Abstract
To cope with water stress, plants must be able to effectively sense, respond to, and adapt to changes in water availability. The Arabidopsis thaliana plasma membrane His kinase ATHK1 has been suggested to act as an osmosensor that detects water stress and initiates downstream responses. Here, we provide direct genetic evidence that ATHK1 not only is involved in the water stress response during early vegetative stages of plant growth but also plays a unique role in the regulation of desiccation processes during seed formation. To more comprehensively identify genes involved in the downstream pathways affected by the ATHK1-mediated response to water stress, we created a large-scale summary of expression data, termed the AtMegaCluster. In the AtMegaCluster, hierarchical clustering techniques were used to compare whole-genome expression levels in athk1 mutants with the expression levels reported in publicly available data sets of Arabidopsis tissues grown under a wide variety of conditions. These experiments revealed that ATHK1 is cotranscriptionally regulated with several Arabidopsis response regulators, together with two proteins containing novel sequences. Since overexpression of ATHK1 results in increased water stress tolerance, our observations suggest a new top-down route to increasing drought resistance via receptor-mediated increases in sensing water status, rather than through genetically engineered changes in downstream transcription factors or specific osmolytes.
INTRODUCTION
Drought or high-salinity conditions adversely affect plant growth and are considered one of the most substantial threats to crop productivity (Ingram and Bartels, 1996; Bray, 1997; Hasegawa et al., 2000; Zhu, 2002; Yamaguchi-Shinozaki and Shinozaki, 2006). The response to water stress resulting from these environmental conditions involves broad physiological and metabolic adaptations in the plant. The hormone abscisic acid (ABA) is an important regulatory component of this stress response and is also involved in various aspects of vegetative and seed development (Finkelstein et al., 2002; Finkelstein and Rock, 2002; Nambara and Marion-Poll, 2003), including acquisition of desiccation tolerance and reserve accumulation during seed maturation (Finkelstein et al., 2002).
In this report, we present evidence that ATHK1, an Arabidopsis thaliana His kinase, plays a novel role in regulating water stress response and seed viability. Two-component His kinase signal transduction pathways are involved in various responses in many prokaryotes, fungi, and plants. In the multistep His kinase system, which is common in eukaryotic systems, there are three proteins that participate in the phosphorelay: the sensor, the phosphotransfer protein, and the response regulator. The sensor protein is a membrane-bound His kinase that often has an additional response regulatory domain at its C terminus. Autophosphorylation of a conserved His residue on the sensor protein occurs in response to an environmental signal, and this phosphate group is then relayed to a conserved aspartate in the response regulatory domain on the same protein. A phosphotransfer protein receives the phosphate group from the His kinase and transfers it to a response regulator, which directly controls the output of this signaling cascade. Arabidopsis contains a family of eight true His kinases, a number of which have been implicated in plant hormone pathways, such as those for ethylene and cytokinin signal transduction (Tena et al., 2001; Hwang et al., 2002; Lohrmann and Harter, 2002; Schaller et al., 2002).
Another Arabidopsis His kinase, ATHK1, has been implicated in some plant stress responses. In particular, ATHK1 has been shown to complement a deletion mutant of the yeast osmosensing His kinase Sln1 and can function as an osmosensor in yeast (Urao et al., 1999). Furthermore, RNA analysis suggests that the ATHK1 transcript is most abundant in roots and is transcriptionally regulated by osmotic changes (Urao et al., 1999), suggesting a role for ATHK1 in osmotic stress. We report here that athk1 null mutants show less tolerance and that 35S:ATHK1 overexpressors show more tolerance to various types of water stress, highlighting an important role for ATHK1 in responding to water stress in the growing plant. Furthermore, hormone levels of ABA and transcription of ABA biosynthetic genes increase during osmotic stress in 35S:ATHK1 overexpressors but fail to increase to wild-type levels in athk1 null mutants, suggesting a pathway between ATHK1 and ABA biosynthesis leading to water stress tolerance. Additionally, we demonstrate a link between water stress tolerance in growing plants and desiccation tolerance in maturing seeds. Thus, we propose a unique role for ATHK1 in sensing or regulating vegetative water stress and desiccation of seeds through regulation of ABA biosynthesis.
RESULTS
ATHK1 Mutants Display Altered Water Stress Sensitivities
To analyze the role of ATHK1, we used four genotypes of plants with altered ATHK1 expression levels: (1) wild-type Wassilewskija (Ws), (2) two independent athk1 T-DNA insertion mutants (athk1-3 and athk1-4), (3) ATHK1-rescued athk1 mutants (athk1/ATHK1), and (4) 35S:ATHK1 overexpressors. athk1-3 contains a T-DNA insertion within the sixth intron, 2465 bases downstream of the translation start site, and athk1-4 contains a T-DNA insertion within the second exon, 593 bases downstream of the translation start site (see Supplemental Figure 1A online). Expression of ATHK1 in both athk1 mutants was undetectable by standard RT-PCR analysis or by quantitative RT-PCR (qRT-PCR; see Supplemental Figure 1B online); thus, it was determined that both alleles represent nulls. Four independently derived lines of ATHK1-rescued athk1-3 or athk1-4 mutants were examined. Levels of ATHK1 RNA in these rescued lines were comparable to that of the wild type (see Supplemental Figure 1B online). Data from a representative ATHK1-rescued athk1-3 line is presented in all subsequent figures. Three independently derived lines of 35S:ATHK1 overexpressors were examined (see Supplemental Figure 1C online). Levels of ATHK1 RNA in these overexpressors were between 2- and 12-fold greater than that of the wild type. Data from an overexpressing line with fivefold increase in ATHK1 expression (see Supplemental Figure 1B online) are presented in all subsequent figures.
To characterize expression patterns of the wild-type ATHK1 transcript in seedlings, we exposed plants to NaCl, ABA, cytokinin, and gibberellin in a time course up to 24 h (see Supplemental Figure 1D online). Consistent with previous results, we observed induction of ATHK1 within 30 min after 100 mM NaCl treatment. ABA treatment (1 μM) induced ATHK1 within 4 h, whereas 1 μM gibberellin did not. Interestingly, 1 μM cytokinin (N6-benzylanimopurine) also rapidly induced ATHK1, indicating a possible overlap in the signal transduction pathways for this and other Arabidopsis His kinases.
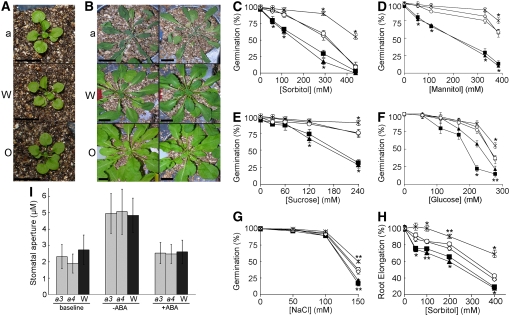
Based on these expression data and previous reports (Urao et al., 1999), we hypothesized that changes in ATHK1 expression might cause altered water stress responses in planta. We found that athk1 null mutants showed significantly more sensitivity to drought (Figures 1A and 1B), osmotic (Figures 1C to 1G), and salinity (Figure 1H) stress than did the wild type. Furthermore, overexpression of ATHK1 increased tolerance to these stresses. However, in the absence of stress, alteration of levels of the ATHK1 transcript did not affect plant growth, as both athk1 null mutants and 35S:ATHK1 overexpressors showed growth comparable to the wild type in nonstress conditions (Figure 1A). The athk1 drought phenotype appears to be a defect related to sensing or responding to levels of water availability in the soil, rather than a stomatal-associated deficiency, as athk1 mutants have stomatal apertures comparable to the wild type (Figure 1I). Furthermore, athk1 stomata respond normally to ABA-mediated inhibition of stomatal opening (Figure 1I). Therefore, our results indicate that loss of ATHK1 gene activity results in plant tissue that is defective in response to general water stress.
Figure 1.
Altered Water Stress Sensitivities and Stomatal Response of ATHK1 Alleles.
For germination and root elongation experiments, wild-type (open diamonds), athk1-3 (closed squares), athk1-4 (closed triangles), athk1/ATHK1 rescued (open circles), and a 35S:ATHK1 overexpressor (asterisks) from matched seed lots were scored for germination on the indicated concentrations of osmolytes. The percentage of germination or root elongation after 5 d of stress treatment is shown. Each value represents the mean percentage of germination for at least four replicates of at least 100 seeds, or the mean percentage of elongation based on the mean length of a nonstressed control root for at least three replicates of at least 20 roots. Error bars represent the se. Stars above data points represent significance based on a two-tailed t test (* P < 0.01, ** P < 0.05).
(A) Fourteen-day-old seedlings of athk1-3 (a), wild-type (W), and 35S:ATHK1 (O) immediately before the onset of drought stress.
(B) athk1-3 (a), wild-type (W), and 35S:ATHK1 (O) after 6 weeks of drought stress.
(C) Percentage of germination on sorbitol-supplemented media.
(D) Percentage of germination on mannitol-supplemented media.
(E) Percentage of germination on sucrose-supplemented media.
(F) Percentage of germination on glucose-supplemented media.
(G) Percentage of germination on NaCl-supplemented media.
(H) Percentage of root elongation on sorbitol-supplemented media.
(I) Average stomatal aperture of mature adult rosette leaves from wild-type (W; dark gray), athk1-3 (a3; light gray), and athk1-4 (a4; light gray). Leaves were initially held in the dark (baseline). Stomata were then induced to open with light in the presence or absence of ABA. Bars represent the mean of the average stomatal aperture for three experiments (two leaves per experiment, with 20 stomatal aperture measurements per leaf). Error indicated is the sd of the experimental means.
ATHK1 Response to Osmotic Stress Acts through an ABA-Dependent Pathway
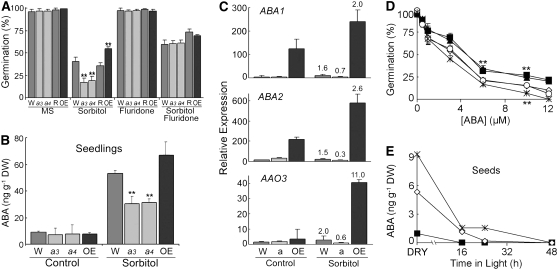
We next sought to explore mechanisms by which ATHK1 might function to mediate water stress tolerance. It is well established that ABA functions to help regulate water stress tolerance in the plant. Water stress leads to ABA accumulation, and external application of ABA induces the expression of genes that function in cellular dehydration tolerance (Finkelstein and Rock, 2002; Schwartz et al., 2003). To determine if the altered water stress response observed in ATHK1 mutants was ABA dependent, endogenous ABA levels were reduced with 100 μM of the ABA biosynthesis inhibitor fluridone, and seeds were treated with 300 mM sorbitol. Fluridone treatment reduced the inhibitory effect of sorbitol on seed germination and also eliminated the differences in sensitivity to sorbitol among all genotypes (Figure 2A). This result suggests that differences in sorbitol sensitivity are due to alterations in ABA accumulation or response to ABA and that the ATHK1-mediated response to osmotic stress acts in an ABA-dependent manner.
Figure 2.
Altered ABA Phenotypes of ATHK1 Alleles.
Wild-type (W; dark-gray bars or open diamonds), athk1-3 (a3; light-gray bars or closed squares), athk1-4 (a4; light-gray bars or closed trianges), athk1/ATHK1 rescued (R; dark-gray bars or open circles), and a 35S:ATHK1 overexpressor (OE; black bars or asterisks) were used for these assays. Error bars represent the se. Stars above data points represent significance based on a two-tailed t test (* P < 0.01; ** P < 0.05).
(A) Effect of the ABA inhibitor fluridone on osmotic sensitivities of ATHK1 alleles. Seeds from matched lots were germinated on Murashige and Skoog (MS) media ± 300 mM sorbitol and ± 100 μM fluridone. Each value represents the mean percentage of germination after 5 d of stress treatment for five replicates of at least 50 seeds.
(B) ABA levels in vegetative tissues of wild-type and ATHK1 mutants. Five-day-old seedlings were exposed to water ± 300 mM sorbitol for 16 h. Each value represents the mean ABA level of three independent biological replicates.
(C) Expression levels of the ABA biosynthetic genes ABA1, ABA2, and AAO3 assayed by qRT-PCR. Five-day-old seedlings were exposed to water ± 300 mM sorbitol for 16 h. All values were normalized to the actin control ACT2 gene. Bars represent the relative mean expression level from five PCR reactions. Printed numbers represent the fold change over a control sample from the same genotype. Error bars represent the se.
(D) Altered ABA sensitivities in germination of ATHK1 alleles. Seeds from matched lots were germinated on MS media ± ABA. Each value represents the mean percentage of germination after 5 d of ABA treatment for four replicates of 100 seeds.
(E) ABA levels in wild-type and ATHK1 mutant seeds. Samples were collected from dry seeds (0 h) and seeds after 16, 24, and 48 h of imbibition. Each value represents the mean ABA level of four independent biological replicates.
We confirmed the ABA dependence of the ATHK1 response through direct measurements of hormone levels and through quantitative measurements of transcript levels of ABA biosynthetic genes. Levels of ABA in untreated control plants were comparable among all genotypes (Figure 2B). However, athk1 null mutants accumulated approximately twofold less ABA in sorbitol stressed tissues than did the wild type, whereas 35S:ATHK1 overexpressors accumulated more ABA than the wild type (Figure 2B). Therefore, consistent with germination data, direct hormone measurements indicate that ATHK1 mediates osmotic stress tolerance by regulating levels of ABA accumulation, either through promotion of ABA biosynthesis or inhibition of ABA degradation.
We sought to further examine the mechanism of ATHK1-regulated ABA accumulation by examining the expression of several ABA biosynthetic genes, which is known to increase upon water stress. We performed qRT-PCR analysis to measure the RNA levels of the ABA biosynthetic genes ABA1 (zeaxanthin epoxidase), ABA2 (xanthoxin dehydrogenase), and AAO3 (aldehyde oxidase), which control the first steps of ABA biosynthesis after zeaxanthin (Marin et al., 1996; Schwartz et al., 1997; Seo and Koshiba, 2002). We observed that ABA1, ABA2, and AAO3 levels correlated well with hormone levels among the three genotypes (Figure 2C). Osmotolerant 35S:ATHK1 overexpressors, which accumulate more ABA during osmotic stress, also upregulate ABA1, ABA2, and AAO3 during osmotic stress. Conversely, osmosensitive athk1 null mutants, which accumulate less ABA during osmotic stress, are unable to upregulate these three ABA biosynthetic genes and actually accumulate less transcript when compared with control levels in athk1 plants. Taken together, these results support a model in which ATHK1 is one component of the ABA regulatory system and that during osmotic stress, ATHK1 functions to increase ABA levels via induction of the ABA biosynthetic genes ABA1, ABA2, and AAO3.
ATHK1 Mutants Display Altered ABA Sensitivity
Since we determined that ATHK1 functions in osmotic stress regulation through an ABA-dependent pathway, it was reasonable to assume that ATHK1 mutants might also have an altered ABA response. To investigate ABA sensitivity, we assayed responsiveness to various concentrations of ABA during germination and root elongation. We found that athk1 null mutants are moderately insensitive to ABA during both germination (Figure 2D) and root elongation (see Supplemental Figure 2 online), suggesting that the mutants are impaired in either their ability to sense or respond to extracellular ABA or that they have reduced endogenous ABA levels in seeds. Conversely, 35S:ATHK1 overexpressors are moderately more sensitive to the inhibition of germination by ABA than are wild-type plants (Figure 2D). The ABA insensitivity in athk1 null mutants is rescued by the introduction of a wild-type copy of ATHK1 (Figure 2D).
To investigate the possibility that observed ABA hyposensitivity in athk1 null mutants and ABA hypersensitivity in 35S:ATHK1 overexpressors was due to differences in endogenous ABA levels, we measured the concentrations of ABA in dry seeds and seeds imbibed in water for 16, 24, or 48 h (Figure 2E). The athk1 null mutants had approximately fivefold less dry seed ABA than wild-type seeds. Furthermore, 35S:ATHK1 overexpressors had approximately twice the level of ABA in dry seeds compared with the wild type. Overexpressors also maintained measurable ABA amounts through 24 h of imbibition, whereas levels of ABA in athk1 and the wild type were undetectable at this time point. Taken together, these results suggest that the observed ABA insensitivity in athk1 null mutants is reflective of the lower levels of endogenous ABA in athk1 seeds and that the ABA hypersensitivity in 35S:ATHK1 overexpressors is reflective of the increased levels of ABA in 35S:ATHK1 seeds. These differences in ABA sensitivity due to endogenous ABA levels are distinct from a mechanism based solely on a defect in ABA perception or response.
ATHK1 Mutants Display Altered Seed Phenotypes
Phenotypic analysis of ATHK1 mutants points to a possible role for ATHK1 in osmotic stress response, a role that is mediated by the phytohormone ABA. However, in addition to its role in vegetative water stress regulation, the hormone ABA also functions during seed maturation (Finkelstein et al., 2002; Nambara and Marion-Poll, 2003). The differences in dry seed ABA levels that we observed in ATHK1 mutants suggested a possible role for ATHK1 during the period of seed maturation when ABA accumulates in the embryo. During this growth stage, cellular water content is reduced from ∼90 to ∼10%. Consequently, an embryo must maintain proper osmotic balance during this desiccation to establish a viable seed (West and Harada, 1993). We examined seeds for phenotypic defects by assaying seed viability and moisture levels in ATHK1 mutants. We observed that athk1 null mutant seeds began to lose the ability to successfully germinate after approximately 8 months of dry storage, conditions that do not affect wild-type seeds. This effect was exacerbated by high temperature and high humidity, which are conditions of a controlled deterioration (CD) test that artificially age seeds (Table 1) (Tesnier et al., 2002). Conversely, 35S:ATHK1 overexpressors had higher rates of survival after deterioration tests. The loss of seed viability over time in athk1 null mutants may be caused by defects in the regulation of embryo desiccation since the mutant athk1 seeds showed a significantly higher amount of moisture than wild-type and ATHK1-rescued seeds (Table 1).
Table 1.
Viability and Moisture Content in Wild-Type and ATHK1 Mutant Seeds
| Ws | athk1-3 | athk1-4 | athk1/ATHK1 | 35S:ATHK1 | |
|---|---|---|---|---|---|
| Survival | 7.9% ± 1.9% | 3.6% ± 2.0% | 3.7% ± 2.8% | 8.5% ± 3.2% | 13.1% ± 2.8% |
| Moisture | 12.5% ± 2.5% | 15.4% ± 2.0% | 14.2% ± 1.5% | 10.4% ± 2.1% | 9.0% ± 2.3% |
For seed survival tests, seeds were subjected to 80% humidity and 60°C for 4 d as a CD treatment. Each value represents the mean percentage of germination for four replicates of at least 100 seeds. For seed moisture tests, ∼200 mg of seeds were dried at 65°C for 16 h, and seed mass before and after drying was measured. Measurements from four independently harvested seed lots were averaged. Error indicated is se. Measurements from athk1-3, athk1-4, and 35S:ATHK1 are significantly different from those of the wild type (P < 0.01, based on two-tailed t test).
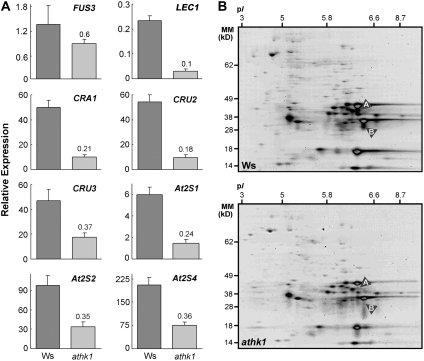
We also examined seed transcript levels of a number of Arabidopsis genes that are active before or during the period of ABA accumulation in the developing seed. RNA levels of FUS3 and LEC1, two seed transcription factors required for embryo maturation and ABA accumulation, were decreased in dry seeds of athk1 null mutants (Figure 3A). Interestingly, mutants in fus3 and lec1 exhibit some of the same seed phenotypes as athk1, including reduced viability, which suggests a possible transcriptional mechanism for the observed athk1 seed defects. FUS3 and LEC1 are known to help to regulate levels of seed storage proteins, which provide the primary source of nitrogen for the growing seed during germination (Pang et al., 1988). Alterations in the accumulation of seed storage mRNAs or proteins have also been linked to germination defects (Koornneef et al., 1989). Accordingly, we examined the seed RNA levels of three Arabidopsis 12S globulin genes (CRA1, CRU2, and CRU3) and three Arabidopsis 2S albumin genes (2S1, 2S2, and 2S4) (Figure 3A) and used two-dimensional protein gel electrophoresis coupled with tandem mass spectrometry to examine seed proteins (Figure 3B). We observed that athk1 knockouts showed decreased levels of the seed proteins CRA1 and CRU3 (Figure 3B), commensurate with a decrease in their mRNA levels (Figure 3A). Taken together, these results suggest that expression of seed maturation genes required for storage protein accumulation is misregulated in ATHK1 mutants, causing decreased storage protein accumulation, and which might lead to the seed defects we have observed. However, ATHK1 most likely functions in a more limited regulatory role during seed maturation, as evidenced by the fact that expression of seed maturation genes is not completely abolished in athk1 nulls and that seed defects are displayed as physiological changes in viability over storage, rather than through gross morphological or developmental defects (see Supplemental Figure 3 online).
Figure 3.
Altered Expression of Seed-Specific Transcripts and of Seed Storage Protein Levels in ATHK1 Mutants.
(A) Expression of FUS3, LEC1, CRA1, CRU2, CRU3, 2S1, 2S2, and 2S4 in wild-type (W; dark gray) and athk1-3 (a; light gray) dry seeds. Expression levels were assayed by qRT-PCR. All values were normalized to the actin control ACT2 gene. Bars represent the relative mean expression level from three PCR reactions. Printed numbers represent the fold change over the wild type. Error bars represent the se.
(B) Relative protein levels in athk1 and wild-type dry seeds. An equal amount (100 μg) of total protein extract from dry mature seeds of wild-type (Ws) and athk1-3 (athk1) was loaded in each gel. The figure shows representative experiments performed at least two times. Two-dimensional gels were stained with SYPRO Ruby and imaged with a UV scanner. Gel images were analyzed using the program PDQuest. Proteins whose abundance was determined to be different among genotypes are labeled: A, CRU3; B, CRA1.
A Survey of Transcriptome Changes Related to ATHK1 Transcript Level
To investigate the role of ATHK1 in gene regulation, we performed full genome transcriptome profiling of athk1-3, an ATHK1-rescued athk1-3 mutant, and an ATHK1 overexpression mutant. Following Robust Multichip Average (RMA) processing of the entire data set, one-way analysis of variance (ANOVA) analysis was performed to identify significant expression differences relating to ATHK1 transcript level. Using a significance level of α = 0.01, with the Benjamini and Hochberg correction, 396 genes were considered significantly differentially expressed. Next, we performed unpaired t tests on this group of 396 genes to identify those for which expression was significantly different compared with the wild type in the athk1 background, in the 35S:ATHK1 background, or in both. Only ATHK1 was significantly differentially regulated in both mutants, and, as expected, ATHK1 is downregulated in the athk1 null background and upregulated in the 35S:ATHK1 background. Of the other genes, 12 were significantly differentially regulated in athk1 compared with the wild type (see Supplemental Table 1 online), and 36 genes were differentially regulated in 35S:ATHK1 compared with the wild type (see Supplemental Table 1 online). We also compared the gene expression profiles of the wild type and the ATHK1-rescued athk1 mutant using an unpaired t test and found that even without additional stringency imposed by corrected P values, no genes were significantly differentially expressed in these two genotypes. This result supports our previous observations that genomic ATHK1 can functionally restore the ATHK1 transcript in athk1 mutants. Because there were no transcriptional differences between wild-type and rescued lines, we did not use these samples in further analysis.
A Survey of Transcriptome Changes Related to Both ATHK1 Transcript Level and Osmotic Stress Conditions
Because ATHK1 is involved in osmotic stress sensing, we also desired to identify genes whose expression was dependent on ATHK1 during osmotic stress. Following RMA processing, two-way ANOVA analysis was performed to identify significant expression differences relating to the interaction of ATHK1 transcript level and sorbitol stress conditions. We identified 233 genes that were significantly (α = 0.01) differentially expressed in these conditions. We further reduced the list to highlight only those genes whose expression was induced compared with the wild type in 35S:ATHK1 plants exposed to sorbitol and whose expression was similar to or reduced compared with the wild type in athk1 plants exposed to sorbitol (Table 2). We classified these genes according to their functional category and gene ontology (GO) annotation and observed a statistical abundance of genes involved in response to stress or abiotic stimulus and binding activity (see Supplemental Figure 4 online). Many are also known to be involved in ABA response. Additionally, some of the genes that we find to be affected by ATHK1 transcript level and osmotic stress condition have also been identified in different studies as responsive to various types of water stress: the transcription factors ATHB12 and ATHB7, the stress responsive genes RD20 and RD29B, the Pro biosynthesis gene P5CS1, and the sucrose biosynthesis gene SUS1. Furthermore, many of the genes on our list (Table 2) are potential downstream targets of both ABA-dependent and ABA-independent transcription factors, suggesting that ATHK1 functions in both ABA-dependent and ABA-independent pathways to control gene expression in response to osmotic stress.
Table 2.
Genes Significantly Differentially Regulated According to Both ATHK1 Transcript Level and Sorbitol Stress Condition
| Log2 FC, Sorbitol Stress (versus Ws Sorbitol)
|
||||||
|---|---|---|---|---|---|---|
| Locus ID | Annotation | FunCata | P Valueb | athk1 | Ws | 35S:ATHK1 |
| At2g39800 | P5CS1δ 1-pyrroline-5-carboxylate synthetase A | 1.7.8.9 | 0.0000026 | 1.30 | 2.10 | 5.40 |
| At4g02280 | Sucrose synthase | 1 | 0.0000005 | 1.70 | 2.00 | 3.40 |
| At3g60140 | DIN2 glycosyl hydrolase family 1 protein | 1,8 | 0.0000467 | 0.31 | 1.06 | 3.08 |
| At5g43840 | AT-HSFA6A heat shock transcription factor | 2,4 | 0.0009559 | 0.76 | 1.00 | 2.99 |
| At3g61890 | ATHB-12 homeobox-leucine zipper 12 | 2,7,8,9 | 0.0025485 | 0.30 | 0.99 | 2.52 |
| At4g19170 | NCED4 9-cis-epoxycarotenoid dioxygenase, putative | 1.9 | 0.0000149 | 1.39 | 2.22 | 2.51 |
| At3g13784 | β-Fructosidase, putative | 1 | 0.0000000 | 0.16 | 0.99 | 2.49 |
| At5g20830 | SUS1 sucrose synthase | 1.7.8 | 0.0001412 | −1.10 | 2.20 | 2.40 |
| At2g46680 | ATHB-7 similar to homeobox-leucine zipper | 2,6,7,8,9 | 0.0002568 | 0.61 | 1.31 | 2.33 |
| At1g17870 | S2P-like putative metalloprotease | 7,8 | 0.0006376 | 0.15 | 1.00 | 2.06 |
| At1g73480 | Hydrolase, α/β fold family protein | 1 | 0.0000073 | 0.39 | 1.06 | 2.00 |
| At2g41190 | Amino acid transporter family protein | 5 | 0.0021325 | 0.43 | 1.12 | 1.93 |
| At2g33380 | RD20 similar to Ca2+ binding EF hand | 4,7,8,9 | 0.0002060 | 0.38 | 1.47 | 1.90 |
| At5g52300 | RD29B stress-responsive protein related | 6,7,8,9 | 0.0038603 | 0.66 | 1.11 | 1.89 |
| At3g02480 | ABA-responsive protein related | 9 | 0.0085394 | 0.17 | 1.24 | 1.86 |
| At1g64110 | AAA-type ATPase family protein | 1,4 | 0.0000178 | 0.33 | 1.30 | 1.81 |
| At1g53540 | 17.6-kD class I small heat shock protein | 7,8 | 0.0002445 | −0.07 | 0.99 | 1.55 |
| At1g07430 | Protein phosphatase 2C, putative | 1,3 | 0.0032986 | 0.18 | 1.09 | 1.53 |
| At1g62510 | Protease inhibitor/seed storage/lipid transfer protein | 4,5 | 0.0081748 | 0.20 | 1.14 | 1.52 |
| At5g12030 | AT-HSP17.6A 1 class II heat shock protein | 3,4,7,8 | 0.0000073 | 0.01 | 1.01 | 1.51 |
| At4g26790 | GDSL-motif lipase/hydrolase | 1 | 0.0000946 | 0.12 | 0.97 | 1.46 |
| At2g21320 | Zinc finger (B-box type) | 2,4 | 0.0028251 | 0.36 | 0.93 | 1.39 |
| At3g62590 | Lipase class 3 | 1 | 0.0021546 | 0.40 | 1.06 | 1.38 |
| At3g09270 | ATGSTU8 glutathione S-transferase, putative | 1 | 0.0011834 | 0.23 | 1.05 | 1.34 |
| At1g80820 | CCR2 cinnamoyl-CoA reductase, putative | 1,7,8 | 0.0002994 | 0.43 | 1.14 | 1.33 |
| At3g09640 | APX2 similar to l-ascorbate peroxidase 1 | 1,7 | 0.0078970 | −0.17 | 1.01 | 1.30 |
| At2g19900 | ATNADP-ME1 malic enzyme | 1,4 | 0.0010296 | 0.17 | 1.15 | 1.26 |
| At5g53870 | Plastocyanin-like domain-containing protein | 4,5 | 0.0019417 | 0.51 | 1.02 | 1.24 |
| At2g46270 | GBF3 G-box binding factor 3 | 2,4,8,9 | 0.0000728 | 0.27 | 1.05 | 1.22 |
| At1g68570 | Proton-dependent oligopeptide transport | 5 | 0.0001645 | 0.26 | 1.06 | 1.20 |
| At3g46230 | ATHSP17.4 class I heat shock protein | 7,8 | 0.0011014 | −0.01 | 1.03 | 1.14 |
| At5g06530 | Similar to ABC transporter family protein | 1,4 | 0.0031217 | 0.17 | 1.02 | 1.14 |
| At4g10250 | ATHSP22.0 ER small heat shock protein | 7,8 | 0.0000051 | 0.02 | 1.00 | 1.12 |
| At5g37500 | GORK guard cell outward rectifying K+ channel | 4,5,7,8,9 | 0.0018003 | 0.24 | 0.94 | 1.12 |
| At3g28270 | Expressed protein | 6 | 0.0002979 | 0.09 | 1.04 | 1.09 |
| At5g09930 | ATGCN2 ABC transporter family protein | 5 | 0.0000105 | 0.07 | 1.04 | 1.06 |
| At2g04160 | AIR3 subtilisin-like protease | 3,8 | 0.0012379 | 0.18 | 0.98 | 1.00 |
| At3g62740 | Glycosyl hydrolase family 1 protein | 1 | 0.0021788 | 0.12 | 0.95 | 0.96 |
The GO annotation of genes in italic text is suggestive of a role in ABA response. The GO annotation of genes in bold type is suggestive of a role in abiotic stress response. Genes were classified by their functional category using MIPS Functional Catalog (FunCat) Database (http://mips.gsf.de/projects/funcat): 1, metabolism; 2, transcription; 3, translation/posttranslation (protein folding, modification, or destination); 4, binding; 5, transport; 6, signal transduction; 7, abiotic stress response; 8, interaction with the environment; 9, ABA response; and 0, unclassified/other.
P values from a two-way ANOVA between log2 RMA-processed expression values were corrected using the method of Benjamini and Hochberg. A significance level of α = 0.01 was used as the cutoff.
As an additional analysis, we attempted to identify enriched elements in the upstream regions of the genes significantly differentially regulated according to water stress and ATHK1 transcript level. We first searched for known cis-elements involved in drought-responsive gene expression, including the ABA-responsive gene element (ABRE) PyACGTGG/TC (Guiltinan et al., 1990; Mundy et al., 1990), the MYB-responsive element TGGTTAG (Abe et al., 1997), the MYC-responsive element CACATG (Abe et al., 1997), and the drought-responsive element TACCGACAT (Yamaguchi-Shinozaki and Shinozaki, 1994). The ABRE ACGTGTC was identified 72 times, usually in tandem with a second ABRE, representing a significant (P = 4.5 e-10) enrichment over the background model. We also identified 43 instances of the MYB-responsive element and 160 instances of the MYC-responsive element, further confirming the ABA dependence of the ATHK1 response. Next, to identify putative novel cis-regulatory elements, we searched for any 6- or 7-bp sequences enriched in the upstream sequences of our 233 interaction significant genes (Table 3). Several of the sequences we identified were identical to ABREs. Another three sequences were highly similar to a conserved cis-acting element called the G-box (CACGTGGC), which has been identified in the promoters of genes responsive to light, ultraviolet radiation, and jasmonic acid (Menkes et al., 1995). A third set of enriched sequences contained the consensus sequence CATATACA, which most likely represents the TATA box core promoter upstream of genes.
Table 3.
Summary of 6- and 7-mer Elements Overrepresented in Upstream Sequences of Interaction Significant Genes
| Sequence | Occurences | Expected Occurences | Expected Frequency | E-Valuea | Known cis-Element?b |
|---|---|---|---|---|---|
| 6-mers | |||||
| ACACGT | 201 | 106.89 | 0.000368898 | 6.80E-13 | ABRE |
| CGTGTC | 102 | 60.72 | 0.000209556 | 1.70E-03 | ABRE |
| TATACA | 407 | 319.81 | 0.001103676 | 3.30E-03 | * |
| ACGTGG | 118 | 76.66 | 0.000264561 | 1.50E-02 | ABRE |
| CATATA | 456 | 369.85 | 0.00127638 | 1.70E-02 | * |
| CACGTG | 95 | 59.28 | 0.000204583 | 2.50E-02 | G-box |
| ATATAC | 389 | 311.29 | 0.001074267 | 2.50E-02 | * |
| 7-mers | |||||
| ACGTGTC | 68 | 28.94 | 9.99642E-05 | 3.70E-06 | ABRE |
| ACACGTG | 71 | 33.43 | 0.000115475 | 8.80E-05 | G-box |
| ACACGTA | 58 | 27.87 | 9.62697E-05 | 3.40E-03 | G-box |
Overrepresented promoter elements were determined using the program RSA Tools (http://rsat.ulb.ac.be/rsat/), with predefined background frequencies as implemented on the website.
E-values were computed as defined on the website.
If the identified upstream element represents a known cis-element, it is listed here; the asterisk indicates consensus sequence of CATATACA.
Confirmation of Microarray Expression Changes
To test whether the transcriptional responses we observed in our microarray experiments were biologically valid and were not artifacts of the array technology, we used qRT-PCR analysis to measure the expression of nine Arabidopsis genes that were significantly differentially regulated in the conditions of our microarrays and which were somewhat well characterized: RD29B, RD20, CCA1, ABI2, MYB2, HB12, KIN1, LEA14, and RAB18 (see Supplemental Figure 5 online). We normalized all values to the ACT2 gene and calculated the linear fold change relative to a wild-type control sample. In almost all cases tested by qRT-PCR, the direction of the fold change was the same in both microarray and qRT-PCR analysis. In cases where the direction of the fold change was not the same, including KIN1 and LEA14 in athk1 compared with the wild type and RD29B, RD20, and ABI2 in athk1 after sorbitol stress compared with the wild type after sorbitol stress, absolute fold change levels were very close to 1.0 (no change) and thus may have been below the limits of detection for the microarray. Similar to what has been previously reported, for genes with large increases or decreases in magnitude, microarrays underestimated the magnitude of the changes (Etienne et al., 2004), implying that qRT-PCR is more accurate when fold changes are large.
A Global Survey of Transcriptome Changes
To expand our analysis of the Arabidopsis transcriptome, we collected 1704 available data files representing full-genome Arabidopsis expression profiling experiments and performed hierarchical clustering. Detailed results of the clustering (referred to as the AtMegaCluster) are presented in Figure 4 and are explained in more detail in the Discussion. Our goal in this analysis was to compile a set of microarray experiments that represented a full spectrum of conditions, genotypes, and other variables to allow for comprehensive coexpression analysis. We used the AtMegaCluster to identify a group of genes coexpressed with ATHK1. This cluster contains ARR4, ARR5, ARR6, ARR8, and ARR9, which are all Arabidopsis response regulators (ARRs) that might function in the final step of the ATHK1 His kinase phosphorelay pathway. The cluster also contains one unknown gene (At4g37080) and a putative 18S ribosomal assembly gene (At1g13650). Notably, like ATHK1, ARR3 is expressed specifically in roots, while ARR8 is also highly, but not exclusively, expressed in roots (Urao et al., 1998). The ATHK1 gene cluster falls into the larger gene cluster D, and its position in the AtMegaCluster is indicated by a plus sign (+) in Figure 4. Overall, cluster D genes were significantly enriched for cell communication and carbohydrate metabolism, suggesting that ATHK1 may also be involved in these types of processes, specifically in communication of a stress-induced signal or in carbohydrate metabolism during osmotic stress.
Figure 4.
The AtMegaCluster Displays Hierarchical Clustering of 444 Experiments and 22,810 Genes in Arabidopsis.
Experiments, represented on the horizontal axis, were grouped into eight clusters according to the fold change values of genes and have been named according to the classification of the majority of experiments in that cluster. Genes, represented on the vertical axis, were grouped into five clusters according to their fold change values and have been named according to the functional category of the majority of genes in that cluster. The ATHK1 gene is represented by a plus sign (+). Induced fold changes are in magenta; repressed fold changes are in green.
We confirmed our list of genes coexpressed with ATHK1 using the ATTED-II database (http://www.atted.bio.titech.ac.jp/), which uses a smaller set of experiments for coexpression analysis. The results of this analysis also identified the ARRs ARR3 and ARR4 as potentially coregulated genes. Finally, to further verify that the ATHK1 gene cluster highlighted a biologically relevant group of genes, we wanted to rule out the possibility that cluster results were due to spurious hybridization events resulting from similar probe sequences among the genes in our cluster. We examined the perfect match probe sequences of all eight genes in the ATHK1 cluster for common sequences using the multiple alignment program ClustalW and found that similarity between probe sequences could not account for the clustering results.
To distinguish experimental conditions that resulted in the most similar expression patterns for the genes in the ATHK1 cluster, and thus contributed to the hierarchical clustering results, we identified experiments in which at least two of the genes in the cluster displayed an average difference in expression of at least twofold. Thus, we were able to highlight experiments in which ATHK1 and the other genes in the cluster share the most expression similarities (see Supplemental Table 2 online). Notably, these experimental conditions were mainly abiotic stress experiments, especially mannitol and salt stress, suggesting that similar transcriptional profiles exist for both ATHK1 and the ARRs specifically during water stresses. The list also included five cytokinin-related experiments, which might be expected since these ARRs are cytokinin responsive (To et al., 2004) and ATHK1 can also be induced by cytokinin (see Supplemental Figure 1D online).
Osmotic Stress Sensitivity of arr Mutants
To characterize the osmotic stress response of the ARRs that clustered with ATHK1, we obtained higher-order mutants of these ARRs, including arr3,4,5,6, arr5,6,8,9, and arr3,4,5,6,8,9, which are described by To et al. (2004). To et al. (2004) examined RNA expression levels for these mutants and concluded that the T-DNA insertions in arr3 and arr8 result in null alleles, whereas the remaining insertions result in hypomorphic alleles. Previous studies have implicated these ARRs in cytokinin signaling and circadian clock regulation (To et al., 2004; Salomé et al., 2006). To assay for osmotic sensitivity, seed germination was measured after 5 d of treatment with various concentrations of the nonmetabolizable sugar sorbitol (see Supplemental Figure 6 online). arr3,4,5,6 null mutants were found to be hypersensitive to osmotic stress during germination (P < 0.01), whereas arr5,6,8,9 null mutants were found to be slightly insensitive to osmotic stress during germination (P < 0.05). The arr3,4,5,6,8,9 hextuple mutant exhibited near wild-type response to osmotic stress, suggesting a complex function for these ARRs in osmotic stress regulation in which arr3,4 and arr8,9 play opposite roles. These results suggest that there may be interactions among the type-A ARRs involving both additive and antagonistic functions. These results are consistent with previous studies in which To et al. (2004) examined the sensitivity of these mutants to red light and found that mutations in ARR3, ARR4, ARR5, and ARR6 increased sensitivity in an additive manner. However, the arr3,4,5,6,8,9 hextuple mutant exhibited a decrease in red light sensitivity compared with arr3,4,5,6, suggesting that arr8 and arr9 may antagonize the effects of the other four arr mutations. We suggest that a similar mechanism exists for ARR3, ARR4, ARR8, and ARR9 during osmotic stress.
DISCUSSION
Novel Phenotypes of ATHK1 Mutants Highlight the Connection between Desiccating Seeds and Osmotically Stressed Seedlings
In this report, we have examined the question of whether ATHK1 has the ability to function as an osmosensor in planta by studying the effects of gene ablation and overexpression on osmotic sensitivity. The results indicate that eliminating the ATHK1 gene results in plant tissue that is defective in the ability to withstand water stress both in vegetative and seed tissues and that constitutive activation of the ATHK1 gene results in plant tissue more resistant to water stress. In addition, phenotypic data suggests that the ATHK1-mediated response to osmotic stress is ABA dependent and functions by positively regulating genes involved in ABA biosynthesis to increase ABA hormone levels. However, these data do not rule out the possibility that the ATHK1 response to osmotic stress is also ABA independent. Indeed, to further characterize the regulatory role of ATHK1 during osmotic stress, we performed whole-genome transcriptional studies and found that genes responsive to ATHK1 during osmotic stress are both ABA dependent and ABA independent. Analysis indicated that many known stress-responsive and ABA-dependent and ABA-independent genes are downregulated in athk1 knockout plants and are upregulated in 35S:ATHK1 plants. Furthermore, statistical enrichment of promoter sequences for known ABA- and stress-responsive transcription factors suggest that ATHK1 functions upstream of these transcription factors to regulate water stress resistance. The increase in expression levels of the Pro synthesis gene P5CS1 and the sucrose synthesis gene SUS1 suggest that the response of ATHK1 helps to regulate osmolyte synthesis as a protective mechanism during water stress. It remains to be determined whether changes in the protein activities of these genes are responsible for the altered osmotic response in athk1 mutants. Similarly, future studies to investigate changes in protein modifications, such as phosphorylation states during the early response to stress, will further clarify the role of ATHK1 in response to water stress.
While this manuscript was under review, Tran et al. (2007) reported similar results for the function of ATHK1. However, Tran et al. (2007) used knockdown mutations located in the upstream region of ATHK1 to show that ATHK1 is involved in drought and salt stress response, whereas we used two true null mutations of athk1 and demonstrate rescue of the mutant phenotypes with a genomic ATHK1 transcript. We also expand on previous work to show that ATHK1 is involved in general water stress, as athk1 is also sensitive to a number of osmotic stressors. These two studies also use different genetic backgrounds and so are complementary to one another. Tran et al. (2007) also report transcriptional analysis of athk1 knockdown mutants after 2.5 or 9 h of dehydration stress.
In this report, we describe the transcriptional network of ATHK1 after 16 h of sorbitol stress. Despite the differences in genotypes and growth conditions, some genes identified in these studies overlap, which further serves to confirm the results of both studies. In their analysis of genes whose expression was altered in athk1, Tran et al. (2007) report 190 genes downregulated with a ratio of >2 after 2.5 h of dehydration stress and 120 genes downregulated after 9 h of dehydration stress. Of these genes, ∼5% were also identified in our study and include the desiccation-responsive gene RD29B, the homeobox Leu zipper genes ATHB12 and ATHB7, the Pro synthesis gene P5CS1, and a sucrose-UDP glucosyltransferase and an AAA-type ATPase family protein. This relatively low overlap between the genes identified in the two studies might not be unexpected, since these two studies used different exposure times (2.5 or 9 h versus 16 h), types of stress (dehydration versus osmotic), microarray platforms (Agilent versus Affymetrix), and methods of identifying interesting genes (fold change cutoff versus statistical criteria). Nonetheless, the commonly identified genes might represent an important biological response toward short- and longer-term general water stress exposure and as such are probably key genes in the whole-plant coordination of water stress response.
Distinct and Interesting Clusters of Genes and Experiments Emerge from the AtMegaCluster
More than half of the genes in the Arabidopsis genome remain unclassified; a major goal of functional genomics studies is to assign putative functional classification to genes on the basis of sequence or expression similarities. Coexpression studies can be valuable when a gene of unknown function clusters next to a gene of known function because genes in biological pathways tend to group together. However, we demonstrate that for a large number of Arabidopsis genes with no known homolog, coexpression analysis using the AtMegaCluster fails to group genes with unknown function near genes with known function. Many of these unknown genes might comprise undiscovered functional gene families whose expression patterns are unique. The AtMegaCluster consists of experiments that represent a full spectrum of conditions, genotypes, and other variables for comprehensive coexpression analysis. By clustering genes, we are able to identify genes with similar expression patterns across diverse conditions whose coregulation might not have been evident in smaller clustering datasets. Furthermore, by clustering experiments in a second dimension, we are able to identify experimental conditions that result in similar global expression patterns.
We first examined five distinct groups of genes that emerged from the AtMegaCluster and identified their functional categories using the GeneBins database. Because these clusters contained thousands of genes, we employed the Bonferroni correction to identify significant functional enrichment in a group (see Supplemental Figure 7 online). Gene cluster A contained 2039 genes, of which ∼55% were unclassified, and another 15% were unclassified with no known homolog in Arabidopsis. All other clusters also contained ∼50% unclassified genes; however, cluster A was significantly (P = 4.64e-08) enriched for these genes. Gene cluster B contained 2023 genes significantly (P = 6.07e-26) enriched for environmental information processing functions, such as signal transduction and ligand–receptor interaction. Gene cluster B was also enriched for carbohydrate metabolism and cell growth genes; however, the vast majority of these were genes with functional category overlap in environmental information processing, including many protein kinases. Gene cluster C contained 2370 genes with less distinct functional classification. However, ∼30% of the genes could be classified as metabolism genes, with an enrichment for energy metabolism, including many photosynthetic genes (P = 3.37e-14), lipid metabolism (P = 1.83e-07), and amino acid metabolism (P = 2.06e-08). Gene cluster D contained 1133 genes, with significant (P = 2.62e-13) enrichment for cellular processes, such as cell communication, cell growth, and cell death. Cluster D was also functionally enriched for carbohydrate metabolism genes (P = 8.54e-07), many of which were cyclins and tubulins, with functional overlap in the cellular processes category. Finally, gene cluster E contained 1357 genes functionally enriched for genetic information processing (P = 4.00e-191), including transcription, translation, and posttranslational processes, such as protein folding, sorting, and degradation, as well as nucleotide metabolism (P = 8.53e-23).
The AtMegaCluster also allowed us to identify groups of experiments with similar global expression patterns. We found that the 444 experiments of the AtMegaCluster could be grouped into eight distinct clusters. The 180 abiotic stress experiments could be grouped into three distinct clusters: Cluster 1 included 45 experiments, mainly wounding, DNA damage, oxidative stress, drought, osmotic stress, and salt stress applied to root tissue; Cluster 2 included 64 experiments, mainly heat, light, and cold stress applied to all tissue types for longer periods of stress (>4 h); and Cluster 8 included 47 experiments, mainly heat, light, and cold stress applied to all tissue types for shorter periods of stress (<4 h). Cluster 3 contained 26 experiments, including all of the 22 hormone treatments; while a short distance away, Cluster 4, containing 20 experiments, was comprised of mainly hormone inhibitor experiments. The 47 biotic stress experiments of the AtMegaCluster were included in Cluster 5, along with ∼20 abiotic stress experiments, mainly 24 h of stress in shoot tissue. The flowering time mutants co, ft, and lfy comprised the 15-experiment Cluster 6. Finally, Cluster 7, which included 59 experiments, was comprised of mostly nutrient starvation, cold stress, and experiments examining cell cycle. The grouping of these experiments in such a manner highlights similarities among broad types of conditions and tissue types.
The Role of ATHK1 in Seed Maturation
Perhaps the most interesting of the findings described in this report is the connection between vegetative osmotic stress and seed maturation. To date, there have been several mutants described that share some of the athk1 phenotypes, but no other mutants that we know of show the unique pattern of reduced seed longevity, osmotic hypersensitivity, and ABA insensitivity of athk1. We have observed various seed defects, including increased moisture content and reduced seed viability, in athk1 seeds, suggesting that ATHK1 may also be involved in sensing or regulating the amount of desiccation that occurs normally during seed maturation. Although this connection between stress sensing and seed maturation has, to our knowledge, not been suggested in prior studies, because osmotic imbalance is a necessary consequence of the massive cellular desiccation that occurs during seed maturation, it seems plausible that a single sensory molecule might be able to control aspects of stress sensing in both seeds and vegetative tissue.
It has generally been accepted that the rise of ABA levels in seeds during maturation correlates directly with a reduction in seed water content. Thus, the reduced ABA levels and increased moisture content in seeds of athk1 null mutants suggests that ATHK1 plays a role in the process of seed desiccation. It is also during this desiccation period in seed maturation when levels of seed storage components accumulate. In a plant with seed phenotypes such as reduced viability, reduced ABA, and increased water content, we might also expect reduced levels of some seed-specific proteins. Indeed, RNA levels of FUS3 and LEC1 are reduced in athk1 nulls, and direct measurements using two-dimensional electrophoresis coupled with tandem mass spectrometry confirmed misexpression of the seed proteins CRA1 and CRU3.
In this study, we also provide evidence of a role for ATHK1 in control of seed levels of ABA. Although we did not measure ABA levels throughout the entire process of seed maturation, we did observe alterations in ABA levels of athk1 and 35S:ATHK1 mature seeds. It is reasonable to hypothesize that if ATHK1 can influence ABA biosynthesis during osmotic stress in the growing plant, ATHK1 might also influence ABA accumulation during desiccation in the maturing seed. Furthermore, processes such as seed moisture level, which are known to be regulated by ABA in the seed, are altered in ATHK1 mutants, leading to further confidence that ATHK1 plays some role in sensing or regulating embryo water loss, most likely through an ABA-dependent pathway. The results are consistent with our hypothesis that ATHK1 plays a sensory or regulatory function during seed maturation and that although not completely eliminated, the exact timing or kinetics of processes such as desiccation, ABA accumulation, or storage protein synthesis are altered in athk1 mutants, leading to the differences in mature seed phenotypes and mature seed levels of certain RNAs.
Mechanisms of ATHK1 Action
An increase in osmolarity often leads to a decrease in turgor pressure at the surface of the cell. Whether ATHK1 directly responds to a change in turgor or is activated by a small molecule or protein that can respond to turgor changes in the lipid bilayer is not known. However, since many prokaryotic osmosensors can respond to pressure changes, it follows that a putative osmosensor in Arabidopsis may also directly respond to changes in turgor pressure. Furthermore, it has been suggested that the yeast osmosensor Sln1 can respond to changes in turgor pressure (Reiser et al., 2003), and so by analogy, ATHK1 might function in a similar manner.
Just as the exact manner in which ATHK1 responds to osmolarity is unknown, downstream members of the ATHK1 signal transduction pathway have also remained elusive. By analogy to other Arabidopsis His kinases, we would expect one or more AHPs and ARRs to be involved in the ATHK1 phosphorelay. Interestingly, our AtMegaCluster identified several ARRs (ARR4, ARR5, ARR6, ARR8, and ARR9) as exhibiting similar expression profiles to ATHK1, indicating that they are cotranscriptionally regulated under a wide variety of conditions. Although these proteins have not previously been shown to function in osmotic stress signal transduction, the ARRs ARR3, ARR4, ARR5, ARR6, ARR8, and ARR9 have been implicated as partially redundant components of cytokinin and light signal transduction and circadian clock regulation (To et al., 2004; Salomé et al., 2006). In particular, To et al. (2004) observed a gradual increase in cytokinin sensitivity with the progressive loss of ARRs, from arr3,4 to arr3,4,5,6,8,9, leading to the hypothesis that type-A ARRs function to negatively regulate cytokinin responses. On the other hand, Salomé et al. (2006) observed altered circadian rhythms in arr3,4 mutants, which seemed to be suppressed by arr8,9 mutants. However, the mechanism of the arr8,9-mediated repression of the arr3,4,5,6 phenotype was not determined in that study.
In this study, we observed a similar phenomenon in regard to osmotic response. Disruption of ARR3, ARR4, ARR5, and ARR6 increases sensitivity to osmotic stress. Additional loss of ARR8 and ARR9 decreases sensitivity to osmotic stress, especially in high concentrations. Because the arr5,6,8,9 mutant has an osmoresponse comparable to the wild type, while the arr3,4,5,6 mutant is extremely sensitive to osmotic stress, we suggest that the arr8,9 mutation can suppress the arr3,4 phenotype during osmotic stress. Taken together, these results suggest a role for ARR3, ARR4, ARR8, and ARR9 in osmotic stress regulation that is complex and may be both positive and negative. Clearly, there exist intricate interactions between these two pairs of ARRs, which may function through differences in the physical interactions between proteins or differences in other phosphorelay signal components. Because it is known that members of two-component systems display complex interactions and can often act in more than one signaling pathway (reviewed in Knight and Knight, 2001), it seems plausible, given prior indirect biochemical information, and the results of our AtMegaCluster as well as phenotyping, that some of these ARRs might function in the ATHK1 osmotic stress signaling cascade.
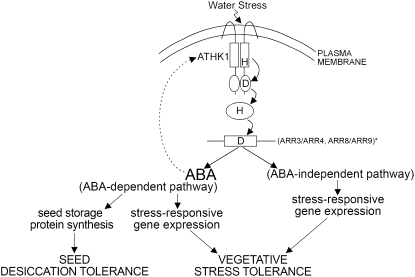
Here, we have described a unique gene encoding a water receptor that is responsible for general water stress tolerance. We have suggested a plausible mechanism in which ATHK1-mediated water stress tolerance functions in both an ABA-dependent and ABA-independent manner and have provided some evidence that certain ARRs function in the ATHK1 phosphorelay. Furthermore, our data indicate an important role for osmosensing during seed development, one that may have important implications for general cell preservation, as well as for agriculture, as society increases crop production in arid areas. A model depicting the potential hierarchy of ATHK1 regulation is shown in Figure 5. In this model, we suggest that the ATHK1 signal transduction cascade is initially activated by the sensing of water stress at the plasma membrane. The ATHK1-mediated water stress signal may be passed through the response regulators ARR3/ARR4 and/or ARR8/9. Activation of the ATHK1 signal transduction pathway induces ABA biosynthesis. The increased levels of ABA then induce the expression of ATHK1, so that the water stress signal may be propagated more rapidly. In the ABA-dependent pathway of ATHK1 response, transcription factors, including ABRE binding proteins, and MYB/MYC induce stress-responsive gene expression leading to vegetative stress tolerance. An ABA-independent pathway of ATHK1 response also exists wherein drought-responsive element binding proteins induce stress-responsive gene expression. During seed maturation, the ATHK1-induced ABA signal can also function to regulate seed storage protein levels and maintain the coordination of seed desiccation.
Figure 5.
Schematic of the Proposed Mechanism of ATHK1 Action in Seeds and Seedlings.
After perception of water stress by ATHK1, a phosphorelay putatively involving ARR3/ARR4 and/or ARR8/ARR9 transmits a signal to increase the transcription of ABA biosynthetic genes, causing a rise in intracellular ABA levels. In vegetative tissues, increased ABA levels initiate the transcription of stress-responsive genes, including ATHK1 itself. An ATHK1-responsive ABA-independent pathway of stress-responsive gene expression also exists. In seeds, ATHK1-mediated increased ABA levels drive the synthesis of seed storage proteins and affect seed desiccation tolerance.
METHODS
T-DNA Mutant Screen and Identification
Using a PCR-based strategy (Krysan et al., 1999), T-DNA–mutagenized populations from the Wisconsin lines (Krysan et al., 1999) were screened for the presence of insertions in ATHK1. The sequences of primers specific for ATHK1 were 5′-AGGAAGGTGTTCGATAAAATGACTGAATG-3′, and 5′-CACATCCAGTATCATCAACCTCAAACCA-3′. The sequences of primers specific for the T-DNA border were 5′-CATTTTATAATAACGCTGCGGACATCTAC-3′ and 5′-TTTCTCCATATTGACCATCATACTCATTG-3′. DNA sequencing of PCR products confirmed the locations of the junctions of genomic and T-DNA sequences. We isolated two independent T-DNA insertions in ATHK1, named athk1-3 and athk1-4 (see Supplemental Figure 1B online).
Molecular Complementation
A 6- and an 8-kb fragment containing the entire coding sequence and putative regulatory sequence of ATHK1 were amplified using high-fidelity PCR. To amplify the 6-kb fragment, the following primers were used: 5′-CCGCTCGACTCTCCATTGGCCATTTTACCTTCTAC-3′ and 5′-ATAAGAATGCGGCCGCTTACCCCCAAAAACCTCATCGTCAA-3′. To amplify the 8-kb fragment, the following primers were used: 5′-CCGCTCGAGTGTTAAATCGCAGTCTATACAGTCATC-3′, and 5′-ATAAGAATGCGGCCGCGGGCTTAAAAATTGTTCCAGAGTTCG-3′. To confirm the presence of full-length wild-type sequence, DNA sequencing was performed on the entire length of all clones used for rescue. Each construct was used to transform six different athk1-3 and athk1-4 plants using an Agrobacterium tumefaciens–mediated floral dip procedure (Clough and Bent, 1998). Transformants were selected with kanamycin (50 μg/mL) (Sigma-Aldrich). Rescued plants were identified as those homozygous for an athk1 T-DNA insertion and homozygous for an ATHK1 transgene, based on PCR genotyping. Multiple rescued lines were identified for both the 6- and 8-kb fragments; data for a representative athk1-3 rescued line are shown in all figures.
Overexpression
A 3.3-kb cDNA fragment containing the entire open reading frame of ATHK1 was overexpressed under the control of the enhanced cauliflower mosaic virus 35S promoter. The tobacco mosaic virus Ω sequence was inserted upstream of the ATHK1 sequence to increase the translational level. Ws and athk1 plants were transformed as described above for molecular complementation. Transformants were selected with hygromycin (30 μg/mL) (Sigma-Aldrich). Multiple overexpressing lines were identified; data for a representative line overexpressed in a wild-type background are shown in all figures.
Plant Materials and Growth Conditions
We used the ecotype Ws for all experiments. For seed propagation, seeds were sown on media, pH 5.7, containing half-strength MS salts (Murashige and Skoog, 1962) (Sigma-Aldrich), 2.5 mM MES, 1% (w/v) sucrose, and 0.8% (w/v) washed agar. Plates were cold treated in the dark at 4°C for 3 d and then transferred to light (42 μmol m−2 s−1). Seedlings were transferred to soil after 10 d of growth on MS plates. Plants were housed under the following growth conditions: 23°C, 24 h light (42 μmol m−2 s−1), and ∼60% humidity. After harvesting and ∼3 weeks of after-ripening, seeds were stored at 4°C.
Sorbitol, mannitol, glucose, and sucrose plates were made with half-strength MS media by adding autoclaved sugar solutions after the medium had cooled to ∼55°C. NaCl plates were made with half-strength MS media by adding solid NaCl directly to media before autoclaving. For ABA assays, ethanolic stock solutions of ABA (Sigma-Aldrich) were made at 1000× strength and added to half-strength MS media after the medium had cooled to ∼55°C. Ethanolic stock solutions of fluridone (Chem Service) were freshly prepared for each experiment.
Water and ABA Stress Assays
For all assays, seeds of simultaneously produced and harvested lots were compared. For drought assays, sterilized seeds were sown on MS plates and were cold treated in the dark at 4°C for 3 d. After 7 d of growth in constant light, seedlings were transferred to soil and grown for an additional 7 d in short-day conditions (8 h light/16 h dark), with water every 3 d. Prior to drought treatment, plants were saturated with water and then transferred to dry conditions. Plants were withheld from water and observed daily for signs of wilting. Plants were photographed after ∼6 weeks when the largest differences between genotypes were apparent.
For osmotic stress germination assays, sterilized seeds were sown on MS plates containing sorbitol, mannitol, sucrose, or glucose and were cold treated in the dark at 4°C for 3 d. Plates were exposed to light (42 μmol m−2 s−1) for 1 h at 23°C and scored after 5 d of growth. At least three replications of 100 seeds per line were tested for all treatments. For ABA germination assays, sterilized seeds were sown on MS plates containing ABA and were cold treated in the dark at 4°C for 3 d. Plates were exposed to light for 1 h at 23°C and scored after 5 d of growth in darkness. Germination was indicated by clear protrusion of the radicle. For root growth assays, sterilized seeds were sown on MS plates and were cold treated at 4°C for 3 d. Plates were grown vertically in constant light for 3 d at 23°C and then transferred to MS plates supplemented with different concentrations of sorbitol and scored after 7 d of additional growth. At least three replications of 20 roots per line were tested for each treatment.
Stomatal Assays
For stomatal assays, sterilized seeds were sown on MS plates and were cold treated in the dark at 4°C for 3 d. After 7 d of growth in constant light, seedlings were transferred to soil and grown for an additional 5 weeks in short-day conditions (8 h light/16 h dark), with water every 3 to 5 d. For inhibition of stomatal opening, adult rosette leaves were harvested immediately prior to onset of light and placed in the dark at room temperature for 2 h in a solution of 10 mM KCl, 7.5 mM iminodiacetatic acid, and 10 mM MES, pH 6.15. ABA was added to the solution to a final concentration of 30 μM from 1000× ethanolic stock solutions, and an equivalent amount of ethanol was added to the controls. Leaves were placed in bright light at room temperature for 2 h. For baseline measurements, stomata were measured after dark treatment. To measure stomata, epidermal peels were taken from leaves and imaged at ×400 magnification under bright-field microscopy. Images were captured by a SPOT Insight CCD camera (Diagnostic Instruments), and stomatal apertures were measured in NIH Image (http://rsb.info.nih.gov/nih-image/).
ABA Measurements
For ABA measurements on sorbitol treated seedlings, plants were prepared as for qRT-PCR measurements. For ABA measurements on seeds, seeds were sown on two layers of chromatography paper moistened with sterile water. Seeds were cold treated in the dark for 3 d at 4°C. Seeds were transferred to light and collected after 16, 24, and 48 h. Dry seeds were used as the control. The extraction procedure was performed as described (Chiwocha et al., 2003), with the following modifications: following extraction in 99% isopropanol and 1% acetic acid, the extract was dried in a SpeedVac. The pellet was resuspended in 15% methanol and 1% acetic acid and then passed through a Sep-Pak C18 column (Waters) as described. The purified extract was dried in a SpeedVac and reconstituted with 30 μL of 15 μM ammonium acetate. For each sample, 10 μL was used for reverse-phase HPLC–electrospray ionization tandem mass spectrometry, and the eluting ions were measured with multiple reaction monitoring. The level of ABA in the samples was quantified in relation to their internal standard using calibration curves that had been generated for each compound. Each experiment was performed twice, with three biological replicates.
CD Test
A CD test simulates maturation of seeds under controlled conditions and can thus reveal the relative storage potential of seeds (Tesnier et al., 2002). Seeds were equilibrated at 85% relative humidity and stored for 4 d at 60°C, for artificial ageing of seeds. Seeds were then allowed to equilibrate at ∼30% humidity. For germination assays, seeds were sown on filter paper moistened with water, stratified for 3 d at 4°C, transferred to light, and scored after 5 d of growth at 23°C. At least three replications of 100 seeds per line were tested.
RNA Isolation, Labeling, and Hybridization
RNA was isolated from 5-d-old plants of four different genotypes in two different conditions as follows: (1) wild-type (Ws) control; (2) wild-type (Ws) + 300 mM sorbitol; (3) a T-DNA knockout (athk1-3) control; (4) a T-DNA knockout (athk1-3) + 300 mM sorbitol; (5) a rescued line (athk1-3/ATHK1) control, (6) a rescued line (athk1-3/ATHK1) + 300 mM sorbitol; (7) an overexpression line (35S:ATHK1) control; and (8) an overexpression line (35S:ATHK1) + 300 mM sorbitol. Three biological replicates of each sample were prepared, for a total of 24 samples. Whole seedlings were ground to a fine powder using a mixer mill (Retsch) in RNase-free conditions. Total RNA was prepared using the RNeasy plant mini kit (Qiagen) according to the manufacturer's instructions. The GeneChip Eukaryotic Poly-A RNA control kit (Affymetrix) was used to provide positive controls to monitor the entire labeling process. Ten micrograms of RNA was used for first- and second-strand cDNA synthesis using the SuperScript Choice system (Invitrogen), with the following modifications to the manufacturer's instructions: first-strand synthesis was performed at 42°C with an oligo(dT) primer containing the T7 RNA polymerase promoter (5′-GGCCAGTGAATTGTAATACGACTCACTATAGGGAGGCGG-T24 -3′). Double-stranded cDNA was purified by phenol-chloroform extraction, precipitated with NH4OAc and ethanol, and resuspended in 12 μL of water. Six microliters of this cDNA was used to produce biotin-labeled cRNA by in vitro transcription using the GeneChip IVT labeling kit (Affymetrix) according to the manufacturer's instructions. The RNeasy kit (Qiagen) was used for cleanup of the in vitro transcription reaction, and labeled cRNA was precipitated with NH4OAc and ethanol at −20°C overnight. The pellet was then dried and resuspended in 11 μL of water. Twenty micrograms of biotin-labeled cRNA was fragmented to a size range of 35 to 200 nucleotides according to Affymetrix recommendations for eukaryotic sample processing. Hybridization to full-genome Arabidopsis Affymetrix ATH1 arrays, scanning, and data extraction were performed at the Gene Expression Center (University of Wisconsin–Madison Biotechnology Center,).
Microarray Processing and Quality Control
For expression analysis, raw probe intensity data were preprocessed using the RMA algorithm implemented in Bioconductor (http://www.bioconductor.org/). The RMA application consisted of background adjustment and quantile normalization, followed by log2 summarization (Bolstad et al., 2003; Irizarry et al., 2003a, 2003b). Several methods of quality assessment were employed to examine our microarray data. As a first method, the program RMAExpress was used to examine pseudo-images of chip residuals (http://rmaexpress.bmbolstad.com/). Examining chip residual images can detect artifacts on arrays and can identify faulty hybridization events. The residual image for one of the wild-type control samples indicated anomalous hybridization and was eliminated from analysis. As a replacement, a new array was hybridized with cRNA from the same tissue sample. Second, the quality of biological replicates was examined by performing a least-square regression analysis. We examined all possible pairwise comparisons of normalized signal intensities within each set of three biological replicates, and r2 values are presented in Supplemental Table 4 online. Following quality control, probe sets corresponding to Affymetrix controls were trimmed from the data set. RMA-processed data from this experiment are available in Supplemental Data Set 1 online.
Microarray Analysis
From the signal intensity values resulting from the application of the RMA algorithm, log2 fold change values were calculated. Statistical tests were applied to the data set using the TIGR MultiExperiment Viewer (http://www.tm4.org/mev.html/) (Saeed et al., 2003). For unpaired t tests, the following parameters were used: Welch approximation for variance assumption, P values based on t-distribution, and α = 0.01. For one- and two-way ANOVA, the following parameters were used: P values based on F-distribution and α = 0.01. Two methods of multiple testing correction (both using α = 0.01) were compared for each statistical test: the Bonferroni method (Bonferroni, 1936) and the Benjamini and Hochberg method (Benjamini and Hochberg, 1995). The Bonferroni correction divides the test-wise significance level by the number of tests and is the most stringent method, tolerating the fewest number of false positives. The Benjamini and Hochberg false discovery rate is much less stringent and will reduce the number of false negatives but also tolerates more false positives. Genes of interest were classified into functional categories using the MIPS Functional Catalog Database (http://mips.gsf.de/projects/funcat) or the GeneBins website (http://bioinfoserver.rsbs.anu.edu.au/utils/GeneBins/) (Goffard and Weiller, 2007). Enrichment of a functional category was determined using the hypergeometric function, as implemented by the websites.
Hierarchical Clustering
A bulk download of Affymetrix ATH1 microarray expression data was obtained from the ftp server on The Arabidopsis Information Resource (TAIR) website (ftp://ftp.arabidopsis.org/home/tair/Miroarrays/analyed_data). At the time when this file was created, there were 1436 RMA-processed microarrays represented in the download. Some of these files were duplicates, and some were from experiments without any replicate chips; these data were removed from the analysis. Additional microarray experiments not contained in this large file were also downloaded individually from the TAIR website (ftp://ftp.arabidopsis.org/home/tair/Microarrays/analyzed_data). An additional 558 CEL files were obtained in this manner and were RMA processed using RMAExpress (http://rmaexpress.bmbolstad.com/). After filtering the microarrays, a total of 1704 Affymetrix microarray expression data files were used for analysis, representing microarray experiments that were performed as part of the AtGenExpress project (http://web.uni-frankfurt.de/fb15/botanik/mcb/AFGN/atgenex.htm) as well as additional experiments from the Nottingham Arabidopsis Stock Centre facility (http://affymetrix.arabidopsis.info/narrays/experimentbrowse.pl). Although every effort was made to ensure data sets were comparably processed, it is impossible to ensure that all were normalized using the exact same parameters. Consequently, any conclusions drawn from the AtMegaCluster should be viewed with a degree of caution.
Expression data from replicate microarrays were identified and averaged. Experimental and control samples were identified, and the expression difference was computed as the log2 fold change of experimental as compared with the control. For some microarray sets, more than one variable was tested; in these cases, all possible combinations of experimental and control pairs were considered. These calculations resulted in 444 experiments (columns in our data file) used for clustering (see Supplemental Data Set 2 online). These experiments represent a wide variety of conditions, including abiotic stress treatment (180 experiments), hormone treatment (22 experiments), mutant genotype (109 experiments), chemical treatment (81 experiments), and biotic stress treatment, mostly pathogens (47 experiments).
This large data file was hierarchically clustered in two dimensions (across 22,810 genes and across 444 experiments) using the average linkage method and the absolute Pearson correlation coefficient as the distance metric (Eisen et al., 1998). Additional hierarchical clustering with a smaller subset of 13 osmotic stress experiments was also performed. The program Cluster (http://rana.lbl.gov/EisenSoftware.htm) was used for clustering, and the program JavaTreeView (http://jtreeview.sourceforge.net/) (Saldanha, 2004) was used for visualization of clustered data. Results of the hierarchical clustering were used to compile a list of genes coexpressed with ATHK1. The ATTED-II database (http://www.atted.bio.titech.ac.jp/) (Obayashi et al., 2007) was used as a comparison to our results.
Promoter Analysis
The set of 233 genes significantly regulated by ATHK1 transcript level during osmotic stress conditions (significance based on two-way ANOVA) was used to identify putative regulatory elements in upstream regions using the motif search and prediction program Regulatory Sequence Analysis (RSA) Tools (http://rsat.ulb.ac.be/rsat/) (van Helden, 2003). Using 1500 bp upstream of our set of 233 genes, we searched for known cis-acting regulatory elements in osmotic stress-responsive genes (reviewed in Yamaguchi-Shinozaki and Shinozaki, 2005). The motif prediction algorithm implemented in RSA Tools uses string-based pattern matching to analyze oligonucleotide occurrences in a set of genes and returns those that are statistically overrepresented. We used predefined background frequencies for Arabidopsis thaliana as provided by the RSA website and searched for statistically overrepresented 6- and 7-mers in the 1500-bp upstream regions of our genes.
qRT-PCR
For RNA analysis on seedlings, plants were grown for 5 d on chromatography paper overlaid on media, pH 5.7, containing half-strength MS, 2.5 mM MES, and 0.8% (w/v) agar. For stress treatment, seedlings were transferred along with the chromatography paper to liquid half-strength MS, 2.5 mM MES ± 300 mM sorbitol, or 1 μM ABA for 16 h, after which time tissue was collected. Whole seedlings were ground to a fine powder using a Mixer Mill (Retsch) in RNase-free conditions. Total RNA was prepared using the RNeasy plant mini kit (Qiagen) according to the manufacturer's instructions. For RNA analysis on seeds, dry seeds were frozen in liquid N2 and ground to a fine powder using a mortar and pestle in RNase-free conditions. Seed RNA was extracted as described by Vicient and Delseny (1999). All RNA samples were DNase treated (Promega) prior to cDNA synthesis. Three micrograms of DNase-treated RNA was used for first-strand cDNA synthesis with Superscript II (Invitrogen) using an oligo(dT) (dT24 + V) primer, with the following modifications to the manufacturer's instructions: the oligo(dT) primer (100 μM) was mixed with RNA in 12 μL of RNase-free water and was heated to 70°C for 10 min prior to addition of first-strand buffer and DTT.
Following first-strand synthesis, optimal cDNA template amount was determined by preparing a dilution series. For final analyses, 2 μL of a 250-fold dilution of the RT reaction (corresponding to 2.4 ng RNA) was used for qRT-PCR amplification with SYBR Premix Ex Taq (Takara), with the following modifications to the manufacturer's instructions: amplifications were performed in 20 μL reaction volumes with 5 pmol of each primer. qRT-PCR reactions were performed with the iCycler Real Time PCR system (Bio-Rad). For each of the genes under study, a primer pair was designed around an intron to obtain a PCR product of ∼100 to 300 bp. The sequences of the primers used for qRT-PCR amplifications are available in Supplemental Table 2 online. Presence of a single PCR product was verified by melt-curve analysis. Relative quantification of gene expression was performed using the Real-Time PCR Miner algorithm (Zhao and Fernald, 2005) to account for differences in PCR efficiencies of different primer pairs used for different genes. All reactions were performed in quadruplicate on separate plates. Expression levels were normalized using values obtained for the housekeeping gene ACT2 (At3g18780).
Seed Protein Extraction
For quantification of soluble proteins, 25 μg of dry seeds were frozen in liquid N2 and ground to a fine powder using a mortar and pestle. Seed powder was combined with 500 μL extraction buffer, pH 8.5, containing 100 mM Tris-HCl and the following protease inhibitors (all from Sigma-Aldrich): 1 mM phenylmethanesulphonyl fluoride, 1 mM pepstatin, 1 mM l-trans-epoxysuccinyl-leucyl amido(4-guanidio)butane (E64), 1 μM 1,10-phenanthroline, and 1 μM bestatin. The extract was gently rocked at 4°C for 20 min to mix the buffer and seed powder. After centrifugation (11,000 g, 10 min, 4°C), the supernatant was used for protein measurements. Protein concentration from 10% of the final extract was measured using a bicinchoninic acid assay kit (Pierce) (Smith et al., 1985). BSA was used as a standard.
Two-Dimensional Gel Electrophoresis
Proteins were analyzed by two-dimensional gel electrophoresis as follows. Protein extract corresponding to 100 μg was precipitated by adding 50% trichloroacetic acid to a final volume of 10% and incubating for 30 min on ice. After centrifugation (11,000g, 10 min, 4°C), the protein pellet was washed six times in cold acetone to remove all traces of trichloroacetic acid. The pellet was resuspended in 300 μL immobilized pH gradient (IPG) sample buffer containing 8 M urea, 4% CHAPS, and 50 mM DTT. Proteins were first separated by electrophoresis according to charge. Isoelectric focusing was performed with 100 μg of protein using gel strips forming an immobilized nonlinear pH gradient from 3 to 10 (ReadyStrip IPG Strips, pH 3 to 10, 11 cm; Bio-Rad). Strips were rehydrated overnight at room temperature with IPG sample buffer and protein extracts. Isoelectric focusing was performed at 20°C in the PROTEAN IEF Cell system (Bio-Rad) with the following program: (1) 1 h at 250 V; (2) 2 h at 3000 V; and (3) 4 h at 8000 V. Prior to the second dimension, the gel strips were equilibrated twice for 45 min each in equilibration solution containing 6 M urea, 20% (v/v) glycerol, 2% (w/v) SDS, and 0.375 M Tris-HCl, pH 8.8. DTT (50 mM) was added to the first equilibration solution, and iodoacetamide (4% [w/v]) was added to the second (Harder et al., 1999). Equilibrated gel strips were placed on top of vertical polyacrylamide gels (Criterion 10 to 20% Tris-HCl precast gels; Bio-Rad). Electrophoresis was performed at room temperature in a buffer, pH 8.3, containing 25 mM Tris, 192 mM glycine, and 0.1% SDS (Bio-Rad), at 20 V for ∼1 h and then at 100 V until proteins had run through the gel (∼3 h). For each genotype analyzed, two-dimensional gels were run in duplicate from three independent protein extractions.
Gels were stained with SYPRO Ruby (Bio-Rad) according to the manufacturer's instructions. Stained gels were scanned with a UV scanner (Fotodyne), and images were captured using the UVP gel documentation system. Image analysis was performed with software (PDQuest; Bio-Rad) according to the manufacturer's instructions. After spot detection and background subtraction, two-dimensional gels were aligned and matched, and the quantitative determination of the spot volumes was performed. For each analysis, the Matching Summary provided by PDQuest indicated a high level of reproducibility between normalized spot volumes of gels (correlation coefficients of replicate samples ranged from 0.878 to 0.763).
Protein Identification by Mass Spectrometry
Spots of interest were excised from two-dimensional gels and were digested with trypsin. Solid phase extraction (SPE) was performed as follows. SPEC Plus PT C18 solid-phase extraction pipette tips were obtained from Ansys Diagnostics. Tips were rinsed by gravity flow with 400 μL of a solution containing 0.1% formic acid in 90% acetonitrile and 10% water, and again with 200 μL of 0.1% formic acid in water. Formic acid was added to the peptide sample to a final concentration of 5%. After centrifugation to remove solid debris, the sample was applied to the SPE unit and allowed to flow through by gravity. The SPE unit was washed with 200 μL of solution, pH 4.5, containing 100 mM ammonium acetate and 1% acetic acid and washed again with 2 mL of 0.1% formic acid in water. Peptides were eluted with 100 μL of a solution containing 0.1% formic acid in 90% acetonitrile and 10% water. The volume of the sample was reduced to ∼10% in a SpeedVac to remove acetonitrile, and 40 μL 0.1% formic acid was added to the remaining solution. Samples were analyzed on a QTOF2 mass spectrometer (Micromass) coupled to an HP 1100 HPLC (Agilent) as described (Nelson et al., 2007). Proteins were identified by searching the Arabidopsis protein databases using MASCOT 2.0 (Perkins et al., 1999) as described (Nelson et al., 2007).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AY093145 (ABA1, At5G67030); AY082344 (ABA2, At1G52340); AC007154 (AAO3, At2G27150); BT029491 (CRA1, At5G44120); AY093005 (CRU2, At1G03880); ATU66916 (CRU3, At4G28520); AY072508 (2S1, At4G27140); BT002073 (2S2, At4G27150); AF446894 (2S4, At4G27170); AY128731 (RD29B, At5G52300); AY062661 (RD20, At2G33380); AY519511 (CCA1, At2G46830); AY136415 (ABI2, At5G57050); BT011656 (MYB2, At2G47190); BT002206 (HB12, At3G61890); AY062849 (KIN1, At5G15960); BT015111 (LEA14, At1G01470); and AY093779 (RAB18, At5G66400). The following genes have no accession numbers currently available: ATHK1 (At2g17820), FUS3 (At3G26790), and LEC1 (At1G21970). Germplasm is available for the following mutants: athk1-3 (CSH10), athk1-4 (CSH17), arr3,4,5,6 (CS25276), arr5,6,8,9 (CS25277), and arr3,4,5,6,8,9 (CS25279).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Locations of athk1 T-DNA Inserts and Expression Levels of ATHK1 RNA.
Supplemental Figure 2. Altered ABA Sensitivities in Root Elongation of athk1 Alleles.
Supplemental Figure 3. athk1 Knockout Mutants Display Normal Embryo Development.
Supplemental Figure 4. Gene Ontology Annotations for Genes Significantly Differentially Regulated According to Both ATHK1 Transcript Level and Sorbitol Stress Conditions.
Supplemental Figure 5. qRT-PCR Measures of Expression Confirm Microarray Expression Changes.
Supplemental Figure 6. Type-A ARR Mutants Display Altered Osmotic Stress Germination Response.
Supplemental Figure 7. Functional Distribution of Gene Clusters from the AtMegaCluster.
Supplemental Table 1. Genes Significantly Differentially Regulated According to ATHK1.
Supplemental Table 2. Common Experimental Conditions in the ATHK1 Gene Cluster.
Supplemental Table 3. Sequences of Primers Used for qRT-PCR Analysis.
Supplemental Table 4. Replicate Quality of Biological Duplicate Microarray Experiments.
Supplemental Data Set 1. RMA-Processed, Log2 Expression Levels Obtained from the Microarray Experiment Described in This Study.
Supplemental Data Set 2. Compilation of 1704 Affymetrix Microarray Experiments Used to Create the AtMegaCluster Described in This Study.
Supplementary Material
Acknowledgments
We gratefully acknowledge funding from the National Science Foundation and the Department of Energy (to M.R.S.), from the National Institutes of Health Graduate Student Genetics Training Grant (to D.J.W.), and from Conselho Nacional de Desenvolvimento Científico e Tecnológico (to B.F.Q.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Michael R. Sussman (msussman@wisc.edu).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Abe, H., Yamaguchi-Shinozaki, K., Urao, T., Iwasaki, T., Hosokawa, D., and Shinozaki, K. (1997). Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 9 1859–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. Roy. Stat. Soc. B 57 289–300. [Google Scholar]
- Bolstad, B.M., Irizarry, R.A., Astrand, M., and Speed, T.P. (2003). A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19 185–193. [DOI] [PubMed] [Google Scholar]
- Bonferroni, C.E. (1936). Teoria statistica delle classi e calcolo delle probabilità. Pubblicazioni del R Istituto Superiore di Scienze Economiche e Commerciali di Firenze 8 3–62. [Google Scholar]
- Bray, E.A. (1997). Plant responses to water deficit. Trends Plant Sci. 2 48–54. [Google Scholar]
- Chiwocha, S.D., Abrams, S.R., Ambrose, S.J., Cutler, A.J., Loewen, M., Ross, A.R., and Kermode, A.R. (2003). A method for profiling classes of plant hormones and their metabolites using liquid chromatography-electrospray ionization tandem mass spectrometry: An analysis of hormone regulation of thermodormancy of lettuce (Lactuca sativa L.) seeds. Plant J. 35 405–417. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Eisen, M.B., Spellman, P.T., Brown, P.O., and Botstein, D. (1998). Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne, W., Meyer, M.H., Peppers, J., and Meyer, R.A., Jr. (2004). Comparison of mRNA gene expression by RT-PCR and DNA microarray. Biotechniques 36 618–620. [DOI] [PubMed] [Google Scholar]
- Finkelstein, R.R., Gampala, S.S., and Rock, C.D. (2002). Abscisic acid signaling in seeds and seedlings. Plant Cell 14(Suppl): S15–S45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein, R.R., and Rock, C.D. (2002). Abscisic acid biosynthesis and response. In The Arabidopsis Book, C.R. Somerville and E.M. Meyerowitz, eds (Rockville, MD: American Society of Plant Biologists), doi/10.1199/tab.0058, http://www.aspb.org/publications/arabidopsis/. [DOI] [PMC free article] [PubMed]
- Goffard, N., and Weiller, G. (2007). GeneBins: A database for classifying gene expression data, with application to plant genome arrays. BMC Bioinformatics 8 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiltinan, M.J., Marcotte, W.R., Jr., and Quatrano, R.S. (1990). A plant leucine zipper protein that recognizes an abscisic acid response element. Science 250 267–271. [DOI] [PubMed] [Google Scholar]
- Harder, A., Wildgruber, R., Nawrocki, A., Fey, S.J., Larsen, P.M., and Gorg, A. (1999). Comparison of yeast cell protein solubilization procedures for two-dimensional electrophoresis. Electrophoresis 20 826–829. [DOI] [PubMed] [Google Scholar]
- Hasegawa, P.M., Bressan, R.A., Zhu, J.K., and Bohnert, H.J. (2000). Plant cellular and molecular responses to high salinity. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51 463–499. [DOI] [PubMed] [Google Scholar]
- Hwang, I., Chen, H.C., and Sheen, J. (2002). Two-component signal transduction pathways in Arabidopsis. Plant Physiol. 129 500–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram, J., and Bartels, D. (1996). The molecular basis of dehydration tolerance in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47 377–403. [DOI] [PubMed] [Google Scholar]
- Irizarry, R.A., Bolstad, B.M., Collin, F., Cope, L.M., Hobbs, B., and Speed, T.P. (2003. a). Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry, R.A., Hobbs, B., Collin, F., Beazer-Barclay, Y.D., Antonellis, K.J., Scherf, U., and Speed, T.P. (2003. b). Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4 249–264. [DOI] [PubMed] [Google Scholar]
- Knight, H., and Knight, M.R. (2001). Abiotic stress signalling pathways: Specificity and cross-talk. Trends Plant Sci. 6 262–267. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., Hanhart, C.J., Hilhorst, H.W., and Karssen, C.M. (1989). In vivo inhibition of seed development and reserve protein accumulation in recombinants of abscisic acid biosynthesis and responsiveness mutants in Arabidopsis thaliana. Plant Physiol. 90 463–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysan, P.J., Young, J.C., and Sussman, M.R. (1999). T-DNA as an insertional mutagen in Arabidopsis. Plant Cell 11 2283–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohrmann, J., and Harter, K. (2002). Plant two-component signaling systems and the role of response regulators. Plant Physiol. 128 363–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin, E., Nussaume, L., Quesada, A., Gonneau, M., Sotta, B., Hugueney, P., Frey, A., and Marion-Poll, A. (1996). Molecular identification of zeaxanthin epoxidase of Nicotiana plumbaginifolia, a gene involved in abscisic acid biosynthesis and corresponding to the ABA locus of Arabidopsis thaliana. EMBO J. 15 2331–2342. [PMC free article] [PubMed] [Google Scholar]
- Menkes, A.E., Schindler, U., and Cashmore, A.R. (1995). The G-box: A ubiquitous regulatory DNA element in plants bound by the GBF family of bZIP proteins. Trends Biochem. Sci. 20 506–510. [DOI] [PubMed] [Google Scholar]
- Mundy, J., Yamaguchi-Shinozaki, K., and Chua, N.H. (1990). Nuclear proteins bind conserved elements in the abscisic acid-responsive promoter of a rice rab gene. Proc. Natl. Acad. Sci. USA 87 1406–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth bioassays with tobacco tissue cultures. Physiol. Plant. 15 473–497. [Google Scholar]
- Nambara, E., and Marion-Poll, A. (2003). ABA action and interactions in seeds. Trends Plant Sci. 8 213–217. [DOI] [PubMed] [Google Scholar]
- Nelson, C.J., Huttlin, E.L., Hegeman, A.D., Harms, A.C., and Sussman, M.R. (2007). Implications of 15N-metabolic labeling for automated peptide identification in Arabidopsis thaliana. Proteomics 7 1279–1292. [DOI] [PubMed] [Google Scholar]
- Obayashi, T., Kinoshita, K., Nakai, K., Shibaoka, M., Hayashi, S., Saeki, M., Shibata, D., Saito, K., and Ohta, H. (2007). ATTED-II: A database of co-expressed genes and cis elements for identifying co-regulated gene groups in Arabidopsis. Nucleic Acids Res. 35 D863–D869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang, P.P., Pruitt, R.E., and Meyerowitz, E.M. (1988). Molecular cloning, genomic organization, expression and evolution of 12S seed storage protein genes of Arabidopsis thaliana. Plant Mol. Biol. 11 805–820. [DOI] [PubMed] [Google Scholar]
- Perkins, D.N., Pappin, D.J., Creasy, D.M., and Cottrell, J.S. (1999). Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20 3551–3567. [DOI] [PubMed] [Google Scholar]
- Reiser, V., Raitt, D.C., and Saito, H. (2003). Yeast osmosensor Sln1 and plant cytokinin receptor Cre1 respond to changes in turgor pressure. J. Cell Biol. 161 1035–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed, A.I., et al. (2003). TM4: A free, open-source system for microarray data management and analysis. Biotechniques 34 374–378. [DOI] [PubMed] [Google Scholar]
- Saldanha, A.J. (2004). Java Treeview – Extensible visualization of microarray data. Bioinformatics 20 3246–3248. [DOI] [PubMed] [Google Scholar]
- Salomé, P.A., To, J.P., Kieber, J.J., and McClung, C.R. (2006). Arabidopsis response regulators ARR3 and ARR4 play cytokinin-independent roles in the control of circadian period. Plant Cell 18 55–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller, E.G., Mathews, D.E., Gribskov, M., and Walker, J.C. (2002). Two-component signaling elements and histidyl-aspartyl phosphorelays. In The Arabidopsis Book, C.R. Somerville and E.M. Meyerowitz, eds (Rockville, MD: American Society of Plant Biologists), doi/10.1199/tab.0086, http://www.aspb.org/publications/arabidopsis/. [DOI] [PMC free article] [PubMed]
- Schwartz, S.H., Leon-Kloosterziel, K.M., Koornneef, M., and Zeevaart, J.A. (1997). Biochemical characterization of the aba2 and aba3 mutants in Arabidopsis thaliana. Plant Physiol. 114 161–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, S.H., Qin, X., and Zeevaart, J.A. (2003). Elucidation of the indirect pathway of abscisic acid biosynthesis by mutants, genes, and enzymes. Plant Physiol. 131 1591–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, M., and Koshiba, T. (2002). Complex regulation of ABA biosynthesis in plants. Trends Plant Sci. 7 41–48. [DOI] [PubMed] [Google Scholar]
- Smith, P.K., Krohn, R.I., Hermanson, G.T., Mallia, A.K., Gartner, F.H., Provenzano, M.D., Fujimoto, E.K., Goeke, N.M., Olson, B.J., and Klenk, D.C. (1985). Measurement of protein using bicinchoninic acid. Anal. Biochem. 150 76–85. [DOI] [PubMed] [Google Scholar]
- Tena, G., Asai, T., Chiu, W.L., and Sheen, J. (2001). Plant mitogen-activated protein kinase signaling cascades. Curr. Opin. Plant Biol. 4 392–400. [DOI] [PubMed] [Google Scholar]
- Tesnier, K., Strookman-Donkers, H.M., van Pijlen, J.G., van der Geest, A.H.M., Bino, R.J., and Groot, S.P.C. (2002). A controlled deterioration test for Arabidopsis thaliana reveals genetic variation in seed quality. Seed Sci. Technol. 30 149–165. [Google Scholar]
- To, J.P., Haberer, G., Ferreira, F.J., Deruere, J., Mason, M.G., Schaller, G.E., Alonso, J.M., Ecker, J.R., and Kieber, J.J. (2004). Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell 16 658–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran, L.S., Urao, T., Qin, F., Maruyama, K., Kakimoto, T., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2007). Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proc. Natl. Acad. Sci. USA 104 20623–20628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urao, T., Yakubov, B., Satoh, R., Yamaguchi-Shinozaki, K., Seki, M., Hirayama, T., and Shinozaki, K. (1999). A transmembrane hybrid-type histidine kinase in Arabidopsis functions as an osmosensor. Plant Cell 11 1743–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urao, T., Yakubov, B., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1998). Stress-responsive expression of genes for two-component response regulator-like proteins in Arabidopsis thaliana. FEBS Lett. 427 175–178. [DOI] [PubMed] [Google Scholar]
- van Helden, J. (2003). Regulatory sequence analysis tools. Nucleic Acids Res. 31 3593–3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicient, C.M., and Delseny, M. (1999). Isolation of total RNA from Arabidopsis thaliana seeds. Anal. Biochem. 268 412–413. [DOI] [PubMed] [Google Scholar]
- West, M., and Harada, J.J. (1993). Embryogenesis in higher plants: An overview. Plant Cell 5 1361–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki, K., and Shinozaki, K. (1994). A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6 251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki, K., and Shinozaki, K. (2005). Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 10 88–94. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki, K., and Shinozaki, K. (2006). Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 57 781–803. [DOI] [PubMed] [Google Scholar]
- Zhao, S., and Fernald, R.D. (2005). Comprehensive algorithm for quantitative real-time polymerase chain reaction. J. Comput. Biol. 12 1047–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J.K. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53 247–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.