is still present with only 50% of CP29 proteins, might play a role in qE. Based on the slower onset of quenching in the double mutant, it can be hypothesized that monomeric Lhcs might transfer conformational information from PsbS to LHCII. The construction of a mutant without minor CPs will allow verification of this hypothesis.
Finally, it has been proposed that CP26 plays a major role in qI, based on its capacity to assume a quenched conformation upon Zea binding that can be isolated from high-light-treated thylakoids (Dall'Osto et al., 2005). Here, we show that koCP26, although having normal levels of qE, has reduced qI (Figure 5), and the double mutant koCP24/26 does not further decrease its qI level, supporting a specific role of CP26 in catalyzing qI type of quenching. This suggests that although Lhcb proteins might have overlapping functions, they each fulfill specific roles in light harvesting and photoprotection.
What Is the Function of CP24 and CP26 in the Organization of PSII?
CP26 is a component of the PSII antenna in the most ancient green algae species in which photoprotection is mainly performed through Zea synthesis (Baroli et al., 2003) independently from qE (Ledford et al., 2007), which is strongly decreased in green algae with respect to higher plants (Finazzi et al., 2004). We propose that CP26, here shown to be largely responsible for qI, is specialized in Zea-mediated photoprotection (Dall'Osto et al., 2005). Unlike CP26 and CP29, CP24 is a recent addition to the PSII antenna system of the green lineage, appearing only in land plants (Rensing et al., 2007). Chloroplasts of land plants are characterized by large grana stacks made up of partition domains with larger diameters than those of green algae (Larkum and Vesk, 2003). Thus, higher plants are expected to experience restriction of PQ diffusion (Lavergne and Joliot, 1991) between PSII reaction centers and cytochrome b6f during linear ET, which has been suggested to occur mainly in grana margins (Joliot and Joliot, 2005). Chlamydomonas reinhardtii lacks CP24 (Teramoto et al., 2001) and forms C2S2 particles (Boekema et al., 2000), which are prone to form regular arrays (Dekker and Boekema, 2005) that further restrict PQ diffusion. The increased size of grana discs in land plants was a result of evolution that separated PSI from PSII, which increased the efficiency of light harvesting for PSII and established a fine-tuning between cyclic and linear electron flow (Finazzi et al., 2001). This might have led to the requirement of an additional monomeric complex for interfacing the PSII core with trimeric LHCII during changes in antenna size induced by acclimation at low light intensities (Ballottari et al., 2007). This implies that the organization of photosystems in the wild type allows for the highest rate of PQ diffusion, reminiscent of grana organization in green algae (A. Alboresi, S. Caffarri, F. Nogué, R. Bassi, and T. Morosinotto, unpublished data).
We conclude that minor chlorophyll proteins function in bridging dimeric PSII core complexes to the major trimeric LHCII antenna both structurally and functionally. This is particularly evident in CP24/CP26-less plants in which PSII-rich and LHCII-rich domains are formed within grana partitions. The tight regular arrays formed by C2S2 supercomplexes also affect ETRs, most likely by restricting PQ diffusion to cytochrome b6f complexes, which yields decreased lumen acidification and reduction of qE. We suggest that CP24, the latest addition to Lhcb proteins during evolution (Rensing et al., 2007), has evolved to overcome limitations in PQ diffusion caused by the increased size of grana stacks in land plants with respect to green algae.
This work showed how a specific antenna protein has, besides a direct involvement in light harvesting, a large effect on ET through its role in thylakoid biogenesis and assembly. This is a clear example of how a complex system like thylakoid membranes functions due to the optimization of all its components and the tuning of their interactions with each other over evolutionary time.
METHODS
Plant Material
Arabidopsis thaliana T-DNA insertion mutants (Columbia ecotype) SALK_077953, with insertion into the Lhcb6 gene, and SALK_014869, with insertion into the Lhcb5 gene, were obtained from NASC collections (Alonso et al., 2003). An additional T-DNA insertion mutant into the Lhcb6 gene (Arabidopsis Gene Trap line GT6248, Landsberg erecta ecotype) was obtained form Cold Spring Harbor Laboratory (Sundaresan et al., 1995). This allele is indicated as koCP24lan, and the corresponding control genotype as WTlan. Homozygous plants were identified by immunoblot analysis. Individual mutants were crossed, and F1 seeds were grown and self-fertilized to obtain the F2 generation. Homozygous double mutant plants were selected in the F2 population by immunoblotting with specific antibodies. Mutants were grown for 4 to 6 weeks at 100 μmol photons m−2 s−1, 21°C, 90% humidity, and 8 h of daylight.
Pigment Analysis
Pigments were extracted from leaf discs, either dark-adapted or light-treated (30 min, 1000 μmol photons m−2 s−1) at room temperature (22°C): samples were frozen in liquid nitrogen, ground in 85% acetone buffered with Na2CO3, and then the supernatant of each sample was recovered after centrifugation (15 min at 15,000g, 4°C); separation and quantification of pigments were performed by HPLC (Gilmore and Yamamoto, 1991) and by fitting of the spectrum of the acetone extract with spectra of individual pigments (Croce et al., 2002) and recorded using an Aminco DW-2000 spectrophotometer (SLM Instruments).
Thylakoid Isolation and Sample Preparation
Unstacked thylakoids were isolated from leaves as previously described (Bassi et al., 1988), while functional chloroplasts for ETR and ΔpH measurements were obtained as described (Casazza et al., 2001).
Gel Electrophoresis and Immunoblotting
SDS-PAGE analysis was performed with the Tris-Tricine buffer system as previously described (Schägger and von Jagow, 1987). For immunotitration, thylakoid samples corresponding to 0.25, 0.5, 0.75, and 1 μg of chlorophyll were loaded for each sample and electroblotted on nitrocellulose membranes. Filters were incubated with antibodies raised against Lhcb1, Lhcb2, Lhcb3, CP29 (Lhcb4), CP26 (Lhcb5), CP24 (Lhcb6), PsbS, or CP47 (PsbB) and were detected with alkaline phosphatase–conjugated antibody, according to Towbin et al. (1979). Signal amplitude was quantified (n = 4) using the GelPro 3.2 software (Bio-Rad). To avoid any deviation between different immunoblots, samples were compared only when loaded in the same gel.
Deriphat PAGE Analysis
Nondenaturing Deriphat-PAGE was performed following the method described previously (Peter et al., 1991), but using 3.5% (w/v) acrylamide (38:1 acrylamide/bisacrylamide) in the stacking gel and in the resolving gel and an acrylamide concentration gradient from 4.5 to 11.5% (w/v) stabilized by a glycerol gradient from 8 to 16%. Thylakoids concentrated at 1 mg/mL chlorophyll were solubilized with a final 0.8% α-DM, and 30 μg of chlorophyll were loaded in each lane. The integrated optical density measured in each band was checked to linearly correlate to the chlorophyll amounts present in each complex.
EM
Intact leaf fragments from wild-type and mutant 3-week-old leaves, grown in control conditions, were fixed, embedded, and observed in thin section as previously described (Sbarbati et al., 2004).
EM on isolated grana membranes was conducted using an FEI Tecnai T12 electron microscope operating at 100 kV accelerating voltage. Samples were applied to glow-discharged carbon-coated grids and stained with 2% uranyl acetate. Images were recorded using a CCD camera (SIS Megaview III).
In Vivo Fluorescence and NPQ Measurements
NPQ of chlorophyll fluorescence and PSII yield (ΦPSII) were measured on whole leaves at room temperature with a PAM 101 fluorimeter (Heinz-Walz). Minimum fluorescence (F0) was measured with a 0.15 μmol m−2 s−1 beam, maximum fluorescence (Fm) was determined with a 0.6-s light pulse (4500 μmol m−2 s−1), and white continuous light (1200 μmol m−2 s−1) was supplied by a KL1500 halogen lamp (Schott). NPQ, ΦPSII, and relative ETR were calculated according to the following equation (Van Kooten and Snel, 1990): NPQ = (Fm-Fm′)/Fm′, rel ETR = ΦPSII · PAR, where Fm is the maximum chlorophyll fluorescence from dark-adapted leaves, Fm′ the maximum chlorophyll fluorescence under actinic light exposure, Fs the stationary fluorescence during illumination, and PAR the photosynthetic active radiations (white light, measured as μmol m−2 s−1).
State transition experiments were performed using whole plants according to established protocols (Jensen et al., 2000). Preferential PSII excitation was provided by illumination with blue light at an intensity of 40 μmol photons m−2 s−1 provided by a KL1500 lamp equipped with a 650-nm interference filter, and excitation of PSI was achieved using far-red light from an LED light source (Heinz-Walz; 102-FR) applied for 15 min simultaneously with red light. Periods of far-red and blue light conditions were used alternately, and the Fm level in State I (Fm′) and State II (Fm″) was determined at the end of each cycle by the application of a saturating light pulse as described above.
Fluorescence induction kinetics was measured with a home-built apparatus. Fluorescence was excited using a green LED with a peak emission at 520 nm and detected in the near infrared. For the antenna size determination, leaf discs were infiltrated with 3.0 10−5 M DCMU, 150 mM sorbitol, and 10 mM HEPES, pH 7.5. Variable fluorescence was induced with a green light of 7 μmol m−2 s−1. The time corresponding to two-thirds of the fluorescence rise (T2/3) was taken as a measure of the functional antenna size of PSII (Malkin et al., 1981). In DCMU-treated leaves, rate of fluorescence rise depends on light intensity and functional antenna size of PSII. Thus, keeping the saturating flash intensity constant, PSII with higher functional antenna size will reduce all the available QA pool more rapidly and will have a lower T2/3 of fluorescence raise. This provides an estimate of the incident photon flux.
The reoxidation kinetics of QA were measured as the decay of chlorophyll a fluorescence using a pulse-amplitude modulated fluorimeter (Heinz-Walz). Saturating single-turnover flashes obtained from a single turnover flash unit (Heinz Walz; XE-ST) were used to convert all QA to QA−. The variable fluorescence decay, reflecting the reoxidation of QA−, was detected at 20-μs resolution. Data from 12 recordings were averaged.
Measure of ΔpH
The kinetics of ΔpH formation across the thylakoid membrane was measured using the method of 9-AA fluorescence quenching, as previously described (Johnson et al., 1994). The reaction buffer composition was as follows: 0.1 M sorbitol, 5 mM MgCl2, 10 mM NaCl, 20 mM KCl, 30 mM Tricine/NaOH, pH 7.8, 100 μM methylviologen, and 2 μM 9-aminoacridine. The chlorophyll concentration in the reaction buffer was adjusted to 20 μg/mL.
ET with Artificial Donors and Acceptor
Linear ET from artificial donors to NADP+ was measured in a dual-wavelength spectrophotometer (Unicam AA; Thermo scientific), while ET from PSII to PBQ and the whole ETR from water to NADP+ were measured following the oxygen evolution. The O2 production was measured on functional thylakoids at 20°C in a Clark-type oxygen electrode system (Hansatech Instruments) under red light illumination (150 μmol photons m−2 s−1). These measurements were performed as described by Casazza et al. (2001). NADP+ reduction rate was measured spectrophotometrically, while oxygen evolution rate was measured with a Clark-type polarographic oxygen electrode system (as described in Casazza et al., 2001) under red light illumination (150 μmol m−2 s−1). Concentrations used were as follows: 0.5 mM NADP+, 300 mM PBQ, 50 μM DPIPH2 (dichlorophenlindophenol), 250 μM TMPDH2 (N, N, N, N-tetramethyl-p-phenylene-diamine, reduced form), and thylakoids to a final chlorophyll concentration of 10 μg/mL.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under accession numbers At4g10340 (LHCB5) and At1g15820 (LHCB6).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Micrograph of Negatively Stained Grana Partition Preparation Obtained by Limited α-DM Solubilization of Stacked Thylakoids.
Supplemental Figure 2. Analysis of Pigment-Protein Complexes of the Wild Type and Mutant.
Supplemental Figure 3. Kinetics of Plastoquinol Reoxidation upon Exposure to Far-Red Light.
Supplemental Figure 4. Characterization of an Additional Allele for koCP24 (koCP24lan) Establishes That This Is the Responsible Mutation for the Observed Phenotype.
Supplemental Figure 5. Chlorophyll Fluorescence Induction Curves Measured on Grana Membrane Preparations from the Wild Type and koCP24 Mutant.
Supplemental Figure 6. QA− Reoxidation Kinetics.
Supplemental Table 1. NPQ Measurements on Intact Chloroplasts of the Wild Type and Mutant Genotypes.
Supplementary Material
Acknowledgments
We thank A. Sbarbati and P. Bernardi for the use of the EM facility at the University of Verona Medical Center. We also thank J. Lavergne (Commissariat à l'Energie Atomique, Cadarache, France) and P. Joliot (Institut de Biologie Physico-Chimique, Paris) for many discussion of PQ diffusion during the early postdoctoral visit of R.B. at Institut de Biologie Physico-Chimique. Financial support for this work was provided by the RBLA0345SF_002 Grant of the Italian Ministry of Research.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Roberto Bassi (bassi@sci.univr.it).
Online version contains Web-only data.
References
- Allen, J.F. (1992). Protein phosphorylation in regulation of photosynthesis. Biochim. Biophys. Acta 1098 275–335. [DOI] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Andersson, J., Walters, R.G., Horton, P., and Jansson, S. (2001). Antisense inhibition of the photosynthetic antenna proteins CP29 and CP26: Implications for the mechanism of protective energy dissipation. Plant Cell 13 1193–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson, J., Wentworth, M., Walters, R.G., Howard, C.A., Ruban, A.V., Horton, P., and Jansson, S. (2003). Absence of the Lhcb1 and Lhcb2 proteins of the light-harvesting complex of photosystem II - Effects on photosynthesis, grana stacking and fitness. Plant J. 35 350–361. [DOI] [PubMed] [Google Scholar]
- Asada, K. (1999). The water-water cycle in chloroplasts: Scavenging of active oxygens and dissipation of excess photons. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50 601–639. [DOI] [PubMed] [Google Scholar]
- Ballottari, M., Dall'Osto, L., Morosinotto, T., and Bassi, R. (2007). Contrasting behavior of higher plant photosystem I and II antenna systems during acclimation. J. Biol. Chem. 282 8947–8958. [DOI] [PubMed] [Google Scholar]
- Barber, J., and Andersson, B. (1992). Too much of a good thing - Light can be bad for photosynthesis. Trends Biochem. Sci. 17 61–66. [DOI] [PubMed] [Google Scholar]
- Baroli, I., Do, A.D., Yamane, T., and Niyogi, K.K. (2003). Zeaxanthin accumulation in the absence of a functional xanthophyll cycle protects Chlamydomonas reinhardtii from photooxidative stress. Plant Cell 15 992–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassi, R., and Dainese, P. (1992). A supramolecular light-harvesting complex from chloroplast Photosystem II membranes. Eur. J. Biochem. 204 317–326. [DOI] [PubMed] [Google Scholar]
- Bassi, R., Giuffra, E., Croce, R., Dainese, P., and Bergantino, E. (1996). Biochemistry and molecular biology of pigment binding proteins. In Light as an Energy Source and Information Carrier in Plant Physiology, R.C. Jennings, G. Zucchelli, F. Ghetti, and G. Colombetti, eds (New York: Plenum Press), pp. 41–63.
- Bassi, R., Rigoni, F., Barbato, R., and Giacometti, G.M. (1988). Light-harvesting chlorophyll a/b proteins (LHCII) populations in phosphorylated membranes. Biochim. Biophys. Acta 936 29–38. [Google Scholar]
- Bennoun, P. (2001). Chlororespiration and the process of carotenoid biosynthesis. Biochim. Biophys. Acta 1506 133–142. [DOI] [PubMed] [Google Scholar]
- Boekema, E.J., van Breemen, J.F., van Roon, H., and Dekker, J.P. (2000). Arrangement of photosystem II supercomplexes in crystalline macrodomains within the thylakoid membrane of green plant chloroplasts. J. Mol. Biol. 301 1123–1133. [DOI] [PubMed] [Google Scholar]
- Boekema, E.J., van Roon, H., van Breemen, J.F., and Dekker, J.P. (1999). Supramolecular organization of photosystem II and its light-harvesting antenna in partially solubilized photosystem II membranes. Eur. J. Biochem. 266 444–452. [DOI] [PubMed] [Google Scholar]
- Bonente, G., Howes, B.D., Caffarri, S., Smulevich, G., and Bassi, R. (2007). Interactions between the photosystem II subunit psbs and xanthophylls studied in vivo and in vitro. J. Biol. Chem., in press. [DOI] [PMC free article] [PubMed]
- Briantais, J.-M. (1994). Light-harvesting chlorophyll a-b complex requirement for regulation of Photosystem II photochemistry by non-photochemical quenching. Photosynth. Res. 40 287–294. [DOI] [PubMed] [Google Scholar]
- Briantais, J.-M., Vernotte, C., Picaud, M., and Krause, G.H. (1980). Chlorophyll fluorescence as a probe for the determination of the photoinduced proton gradient in isolated chloroplasts. Biochim. Biophys. Acta 591 198–202. [DOI] [PubMed] [Google Scholar]
- Butler, W.L., and Strasser, R.J. (1978). Effect of divalent cations on energy coupling between the light harvesting chlorophyll a/b complex and photosystem II. In Proceedings of the 4th International Congress on Photosynthesis, D.O. Hall, J. Coombs, and T. Goodwin, eds (London: Biochemical Society), pp. 9–20.
- Caffarri, S., Croce, R., Breton, J., and Bassi, R. (2001). The major antenna complex of photosystem II has a xanthophyll binding site not involved in light harvesting. J. Biol. Chem. 276 35924–35933. [DOI] [PubMed] [Google Scholar]
- Casazza, A.P., Tarantino, D., and Soave, C. (2001). Preparation and functional characterization of thylakoids from Arabidopsis thaliana. Photosynth. Res. 68 175–180. [DOI] [PubMed] [Google Scholar]
- Crimi, M., Dorra, D., Bosinger, C.S., Giuffra, E., Holzwarth, A.R., and Bassi, R. (2001). Time-resolved fluorescence analysis of the recombinant photosystem II antenna complex CP29. Effects of zeaxanthin, pH and phosphorylation. Eur. J. Biochem. 268 260–267. [DOI] [PubMed] [Google Scholar]
- Croce, R., Canino, G., Ros, F., and Bassi, R. (2002). Chromophore organization in the higher-plant photosystem II antenna protein CP26. Biochemistry 41 7334–7343. [DOI] [PubMed] [Google Scholar]
- Dall'Osto, L., Caffarri, S., and Bassi, R. (2005). A mechanism of nonphotochemical energy dissipation, independent from Psbs, revealed by a conformational change in the antenna protein CP26. Plant Cell 17 1217–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker, J.P., and Boekema, E.J. (2005). Supramolecular organization of thylakoid membrane proteins in green plants. Biochim. Biophys. Acta 1706 12–39. [DOI] [PubMed] [Google Scholar]
- Di Paolo, M. L., Peruffo dal Belin, A., and Bassi, R. (1990). Immunological studies on chlorophyll-a/b proteins and their distribution in thylakoid membrane domains. Planta 181 275–286. [DOI] [PubMed] [Google Scholar]
- Dominici, P., Caffarri, S., Armenante, F., Ceoldo, S., Crimi, M., and Bassi, R. (2002). Biochemical properties of the PsbS subunit of photosystem II either purified from chloroplast or recombinant. J. Biol. Chem. 277 22750–22758. [DOI] [PubMed] [Google Scholar]
- Durnford, D.G. (2003). Structure and regulation of algal light-harvesting complex genes. In Photosynthesis in Algae, A.W.D. Larkum, S.E. Douglas, and J.A. Raven, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 63–82.
- Finazzi, G., Barbagallo, R.P., Bergo, E., Barbato, R., and Forti, G. (2001). Photoinhibition of Chlamydomonas reinhardtii in State 1 and State 2: Damages to the photosynthetic apparatus under linear and cyclic electron flow. J. Biol. Chem. 276 22251–22257. [DOI] [PubMed] [Google Scholar]
- Finazzi, G., Johnson, G.N., Dall'Osto, L., Joliot, P., Wollman, F.A., and Bassi, R. (2004). A zeaxanthin-independent nonphotochemical quenching mechanism localized in the photosystem II core complex. Proc. Natl. Acad. Sci. USA 101 12375–12380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganeteg, U., Kulheim, C., Andersson, J., and Jansson, S. (2004). Is each light-harvesting complex protein important for plant fitness? Plant Physiol. 134 502–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore, A.M., and Yamamoto, H.Y. (1991). Zeaxanthin formation and energy-dependent fluorescence quenching in pea chloroplasts under artificially mediated linear and cyclic electron transport. Plant Physiol. 96 635–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, J.W., Guo, J.K., Zhao, Y., and Du, L.F. (2007). Changes of Photosystem II electron transport in the chlorophyll-deficient oilseed rape mutant studied by chlorophyll fluorescence and thermoluminescence. J. Integr. Plant Biol. 49 689–705. [Google Scholar]
- Harrer, R., Bassi, R., Testi, M.G., and Schäfer, C. (1998). Nearest-neighbor analysis of a photosystem II complex from Marchantia polymorpha L.(liverwort), which contains reaction center and antenna proteins. Eur. J. Biochem. 255 196–205. [DOI] [PubMed] [Google Scholar]
- Havaux, M., Dall'Osto, L., Cuine, S., Giuliano, G., and Bassi, R. (2004). The effect of zeaxanthin as the only xanthophyll on the structure and function of the photosynthetic apparatus in Arabidopsis thaliana. J. Biol. Chem. 279 13878–13888. [DOI] [PubMed] [Google Scholar]
- Horton, P., Ruban, A.V., and Walters, R.G. (1996). Regulation of light harvesting in green plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47 655–684. [DOI] [PubMed] [Google Scholar]
- Jansson, S. (1999). A guide to the Lhc genes and their relatives in Arabidopsis. Trends Plant Sci. 4 236–240. [DOI] [PubMed] [Google Scholar]
- Jensen, P.E., Gilpin, M., Knoetzel, J., and Scheller, H.V. (2000). The PSI-K subunit of photosystem I is involved in the interaction between light-harvesting complex I and the photosystem I reaction center core. J. Biol. Chem. 275 24701–24708. [DOI] [PubMed] [Google Scholar]
- Johnson, G.N., Young, A.J., and Horton, P. (1994). Activation of non-photochemical quenching in thylakoids and leaves. Planta 194 550–556. [Google Scholar]
- Joliot, P., and Joliot, A. (1977). Evidence for a double hit process in photosystem II based on fluorescence studies. Biochim. Biophys. Acta 462 559–574. [DOI] [PubMed] [Google Scholar]
- Joliot, P., and Joliot, A. (2005). Quantification of cyclic and linear flows in plants. Proc. Natl. Acad. Sci. USA 102 4913–4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs, L., Damkjaer, J., Kereiche, S., Ilioaia, C., Ruban, A.V., Boekema, E.J., Jansson, S., and Horton, P. (2006). Lack of the light-harvesting complex CP24 affects the structure and function of the grana membranes of higher plant chloroplasts. Plant Cell 18 3106–3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkum, A.W.D., and Vesk, M. (2003). Algal plastids: Their fine structure and properties. In Photosynthesis in Algae, A.W.D. Larkum, S.E. Douglas, and J.A. Raven, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 11–28.
- Lavergne, J., and Joliot, P. (1991). Restricted diffusion in photosynthetic membranes. Trends Biochem. Sci. 16 129–134. [DOI] [PubMed] [Google Scholar]
- Ledford, H.K., Chin, B.L., and Niyogi, K.K. (2007). Acclimation to singlet oxygen stress in Chlamydomonas reinhardtii. Eukaryot. Cell 6 919–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X.P., Bjorkman, O., Shih, C., Grossman, A.R., Rosenquist, M., Jansson, S., and Niyogi, K.K. (2000). A pigment-binding protein essential for regulation of photosynthetic light harvesting. Nature 403 391–395. [DOI] [PubMed] [Google Scholar]
- Li, X.P., Gilmore, A.M., Caffarri, S., Bassi, R., Golan, T., Kramer, D., and Niyogi, K.K. (2004). Regulation of photosynthetic light harvesting involves intrathylakoid lumen pH sensing by the PsbS protein. J. Biol. Chem. 279 22866–22874. [DOI] [PubMed] [Google Scholar]
- Li, X.P., Gilmore, A.M., and Niyogi, K.K. (2002. a). Molecular and global time-resolved analysis of a psbS gene dosage effect on pH- and xanthophyll cycle-dependent nonphotochemical quenching in photosystem II. J. Biol. Chem. 277 33590–33597. [DOI] [PubMed] [Google Scholar]
- Li, X.P., Muller-Moule, P., Gilmore, A.M., and Niyogi, K.K. (2002. b). PsbS-dependent enhancement of feedback de-excitation protects photosystem II from photoinhibition. Proc. Natl. Acad. Sci. USA 99 15222–15227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkin, S., Armond, P.A., Mooney, H.A., and Fork, D.C. (1981). Photosystem II photosynthetic unit sizes from fluorescence induction in leaves. Correlation to photosynthetic capacity. Plant Physiol. 67 570–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morosinotto, T., Baronio, R., and Bassi, R. (2002). Dynamics of chromophore binding to Lhc proteins in vivo and in vitro during operation of the xanthophyll cycle. J. Biol. Chem. 277 36913–36920. [DOI] [PubMed] [Google Scholar]
- Morosinotto, T., Bassi, R., Frigerio, S., Finazzi, G., Morris, E., and Barber, J. (2006). Biochemical and structural analyses of a higher plant photosystem II supercomplex of a photosystem I-less mutant of barley. Consequences of a chronic over-reduction of the plastoquinone pool. FEBS J. 273 4616–4630. [DOI] [PubMed] [Google Scholar]
- Morosinotto, T., Caffarri, S., Dall'Osto, L., and Bassi, R. (2003). Mechanistic aspects of the xanthophyll dynamics in higher plant thylakoids. Physiol. Plant. 119 347–354. [Google Scholar]
- Munekage, Y., Hojo, M., Meurer, J., Endo, T., Tasaka, M., and Shikanai, T. (2002). PGR5 is involved in cyclic electron flow around photosystem I and is essential for photoprotection in Arabidopsis. Cell 110 361–371. [DOI] [PubMed] [Google Scholar]
- Munekage, Y., Takeda, S., Endo, T., Jahns, P., Hashimoto, T., and Shikanai, T. (2001). Cytochrome b(6)f mutation specifically affects thermal dissipation of absorbed light energy in Arabidopsis. Plant J. 28 351–359. [DOI] [PubMed] [Google Scholar]
- Niyogi, K.K. (1999). Photoprotection revisited: Genetic and molecular approaches. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50 333–359. [DOI] [PubMed] [Google Scholar]
- Niyogi, K.K. (2000). Safety valves for photosynthesis. Curr. Opin. Plant Biol. 3 455–460. [DOI] [PubMed] [Google Scholar]
- Pesaresi, P., Sandona, D., Giuffra, E., and Bassi, R. (1997). A single point mutation (E166Q) prevents dicyclohexylcarbodiimide binding to the photosystem II subunit CP29. FEBS Lett. 402 151–156. [DOI] [PubMed] [Google Scholar]
- Peter, G.F., Takeuchi, T., and Thornber, J.P. (1991). Solubilization and two-dimensional electrophoretic procedures for studying the organization and composition of photosynthetic membrane polypeptides. Methods 3 115–124. [Google Scholar]
- Rensing, S.A., et al. (2007). The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319 64–69. [DOI] [PubMed] [Google Scholar]
- Ruban, A.V., Walters, R.G., and Horton, P. (1992). The molecular mechanism of the control of excitation energy dissipation in chloroplast membranes - Inhibition of ΔpH-dependent quenching of chlorophyll fluorescence by dicyclohexylcarbodiimide. FEBS Lett. 309 175–179. [DOI] [PubMed] [Google Scholar]
- Ruban, A.V., Wentworth, M., Yakushevska, A.E., Andersson, J., Lee, P.J., Keegstra, W., Dekker, J.P., Boekema, E.J., Jansson, S., and Horton, P. (2003). Plants lacking the main light-harvesting complex retain photosystem II macro-organization. Nature 421 648–652. [DOI] [PubMed] [Google Scholar]
- Sane, P.V., Ivanov, A.G., Hurry, V., Huner, N.P., and Oquist, G. (2003). Changes in the redox potential of primary and secondary electron-accepting quinones in photosystem II confer increased resistance to photoinhibition in low-temperature-acclimated Arabidopsis. Plant Physiol. 132 2144–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sbarbati, A., Merigo, F., Benati, D., Tizzano, M., Bernardi, P., and Osculati, F. (2004). Laryngeal chemosensory clusters. Chem. Senses 29 683–692. [DOI] [PubMed] [Google Scholar]
- Schägger, H., and von Jagow, G. (1987). Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166 368–379. [DOI] [PubMed] [Google Scholar]
- Simpson, D.J. (1979). Freeze-fracture studies on barley plastid membranes. 3. Location of the light-harvesting chlorophyll-protein. Carlsberg Res. Commun. 44 305–336. [Google Scholar]
- Simpson, D.J. (1983). Freeze-fracture studies on barley plastid membranes. VI. Location of the P700-chlorophyll a-protein 1. Eur. J. Cell Biol. 31 305–314. [DOI] [PubMed] [Google Scholar]
- Strasser, R.J., Srivastava, A., and Govindjee. (1995). Polyphasic chlorophyll a fluorescence transient in plants and cyanobacteria. Photochem. Photobiol. 61: 32–42.
- Sundaresan, V., Springer, P., Volpe, T., Haward, S., Jones, J.D., Dean, C., Ma, H., and Martienssen, R. (1995). Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev. 9 1797–1810. [DOI] [PubMed] [Google Scholar]
- Teardo, E., De Laureto, P.P., Bergantino, E., Dalla, V.F., Rigoni, F., Szabo, I., and Giacometti, G.M. (2007). Evidences for interaction of PsbS with photosynthetic complexes in maize thylakoids. Biochim. Biophys. Acta 1767 703–711. [DOI] [PubMed] [Google Scholar]
- Teramoto, H., Ono, T., and Minagawa, J. (2001). Identification of Lhcb gene family encoding the light-harvesting chlorophyll-a/b proteins of photosystem II in Chlamydomonas reinhardtii. Plant Cell Physiol. 42 849–856. [DOI] [PubMed] [Google Scholar]
- Towbin, H., Staehelin, T., and Gordon, J. (1979). Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc. Natl. Acad. Sci. USA 76 4350–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremmel, I.G., Kirchhoff, H., Weis, E., and Farquhar, G.D. (2003). Dependence of plastoquinol diffusion on the shape, size, and density of integral thylakoid proteins. Biochim. Biophys. Acta 1607 97–109. [DOI] [PubMed] [Google Scholar]
- Tremolieres, A., Dainese, P., and Bassi, R. (1994). Heterogenous lipid distribution among chlorophyll-binding proteins of photosystem II in maize mesophyll chloroplasts. Eur. J. Biochem. 221 721–730. [DOI] [PubMed] [Google Scholar]
- Vallon, O., Bulte, L., Dainese, P., Olive, J., Bassi, R., and Wollman, F.A. (1991). Lateral redistribution of cytochrome b6/f complexes along thylakoid membranes upon state transitions. Proc. Natl. Acad. Sci. USA 88 8262–8266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Kooten, O., and Snel, J.F.H. (1990). The use of chlorophyll fluorescence nomenclature in plant stress physiology. Photosynth. Res. 25 147–150. [DOI] [PubMed] [Google Scholar]
- Walters, R.G., Ruban, A.V., and Horton, P. (1996). Identification of proton-active residues in a higher plant light-harvesting complex. Proc. Natl. Acad. Sci. USA 93 14204–14209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wentworth, M., Ruban, A.V., and Horton, P. (2003). Thermodynamic investigation into the mechanism of the chlorophyll fluorescence quenching in isolated photosystem II light-harvesting complexes. J. Biol. Chem. 278 21845–21850. [DOI] [PubMed] [Google Scholar]
- Yakushevska, A.E., Keegstra, W., Boekema, E.J., Dekker, J.P., Andersson, J., Jansson, S., Ruban, A.V., and Horton, P. (2003). The structure of photosystem II in Arabidopsis: Localization of the CP26 and CP29 antenna complexes. Biochemistry 42 608–613. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.