Abstract
Regulated cell expansion allows plants to adapt their morphogenesis to prevailing environmental conditions. Cell expansion is driven by turgor pressure created by osmotic water uptake and is restricted by the extensibility of the cell wall, which in turn is regulated by the synthesis, incorporation, and cross-linking of new cell wall components. The vacuolar H+-ATPase (V-ATPase) could provide a way to coordinately regulate turgor pressure and cell wall synthesis, as it energizes the secondary active transport of solutes across the tonoplast and also has an important function in the trans-Golgi network (TGN), which affects synthesis and trafficking of cell wall components. We have previously shown that det3, a mutant with reduced V-ATPase activity, has a severe defect in cell expansion. However, it was not clear if this is caused by a defect in turgor pressure or in cell wall synthesis. Here, we show that inhibition of the tonoplast-localized V-ATPase subunit isoform VHA-a3 does not impair cell expansion. By contrast, inhibition of the TGN-localized isoform VHA-a1 is sufficient to restrict cell expansion. Furthermore, we provide evidence that the reduced hypocotyl cell expansion in det3 is conditional and due to active, hormone-mediated growth inhibition caused by a cell wall defect.
INTRODUCTION
Plant cells are embedded in an extracellular matrix that prevents cell migration. In the absence of cell movement, plants rely on a tightly regulated interplay of cell division and cell expansion to achieve their unique developmental plasticity that enables adaptive morphogenesis and compensates for the lack of mobility. Cell expansion is achieved by osmotic water uptake into the vacuole, creating the turgor pressure necessary for the irreversible extension of the cell wall caused by the synthesis, incorporation, and cross-linking of new cell wall components (Cosgrove, 2005). The primary walls of young, growing cells are composed of cellulose fibers embedded in a matrix of pectins and hemicelluloses, two classes of complex polysaccharides. Whereas cellulose is produced at the plasma membrane by cellulose synthase (CESA) complexes (Somerville, 2006), synthesis of the matrix polysaccharides takes place in the Golgi apparatus (Lerouxel et al., 2006). Plant cell growth thus involves processes and forces acting in different cellular compartments, including the vacuole, Golgi, and plasma membrane. One of the key questions in plant biology is how integration of the individual components is achieved to allow adaptive cell expansion.
The Arabidopsis thaliana hypocotyl has been widely used as a model system, as its growth relies exclusively on cell expansion (Gendreau et al., 1997) and is regulated by light (Chen et al., 2004) and by several plant hormones (Collett et al., 2000; Achard et al., 2007). Accordingly, mutants with reduced hypocotyl growth can be classified as either being involved in light or hormone signaling or having defects in the actual cell expansion machinery (Vandenbussche et al., 2005). The vast majority of the latter class corresponds to mutants with defects in the biosynthesis or assembly of cell wall components, including cellulose synthase mutants (Arioli et al., 1998; Taylor et al., 1999), pectin-deficient mutants (Bouton et al., 2002), and cell wall remodeling mutants (Nicol et al., 1998).
One of the few candidates potentially involved in creating or regulating turgor is vacuolar H+-ATPase (V-ATPase). V-ATPases are highly conserved, multisubunit endomembrane proton pumps that consist of two subcomplexes. The peripheral V1 complex, which consists of eight subunits (subunits A to H), is responsible for ATP hydrolysis, whereas the membrane-integral V0 complex (subunits a, c, c', c'', d, and e) is responsible for proton translocation from the cytosol into the lumen of endomembrane compartments. V-ATPases represent a major fraction of the total tonoplast protein, and their most prominent function in plants is to maintain ion and metabolite homeostasis by energizing secondary active transport across the tonoplast.
V-ATPase function is impaired in the det3 mutant. det3 was originally identified as a potential negative regulator of photomorphogenesis (Cabrera y Poch et al., 1993). Schumacher et al. (1999) showed that DET3 encodes V-ATPase subunit C, and the det3 mutant carries a weak allele of VHA-C that leads to a conditional defect in hypocotyl elongation in dark-grown seedlings. One hypothesis to explain the growth defect in det3 is reduced secondary active vacuolar solute uptake, resulting in inadequate turgor pressure for cell expansion. However, the severity of the det3 phenotype, which encompasses cell death (Schumacher et al., 1999) and ectopic lignification (Cano-Delgado et al., 2003), makes it difficult to identify the primary defects. It has been reported that overexpression of the transcription factor MYB61 is responsible for the dark photomorphogenic phenotype of det3, but the causal relation between reduced V-ATPase activity and MYB61 overexpression was unclear (Newman et al., 2004).
We have shown that the Arabidopsis V-ATPase is essential for Golgi structure and function (Dettmer et al., 2005, 2006) and therefore propose the alternative hypothesis that reduced growth could be the consequence of a defect in a Golgi-dependent process, such as cell wall synthesis or protein trafficking. Moreover, V-ATPase might provide a way to coordinately regulate turgor pressure and cell wall synthesis. Arabidopsis encodes three isoforms of VHA-a, and Dettmer et al. (2006) showed that VHA-a1 is localized to the trans-Golgi network (TGN), whereas VHA-a2 and VHA-a3 are found in the tonoplast. Here, we analyze the respective functions of the TGN and tonoplast V-ATPases during cell expansion using plants compromised for VHA-a1 or VHA-a3 activity. Furthermore, we characterized the conditional nature of the det3 hypocotyl phenotype and show that reduced hypocotyl cell expansion in mutant seedlings is due to 12-oxophytodienoic acid (OPDA) and ethylene- mediated changes in gene expression caused by a defect in cell wall integrity.
RESULTS
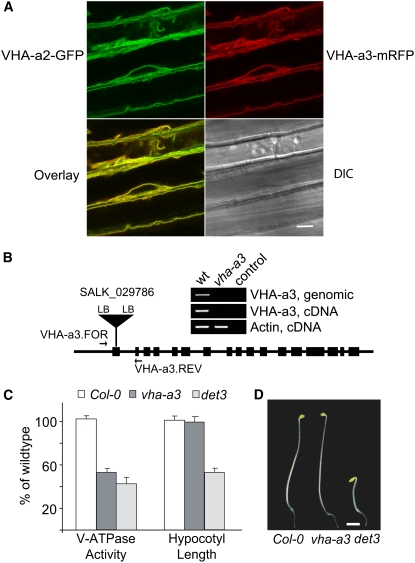
Cell Expansion Is Not Affected in the vha-a3 Mutant
We previously reported that green fluorescent protein (GFP) fusion proteins of VHA-a2 (At2g21410) and VHA-a3 (At4g39080) are localized to the tonoplast of Arabidopsis root tip cells (Dettmer et al., 2006). To determine the function of the tonoplast V-ATPase during cell expansion, we first characterized the subcellular localization and expression levels of VHA-a2 and VHA-a3 in etiolated seedling hypocotyls. Transgenic seedlings coexpressing genomic fusions of VHA-a2 protein with GFP and VHA-a3 with monomeric red fluorescent protein (mRFP) were established and analyzed by confocal laser scanning microscopy (CLSM). GFP and mRFP signals were identical, demonstrating that VHA-a2 and VHA-a3 colocalize at the tonoplast of rapidly growing etiolated hypocotyl cells (Figure 1A). According to publicly available microarray data, VHA-a2 and VHA-a3 are expressed ubiquitously, with VHA-a3 more strongly expressed in most tissues and organs. However, based on our own Affymetrix ATH1 microarray analysis (see below), VHA-a2 and VHA-a3 are expressed to very similar levels in 4-d-old etiolated seedlings (average raw intensities: VHA-a2, 1038 ± 73; VHA-a3, 1000 ± 103), suggesting that both isoforms contribute equally to tonoplast V-ATPase activity in rapidly growing hypocotyl cells. A T-DNA line annotated as carrying an insertion in exon 1 of VHA-a3 (SALK_29786; Alonso et al., 2003) was obtained from the SALK collection, and sequencing of both left border–derived PCR products was used to determine the exact insertion site at chromosome 4 position 18209630 (Figure 1B). Homozygous vha-a3 individuals were identified by PCR. RT-PCR showed that vha-a3 is a null allele, as no full-length VHA-a3 transcript could be detected (Figure 1B). We next determined total V-ATPase activity in microsomal extracts from etiolated seedlings of Columbia-0 (Col-0), vha-a3, and det3 as Concanamycin A (ConcA)-sensitive ATP hydrolysis. In vha-a3, total V-ATPase activity was reduced to a similar extent as in the det3 mutant (Figure 1C). However, in contrast with det3, the hypocotyl length of etiolated vha-a3 seedlings was indistinguishable from Col-0 (Figures 1C and 1D), indicating that the reduced cell expansion of det3 seedlings was not caused by a lack of tonoplast V-ATPase.
Figure 1.
Reduced Tonoplast V-ATPase Activity Does Not Limit Cell Expansion.
(A) Etiolated hypocotyl cells coexpressing VHA-a2-GFP and VHA-a3-mRFP. Green and red fluorescence signals represents the tonoplast of the large central vacuoles. Bar = 10 μm. DIC, differential interference contrast.
(B) Structure and expression of the vha-a3 T-DNA allele; the indicated primers were used to identify homozygous plants and to demonstrate the absence of the VHA-a3 transcript. The control PCR reactions contain no input DNA. LB, left border.
(C) V-ATPase activity and hypocotyl length of etiolated Col-0, vha-a3, and det3 seedlings grown on MS plates. V-ATPase activity of total microsomal extracts was measured as ConcA-sensitive ATP hydrolysis. Data from at least three independent measurements were averaged, and the value obtained for Col-0 was set to 100%. Error bars indicate sd for the enzyme activity and se for hypocotyl length.
(D) Four-day-old etiolated seedlings of Col-0, vha-a3, and det3. Bar = 2 mm.
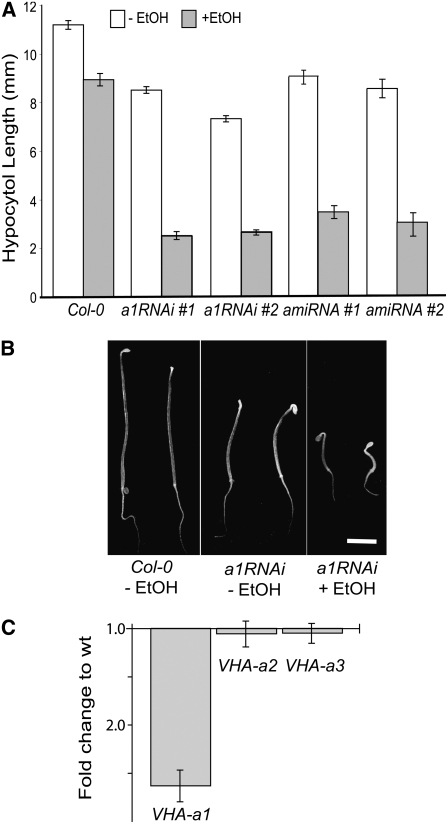
Inhibition of V-ATPase in the TGN Is Sufficient to Reduce Cell Expansion
Assuming that a null allele of the TGN-localized (Dettmer et al., 2006) isoform VHA-a1 (At2g28520) is lethal, we employed inducible RNA interference (RNAi) and artificial microRNA (amiRNA) constructs to reduce V-ATPase activity specifically in the TGN. For the RNAi construct, a 203-bp fragment, derived from the 5′ region of VHA-a1, that has only 30% sequence identity to VHA-a2 and VHA-a3 was cloned into an RNAi vector.
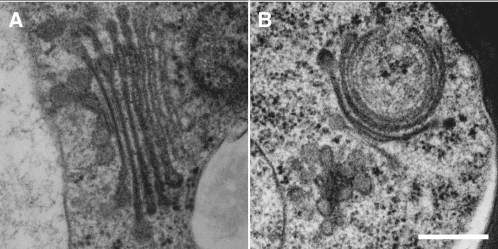
An amiRNA specific for VHA-a1 was designed via the Web MicroRNA Designer (see Methods) and cloned into the MIR319a precursor (Schwab et al., 2006). Both the RNAi cassette and amiRNA were expressed under the control of the ethanol-inducible promoter AlcA (Roslan et al., 2001). Several T2 lines showed mild reductions in hypocotyl length when grown in the absence of ethanol, probably due to leakiness of the AlcA promoter, but were strongly impaired when grown on plates containing 0.2% ethanol (Figure 2A). Quantitative RT-PCR of one of the RNAi lines confirmed that the expression of VHA-a2 and VHA-a3 was not inhibited by ethanol, whereas VHA-a1 transcript was reduced (Figure 2C). Therefore, we concluded that inhibition of the V-ATPase in the TGN is sufficient to cease cell expansion. Electron microscopy of high-pressure frozen and freeze-substituted seedlings was used to examine the ultrastructural effects of VHA-a1 inhibition. In contrast with the normal Golgi morphology of control seedlings (Figure 3A), Golgi stacks of seedlings grown in the presence of ethanol displayed bending and swelling of cisternae (Figure 3B).
Figure 2.
V-ATPase Activity in the TGN Is Necessary for Cell Expansion.
(A) Hypocotyl length of 4-d-old etiolated seedlings grown in the absence or presence of ethanol. Shown are the results for Col-0 and two independent transgenic lines expressing ethanol-inducible constructs with either an RNAi or an amiRNA targeted against VHA-a1, the TGN-localized isoform of VHA-a. At least 30 seedlings of each line were measured. Error bars represent se.
(B) Four-day-old etiolated seedlings of Col-0 and a homozygous, ethanol-inducible VHA-a1 RNAi line grown in the absence or the presence of 0.2% ethanol. Bar = 2 mm.
(C) Quantitative RT-PCR analysis of VHA-a1, VHA-a2, and VHA-a3 transcript levels in VHA-a1 RNAi seedlings grown in the presence of ethanol. Shown is the fold downregulation in comparison to Col-0. Error bars represent sd of three technical replicates.
Figure 3.
V-ATPase Inhibition Affects Golgi Morphology.
Electron micrographs of Golgi stacks and their associated TGNs in high-pressure frozen and freeze-substituted root tip cortex cells of uninduced (A) and induced VHA-a1 RNAi seedlings (B). Bar = 0.25 μm.
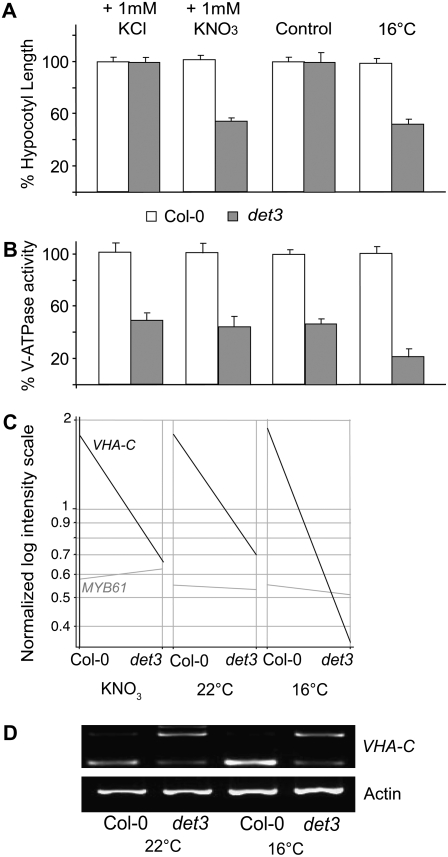
The det3 Hypocotyl Elongation Defect Is Conditional, Depending on Nitrate and Temperature
To gain further insight into the events that underlie growth inhibition caused by a lack of V-ATPase activity, we made use of the conditional nature of the det3 phenotype. We previously reported that the hypocotyl growth of etiolated det3-seedlings is normal when the gravitropic response causes roots to grow away from the plate (Schumacher et al., 1999). Therefore, we investigated whether growth inhibition is affected by nutrient uptake. Indeed, in the absence of external nutrients, etiolated seedlings of det3 were indistinguishable from the wild type, whereas 1 mM KNO3 inhibited hypocotyl length to 50% of the wild type (Figure 4A). The addition of 1 mM KCl (Figure 4A) or other potassium salts (see Supplemental Figure 1 online) did not cause growth inhibition, confirming that the observed defect is not a general anion effect and is not caused by potassium. Growth inhibition was also not detected when nitrogen was supplied as ammonium, whereas the nitrate analog chlorate inhibited growth efficiently, indicating that the observed effect is not a general nitrogen effect (see Supplemental Figure 1 online).
Figure 4.
Hypocotyl Growth Inhibition in det3 Can Be Induced by Nitrate and Temperature.
(A) Hypocotyl length of 4-d-old etiolated seedlings grown at 22°C in the absence of external nutrients (control), in the presence of 1 mM KNO3 or 1 mM KCl, or at 16°C in the absence of external nutrients. At least 30 seedlings from each of three independent experiments were measured. Col-0 length was set to 100%. Absolute values are presented in Supplemental Table 2 online. Error bars represent se.
(B) V-ATPase activity of total microsomal extracts of 4-d-old etiolated seedlings was measured as ConcA-sensitive ATP hydrolysis (see Methods). Data from at least three independent measurements were averaged, and the values obtained for Col-0 were set to 100%. Absolute values are presented in Supplemental Table 2 online. Error bars indicate sd.
(C) Expression of VHA-C and MYB61 on a normalized log intensity scale. cDNA of 4-d-old etiolated Col-0 and det3 seedlings grown under permissive (22°C, 0 mM KNO3) and restrictive conditions (22°C, 1 mM KNO3; 16°C, 0 mM KNO3) was used for hybridization of the Affymetrix ATH1 microarray.
(D) RT-PCR on cDNA of 4-d-old etiolated seedlings using primers flanking intron 1 of VHA-C. Primers for actin2 (see Supplemental Table 1 online) served as a control. The lower band is derived from the spliced transcripts; the upper band represents transcripts in which intron 1 is present.
Nitrate is an inhibitor of the V-ATPase in vitro (Dschida and Bowman, 1995) and is transported into the vacuole of Arabidopsis cells by ClC-a, an anion-H+ exchanger of the chloride channel family ClC (De Angeli et al., 2006). On the premise that reduced V-ATPase activity in det3 could lead to hyperaccumulation of nitrate in the cytosol, we next measured total V-ATPase activity as ConcA-sensitive ATP hydrolysis in the presence and absence of nitrate. However, growth in the presence of nitrate did not further reduce V-ATPase activity (Figure 4B). In addition, the det3 phenotype is temperature sensitive. Incubation at 16°C in the absence of nitrate also leads to a 50% reduction in hypocotyl growth (Figure 4A). Under these conditions total V-ATPase activity was reduced to ∼20% of Col-0 (Figure 4B). However, V-ATPase activity did not appear to be strictly correlated with the det3 phenotype under all conditions.
The det3 Mutant Shows a Temperature-Sensitive Reduction in VHA-C Splicing Efficiency
By microarray analysis (discussed below), we found that at 16°C VHA-C mRNA levels were reduced to 18% (average raw intensities: wild type, 1478; det3, 270) compared with 40% (average raw intensities: wild type, 1378; det3, 531) of Col-0 at 22°C, whereas the presence of 1 mM KNO3 did not affect VHA-C transcript levels (average raw intensities: wild type, 1472; det3, 512; Figure 4C). The det3 allele carries a point mutation in the branch point consensus sequence of the first intron, which causes 50% of the mRNA to be degraded, whereas the remaining mRNA is correctly spliced, probably due to the existence of an alternative branch point sequence (Schumacher et al., 1999). RT-PCR showed that cDNAs containing intron 1 are readily detected in det3 but not in Col-0 seedlings (Figure 4D). Taken together, these data indicate that although correctly spliced VHA-C transcripts are found under all conditions, splicing efficiency is reduced at 16°C, leading to variable levels of VHA-C mRNA degradation in the det3 mutant.
MYB61 Is Not Responsible for the Phenotype of det3
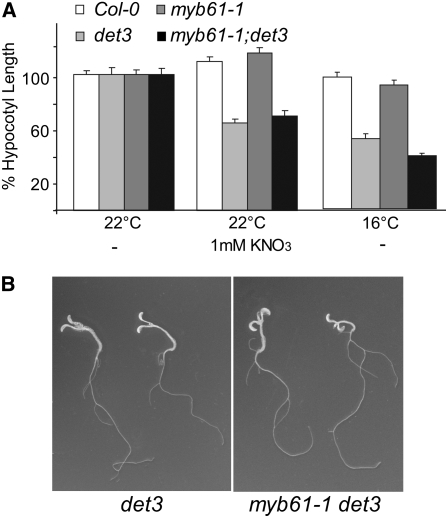
It was previously reported that the deetiolated phenotype of det3 is caused by the misexpression of the transcription factor MYB61 (Newman et al., 2004). As we could not detect overexpression of MYB61 in the microarray analysis (Figure 4C), we created the double mutant with the null allele myb61-1 (Penfield et al., 2001) to clarify the contribution of MYB61 to the det3 phenotype. Hypocotyl length of myb61-1det3 was not increased (Figure 5A), and extended growth in the dark in the presence of sucrose revealed that the phenotype of the double mutant was fully deetiolated (Figure 5B), indicating that misexpression of MYB61 cannot be responsible for the dark photomorphogenic phenotype of det3.
Figure 5.
Phenotype of the myb61 det3 Double Mutant.
(A) Hypocotyl length of 4-d-old etiolated seedlings. At least 30 seedlings from each of three independent experiments were measured. Col-0 length was set to 100%. Absolute values are presented in Supplemental Table 2 online. Error bars represent se.
(B) Phenotype of 14-d-old etiolated seedlings grown on MS plates with 1% sucrose.
Expression Profiling of det3 Mutants under Restrictive and Permissive Conditions Implicates Oxylipin Signaling
To investigate the molecular basis for the observed conditional defect, the expression profiles of etiolated Col-0 and det3 seedlings grown under permissive (22°C, 0 mM KNO3) and two restrictive conditions (22°C, 1 mM KNO3 and 16°C, 0 mM KNO3) were analyzed using the Affymetrix ATH1 chip. Two chips were hybridized for each genotype in each condition. Each chip was hybridized with cRNA pools derived from two biological replicates. The obtained data were normalized using gcRMA, and statistical significance of differential expression was determined by Logit-T (Lemon et al., 2003).
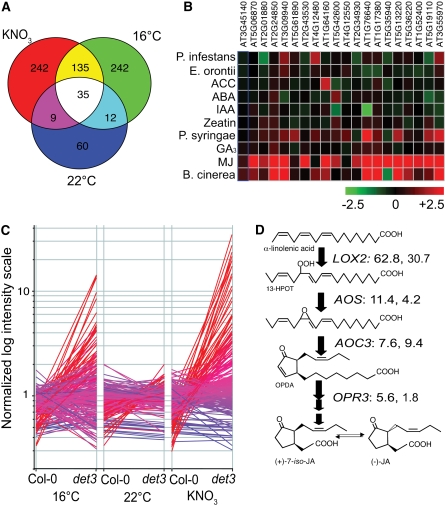
The Logit-T scores identified the following numbers of genes as differentially expressed (Logit-T scores of >29,000) between Col-0 and det3: 710 under permissive conditions of 22°C, 0 mM KNO3 (see Supplemental Data Set 1 online), 1639 at 22°C, 1 mM KNO3 (see Supplemental Data Set 2 online), and 2052 at 16°C, 0 mM KNO3 (see Supplemental Data Set 3 online). For further downstream analysis, we increased the stringency of these tests by applying an additional filter of genes showing fold change in expression greater than two or three (see below).
Among the differentially expressed genes, 116 showed at least a threefold difference in expression levels between Col-0 and det3 under permissive conditions (22°C, 0 mM KNO3; see Supplemental Data Set 1 online), and 421 (NO3; see Supplemental Data Set 2 online) and 424 (16°C; see Supplemental Data Set 3 online) showed a difference of this magnitude under restrictive conditions (Figure 6A). Quantitative RT-PCR was used to confirm the differential expression of two genes (see Supplemental Figure 2 online). Among the Gene Ontology terms in the category “biological process” associated with the genes misregulated in det3 under both restrictive conditions (170 genes, yellow and white in Figure 6A), the following terms were overrepresented with an e-score of <0.05 probability that the overrepresentation is due to chance: “response to oxidative stress” (7.77E-05), “response to pest, pathogen, or parasite” (0.028), “response to jasmonic acid stimulus” (0.047), and “jasmonic acid biosynthesis” (0.0004). Comparison with the AtGenExpress data sets for biotic stress and hormone treatments (http://www.arabidopsis.org/info/expression/ATGenExpress.jsp) using Genevestigator (Zimmermann et al., 2004) revealed that many of the genes deregulated in det3 are responsive to methyl jasmonate (MeJA) and/or the necrotrophic fungus Botrytis cinerea (Figure 6B). Five percent of all genes represented on the ATH1 array are MeJA responsive; compared with this, 35% of the genes deregulated in det3 by both nitrate and temperature are classified as MeJA responsive (Nemhauser et al., 2006). Moreover, of the 243 genes classified as MeJA specific (Nemhauser et al., 2006), 30% are more than twofold deregulated by nitrate and 26% by temperature (Figure 6C; see Supplemental Data Set 4 online).
Figure 6.
Gene Expression in det3 under Permissive and Restrictive Conditions.
(A) Venn diagram showing intersections between the sets of genes with at least threefold differences in expression level between det3 and Col-0.
(B) Heat map displaying the responses to phytohormones and biotic stress for 20 genes showing strong deregulation in det3. ACC, 1-aminocyclopropane-1-carboxylic acid; ABA, abscisic acid; IAA, indole-3-acetic acid; MJ, MeJA.
(C) Expression of genes classified as MeJA specific in 4-d-old etiolated Col-0 and det3 seedlings grown under permissive (22°C, 0 mM KNO3) and restrictive conditions (22°C, 1 mM KNO3; 16°C, 0 mM KNO3) as determined by Affymetrix ATH1 hybridization and displayed on a normalized log intensity scale. Coloring represents expression in det3 at 16°C ranging from red (normalized intensity > 5.0) to blue (normalized intensity < 0.2).
(D) Schematic representation of the JA biosynthetic pathway including the fold changes in expression level between Col-0 and det3 under the two restrictive conditions used: the first number in the KNO3 condition and the second number in the 16°C condition.
Taking into account that our microarray analysis used dark-grown seedlings, but MJ-responsive genes have been identified in experiments with light-grown seedlings and that light is required for the expression of certain JA-responsive genes, such as VSP and PDF1.2, the overlap is likely to be an underestimation. Strikingly, the genes encoding the enzymes responsible for the first four steps of jasmonate (JA) biosynthesis (LOX2, AOS, AOC3, and OPR3) were highly to moderately overexpressed in det3 mutants under restrictive conditions (Figure 6D).
Oxylipin and Ethylene Signaling Are Necessary and Sufficient for Hypocotyl Growth Inhibition in det3
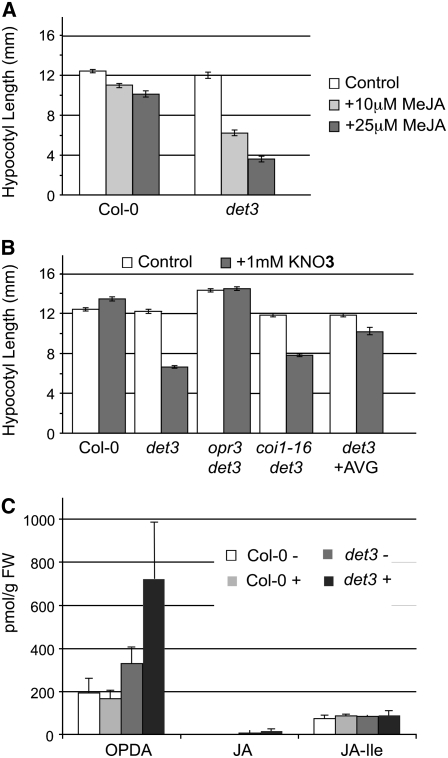
Given the strong overrepresentation of JA biosynthesis and JA-regulated genes among the genes affected by det3, we next addressed the causal relation between JA signaling and growth inhibition. When wild-type and det3 seedlings were grown under permissive conditions (22°C, 0 mM KNO3) in the presence of MeJA, hypocotyl growth in det3 was strongly inhibited, whereas wild-type seedlings showed only weak inhibition (Figure 7A). Thus, JA signaling is sufficient for hypocotyl growth inhibition in det3. To determine if JA signaling is also necessary, double mutants between det3 and opr3, a mutant that lacks activity of a OPDA reductase required for JA synthesis (Stintzi and Browse, 2000), and the JA-insensitive coi1-16 mutant (Ellis and Turner, 2002) were generated. Nitrate-mediated growth inhibition was abolished in the det3 opr3 double mutant, demonstrating that JA signaling is not only sufficient but also necessary for growth inhibition. It is important to note that this effect is limited to nitrate concentrations up to 5 mM, whereas the severe phenotype caused by growth on Murashige and Skoog (MS) medium, which contains almost 40 mM nitrate, is not affected in the det3 opr3 double mutant. Surprisingly, the det3 coi1-16 double mutant showed little difference when compared with the det3 single mutant (Figure 7B), suggesting that the signal leading to growth inhibition is not perceived by the central JA-signaling component SCFCOI1 (Turner et al., 2002; Wasternack, 2006) that has recently been shown to interact with JAZ repressor proteins in the presence of the jasmonoyl-isoleucine conjugate (JA-Ile) (Thines et al., 2007).
Figure 7.
Oxylipins Are Necessary and Sufficient for Hypocotyl Growth Inhibition.
(A) and (B) Hypocotyl length of 4-d-old etiolated seedlings; genotype and growth conditions are indicated. At least 30 seedlings from each of three independent experiments were measured. Wild-type length was set to 100%. Error bars represent se.
(C) Amount of OPDA, JA, and JA-Ile in 3-d-old etiolated Col-0 and det3 seedlings grown with (+) or without 2 mM KNO3 as analyzed by gas chromatography–mass spectrometry. Error bars indicate sd of three independent experiments. FW, fresh weight.
When endogenous levels of JA-Ile, JA, and the biosynthetic intermediate OPDA were determined in 3-d-old seedlings grown on KNO3, levels of JA-Ile were found to be comparable in Col-0 and det3 (Figure 6C). Very low levels of JA were detected in det3 but showed no difference between seedlings grown in the presence or absence of nitrate. By contrast, the level of OPDA was already slightly increased in det3 grown in the absence of nitrate and was increased approximately threefold in det3 seedlings grown in the presence of nitrate (Figure 7C).
Although the gene expression profiles did not indicate an involvement of ethylene, hypocotyl growth inhibition is a classical ethylene response and we therefore addressed the contribution of this phytohormone using the biosynthesis inhibitor aminovinylglycine (AVG) (Yu and Yang, 1979). The presence of AVG strongly reduced the growth-inhibiting effect of nitrate (Figure 7B); however, in the same samples used to determine the oxylipins, ethylene could not be detected by gas chromatography–mass spectroscopy.
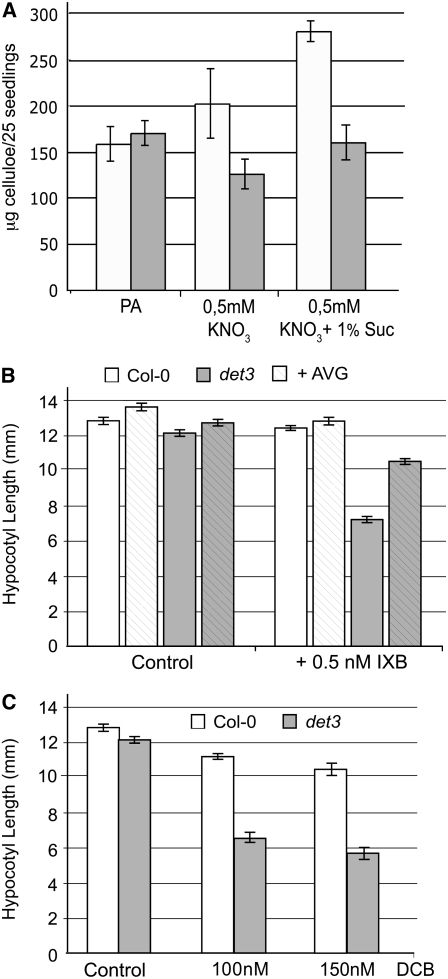
Cellulose Synthesis Is Reduced in det3
OPDA, JA, and ethylene levels are elevated in mutants with defects in cellulose biosynthesis (Ellis et al., 2002; Cano-Delgado et al., 2003), and it has been shown that cellulose biosynthesis is reduced in severely dwarfed light-grown det3 seedlings (Cano-Delgado et al., 2003). To determine if cellulose biosynthesis is also affected in det3 seedlings showing only a mild growth reduction, we first measured the cellulose content of 4-d-old etiolated seedlings. Whereas no difference was found between Col-0 and det3 seedlings grown in the absence of nitrate, cellulose content of det3 was less than that of wild-type seedlings when grown in the presence of 1 mM KNO3 (Figure 8A). The presence of sucrose strongly increased the cellulose content of Col-0 but not of det3 seedlings (Figure 8A). Moreover, det3 displays strong hypersensitivity when grown under permissive conditions in the presence of the cellulose synthase inhibitors isoxaben (IXB; Figure 8B) and dichlorobenzonitrile (DCB; Figure 8C). Again, the presence of AVG strongly ameliorated the phenotype, confirming that hormone signaling is necessary for hypocotyl growth inhibition.
Figure 8.
det3 Has Reduced Amounts of Cellulose and Is Hypersensitive to Inhibitors of Cellulose Synthase.
(A) Amount of cellulose in 4-d-old etiolated Col-0 and det3 seedlings grown under the indicated conditions. Dry weight of etiolated wild-type and det3 seedlings is not significantly different (Col-0, 43.5 ± 1.2 mg/1000 seedlings; det3, 42.9 ± 1.5 mg). Error bars indicate sd of three independent experiments. PA, phytagar.
(B) Hypocotyl length of 4-d-old etiolated Col-0 and det3 seedlings grown in the absence or presence of the indicated concentrations of IXB and AVG. At least 30 seedlings from each of three independent experiments were measured. Error bars indicate se.
DISCUSSION
Reduced Tonoplast V-ATPase Activity Does Not Interfere with Cell Expansion
The activity of the V-ATPase is strongly correlated with cell expansion, and the genetic inhibition of the V-ATPase caused either by antisense (Gogarten et al., 1992), RNAi (Padmanaban et al., 2004), or a weak mutant allele (Schumacher et al., 1999) reduces cell expansion. In all these cases both, the Tonoplast and the TGN isoforms of the V-ATPase were affected, and the cellular basis of the observed growth defects has not been determined. We showed here that cell expansion is not affected in a mutant that lacks VHA-a3, one of two tonoplast-localized isoforms of the membrane-integral subunit VHA-a, although total V-ATPase activity is limited to a similar extent as in the severely dwarfed det3 mutant. Similarly, for reasons yet to be determined, tonoplast V-ATPase activity is reduced in the cax1 mutant deficient in vacuolar Ca2+/H+-antiport, but cell expansion is normal (Cheng et al., 2003). In det3, the expression of the single-copy gene VHA-C is reduced so that not only the V-ATPase at the tonoplast but also in the TGN is affected. Taken together, these results indicate that in VHA-a3 mutants, the remaining tonoplast V-ATPase activity together with the second vacuolar proton pump, the vacuolar H+-pyrophosphatase (Schumacher, 2006), is sufficient to create the necessary turgor pressure. By contrast, in det3 mutants and VHA-a1 mutants, reduced V-ATPase activity in the TGN restricts cell expansion. Indeed, we showed that inhibition of the TGN-localized isoform VHA-a1 leads to reduced hypocotyl growth. On the subcellular level, RNAi inhibition of VHA-a1 causes the same characteristic changes in Golgi morphology previously described for cells treated with the V-ATPase inhibitor ConcA (Dettmer et al., 2005, 2006). Hypocotyl growth is inhibited in a dose-dependent manner by ConcA, and this effect is enhanced in a mutant lacking the TGN-localized anion transporter ClC-d (von der Fecht-Bartenbach et al., 2007), again confirming that reduced acidification of the TGN causes cell expansion to cease.
The Conditional Phenotype of det3
The temperature dependence of det3 mRNA splicing provides a molecular explanation for the reduced cell expansion at 16°C. It has recently been proposed that nonsense-mediated mRNA decay can compensate for suboptimal splicing efficiency (Jaillon et al., 2008), and it remains to be determined if the two in-frame stop codons found in intron 1 induce nonsense-mediated mRNA decay of unspliced VHA-C. By contrast, the nitrate sensitivity of det3 is more difficult to explain as nitrate does not cause a further reduction of VHA-C transcript or ATP hydrolysis. The limited amounts of VHA-C present in det3 might be sufficient to maintain activity in the TGN in the absence of nitrate, whereas the presence of nitrate would increase the demand for V1 subcomplexes at the tonoplast, thus limiting the activity in the TGN. We have not provided experimental evidence for this hypothesis, but it is in agreement with the observation that nitrate can cause changes in V-ATPase structure and composition (Ratajczak, 2000; Drobny et al., 2002). Regardless of the molecular mechanisms, the conditional nature of the det3 phenotype provided a unique opportunity to study the chain of events leading from reduced V-ATPase activity to reduced cell expansion.
Hormone Signaling and Cell Expansion
Expression profiling pointed to an involvement of oxylipin signaling, and we could show that oxylipins are necessary and sufficient for hypocotyl elongation inhibition in det3, at least under conditions that cause a mild inhibition of hypocotyl growth. Surprisingly, OPDA levels (but not JA or JA-IIe levels) are increased in det3 seedlings, and hypocotyl growth inhibition is independent of the SCFCOI1 complex. The phenotype of det3 seedlings is similar to that of cev1, a mutant affected in the cellulose synthase subunit CESA3 that also has elevated levels of OPDA (Ellis et al., 2002). This could mean that OPDA is the active molecule mediating the growth inhibition and that the response to MeJA is due to the well-documented positive feedback in JA biosynthesis (Turner et al., 2002; Wasternack 2006) leading to enhanced synthesis of OPDA. The restored hypocotyl growth of the opr3 det3 double mutant, at first glance, argues against OPDA as the active molecule, as OPR3 catalyzes the conversion of OPDA and not its synthesis. However, OPDA levels are reduced in the opr3 mutant (Stintzi et al., 2001) most likely due to the reduced positive feedback regulation of its biosynthesis. It has been shown that OPDA has JA-independent signaling functions in plant defense reactions (Stintzi et al., 2001) and OPDA-specific responses may occur by the OPDA-specific expression of genes coding for cell wall modifying enzymes (Taki et al., 2005). Although JA is well known to cause inhibition of root growth, no member of the oxylipin family has so far been implicated in the control of hypocotyl elongation. Our data therefore point to OPDA as a new component of the well-studied regulatory network controlling hypocotyl growth.
Ethylene is the classic negative regulator of hypocotyl growth (Guzman and Ecker, 1990), and although expression profiling did not point to a strong involvement of ethylene signaling, inhibition of ethylene biosynthesis was able to restore hypocotyl elongation in det3. One possibility would be that ethylene biosynthesis might be necessary for oxylipin synthesis in etiolated seedlings, but the exact mode of interaction between the two pathways remains to be determined. Interestingly, it has recently been shown that the ethylene receptor ETR1 and the interacting protein RTE1 (Resnick et al., 2006; Zhou et al., 2007) colocalize in the Golgi apparatus (Dong et al., 2008), and it thus seems possible that ethylene signaling is linked to Golgi function and that cell wall synthesis could be directly regulated by ethylene.
Crosstalk between JA and ethylene signaling is well documented (Xu et al., 1994; Penninckx et al., 1998) in particular during defense responses against necrotrophic pathogens. Indeed, the expression profile of det3 is quite similar to the one of plants infected with the necrotrophic fungus B. cinerea. Many of the genes encode typical defense components like peroxidases, protease inhibitors, or myrosinase-associated proteins. Therefore, it is of interest if these changes in gene expression reflect an accidental defense reaction or if they simply reflect a program for growth inhibition that can be triggered by different stimuli leading to cell wall crosslinking and growth inhibition.
det3: A Cellulose-Deficient Mutant
Activation of oxylipin and ethylene signaling is a feature that det3 shares with mutants like cev1 and eli1, which are cellulose deficient due to mutations in cellulose synthase genes (Ellis et al., 2002; Cano-Delgado et al., 2003). Light-grown det3 seedlings were previously shown to have reduced cellulose synthesis (Cano-Delgado et al., 2003), but it was not clear if this is a primary defect or a secondary facet of the severe and pleiotropic dwarf phenotype. We have shown here that etiolated det3 seedlings, displaying only a mild growth reduction, are also cellulose deficient and are hypersensitive to the cellulose synthesis inhibitors IXB and DCB. IXB targets the cellulose synthase catalytic subunits CESA3 and CESA6 (Scheible et al., 2001; Desprez et al., 2002) and has recently been shown to affect the trafficking of yellow fluorescent protein-CESA6 (Paredez et al., 2006) and of KOR, an endo-1,4-β-d-glucanase involved in cellulose synthesis that undergoes cycling between intracellular compartments, including Golgi, early endosomes, and the vacuole (Robert et al., 2005). The activity of the V-ATPase in the TGN is required for endocytic and secretory trafficking (Dettmer et al., 2006), and the trafficking of proteins involved in cellulose biosynthesis could be sensitive to changes in V-ATPase activity. Furthermore, the assembly of CesA complexes takes place in the Golgi/TGN (Haigler and Brown, 1986) and could be pH dependent. However, we cannot exclude that the primary defect lies in the synthesis of other cell wall components in the Golgi and that the reduced cellulose amount is only a secondary effect. More detailed studies of det3 cell walls may elucidate the connection between Golgi/TGN acidification and cell wall synthesis. Dual-function proteins like KAM1/MUR3, a xyloglucan galactosyltransferase (Madson et al., 2003) that also plays an important role in Golgi organization (Tamura et al., 2005), already provide evidence for a close connection between cell wall biosynthesis and endomembrane organization.
In summary, we have shown here that a subtle cell wall defect caused by inhibition of V-ATPase in the TGN leads to oxylipin- and ethylene-mediated changes in gene expression that in turn prohibit further cell expansion, possibly by increased cell wall crosslinking. Our results support the existence of a mechanism that monitors cell wall integrity and actively prevents further growth if either composition or physical properties of the cell wall deviate. A receptor-like kinase that may act as a cell wall integrity sensor has recently been identified (Hematy et al., 2007), and it will be of great interest to see if this receptor is also responsible for the activation of the signaling cascade that prohibits cell expansion in the det3 mutant.
METHODS
Fluorescent Protein Constructs and Fluorescence Microscopy
VHA-a2-GFP and VHA-a3-GFP were described previously (Dettmer et al., 2006). To obtain a fusion construct that allows colocalization of both isoforms, the GFP coding sequence was replaced by mRFP. Seedlings expressing the resulting construct VHA-a3-mRFP were crossed to homozygous VHA-a2-GFP plants, and the segregating F2 was inspected by microscopy. Fluorescence microscopy was performed using a Leica TCS SP2 CLSM. All CLSM images were obtained using the Leica Confocal Software and a ×63 water immersion objective. The excitation wavelength was 488 nm, and emission was detected for GFP between 500 and 530 nm and for RFP between 565 and 600 nm. Images were processed using Adobe Photoshop.
PCR and RT-PCR
Genomic DNA and RNA isolation and cDNA synthesis were performed as described previously (Dettmer et al., 2005). Quantitative RT-PCR amplification was performed in the presence of the double-stranded DNA binding dye SYBR Green (Molecular Probes) and monitored in real time with the Opticon continuous fluorescence detection system (MJ Research). Amplification of TUBULIN BETA-2 served as cDNA loading control. A complete list of oligonucleotides used for PCR, RT-PCR, and quantitative RT-PCR reactions is provided in Supplemental Table 1 online.
V-ATPase Activity Measurements
V-ATPase activity was measured using a modification of the protocol described by Palmgren (1990) in which ATP hydrolysis is coupled enzymatically to the oxidation of NADH, which can be followed by an absorbance decrease at 340 nm. In brief, 10 μg of microsomal protein was added to a master mix containing 37.5 mM MOPS, 4 mM MgSO4, 50 mM KCl, 3 mM orthovanadate, 1 mM phosphoenolpyruvate, 0.3 mM NADH, 10 mg/mL pyruvate kinase, and 10 mg/mL lactate dehydrogenase. Reactions with and without the V-ATPase–specific inhibitor ConcA (100 nM) were started by addition of ATP to a final concentration of 2 mM in a microtiter plate, and absorbance at 340 nm was monitored for 20 min in a Tecan Safire plate reader. All measurements were performed in triplicate, and V-ATPase activity was calculated as ConcA-inhibited ATP hydrolysis.
Plant Growth and Hypocotyl Measurements
Etiolated Arabidopsis thaliana seedlings were grown in Petri dishes on either PA medium (water solidified with 1% phytagar adjusted to pH 5.8 with 20 mM MES) or on MS medium (1× MS salts in water solidified with 0.7% phytagar, pH 5.8). Seeds were surface sterilized and treated for 48 h at 4°C before planting. After exposure to 100 μmol m−2 s−1 of fluorescent white light for 4 h, plates were wrapped in two layers of aluminum foil and incubated for 4 d at 22 or 16°C. All plants for microarray analysis and enzyme assays were grown in this manner. For hypocotyl length measurements, seedlings were sandwiched between two sheets of acetate and scanned in a flatbed scanner at a resolution of 300 points per inch. The digitized images were analyzed using the NIH image software.
Ethanol-Inducible RNAi and amiRNA
A 203-bp fragment derived from the 5′ region of VHA-a1 was amplified from cDNA using oligonucleotides VHA-a1RNAi.FOR and VHA-a1RNAi.REV (see Supplemental Table 1 online) and cloned as an inverted repeat into a derivative of pHANNIBAL (Wesley et al., 2001) in which the 35S promoter has been replaced by the ethanol-inducible promoter pAlcA (Roslan et al., 2001). The pAlcA:VHA-a1RNAi cassette was then cloned into the NotI site of the plant transformation vector pBart_AlcR that contains the coding sequence of the alcR transcription factor required to activate the pAlcA promoter in the presence of ethanol.
The amiRNA for VHA-a1 was PCR amplified (see Supplemental Table 1 online) according to the protocol provided via WMD2 Web MicroRNA Designer (http://wmd2.weigelworld.org) and cloned into the pCR2.1-TOPO vector. The VHA-a1 amiRNA was then cloned into pBJ36_AlcA, from which it was subcloned into pBART_AlcR. Transgenic plants were selected based on the phosphinothricin (BASTA) resistance conferred by the bar gene contained in pBART_AlcR. Homozygous lines were established, and hypocotyl length of etiolated seedlings grown on MS plates containing 0.2% ethanol was compared with seedlings grown in the absence of ethanol.
Transmission Electron Microscopy
Seedling root tips were high pressure frozen (Bal-Tec HPM 010; Balzers) in hexadecane (Merck Sharp and Dohme), freeze substituted (72 h, −90°C; 8 h, −60°C; 8 h, −35°C; 4 h, 0°C) in acetone containing 2% osmium tetroxide and 0.5% uranyl acetate, washed at 0°C, and embedded in Epon, then sectioned as described by Dettmer et al. (2006).
Microarray Analysis
Four-day-old etiolated Col-0 and det3 seedlings were grown on PA medium at 16°C (Condition 1), at 22°C (Condition 2), and at 22°C with 1 mM KNO3 added (Condition 3). RNA was extracted from pools of seedlings (50 to 100 mg fresh weight) grown in four independent biological replicates using the Plant RNeasy Mini kit (Qiagen). After quality control by agarose gel electrophoresis, 5 μg total RNA from two biological replicates were pooled to reduce replicate number from four to two. These pools were used as starting material for Affymetrix probe synthesis following the AtGenExpress protocol (Schmid et al., 2005). Two Affymetrix ATH1 high-density oligonucleotide arrays were used for hybridization of the two independent probes per genotype per condition, for a total of 12 chips (two genotypes × two replicate chips per genotype × three conditions). Hybridization, washing, and scanning were performed according to the manufacturer's instructions (Affymetrix). Arrays were scanned using Affymetrix Microarray Suite version 5.0, and expression estimates were calculated by gcRMA implemented in R (Irizarry et al., 2006). Expression estimates were imported into Genespring software (version 7.2; Agilent Technologies) for cluster analysis and visual inspection. Differentially expressed genes were identified using Logit-T (Lemon et al., 2003) on raw CEL files with a probability cutoff set to P < 0.05. Logit-T employs a logit transformation for normalization followed by statistical testing and multiple testing correction. In contrast with other methods for detecting differentially expressed genes that are based on expression estimates, Logit-T works at the probe level. In the case of the ATH1 array used, every transcript is represented by 11 probes, and those 11 probes are used for independent testing by Logit-T. The Logit-T score is defined as the median t-value found among all the perfect match probes in the set across the experiments compared. The Logit-T score corresponding to the desired P value cutoff was defined using a standard t-table at 2 degrees of freedom. By this measure, Logit-T scores of >29,000 indicate a significant difference (positive and negative scores being associated with a decrease or an increase, respectively, in det3 relative to the wild type). Since Logit-T is independent of expression values, some of the transcripts detected as differentially expressed only show marginal fold changes between experiment and control. Therefore, we used fold change filtering based on gcRMA expression estimates to facilitate downstream analysis in addition to statistical testing. Analysis of associated Gene Ontology terms was performed with the Genemerge program (Castillo-Davis and Hartl, 2003).
Double Mutant Analysis
The det3 mutant (Col-0) was used to pollinate the male-sterile opr3 (Wassilewskija [Ws]; Stintzi and Browse, 2000) and coi1-16 (Col-0; Ellis and Turner, 2002) mutants. Among the homozygous det3 plants identified in the segregating F2 by their characteristic dwarf phenotype, potential opr3 det3 and coi1-16 det3 double mutants were identified based on their sterility, which was cured either by spraying with 450 μM MeJA (opr3) or shifting to the permissive temperature of 16°C (coi1-16). Homozygosity was confirmed using the segregation of kanamycin resistance for opr3 or a cleaved-amplified polymorphic sequence marker (MboI) in the case of coi1-16. To exclude the influence of modifier genes in the mixed Ws/Col-0, we confirmed that the det3 phenotype was fully penetrant in the Ws background (data not shown), and pooled seeds of several opr3 det3 double mutants identified in the F2 were used for hypocotyl measurements.
Hormone Measurements
JA, JA-Ile, and OPDA in 4-d-old etiolated Col-0 and det3 seedlings grown in the presence of 1 mM KNO3 were determined as described previously (Hause et al., 2000). For ethylene measurements, seedlings were grown for 4 d on MS medium in erlenmeyer flasks closed with a SUBA-seal. Duplicate head space samples were analyzed using a gas chromatograph (Hewlett Packard 5890 Series II). Seedling fresh weight was determined, and ethylene production per seedling was calculated. All hormone measurements were performed in triplicate using independent seed batches.
Determination of Cell Wall Components
Cellulose content was determined according to Updegraff (1969) with slight modifications. Four-day-old etiolated seedlings were incubated for 30 min in 90% ethanol at 65°C to inactivate enzymes. After seedlings were dried overnight at 80°C, hemicellulose and pectin were removed by boiling for 30 min in 73% acetic acid and 9% nitric acid. After centrifugation, the remaining pellet was washed with water followed by acetone. Cellulose was dissolved in 72% sulfuric acid, and the resulting glucose concentration was determined photometrically at 650 nm after addition of 3% anthrone in sulfuric acid.
Accession Numbers
Arabidopsis Genome Initiative locus identifiers are as follows: VHA-a1 (AT2G28520), VHA-a2 (AT2G21410), VHA-a3 (AT4G39080), MYB61 (AT1G09540), LOX2 (AT3G45140), AOC3 (AT3G25780), AOS (AT5G42650), OPR3 (AT2G06050), and TUBULIN BETA-2 (AT5G62690). The microarray data set has been deposited at ArrayExpress (EMBL-EBI) under accession number E-MEXP-1468.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. MYB61 RT-PCR and Phenotype of the myb61-1 det3 Double Mutant.
Supplemental Figure 2. RT-PCR of Genes Misregulated in det3.
Supplemental Table 1. List of Oligonucleotides Used in This Study.
Supplemental Table 2. Absolute Values for Figures 4 and 5.
Supplemental Data Set 1. Genes Deregulated in det3 under Permissive Conditions.
Supplemental Data Set 2. Genes Deregulated in det3 under Restrictive Conditions (Nitrate).
Supplemental Data Set 3. Genes Deregulated in det3 under Restrictive Conditions (Temperature).
Supplemental Data Set 4. MeJA-Specific Genes Deregulated in det3 under Both Restrictive Conditions.
Supplementary Material
Acknowledgments
We thank Anne Hong-Hermesdorf and Felicity de Courcy for critical reading of the manuscript. We also thank Annik Stintzi for providing opr3 and coi1-16 seeds and Mike Bevan for providing myb61-1 seeds. We are grateful to Mathias Grauer, Zhao-Xin Wang, Elke Sauberzweig, and Dagmar Ripper for excellent technical assistance. This work was supported by the Deutsche Forschungsgemeinschaft (SFB 446: A20, Z2, B22) and an EMBO Young Investigator Award to J.U.L.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Karin Schumacher (karin.schumacher@hip.uni-heidelberg.de)
Online version contains Web-only data.
References
- Achard, P., Liao, L., Jiang, C., Desnos, T., Bartlett, J., Fu, X., and Harberd, N.P. (2007). DELLAs contribute to plant photomorphogenesis. Plant Physiol. 143 1163–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Arioli, T., et al. (1998). Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 279 717–720. [DOI] [PubMed] [Google Scholar]
- Bouton, S., Leboeuf, E., Mouille, G., Leydecker, M.T., Talbotec, J., Granier, F., Lahaye, M., Hofte, H., and Truong, H.N. (2002). QUASIMODO1 encodes a putative membrane-bound glycosyltransferase required for normal pectin synthesis and cell adhesion in Arabidopsis. Plant Cell 14 2577–2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera y Poch, H., Peto, C., and Chory, J. (1993). A mutation in the Arabidopsis DET3 gen uncouples photoregulated leaf development from gene expression and chloroplast biogenesis. Plant J. 4 671–682. [Google Scholar]
- Cano-Delgado, A., Penfield, S., Smith, C., Catley, M., and Bevan, M. (2003). Reduced cellulose synthesis invokes lignification and defense responses in Arabidopsis thaliana. Plant J. 34 351–362. [DOI] [PubMed] [Google Scholar]
- Castillo-Davis, C.I., and Hartl, D.L. (2003). GeneMerge – Post-genomic analysis, data mining, and hypothesis testing. Bioinformatics 19 891–892. [DOI] [PubMed] [Google Scholar]
- Chen, M., Chory, J., and Fankhauser, C. (2004). Light signal transduction in higher plants. Annu. Rev. Genet. 38 87–117. [DOI] [PubMed] [Google Scholar]
- Cheng, N.H., Pittman, J.K., Barkla, B.J., Shigaki, T., and Hirschi, K.D. (2003). The Arabidopsis cax1 mutant exhibits impaired ion homeostasis, development, and hormonal responses and reveals interplay among vacuolar transporters. Plant Cell 15 347–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collett, C.E., Harberd, N.P., and Leyser, O. (2000). Hormonal interactions in the control of Arabidopsis hypocotyl elongation. Plant Physiol. 124 553–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove, D.J. (2005). Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 6 850–861. [DOI] [PubMed] [Google Scholar]
- De Angeli, A., Monachello, D., Ephritikhine, G., Frachisse, J.M., Thomine, S., Gambale, F., and Barbier-Brygoo, H. (2006). The nitrate/proton antiporter AtCLCa mediates nitrate accumulation in plant vacuoles. Nature 442 939–942. [DOI] [PubMed] [Google Scholar]
- Desprez, T., Vernhettes, S., Fagard, M., Refregier, G., Desnos, T., Aletti, E., Py, N., Pelletier, S., and Hofte, H. (2002). Resistance against herbicide isoxaben and cellulose deficiency caused by distinct mutations in same cellulose synthase isoform CESA6. Plant Physiol. 128 482–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettmer, J., Hong-Hermesdorf, A., Stierhof, Y.D., and Schumacher, K. (2006). Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell 18 715–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettmer, J., Schubert, D., Calvo-Weimar, O., Stierhof, Y.D., Schmidt, R., and Schumacher, K. (2005). Essential role of the V-ATPase in male gametophyte development. Plant J. 41 117–124. [DOI] [PubMed] [Google Scholar]
- Dong, C.H., Rivarola, M., Resnick, J.S., Maggin, B.D., and Chang, C. (2008). Subcellular co-localization of Arabidopsis RTE1 and ETR1 supports a regulatory role for RTE1 in ETR1 ethylene signaling. Plant J. 53 275–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drobny, M., Schnolzer, M., Fiedler, S., Luttge, U., Fischer-Schliebs, E., Christian, A.L., and Ratajczak, R. (2002). Phenotypic subunit composition of the tobacco (Nicotiana tabacum L.) vacuolar-type H(+)-translocating ATPase. Biochim. Biophys. Acta 1564 243–255. [DOI] [PubMed] [Google Scholar]
- Dschida, W.J., and Bowman, B.J. (1995). The vacuolar ATPase: Sulfite stabilization and the mechanism of nitrate inactivation. J. Biol. Chem. 270 1557–1563. [DOI] [PubMed] [Google Scholar]
- Ellis, C., Karafyllidis, I., Wasternack, C., and Turner, J.G. (2002). The Arabidopsis mutant cev1 links cell wall signaling to jasmonate and ethylene responses. Plant Cell 14 1557–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis, C., and Turner, J.G. (2002). A conditionally fertile coi1 allele indicates cross-talk between plant hormone signalling pathways in Arabidopsis thaliana seeds and young seedlings. Planta 215 549–556. [DOI] [PubMed] [Google Scholar]
- Gendreau, E., Traas, J., Desnos, T., Grandjean, O., Caboche, M., and Hofte, H. (1997). Cellular basis of hypocotyl growth in Arabidopsis thaliana. Plant Physiol. 114 295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogarten, J.P., Fichmann, J., Braun, Y., Morgan, L., Styles, P., Taiz, S.L., DeLapp, K., and Taiz, L. (1992). The use of antisense mRNA to inhibit the tonoplast H+ ATPase in carrot. Plant Cell 4 851–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman, P., and Ecker, J.R. (1990). Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 2 513–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigler, C.H., and Brown, R.M., Jr. (1986). Transport of rosettes from the Golgi apparatus to the plasma membrane in isolated mesophyll cells of Zinnia elegans during differentiation. Protoplasma 134 111–120. [Google Scholar]
- Hause, B., Stenzel, I., Miersch, O., Maucher, H., Kramell, R., Ziegler, J., and Wasternack, C. (2000). Tissue-specific oxylipin signature of tomato flowers: Allene oxide cyclase is highly expressed in distinct flower organs and vascular bundles. Plant J. 24 113–126. [DOI] [PubMed] [Google Scholar]
- Hematy, K., Sado, P.E., Van Tuinen, A., Rochange, S., Desnos, T., Balzergue, S., Pelletier, S., Renou, J.P., and Hofte, H. (2007). A receptor-like kinase mediates the response of Arabidopsis cells to the inhibition of cellulose synthesis. Curr. Biol. 17 922–931. [DOI] [PubMed] [Google Scholar]
- Irizarry, R.A., Wu, Z., and Jaffee, H.A. (2006). Comparison of Affymetrix GeneChip expression measures. Bioinformatics 22 789–794. [DOI] [PubMed] [Google Scholar]
- Jaillon, O., et al. (2008). Translational control of intron splicing in eukaryotes. Nature 451 359–362. [DOI] [PubMed] [Google Scholar]
- Lemon, W.J., Liyanarachchi, S., and You, M. (2003). A high performance test of differential gene expression for oligonucleotide arrays. Genome Biol. 4 R67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerouxel, O., Cavalier, D.M., Liepman, A.H., and Keegstra, K. (2006). Biosynthesis of plant cell wall polysaccharides - A complex process. Curr. Opin. Plant Biol. 9 621–630. [DOI] [PubMed] [Google Scholar]
- Madson, M., Dunand, C., Li, X., Verma, R., Vanzin, G.F., Caplan, J., Shoue, D.A., Carpita, N.C., and Reiter, W.D. (2003). The MUR3 gene of Arabidopsis encodes a xyloglucan galactosyltransferase that is evolutionarily related to animal exostosins. Plant Cell 15 1662–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser, J.L., Hong, F., and Chory, J. (2006). Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126 467–475. [DOI] [PubMed] [Google Scholar]
- Newman, L.J., Perazza, D.E., Juda, L., and Campbell, M.M. (2004). Involvement of the R2R3-MYB, AtMYB61, in the ectopic lignification and dark-photomorphogenic components of the det3 mutant phenotype. Plant J. 37 239–250. [DOI] [PubMed] [Google Scholar]
- Nicol, F., His, I., Jauneau, A., Vernhettes, S., Canut, H., and Hofte, H. (1998). A plasma membrane-bound putative endo-1,4-beta-D-glucanase is required for normal wall assembly and cell elongation in Arabidopsis. EMBO J. 17 5563–5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanaban, S., Lin, X., Perera, I., Kawamura, Y., and Sze, H. (2004). Differential expression of vacuolar H+-ATPase subunit c genes in tissues active in membrane trafficking and their roles in plant growth as revealed by RNAi. Plant Physiol. 134 1514–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmgren, M.G. (1990). An H-ATPase assay: Proton pumping and ATPase activity determined simultaneously in the same sample. Plant Physiol. 94 882–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredez, A.R., Somerville, C.R., and Ehrhardt, D.W. (2006). Visualization of cellulose synthase demonstrates functional association with microtubules. Science 312 1491–1495. [DOI] [PubMed] [Google Scholar]
- Penfield, S., Meissner, R.C., Shoue, D.A., Carpita, N.C., and Bevan, M.W. (2001). MYB61 is required for mucilage deposition and extrusion in the Arabidopsis seed coat. Plant Cell 13 2777–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx, I.A., Thomma, B.P., Buchala, A., Metraux, J.P., and Broekaert, W.F. (1998). Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 10 2103–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratajczak, R. (2000). Structure, function and regulation of the plant vacuolar H(+)-translocating ATPase. Biochim. Biophys. Acta 1465 17–36. [DOI] [PubMed] [Google Scholar]
- Resnick, J.S., Wen, C.K., Shockey, J.A., and Chang, C. (2006). REVERSION-TO-ETHYLENE SENSITIVITY1, a conserved gene that regulates ethylene receptor function in Arabidopsis. Proc. Natl. Acad. Sci. USA 103 7917–7922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert, S., Bichet, A., Grandjean, O., Kierzkowski, D., Satiat-Jeunemaitre, B., Pelletier, S., Hauser, M.T., Hofte, H., and Vernhettes, S. (2005). An Arabidopsis endo-1,4-beta-D-glucanase involved in cellulose synthesis undergoes regulated intracellular cycling. Plant Cell 17 3378–3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roslan, H.A., Salter, M.G., Wood, C.D., White, M.R., Croft, K.P., Robson, F., Coupland, G., Doonan, J., Laufs, P., Tomsett, A.B., and Caddick, M.X. (2001). Characterization of the ethanol-inducible alc gene-expression system in Arabidopsis thaliana. Plant J. 28 225–235. [DOI] [PubMed] [Google Scholar]
- Scheible, W.R., Eshed, R., Richmond, T., Delmer, D., and Somerville, C. (2001). Modifications of cellulose synthase confer resistance to isoxaben and thiazolidinone herbicides in Arabidopsis Ixr1 mutants. Proc. Natl. Acad. Sci. USA 98 10079–10084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid, M., Davison, T.S., Henz, S.R., Pape, U.J., Demar, M., Vingron, M., Scholkopf, B., Weigel, D., and Lohmann, J.U. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37 501–506. [DOI] [PubMed] [Google Scholar]
- Schumacher, K. (2006). Endomembrane proton pumps: Connecting membrane and vesicle transport. Curr. Opin. Plant Biol. 9 595–600. [DOI] [PubMed] [Google Scholar]
- Schumacher, K., Vafeados, D., McCarthy, M., Sze, H., Wilkins, T., and Chory, J. (1999). The Arabidopsis det3 mutant reveals a central role for the vacuolar H(+)-ATPase in plant growth and development. Genes Dev. 13 3259–3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab, R., Ossowski, S., Riester, M., Warthmann, N., and Weigel, D. (2006). Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18 1121–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville, C. (2006). Cellulose synthesis in higher plants. Annu. Rev. Cell Dev. Biol. 22 53–78. [DOI] [PubMed] [Google Scholar]
- Stintzi, A., and Browse, J. (2000). The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc. Natl. Acad. Sci. USA 97 10625–10630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stintzi, A., Weber, H., Reymond, P., Browse, J., and Farmer, E.E. (2001). Plant defense in the absence of jasmonic acid: The role of cyclopentenones. Proc. Natl. Acad. Sci. USA 98 12837–12842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taki, N., et al. (2005). 12-Oxo-phytodienoic acid triggers expression of a distinct set of genes and plays a role in wound-induced gene expression in Arabidopsis. Plant Physiol. 139 1268–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, K., Shimada, T., Kondo, M., Nishimura, M., and Hara-Nishimura, I. (2005). KATAMARI1/MURUS3 Is a novel Golgi membrane protein that is required for endomembrane organization in Arabidopsis. Plant Cell 17 1764–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, N.G., Scheible, W.R., Cutler, S., Somerville, C.R., and Turner, S.R. (1999). The irregular xylem3 locus of Arabidopsis encodes a cellulose synthase required for secondary cell wall synthesis. Plant Cell 11 769–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines, B., Katsir, L., Melotto, M., Niu, Y., Mandaokar, A., Liu, G., Nomura, K., He, S.Y., Howe, G.A., and Browse, J. (2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448 661–665. [DOI] [PubMed] [Google Scholar]
- Turner, J.G., Ellis, C., and Devoto, A. (2002). The jasmonate signal pathway. Plant Cell 14 (suppl.): S153–S164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updegraff, D. (1969). Semimicro determination of cellulose in biological materials. Anal. Biochem. 32 420–424. [DOI] [PubMed] [Google Scholar]
- Vandenbussche, F., Verbelen, J.P., and Van Der Straeten, D. (2005). Of light and length: regulation of hypocotyl growth in Arabidopsis. Bioessays 27 275–284. [DOI] [PubMed] [Google Scholar]
- von der Fecht-Bartenbach, J., Bogner, M., Krebs, M., Stierhof, Y.D., Schumacher, K., and Ludewig, U. (2007). Function of the anion transporter AtCLC-d in the trans-Golgi network. Plant J. 50 466–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack, C. (2006). Oxilipins: Biosynthesis, signal transduction and action. In Plant Hormone Signaling, P.T. Hedden, ed (Oxford, UK: Blackwell), pp. 185–228.
- Wesley, S.V., et al. (2001). Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 27 581–590. [DOI] [PubMed] [Google Scholar]
- Xu, Y., Chang, P., Liu, D., Narasimhan, M.L., Raghothama, K.G., Hasegawa, P.M., and Bressan, R.A. (1994). Plant defense genes are synergistically induced by ethylene and methyl jasmonate. Plant Cell 6 1077–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Y.B., and Yang, S.F. (1979). Auxin-induced ethylene production and its inhibition by aminoethyoxyvinylglycine and cobalt ion. Plant Physiol. 64 1074–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, X., Liu, Q., Xie, F., and Wen, C.K. (2007). RTE1 is a Golgi-associated and ETR1-dependent negative regulator of ethylene responses. Plant Physiol. 145 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann, P., Hirsch-Hoffmann, M., Hennig, L., and Gruissem, W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.