Abstract
Indole acetic acid (auxin) is a key regulator of wood formation, and an observed overlap between auxin concentration gradient and developing secondary xylem cells has led to the hypothesis that auxin regulates wood formation by acting as a morphogen. We dissected the role of auxin in wood formation by identifying the auxin-responsive transcriptome in wood-forming tissues and investigating alterations in wood formation in transgenic hybrid aspen plants (Populus tremula × Populus tremuloides) with perturbed auxin signaling. We showed that auxin-responsive genes in wood-forming tissues respond dynamically to changes in cellular auxin levels. However, the expression patterns of most of the auxin-responsive genes displayed limited correlation with the auxin concentration across this developmental zone. Perturbing auxin signaling by reducing auxin responsiveness reduced the cambial cell division activity, caused spatial deregulation of cell division of the cambial initials, and led to reductions in not only radial but also axial dimensions of fibers and vessels. We propose that, instead of acting as a morphogen, changes in auxin concentration in developing secondary xylem cells may provide important regulatory cues that modulate the expression of a few key regulators; these, in turn, may control the global gene expression patterns that are essential for normal secondary xylem development.
INTRODUCTION
In woody plants, the development of secondary xylem cells involves the sequential processes of cell division, expansion, secondary cell wall formation, and cell death. This leads to the organization of secondary xylem cells into a developmental gradient consisting of cells at distinct phases of development (Larson, 1994). The mechanism regulating the sequential development of secondary xylem cells is not well understood, but the plant growth regulator indole acetic acid (IAA/auxin [AUX]) has been proposed to play a key role in it (Sundberg et al., 2000). Auxin is distributed in a radial concentration gradient, with high levels in dividing secondary xylem cells that decline gradually over the regions containing expanding and secondary cell wall–forming cells (Uggla et al., 1996; Tuominen et al., 1997). The coincidence of an auxin concentration gradient with the developmental gradient in secondary xylem has prompted the suggestion that it could be the primary regulator of the sequential patterning of secondary xylem development, with auxin acting in a manner similar to a morphogen (Uggla et al., 1996; Tuominen et al., 1997; Sundberg et al., 2000).
A central tenet of the hypothesis that the auxin concentration gradient regulates secondary xylem development is the concept that integration of the intracellular auxin concentration over the time during which cells experience a particular concentration may control the pattern of secondary xylem development. While attractive as a hypothesis, experimental validation of this model has proven difficult. Attempts to disrupt the in vivo distribution of auxin in secondary xylem by overexpressing auxin biosynthesis genes in transgenic poplar plants (Populus spp) have been largely unsuccessful (Tuominen et al., 1997). Furthermore, the nature of auxin-regulated transcriptional programs underlying secondary xylem development remains largely uninvestigated. Importantly, no auxin response mutants in tree species have been reported that could be used to analyze the role of auxin in secondary xylem development. As a result, our understanding of how auxin could regulate secondary xylem development at the molecular level is rudimentary.
To improve our understanding of the molecular mechanisms involved in auxin-mediated regulation of secondary xylem development, we further characterized the auxin-responsive transcriptome of hybrid aspen plants (Populus tremula × Populus tremuloides). In contrast with earlier studies (Moyle et al., 2002; Schrader et al., 2003), we investigated the dynamic responses of global auxin-responsive gene expression to changes in endogenous auxin levels in wood-forming tissues of these plants. Furthermore, we examined the correlations between patterns of expression of auxin-responsive genes, the transitions between distinct stages of secondary xylem development, and their positions within the radial auxin concentration gradient. The transcriptomic analysis was complemented by an investigation of secondary xylem development in transgenic hybrid aspen plants with altered auxin responses due to the overexpression of a mutant form of the poplar AUX/IAA gene Ptt IAA3, which is a key component in the auxin signaling pathway. (Here, Ptt IAA3 refers to the wild-type gene and Ptt IAA3m refers to the mutant gene; for the corresponding proteins, Ptt IAA3 indicates the wild-type protein and Ptt IAA3m refers to mutant protein.) In this mutant, a conserved Pro residue has been changed to a Ser in the degron domain involved in the auxin-regulated degradation of AUX/IAA proteins (Gray et al., 2001; Ramos et al., 2001; Tiwari et al., 2001). In several Arabidopsis thaliana mutants, similar mutations in the degron domain have been shown to lead to aberrant auxin responses (Rouse et al., 1998; Nagpal et al., 2000; Fukaki et al., 2002; Yang et al., 2004). Combining these approaches has allowed us to dissect the molecular basis of the auxin concentration–mediated regulation of secondary xylem development.
RESULTS
The Auxin-Responsive Transcriptome of Hybrid Aspen Stem Tissues
To elucidate how auxin regulates secondary xylem development, we used transcript profiling to identify global changes in gene expression in hybrid aspen stem tissue in response to changes in auxin levels. Auxin levels were modulated in the stem tissues of hybrid aspen plants by depleting them of auxin for 16 h and then replenishing the levels through the external application of auxin for a period of 30 min to 4 h. Auxin levels were reduced to about half of the starting levels after 16 h of depletion (Figure 1A and inset); then, following the addition of auxin, the levels increased by 10-fold that of the starting levels after 30 min and stabilized at four times the starting levels. We then characterized the global changes in gene expression that occurred in response to the modulation of auxin levels using the poplar POP1 cDNA microarray spotted with 13,000 probes (Sterky et al., 2004). Genes whose expression was altered at 30 min or 2 or 4 h after adding auxin following depletion for 16 h were classified as auxin-responsive genes. The data indicate that 632 of the >13,000 cDNAs interrogated were auxin-responsive (Figure 1B; summarized in Supplemental Data Set 1 online); 530 of these were upregulated and 102 were downregulated. As expected, the expression of known auxin-regulated genes, such as members of the AUX/IAA and auxin transporter families, appeared to be regulated by auxin, in agreement with earlier data (Moyle et al., 2002; Schrader et al., 2003). However, in addition to these well-known candidates, various other genes expressed in developing secondary xylem cells were also found to respond to changes in auxin levels (see Supplemental Data Set 1 online). Subsequent assessment of the auxin responsiveness of five candidate genes by RT-PCR (see Supplemental Figure 1 online) confirmed the findings from the microarray experiment.
Figure 1.
Altered Auxin Levels and Corresponding Gene Expression.
(A) Modulation of auxin levels in the stem. Stem samples were placed in half-strength MS medium for 16 h to deplete their auxin levels, then they were replenished by transfer to the same medium with 20 μM IAA. The graph shows changes in IAA levels at various time points: after 0 h (d0), 4 h (d4), and 16 h (d16) of depletion, and 30 min (i0.5), 2 h (i2), and 4 h (i4) after the addition of IAA. The inset shows a magnification covering the depletion step. n = 3; error bars represent sd. t tests indicated that there were significant differences in auxin levels between d0 and d16 (P = 0.05) and between d16 and both i0.5 and i2 (P ≤ 0.01). FW, fresh weight.
(B) Responses of gene expression in stem tissues to changes in auxin levels. The expression patterns of auxin-responsive genes could be divided into nine clusters. The lines in the graphs represent the average expression profile of all genes in each cluster. The number of auxin-responsive cDNA clones identified by microarray analysis belonging to each cluster is indicated. Supplemental Data Set 1 online provides details of the expression clusters.
The Auxin Concentration Gradient Indirectly Regulates Global Gene Expression in Developing Secondary Xylem Cells
The global expression pattern of a large number of genes in developing secondary xylem mirrors the graded distribution of auxin, with high levels in the cambial region that decline gradually in secondary wall–forming secondary xylem cells (Hertzberg et al., 2001). Therefore, we investigated whether the expression patterns of these genes could be directly regulated by the auxin concentration gradient. To address this possibility, we identified genes expressed in developing secondary xylem (Hertzberg et al., 2001; Schrader et al., 2004) whose expression patterns are highly correlated with the auxin distribution pattern. Partial least squares (PLS) analysis indicated that the expression patterns of 250 genes expressed in the developing secondary xylem correlated highly with the auxin distribution pattern. However, as shown in Figure 2, only 26 of this set of 250 genes were present in the set of 632 auxin-responsive genes identified in the transcriptomic analysis (see Supplemental Data Set 1 online). Thus, the vast majority of genes whose expression pattern mirrors the graded distribution of auxin do not appear to be regulated directly by auxin.
Figure 2.
PLS Analysis.
Analysis of correlations between the auxin concentration gradient and the expression pattern of auxin-responsive genes in wood-forming tissues. The Venn diagram shows the overlap (orange area) between the auxin-responsive transcriptome (dark gray area) and the genes showing expression patterns that were highly correlated to the auxin concentration gradient (light gray area).
Expression of Auxin-Responsive Genes Responds Dynamically to Changes in Cellular Auxin Content
Most of the IAA in the cambium and developing secondary xylem is supplied by polar auxin transport (Sundberg et al., 1994). Thus, as cells divide during the course of secondary xylem development, their auxin levels decrease as they move away from the source of auxin. These dynamic changes in auxin concentration in developing secondary xylem cells could play an important role in the regulation of gene expression and thereby influence secondary xylem development. Therefore, we investigated whether the transcript levels of auxin-responsive genes respond dynamically to changes in cellular auxin content, and we found that the expression of a large number of genes responds to changes in auxin levels (Figure 1B). For several of these genes, the transcript levels declined as cellular auxin levels were reduced following 16 h of depletion (Figure 1A, inset), and the transcript levels of these genes increased as auxin levels increased after both 30 min and 2 h of auxin treatment (Figure 1B, clusters D, E, and F). By contrast, for several genes, the opposite trend was observed: their transcript levels increased as auxin levels declined and then declined as auxin levels increased (Figure 1B, clusters G, H, and I). However, it should be noted that while auxin levels were significantly higher than the predepletion levels at the end of the experimental period, the steady state levels of transcripts of auxin upregulated genes did not always show corresponding increases.
We also clustered the auxin-responsive genes according to their expression profiles during secondary xylem development (Figure 3). The results indicated that a large proportion of auxin-responsive genes were expressed at higher levels in expanding and secondary cell wall–forming cells than in the cambium, although these cells have lower auxin levels than the cambium. These findings may reflect the possibility that gene expression is less sensitive to changes in auxin levels in the cambial meristem than in the expanding and secondary cell wall–forming cells or, alternatively, that our experimental treatment did not cause increases in the levels of auxin in the cambium. We believe that we can discount the latter possibility, since application of auxin to the cambium via the polar auxin stream using lanolin supplemented with auxin gives very similar results to those described here (Björklund et al., 2007; A. Karlberg, unpublished results). Furthermore, only minor fluctuations in auxin levels in the cambial meristem have been found throughout the growing season (Uggla et al., 2001). These findings suggest that the expression of auxin-responsive genes may be less sensitive to changes in auxin levels in the cambium than in expanding xylem cells, which experience greater fluctuations in auxin levels. Importantly, these results indicate that the expression of a large proportion of auxin-responsive genes responds dynamically to transient changes in auxin levels rather than being governed by steady state auxin levels.
Figure 3.
Cluster Analysis.
Expression of auxin-responsive genes versus auxin levels in the wood-forming tissues of poplar. The auxin-responsive cDNA clones identified by microarray analysis (see Supplemental Data Set 1 online) were clustered according to their expression across the developing wood cells derived from previous expression data (Hertzberg et al., 2001). The red color gradient reflects auxin levels in the boundary between meristematic cells of the cambium and early expanding cells.
Expression of a Mutated Version of Ptt IAA3 Alters Auxin Responsiveness in Hybrid Aspen
To understand how auxin regulates secondary xylem development, we altered auxin response in transgenic hybrid aspen plants by expressing a mutated version of the poplar AUX/IAA gene Ptt IAA3, referred to as Ptt IAA3m, that is normally expressed in the cambial meristem and young secondary xylem cells (Moyle et al., 2002). In Ptt IAA3m, a conserved Pro residue, Pro-62, was changed to a Ser in the domain responsible for auxin-mediated degradation of AUX/IAA proteins (Rouse et al., 1998; Gray et al., 2001; Ramos et al., 2001; Tiwari et al., 2001) (Figure 4A). Similar mutations in AUX/IAA proteins have been shown to stabilize the mutant proteins in Arabidopsis (Gray et al., 2001). Several transgenic hybrid aspen lines were generated expressing the mutated version of Ptt IAA3 cDNA (Ptt IAA3m) under the control of the 35S cauliflower mosaic virus (CaMV) promoter, and the Ptt IAA3m transcript levels were tested (Figure 4B). Although our RT-PCR primers cannot distinguish between the wild-type Ptt IAA3 and the mutant Ptt IAA3m expression, in all of the transgenic lines tested, the transcript levels of Ptt IAA3 were higher than in the wild type. These data suggest that the increased levels of Ptt IAA3 transcripts in transgenic lines stem from the overexpression of the Ptt IAA3m transgene. The auxin responsiveness of the Ptt IAA3m-expressing plants was investigated by comparing their root growth with that of wild-type (T89) hybrid aspen plants on Murashige and Skoog (MS) medium supplemented with 0.5 μM indole butyric acid (IBA) (Figure 4D). While 0.5 μM IBA was sufficient to inhibit root elongation in wild-type plants, no significant inhibition was observed in the transgenic plants at this concentration of IBA.
Figure 4.
Molecular Analysis of Transgenic Hybrid Aspen Lines with Altered Auxin Signaling.
(A) Amino acid sequence of mutated Ptt IAA3m protein. In the mutated Ptt IAA3m cDNA, the codon encoding the conserved Pro residue (highlighted in gray) was changed to a codon encoding a Ser residue.
(B) Transcript levels of the mutated version of Ptt IAA3 cDNA (Ptt IAA3m) under the control of the 35S CaMV promoter in three independent transgenic lines (Ptt IAA3m) and the wild type by RT-PCR. A total of three replicates were used for each RT-PCR experiment. Bars shows averages (n = 3) of the ratio of the intensity of the fragment of interest relative to that of 18S. Error bars represent sd.
(C) Comparison of the height of transformed and wild-type plants.
(D) Transgenic hybrid aspen plants expressing mutated Ptt IAA3 cDNA display auxin-resistant root growth. Root growth of wild-type hybrid aspen shoots (T89) was compared with that of three independent transgenic lines (35S:PttIAA3m) expressing mutated Ptt IAA3 cDNA (lines 6, 12, and 15) on a medium supplemented with 0.5 μM IBA.
(E) and (F) Auxin inducibility of Ptt IAA6 and Ptt PIN1 transcripts in wild-type (T89) and 35S:Ptt IAA3m transgenic trees. Stem tissues of T89 and three independent transgenic lines were depleted of auxin for 16 h before the addition of auxin. Ptt IAA6 and Ptt PIN1 are auxin-inducible in wild-type plants, but their auxin induction is reduced in the three independent lines expressing Ptt IAA3m cDNA. The difference in transcript levels between the wild type and the Ptt IAA3m lines was significant (P < 0.01) according to t tests.
In addition, the auxin responsiveness of the Ptt IAA6 and Ptt PIN1 genes in stem tissues of all of the Ptt IAA3m–expressing lines was weaker than that of the corresponding wild-type tissues (Figures 4E and 4F). However, the three tested lines differed from each other: auxin induction of Ptt IAA6 and Ptt PIN1 was more severely reduced in transgenic line 6 than in lines 12 and 15. These lines also differed in the severity of other analyzed traits. For example, both reductions in height (Figure 4C) and leaf curling (see Supplemental Figure 2 online) were more severe in line 6 than in the other two lines. Furthermore, differences in the level of reduction of auxin responsiveness in the transgenic lines correlated well with their levels of Ptt IAA3m transcripts, which were higher in the severely affected line 6 than in lines 12 and 15 (Figure 4B). From these results, we conclude that expression of Ptt IAA3m leads to altered auxin response in the transgenic hybrid aspen plants. Based on these findings, we chose the severely affected line 6 and the more moderately affected line 15 for further analysis.
Secondary Xylem Development Is Altered in Ptt IAA3m–Expressing Trees
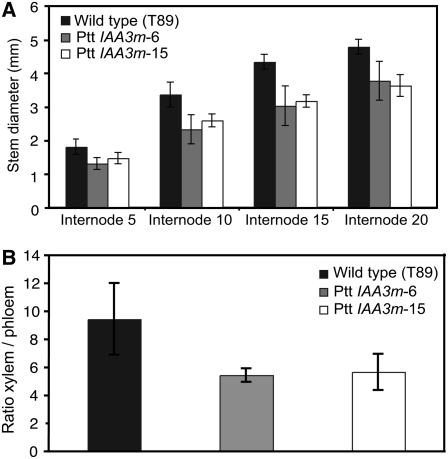
We found the stem diameter to be significantly (P < 0.05) smaller in the Ptt IAA3m than in the wild-type plants, at all measured internodes and spanning early to late stages of wood development (Figure 5A). We then examined phloem and xylem cells (both of which are derived from the cambial meristem) in the transgenic and wild-type plants to determine whether both of these types of cells were affected by the overexpression of Ptt IAA3m. The results showed that the xylem-to-phloem ratios were significantly lower in the Ptt IAA3m lines than in the wild-type plants (Figure 5B). However, anatomical data (see Supplemental Figure 4 online) indicated that there were no significant differences in phloem width between the wild-type and transgenic lines; thus, the overexpression of Ptt IAA3m appeared to affect xylem development much more than phloem development.
Figure 5.
Anatomical Analysis of Ptt IAA3m–Expressing Lines.
(A) Comparison of the stem diameter of the wild type (T89) and transgenic lines 6 and 15 expressing Ptt IAA3m cDNA. The diameter of three plants per clone of the wild type and transgenic lines expressing Ptt IAA3m cDNA was compared at different internodes. Transgenic lines display reductions in diameter compared with the wild type at all stages of development. Bars show average diameters (n = 5; error bars represent sd). The stem diameter of the Ptt IAA3m lines was significantly (P < 0.05) smaller than that of the wild type at all measured internodes covering early to late stages of wood development. All Ptt IAA3m–expressing lines showed statistically significant differences from the wild type (t test, P ≤ 0.01), apart from line 15 at internode 5 (P = 0.015).
(B) Comparison of the xylem-to-phloem ratio between T89 and lines expressing Ptt IAA3m cDNA. Xylem and phloem width was measured in anatomical sections from internode 25. Bars show average xylem:phloem ratios (n = 3; error bars represent sd). The differences between the wild type and Ptt IAA3m lines were found to be significant according to t tests (P < 0.1).
Reduced Auxin Responsiveness Alters Cambial Cell Division Activity
Since the width of the secondary xylem was reduced in the Ptt IAA3m lines, we investigated whether cambial cell division activity was also reduced in these plants. Two types of cell divisions occur during secondary xylem development: periclinal and anticlinal. The former determines the number of secondary xylem cells in each radial file (Larson, 1994), while the latter occurs in cambial initials and determines the number of radial files of secondary xylem cells (Larson, 1994; Schrader et al., 2004). The frequency of periclinal divisions was significantly reduced in line 6, in which auxin responsiveness was most severely affected (Figure 6A), but the reduction of cell division activity was not as pronounced in the more moderately affected line 15. In contrast with periclinal divisions, the effect of altered auxin signaling on the number of anticlinal divisions was not as prominent; nevertheless, they also tended to be less frequent in plants of the severely affected line 6 (Figure 6B). These findings show that auxin plays an important role in regulating cell divisions of secondary xylem cells.
Figure 6.
Analysis of Cell Division Activity in Secondary Xylem Cells in Ptt IAA3m–Expressing Plants.
(A) Periclinal cell divisions. Total numbers of divisions were determined by counting mitotic cell divisions in samples of fusiform cambial cells of three different trees per transformed line and the wild type. For each tree, 500 cambial fusiform cells were examined. Bars represent the ratio of dividing cells and the total number of fusiform cells counted in one tree. The difference in periclinal divisions between the wild type and Ptt IAA3m line 6 was significant according to a Wilcoxon test (P = 0.0496).
(B) Comparison of anticlinal cell division activity in the wild type (T89) and transgenic hybrid aspen lines (6 and 15) expressing Ptt IAA3m cDNA. Cell divisions were counted in each of three individual plants per transformed line and the wild type. Each bar represents one tree and shows the number of anticlinal divisions in relation to the total number of cell files per section studied. A minimum of four sections were examined per tree. Ptt IAA3m line 6 was significantly different from the wild type according to a Wilcoxon test (P < 0.1).
Auxin Signaling Is Necessary for Spatial Regulation of Cambial Initial Cell Division
The anticlinal divisions associated with the cambial initials coincide with high auxin levels in the cambium (Sabatini et al., 1999; Schrader et al., 2004); thus, auxin may play a key role in the spatial regulation of these anticlinal divisions. Therefore, we investigated whether the spatial regulation of anticlinal cell divisions is affected in plants with reduced auxin responsiveness. As can be seen from Figure 7, the zone of anticlinal divisions was broader in plants with altered auxin responses than in the wild-type plants. Interestingly, the pattern of anticlinal divisions was affected not only in line 6 but also in line 15, in which the reduction in auxin responsiveness is less severe. These findings suggest that auxin signaling could play an important role in the spatial regulation of anticlinal divisions of the cambial initials. Furthermore, the mechanism involved in the spatial regulation of anticlinal divisions of the cambial initials appears to be much more sensitive to changes in auxin responsiveness than the mechanism responsible for periclinal divisions, since defects in the pattern of anticlinal divisions were observed in weakly as well as strongly affected lines, whereas periclinal divisions appeared to be normal in the weakly affected line 15.
Figure 7.
Spatial Pattern of Anticlinal Divisions in the Cambium.
The positions of anticlinal divisions are altered in hybrid aspen plants expressing Ptt IAA3m cDNA. Positions of dividing cells within the cambial region were noted manually in relation to the phloem. Position 1 equals the cambial cell closest to the phloem. The graph shows the proportions of anticlinal divisions at different positions (divisions at the respective position per total number of anticlinal divisions). The differences in anticlinal divisions between the wild type and the two Ptt IAA3m lines were significant at positions 3, 4, 5, 6, and 8 (indicated by arrows) according to t tests (P < 0.05), indicating that the zone of anticlinal divisions was wider in the Ptt IAA3m lines than in the wild type.
Reduced Auxin Responsiveness Affects Both Fiber and Vessel Development
It has been suggested that auxin plays an important role in regulating both vessel and fiber development (Sundberg et al., 2000). Data obtained from studies of the effects of both applying auxin to wild-type plants and altering auxin metabolism transgenically support this hypothesis, although the results have often been conflicting (Doley and Leyton, 1968; Aloni and Zimmermann, 1983; Aloni, 1987). However, in contrast with the abundance of data obtained by altering auxin levels, there is little information regarding the role of auxin-sensing in the regulation of fiber and vessel development. To address this issue, we compared the patterns of fiber and vessel development in wild-type and transgenic plants with reduced auxin responsiveness, and found that the width of both fibers and vessel elements was reduced in the transgenic lines (Figures 8A and 8C). In addition, we found that both the fiber and vessel lengths were reduced in the Ptt IAA3m lines, compared with the wild-type trees (Figures 8B and 8D), and these effects were particularly severe in line 6. Together, these findings suggest that auxin plays a key role in fiber and vessel development and, importantly, that auxin is involved in regulating not only the radial expansion but also the length of fibers and vessels.
Figure 8.
Length and Width of Fibers and Vessel Elements.
Measurements of fibers and vessels in plants expressing Ptt IAA3m cDNA from internode 25 were performed manually.
(A) Frequency distribution of fiber widths.
(B) Frequency distribution of fiber lengths. Arrowheads indicate peak size range or size ranges found only in transgenic lines or the wild type. For each line (wild-type T89 and the two transgenic lines), ≥180 fibers originating from three different trees were measured. The differences in mean fiber length and width were significant between the wild type and the Ptt IAA3m lines according to t tests (P < 0.01).
(C) Frequency distribution of vessel widths.
(D) Frequency distribution of vessel lengths. Arrowheads indicate peak size range or size ranges found only in transgenic lines or wild-type vessels. A total of 180 vessels originating from three different trees were measured for each of the wild-type and transgenic lines. The differences in mean vessel width were significant between the wild type and the Ptt IAA3m lines according to t tests (P < 0.01). There was also a significant difference in vessel length between line 6 (P = 0.025), but not line 15, and wild-type plants.
DISCUSSION
Auxin has been implicated in regulating various aspects of secondary xylem development in tree species (Sundberg et al., 2000). However, the lack of suitable tools, such as auxin response mutants in tree species, has hindered molecular genetic analysis of the role of auxin in secondary xylem development. Therefore, we applied two alternative approaches: transcript profiling to explore the auxin-modulated transcriptional networks underlying secondary xylem development, and an investigation of secondary xylem development in hybrid aspen trees in which auxin responsiveness was transgenically impaired.
An important mode of auxin action is through its effects on gene expression (Teale et al., 2006). We thus decided to investigate the auxin-responsive transcriptome in trees and the behavior of auxin-regulated transcriptional networks in wood-forming tissues during secondary xylem development. One of the most interesting findings of our transcript profiling experiment is that for a large number of the genes found to be auxin-responsive in stem tissues of poplar (see Supplemental Data Set 1 online), their potential Arabidopsis orthologs have not been reported as being auxin-regulated. There are several possible explanations for this finding. It is possible that the experimental setup used in this study to detect auxin-responsive genes may lead to artifacts (e.g., the wounding response from the use of cut stem pieces can potentially interfere with the investigated gene expression profiles). Moreover, auxin application, as performed here, may not accurately reflect the in planta changes in auxin occurring in the wood-forming tissues, and cell types such as the cambium may not have been exposed to the applied auxin. In addition, we may not have detected some auxin-responsive genes, since their response times may have been outside the experimental time frame.
Although it is possible that we may not be able to detect auxin-responsive genes that require longer periods to respond (such as the 24 h or more required for the induction of PLETHORA genes [Aida et al., 2004]), our experimental system nevertheless correctly identified several well-known auxin-responsive genes. Moreover, comparing gene expression profiles of stems following auxin application with those obtained after the 16-h auxin depletion period, rather than predepletion, should largely overcome potential problems associated with wounding responses. Thus, in spite of the above caveats, the likelihood of the differences in sets of auxin-responsive genes between poplar stem tissues and Arabidopsis being entirely due to differences in the experimental setups used to detect them is probably low. Another factor to consider is that poplar has undergone full genome duplication and there is evidence that subfunctionalization and neofunctionalization of the duplicated genes has occurred (Tuskan et al., 2006; Yang et al., 2006); this may account for some of the differences between poplar and Arabidopsis in the regulation of various members of gene families. However, Arabidopsis is an herbaceous annual, and to date, most microarray profiling of auxin-responsive gene expression in plants has been performed using Arabidopsis seedlings, which lack secondary xylem tissue (Sawa et al., 2002; Tian et al., 2002; Goda et al., 2004). Thus, it is possible that the set of auxin-responsive genes detected here in the stem tissues of hybrid aspen may be involved in specific aspects of secondary xylem development; thus, their putative orthologs may not have been identified as auxin-responsive in the previous studies of Arabidopsis.
Having identified auxin-regulated genes, we investigated whether the expression of these genes correlated well with the auxin concentration gradient. Such a correlation would suggest that the auxin concentration gradient plays a direct role in regulating secondary xylem development via the regulation of gene expression and would support the hypothesis that auxin acts in a morphogen-like manner in this process (Uggla et al., 1996; Tuominen et al., 1997; Sundberg et al., 2000; Bhalerao and Bennett, 2003). However, the expression of only a few auxin-responsive genes correlated well with the auxin concentration gradient, and few genes whose expression mirrored the auxin distribution pattern in secondary xylem cells were among the set of auxin-responsive genes we had identified. Furthermore, the transcripts of auxin-responsive genes responded dynamically to the changes in auxin levels and did not appear to be dependent on the steady state auxin levels. Together, these results raise doubts about the validity of the proposed mode of auxin action in the regulation of secondary xylem development.
In addition to our data, some results from Arabidopsis also argue against auxin displaying morphogen-like activity. In embryo development, auxin accumulation leads first to the specification of apical fate, followed (almost immediately) by auxin-mediated specification of basal fate (Friml et al., 2002). Moreover, in Arabidopsis embryos, auxin affects cell fate indirectly, by eliciting a primary response in the cell adjacent to the one whose fate it actually affects (Weijers et al., 2006). These findings, in addition to previous results that show difficulties in correlating physiological responses such as the development of vascular cell types with specific auxin concentration (Aloni, 1987), indicate the difficulties involved in establishing simple relationships between auxin concentrations and specific auxin-regulated cellular responses. Consequently, Berleth and Sachs (2001) have argued against a simple mechanism based on passive responses of cells to specific auxin concentrations in a rigid auxin concentration gradient.
Interestingly, we found that the few auxin-responsive genes whose expression patterns correlate well with the auxin concentration gradient include transcription factors such as Ptt HB8 and those of the AUX/IAA gene family. Thus, we cannot discount the possibility that auxin could directly regulate the expression of a few genes, such as the abovementioned transcription factors, including Ptt HB8 (whose Arabidopsis orthologs have been implicated in vascular development [Baima et al., 2001]), and that these may in turn regulate the expression of downstream genes. However, without knowledge of the downstream targets of Ptt HB8, this suggestion remains speculative. Alternatively, auxin may exert its regulatory effects on secondary xylem development via posttranscriptional mechanisms. In support of this hypothesis, changes in auxin levels are known to affect the stability of AUX/IAA transcriptional repressors (Gray et al., 2001; Tiwari et al., 2001), several of which are expressed in secondary xylem cells (Moyle et al., 2002). Thus, auxin could regulate secondary xylem development posttranscriptionally by changing the stability of transcription factors and, thus, the gene expression profiles of the developing cells.
Our transcriptional analysis identified a number of genes that responded dynamically to changes in auxin levels and that could potentially be involved in secondary xylem development. We reasoned that further elucidation of the role of auxin in secondary xylem development could be greatly facilitated by analyzing this process in plants in which either the auxin distribution or auxin responses had been changed. However, earlier attempts to disrupt the auxin concentration gradient in secondary xylem had been largely unsuccessful, with overexpression of auxin biosynthetic genes having a relatively minor impact on the auxin concentration gradient in hybrid aspen plants (Tuominen et al., 1997). Therefore, we used an alternative approach by analyzing secondary xylem in trees in which auxin signaling had been perturbed by the expression of a dominant mutant version of Ptt IAA3, a poplar AUX/IAA gene.
Ptt IAA3m lines, with altered auxin responsiveness, displayed clear modifications in secondary xylem development, with little effect on the phloem. These findings were surprising, since we expected changes in auxin responsiveness to affect both of these tissues. However, an earlier study of the effects of reductions in auxin levels in both xylem and phloem in hybrid aspen plants found the effects to be much greater in the xylem than in the phloem (Tuominen et al., 1997). One possible explanation for these observations is that there are fewer cell divisions on the phloem side than on the xylem side of the cambium; thus, effects on the xylem may be more readily apparent. Moreover, based on earlier data (Tuominen et al., 1995) and our data showing the expression of Ptt IAA3m in the phloem (see Supplemental Figure 3 online), we can rule out the possibility that the lack of alteration of the phloem is due to the absence of expression of mutated Ptt IAA3m in this tissue.
Perturbing the auxin responsiveness resulted in substantial reductions in periclinal cell division in the secondary xylem and less pronounced reductions in anticlinal divisions, extending previous findings that reducing cambial auxin levels causes reductions in xylem divisions (Tuominen et al., 1997). Interestingly, our data uncovered a role for auxin signaling in spatially restricting anticlinal divisions of cambial initials to a narrow zone. The exact mechanisms by which auxin may regulate and position the cambial anticlinal divisions are not clear, but studies of root development in Arabidopsis provide clues concerning a potential mechanism. In Arabidopsis roots, the position of the auxin response maxima plays an important role in regulating the fates of distal cells, and disruption of either the auxin response maxima or auxin distribution in root apices alters the patterning and positions of the cell types within them (Sabatini et al., 1999). The polar auxin transport components play key roles in positioning the auxin maxima (Friml et al., 2002; Blilou et al., 2005), and the graded expression of the auxin-responsive PLETHORA transcription factors (Aida et al., 2004) regulates the expression of polar auxin transport components, thereby regulating patterning in the roots (Blilou et al., 2005; Galinha et al., 2007). Indeed, we do find that PLETHORA-related genes are expressed in the cambial zone of poplar plants, and it will be interesting to investigate their role in the spatial regulation of cambial initials.
Auxin plays important roles not only in regulating divisions of secondary xylem cells but also in their subsequent expansion and development. Tuominen et al. (1997) found that transgenic plants with reduced auxin levels and wider distributions of auxin had wider fibers than their wild-type counterparts. However, the effects of auxin on cell division and expansion cannot be readily determined from these data, since the fibers may have been wider due to the reduced rate of cell division leading to a longer expansion phase. Therefore, it is interesting that even weak alterations in auxin responsiveness that had little effect on rates of cell division (as seen in Ptt IAA3m line 15) still had significant effects on fiber expansion. Thus, the effects of auxin on cell division and cell expansion can be at least partially distinguished using our data, and the results indicate that cell expansion is more sensitive to alterations in auxin responsiveness than is cell division during secondary xylem development.
It has been shown previously that reducing auxin levels has much more pronounced effects on the radial expansion of fibers than of vessels (Tuominen et al., 1997). Furthermore, secondary wall formation is initiated earlier in vessels than in fibers (Doley and Leyton, 1968; Ridoutt and Sands, 1994), suggesting that several aspects of fiber and vessel development differ substantially. Thus, it has been unclear whether auxin plays a role in vessel development. However, our results show that radial expansion of not only fibers but also of vessels is affected in Ptt IAA3m–expressing plants. Moreover, not only the radial expansion but also the axial elongation of fibers and vessels is reduced in these plants. Thus, our approach using plants expressing a mutated form of Ptt IAA3 to perturb auxin responsiveness has revealed a role for auxin in vessel development in addition to the regulation of fibers.
In summary, the studies presented here provide information on several aspects of the molecular regulation of secondary xylem development by auxin in trees. Our results suggest that dynamic changes in gene expression orchestrated by transient alterations of auxin levels may play a key role in the transcriptional regulation of secondary xylem development by auxin. While transcript profiling identified auxin-responsive networks and their potential role in secondary xylem development, perturbing auxin responsiveness by expressing dominant Ptt IAA3m provided important insights into the role of auxin signaling in various aspects of wood formation (e.g., spatial regulation of the anticlinal divisions of the cambial initials, and the subsequent development of fibers and vessels). Although the results of expressing dominant mutants should be treated with caution, the use of dominant AUX/IAA proteins as tools to disrupt auxin signaling in Arabidopsis has yielded important information concerning the roles of auxin in various processes, such as root hair development, gravitropic responses, and lateral root development (Knox et al., 2003; Fukaki et al., 2005; Swarup et al., 2005). In addition, the Ptt IAA3 used here to alter auxin responsiveness is expressed in developing secondary xylem (Moyle et al., 2002). Therefore, in spite of the caveats associated with the experimental use of dominant mutants, the dominant Ptt IAA3m provides a useful tool for elucidating the role of auxin in secondary xylem development in organisms, such as trees, in which forward genetic approaches like those used in model herbaceous plants, such as Arabidopsis, are not currently feasible.
While this work focused mainly on elucidating the molecular basis of the regulation of secondary xylem development by auxin, other hormones also play important roles in this process. For example, both gibberellin and ethylene affect secondary xylem development (Eriksson et al., 2000; Junghans et al., 2004). Moreover, crosstalk between these two hormones and auxin is well documented. For example, in Arabidopsis, AXR1 is implicated in the degradation of RGA (Fu and Harberd, 2003), a DELLA domain protein, and ethylene and auxin crosstalk plays an important role in root elongation (Ruzicka et al., 2007; Stepanova et al., 2007; Swarup et al., 2007). Similar interactions also probably occur in secondary xylem development; a hypothesis supported by data gathered in this study showing the induction of ethylene biosynthetic genes by auxin (see Supplemental Data Set 1 online) and other recent results indicating that a common set of genes responds to auxin and gibberellin (Björklund et al., 2007). In the future, analysis of the crosstalk between signaling pathways for auxin and various hormones using response mutants in trees will be necessary to acquire a more comprehensive understanding of the molecular control of wood formation.
METHODS
Plant Material and Growth Conditions
For the IAA microarray analysis, wild-type hybrid aspen T89 (Populus tremula × Populus tremuloides) plants were grown in sterile MS medium (Duchefa). Stem tissues from soil-grown P. tremula trees growing under natural conditions in the first week of July 2004 were used for sectioning cambial tissue for high-resolution auxin measurements. For comparisons of wild-type T89 and Ptt IAA3m–expressing lines, plants grown in soil at 23°C with 16-h daylengths were used.
Mutagenesis and Cloning of Ptt IAA3 cDNA in a Binary Vector and Hybrid Aspen Transformation
Ptt IAA3 cDNA (Moyle et al., 2002) was cloned in vector pGEM-T (Promega) and a Pro-to-Ser mutation was introduced using the GeneEditor kit (Promega) according to the manufacturer's instructions. Following mutagenesis, the mutated Ptt IAA3 cDNA, termed Ptt IAA3m, was cloned into the pCAMBIA 1305.1 binary vector (www.cambia.org) and introduced into Agrobacterium tumefaciens GV3101pmp90 (Koncz and Schell, 1986). Hybrid aspen was transformed with the mutated Ptt IAA3m cDNA as described earlier (Nilsson et al., 1992). Primers used to test the transgenic lines for the expression of Ptt IAA3m under the control of the 35S CaMV promoter were 3′-CTGCCTCTAGCTCTTCATCATCT-5′ and 3′-CGCACAGAAGGTAAGAACACC-5′.
Auxin Treatments for Microarray Analysis
Five-centimeter-long stem segments with fully elongated internodes from T89 hybrid aspen plants grown in vitro were cut and defoliated. Six stem segments from six individual trees were frozen immediately after cutting and defoliating, thus serving as the pooled control (d0). All of the remaining stems were then placed in auxin-free half-strength MS medium in the same container to deplete them of auxin under identical conditions; two further sets of six stem segments, each representing six individual trees, were randomly selected and pooled into single samples 4 h (d4) and 16 h (d16) later. The remaining stem segments were carefully moved to fresh half-strength MS medium supplemented with 20 μM IAA, from which further pooled sets of six samples were taken after 30 min (i0.5), 2 h (i2), and 4 h (i4).
Sampling for IAA Measurements
To measure IAA levels in the samples and verify that expected changes in the levels had occurred, tissues were collected at each sampling point during the course of the microarray experiment. For each sample, three individual stems were pooled. To remove any possible excess of IAA from the surface of stems exposed to IAA during the experiment, a series of three MS media was used. Stems were briefly dipped once in half-strength MS and 0.5% Triton X-100 and twice into two separate buffers of half-strength MS medium, before freezing in liquid nitrogen. In order to compare the auxin responsiveness of wild-type T89 and the three transgenic 35S:PttIAA3m lines, similar treatments were applied to stem tissues of plants from each of the lines, grown in vitro as described above.
Tissue Sampling for High-Resolution Auxin Measurements
For high-resolution measurements of auxin levels in the wood-forming zone, five series of 20-μm-thick tangential sections were obtained (all from the same tree) by cryosectioning frozen stem tissues across the early phloem, active cambium, and expanding xylem, following the technique described by Uggla et al. (1996).
IAA Measurements
For measurements of IAA, samples were extracted (after adding 50 pg [13C6]IAA/mg fresh weight of sample), purified, and analyzed by combined gas chromatography-selected reaction monitoring-mass spectrometry as described by Edlund et al. (1995). IAA levels in the samples were then quantified using 13C6:12C6 isotopic dilution ratios of the monitored daughter ions. For auxin measurements in thin sections, endogenous auxin levels were measured separately in individual sections using the technique described above, but with isotopic dilution factors based on the addition of 500 pg [13C6]IAA/sample.
cDNA Microarray Analysis
The POP1 cDNA microarrays used in the hybridization experiments represent a double spotted unigene set of 13,526 clones derived from the poplar EST sequencing project and various controls (Sterky et al., 2004). For information regarding plant material, experimental setup, and hybridization and normalization procedures for high-resolution analysis of expression across the wood-forming zone, see Schrader et al. (2004) and Hertzberg et al. (2001).
Microarray Experimental Design
In an attempt to obtain a satisfactory compromise between minimizing the number of hybridizations for each sample and ensuring that the results were statistically reliable, we chose to use an all-versus-all design. In such designs (in contrast with common reference designs, in which a common reference sample is included), data from all samples are directly compared, pairwise, using an in silico–based reference (Kerr and Churchill, 2001; Churchill, 2002). In the hybridization design, all samples were labeled six times (three times Cy3 and respective Cy5), and three technical replicates with dye swaps were used for the analysis of gene expression at time points d0 versus i0.5, d4 versus i2, and d16 versus i4.
RNA Preparation and Hybridization
For hybridizations in the global auxin response study, total RNA was prepared from the pooled sample of six individual stems according to Chang et al. (1993). For analysis of Ptt IAA3m expression in phloem, bark was separated from stem, tissue was scraped from inner bark to obtain phloem-enriched tissue, and total RNA was isolated as described above. After LiCl precipitation, RNA was resuspended and purified using an RNeasy mini kit according to the manufacturer's (Qiagen) recommendations. cDNA synthesis, labeling, and hybridizations were performed according to Andersson-Gunnerås et al. (2006), except that 20 μg of total RNA was used for reverse transcription.
For RT-PCR analysis, first-strand cDNA was prepared from RNA using a first-strand cDNA synthesis kit (Amersham-Pharmacia Biotech). RT-PCR was performed using a Quantum 18S RNA internal standard kit with Sybergreen dye (Ambion; http://www.ambion.com) using first-strand cDNA as template, ensuring that the amount of amplified product remained in linear proportion to the initial template present in the reaction. All RT-PCR experiments were replicated three times, and the ratio of the intensity of the fragment for the gene of interest was obtained relative to that for 18S for each PCR. Primers used for RT-PCR analysis are described in Supplemental Table 1 online, with the exception of primers for Ptt PIN1, which have been described previously by Schrader et al. (2003).
Quality Control, Normalization, and Statistical Analysis
Hybridization intensities were quantified using GenePixPRo 4.1 software (Axon Instruments). For the IAA induction data set, spots with high background, uneven morphology, or dust speckles were manually flagged as bad. Mean foreground intensity minus median background values were used for subsequent analysis. The linear regressions and normalization were calculated using the Linear Models for Microarray Data package (Smyth, 2004) in the statistical software R (Ihaka and Gentleman, 1996). Data were background corrected by subtracting background intensities. To remove systematic intensity- and space-dependent artifacts, a global intensity- and space-dependent (print-tip Loess) normalization with the smooth function was used, and multiple correction was done by the false discovery rate of Benjamini and Hochberg (1995). Furthermore, in order to reduce the number of false-positives as effectively as possible, B-statistics were used to rank significantly expressed clones (Lönnstedt, 2005). The cutoff value for B, corresponding to a 95% probability of differential expression, was determined (from the properties of the ranked data set and the experimental design) to be 3. Clones were considered to be IAA-inducible if they displayed upregulation at either the first (i0.5) or second (i2) time point after IAA application relative to 16 h of depletion (d16), but not between 4 h (d4) and 16 h of depletion (d16). The reverse criteria were set for the clones to be considered IAA-repressed. Normalized data were imported into GeneSpring software (version 6; Silicon Genetics), and expression profiles were clustered through self-organizing maps using default settings. All data reported here are log2 ratios of Cy3/Cy5 signals.
PLS Analysis
Prior to PLS analysis, the normalized microarray data from a data set with high-resolution transcript profiles over the wood-forming zone (Schrader et al., 2004) were mean-centered by subtracting the average for each individual gene expression variable over all samples; this is commonly done prior to projection-based modeling to reduce the complexity of the resulting model. A PLS model was then calculated to assess the correlations between the 11,058 gene expression variables from the microarray data set (X) and the auxin concentration gradient (Y). The goodness of fit, R2, and the predictive ability based on sevenfold partial cross-validation, Q2, was used to judge the quality of the model. The PLS variable weights were used to identify the most strongly correlating genes for further evaluation. All PLS calculations were performed using SIMCA-P+ 11 software (Umetrics). The resultant PLS model described 71.1% of the variation in X (R2X = 0.711), 98% of the variation in Y (R2Y = 0.98), and could predict 95.3% of the variation in Y according to sevenfold partial cross-validation (Q2 = 0.953), indicating a very strong correlation between the gene expression and auxin concentration data sets, since the maximum value for Q2 (indicating perfect correlation between X and Y) is 1. The list of 250 clones showing the highest correlations to the auxin gradient was compared manually with the list of IAA-regulated genes.
Root Growth Assay
Top shoots of wild-type T89 hybrid aspen plants grown in vitro and three independent clones of 35S:PttIAA3m transgenic plants were cut and allowed to root in sterile half-strength MS medium (control) or half-strength MS medium supplemented with 0.5 μM IBA. The plantlets were propagated under controlled conditions as described above, and after a period of 5 weeks, the roots were examined.
Anatomical Analysis
Stem samples were taken from trees grown in soil under controlled conditions (16-h/8-h light/dark cycles, 20°C, humidity 70%) for 6 weeks. The stem diameter of each of three trees of each transgenic line and the wild-type T89 was measured at internodes 5, 10, 15, and 20 using a digital caliper. To analyze cell division activities, 1-cm stem samples cut from internodes 10 and 25 were fixed, for 24 h, in FAA solution containing 5% formaldehyde, 5% acetic acid, and 50% ethanol. Samples were then sequentially dehydrated, gradually infiltrated, and embedded in LR White (TAAB) in polypropylene capsules (TAAB), then 3-μm thin sections were obtained by sectioning using a Microm HM350 microtome (Microm International). The sections were floated on water, heat-fixed to glass slides, stained with toluidine blue, and mounted in Entellan neu (Merck). Sections were then visualized with a Zeiss Axioplan light microscope, and images were captured using an Axiocam digital camera and Axiovision 4.5 software (Zeiss). Xylem and phloem widths were measured using the Axiovision 4.5 software (Zeiss) as described earlier (Tuominen et al., 1997).
Fiber and Vessel Element Measurements
For fiber and vessel measurements, samples from internode 25 of plants representing the wild-type T89 and the 35S:PttIAA3m transgenic lines were fixed in FAA and placed in a solution of 7.5% H2O2 and glacial acetic acid (1:1), then boiled at 95°C for 6 h. After rinsing and mechanical breakage, the xylem cell suspension was placed on a slide and visualized with a Zeiss Axioplan light microscope, and images were captured using a Axiocam digital camera (Zeiss). The width and length of fibers and vessel elements were measured manually using the Axiovision 4.5 software (Zeiss).
Analysis of Cell Division Activity
The cell division activity of fusiform cells was analyzed according to Espinosa-Ruiz et al. (2004). Stem segments from internode 10 of plants representing the wild-type T89 and the two transgenic 35S:PttIAA3m lines were fixed in FAA and treated with 5% pectinase and 1% peptone overnight. Following the treatment, the bark was separated from the stem and cambial cells were lifted from the exposed xylem and bark sides of the stem with a fine needle. The isolated cells were placed in a drop of water on a gelatin-coated slide and dispersed by vibrating the needle in the drop. The slides were air-dried on a slide warmer at ∼40 to 50°C, stained in toluidine blue, rinsed in water, and observed directly. The number of fusiform cells in mitotic phase was counted manually using a light microscope and expressed as a percentage of the total number of fusiform cells. A total of 500 cells were examined for each sample. Anticlinal cell divisions were analyzed according to Schrader et al. (2004). Briefly, stem samples from internode 10 of the wild type and the two transgenic lines were fixed, embedded in plastic, stained, and sectioned as described above. Anticlinal divisions in the cambial zone were recorded manually with a Zeiss Axioplan light microscope (Zeiss).
Accession Numbers
Microarray data used in this study can be found in the public database http://www.upscbase.umu.se (Sjödin et al., 2006) under accession numbers UMA 0007, 0021, and 0026. Information regarding annotations and clone identifiers (PUIDs) reported here and in Supplemental Data Set 1 has been published (Sterky et al., 2004; Sjödin et al., 2006) and deposited in the public Populusdb database (http://www.populus.db.umu.se).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Real-Time PCR Verification of Auxin Responsiveness of Selected Candidate Genes.
Supplemental Figure 2. Leaf Curling in the Ptt IAA3m–Expressing Lines.
Supplemental Figure 3. Expression of Ptt IAA3m in Phloem-Enriched Tissues.
Supplemental Figure 4. Anatomical Sections of the Stem from Ptt IAA3m–Expressing Lines.
Supplemental Table 1. RT-PCR Primers.
Supplemental Data Set 1. Microarray Data and Cluster Identities for Reported Clones.
Supplementary Material
Acknowledgments
We thank Brian Ellis, Catherine Bellini, Alan Marchant, Urs Fischer, and Ben Scheres for helpful comments; Patrik Rydén for statistical help; Jarmo Schrader for assistance with microarray analysis; and Richard Moyle for his gift of Ptt IAA3 cDNA. We also acknowledge technical support from Ingela Sandström, Roger Granblom, David Sandström, and Veronica Bourquin. This work was funded by grants from Formas, Vetenskapsrådet, and the European Union project POPWOOD to R.P.B. and G.S.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Rishikesh P. Bhalerao (rishi.bhalerao@genfys.slu.se).
Online version contains Web-only data.
References
- Aida, M., Beis, D., Heidstra, R., Willemsen, V., Blilou, I., Galinha, C., Nussaume, L., Noh, Y.S., Amasino, R., and Scheres, B. (2004). The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119 109–120. [DOI] [PubMed] [Google Scholar]
- Aloni, R. (1987). Differentiation of vascular tissues. Annu. Rev. Plant Physiol. 38 179–204. [Google Scholar]
- Aloni, R., and Zimmermann, M. (1983). The control of vessel size and density along the plant axis: A new hypothesis. Differentiation 24 203–208. [Google Scholar]
- Andersson-Gunnerås, S., Mellerowicz, E., Love, J., Segerman, B., Ohmiya, Y., Coutinho, P.M., Nilsson, P., Henrissat, B., Moritz, T., and Sundberg, B. (2006). Biosynthesis of cellulose-enriched tension wood in Populus: Global analysis of transcripts and metabolites identifies biochemical and developmental regulators in secondary wall biosynthesis. Plant J. 45 144–165. [DOI] [PubMed] [Google Scholar]
- Baima, S., Possenti, M., Matteucci, A., Wisman, E., Altamura, M., Ruberti, I., and Morelli, G. (2001). The Arabidopsis ATHB-8 HD-zip protein acts as a differentiation-promoting transcription factor of the vascular meristems. Plant Physiol. 126 643–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Statist. Soc. Ser. B. Methodological 57 289–300. [Google Scholar]
- Berleth, T., and Sachs, T. (2001). Plant morphogenesis: Long-distance coordination and local patterning. Curr. Opin. Plant Biol. 4 57–62. [DOI] [PubMed] [Google Scholar]
- Bhalerao, R.P., and Bennett, M.J. (2003). The case for morphogens in plants. Nat. Cell Biol. 5 939–943. [DOI] [PubMed] [Google Scholar]
- Björklund, S., Antti, H., Uddestrand, I., Moritz, T., and Sundberg, B. (2007). Cross-talk between gibberellin and auxin in development of Populus wood: Gibberellin stimulates polar auxin transport and has a common transcriptome with auxin. Plant J. 3 499–511. [DOI] [PubMed] [Google Scholar]
- Blilou, I., Xu, J., Wildwater, M., Willemsen, V., Paponov, I., Friml, J., Heidstra, R., Aida, M., Palme, K., and Scheres, B. (2005). The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433 39–44. [DOI] [PubMed] [Google Scholar]
- Chang, S., Puryear, J., and Cairney, J. (1993). A simple and efficient method for isolating RNA from pine. Plant Mol. Biol. Rep. 11 113–116. [Google Scholar]
- Churchill, G.A. (2002). Fundamentals of experimental design for cDNA microarrays. Nat. Genet. 37 490–495. [DOI] [PubMed] [Google Scholar]
- Doley, D., and Leyton, L. (1968). Effects of growth regulating substances and water potential on the development of secondary xylem in Fraxinus. New Phytol. 67 579–594. [Google Scholar]
- Edlund, A., Eklöf, S., Sundberg, B., Moritz, T., and Sandberg, G. (1995). A microscale technique for gas chromatography-mass spectrometry measurements of picogram amounts of indole-3-acetic acid in plant tissues. Plant Physiol. 108 1043–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson, M.E., Israelsson, M., Olsson, O., and Moritz, T. (2000). Increased gibberellin biosynthesis in transgenic trees promotes growth, biomass production and xylem fiber length. Nat. Biotechnol. 18 784–788. [DOI] [PubMed] [Google Scholar]
- Espinosa-Ruiz, A., Saxena, S., Schmidt, J., Mellerowicz, E., Miskolczi, P., Bakó, L., and Bhalerao, R.P. (2004). Differential stage-specific regulation of cyclin-dependent kinases during cambial dormancy in hybrid aspen. Plant J. 38 603–615. [DOI] [PubMed] [Google Scholar]
- Friml, J., Benkova, E., Blilou, I., Wisniewska, J., Hamann, T., Ljung, K., Woody, S., Sandberg, G., Scheres, B., Jurgens, G., and Palme, K. (2002). AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell 108 661–673. [DOI] [PubMed] [Google Scholar]
- Fu, X., and Harberd, N.P. (2003). Auxin promotes Arabidopsis root growth by modulating gibberellin response. Nature 421 740–743. [DOI] [PubMed] [Google Scholar]
- Fukaki, H., Nakao, Y., Okushima, Y., Theologis, A., and Tasaka, M. (2005). Tissue-specific expression of stabilized SOLITARY-ROOT/IAA14 alters lateral root development in Arabidopsis. Plant J. 44 382–395. [DOI] [PubMed] [Google Scholar]
- Fukaki, H., Tameda, S., Masuda, H., and Tasaka, M. (2002). Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J. 29 153–168. [DOI] [PubMed] [Google Scholar]
- Galinha, C., Hofhuis, H., Luijten, M., Willemsen, V., Blilou, I., Heidstra, R., and Scheres, B. (2007). PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449 1053–1057. [DOI] [PubMed] [Google Scholar]
- Goda, H., Sawa, S., Asami, T., Fujioka, S., Shimada, Y., and Yoshida, S. (2004). Comprehensive comparison of auxin-regulated and brassinosteroid-regulated genes in Arabidopsis. Plant Physiol. 134 1555–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, W.M., Kepinski, S., Rouse, D., Leyser, O., and Estelle, M. (2001). Auxin regulates SCF (TIR1)-dependent degradation of AUX/IAA proteins. Nature 414 271–276. [DOI] [PubMed] [Google Scholar]
- Hertzberg, M., et al. (2001). A transcriptional roadmap to wood formation. Proc. Natl. Acad. Sci. USA 98 14732–14737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihaka, R., and Gentleman, R. (1996). R: A language for data analysis and graphics. J. Comput. Graph. Statist. 5 299–314. [Google Scholar]
- Junghans, U., Langenfeld-Heyser, R., Polle, A., and Teichmann, T. (2004). Effect of auxin transport inhibitors and ethylene on the wood anatomy of poplar. Plant Biol. 6 22–29. [DOI] [PubMed] [Google Scholar]
- Kerr, M.K., and Churchill, G.A. (2001). Experimental design for gene expression microarrays. Biostatistics 2 183–201. [DOI] [PubMed] [Google Scholar]
- Knox, K., Grierson, C.S., and Leyser, O. (2003). AXR3 and SHY2 interact to regulate root hair development. Development 130 5769–5777. [DOI] [PubMed] [Google Scholar]
- Koncz, C., and Schell, J. (1986). The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 204 383–396. [Google Scholar]
- Larson, P. (1994). The Vascular Cambium, Development and Structure. (Berlin: Springer-Verlag).
- Lönnstedt, I. (2005). Empirical Bayes Methods for DNA Microarray Data. (Uppsala, Sweden: Department of Mathematics, Uppsala University).
- Moyle, R., Schrader, J., Stenberg, A., Olsson, O., Saxena, S., Sandberg, G., and Bhalerao, R.P. (2002). Environmental and auxin regulation of wood formation involves members of the Aux/IAA gene family in hybrid aspen. Plant J. 31 675–685. [DOI] [PubMed] [Google Scholar]
- Nagpal, P., Walker, L.M., Young, J.C., Sonawala, A., Timpte, C., Estelle, M., and Reed, J.W. (2000). AXR2 encodes a member of the Aux/IAA protein family. Plant Physiol. 123 563–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson, O., Aldén, T., Sitbon, F., Little, C.H.A., Chalupa, V., Sandberg, G., and Olsson, O. (1992). Spatial pattern of cauliflower mosaic virus 35S promoter-luciferase expression in transgenic hybrid aspen trees monitored by enzymatic assay and non-destructive imaging. Transgenic Res. 1 209–220. [Google Scholar]
- Ramos, J.A., Zenser, N., Leyser, O., and Callis, J. (2001). Rapid degradation of auxin/indoleacetic acid proteins requires conserved amino acids of domain II and is proteasome dependent. Plant Cell 13 2349–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridoutt, B., and Sands, R. (1994). Quantification of the processes of secondary xylem fiber development in Euclayptus globules at two height levels. IAWA J. 15 417–424. [Google Scholar]
- Rouse, D., Mackay, P., Stirnberg, P., Estelle, M., and Leyser, O. (1998). Changes in auxin response from mutations in an AUX/IAA gene. Science 279 1371–1373. [DOI] [PubMed] [Google Scholar]
- Ruzicka, K., Ljung, K., Vanneste, S., Podhorska, R., Beeckman, T., Friml, J., and Benkova, E. (2007). Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. Plant Cell 19 2197–2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini, S., Beis, D., Wolkenfelt, H., Murfett, J., Guilfoyle, T., Malamy, J., Benfey, P., Leyser, O., Bechtold, N., Weisbeek, P., and Scheres, B. (1999). An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99 463–472. [DOI] [PubMed] [Google Scholar]
- Sawa, S., Ohgishi, M., Goda, H., Higuchi, K., Shimada, Y., Yoshida, S., and Koshiba, T. (2002). The HAT2 gene, a member of the HD-Zip gene family, isolated as an auxin inducible gene by DNA microarray screening, affects auxin response in Arabidopsis. Plant J. 32 1011–1022. [DOI] [PubMed] [Google Scholar]
- Schrader, J., Baba, K., May, S.T., Palme, K., Bennett, M., Bhalerao, R.P., and Sandberg, G. (2003). Polar auxin transport in the wood-forming tissues of hybrid aspen is under simultaneous control of developmental and environmental signals. Proc. Natl. Acad. Sci. USA 100 10096–10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader, J., Nilsson, J., Mellerowicz, E., Berglund, A., Nilsson, P., Hertzberg, M., and Sandberg, G. (2004). A high-resolution transcript profile across the wood-forming meristem of poplar identifies potential regulators of cambial stem cell identity. Plant Cell 16 2278–2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjödin, A., Bylesjö, M., Skogström, O., Eriksson, D., Nilsson, P., Rydén, P., Jansson, S., and Karlsson, J. (2006). UPSC-BASE—Populus transcriptomics online. Plant J. 48 806–817. [DOI] [PubMed] [Google Scholar]
- Smyth, G.K. (2004). Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3 Article 3. [DOI] [PubMed] [Google Scholar]
- Stepanova, A.N., Yun, J., Likhacheva, A.V., and Alonso, J.M. (2007). Multilevel interactions between ethylene and auxin in Arabidopsis roots. Plant Cell 19 2169–2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterky, F., et al. (2004). A Populus EST resource for plant functional genomics. Proc. Natl. Acad. Sci. USA 101 13951–13956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundberg, B., Tuominen, H., and Little, C. (1994). Effects of the indole-3-acetic acid (IAA) transport inhibitors N-1-naphthylphthalamic acid and morphactin on endogenous IAA dynamics in relation to compression wood formation in 1-year-old Pinus sylvestris (L.) shoots. Plant Physiol. 106 469–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundberg, B., Uggla, C., and Tuominen, H. (2000). Cambial growth and auxin gradients. In Cell and Molecular Biology of Wood Formation, R. Savidge, J. Barnett, and R. Napier, eds (Oxford, UK: BIOS Scientific Publishers), p. 169–188.
- Swarup, R., Kramer, E.M., Perry, P., Knox, K., Leyser, H.M., Haseloff, J., Beemster, G.T., Bhalerao, R., and Bennett, M.J. (2005). Root gravitropism requires lateral root cap and epidermal cells for transport and response to a mobile auxin signal. Nat. Cell Biol. 7 1057–1065. [DOI] [PubMed] [Google Scholar]
- Swarup, R., Perry, P., Hagenbeek, D., Van Der Straeten, D., Beemster, G.T., Sandberg, G., Bhalerao, R., Ljung, K., and Bennett, M.J. (2007). Ethylene upregulates auxin biosynthesis in Arabidopsis seedlings to enhance inhibition of root cell elongation. Plant Cell 19 2186–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teale, W.D., Paponov, I.A., and Palme, K. (2006). Auxin in action: Signalling, transport and the control of plant growth and development. Nat. Rev. Mol. Cell Biol. 7 847–859. [DOI] [PubMed] [Google Scholar]
- Tian, Q., Uhlir, N.J., and Reed, J.W. (2002). Arabidopsis SHY2/IAA3 inhibits auxin-regulated gene expression. Plant Cell 14 301–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari, S.B., Wang, X.J., Hagen, G., and Guilfoyle, T.J. (2001). AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell 13 2809–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuominen, H., Puech, L., Fink, S., and Sundberg, B. (1997). A radial concentration gradient of indole-3-acetic acid is related to secondary xylem development in hybrid aspen. Plant Physiol. 115 577–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuominen, H., Sitbon, F., Jacobsson, C., Sandberg, G., Olsson, O., and Sundberg, B. (1995). Altered growth and wood characteristics in transgenic hybrid aspen expressing Agrobacterium tumefaciens T-DNA indoleacetic-acid biosynthetic genes. Plant Physiol. 109 1179–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan, G., et al. (2006). The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313 1596–1604. [DOI] [PubMed] [Google Scholar]
- Uggla, C., Magel, E., Moritz, T., and Sundberg, B. (2001). Function and dynamics of auxin and carbohydrates during earlywood/latewood transition in Scots pine. Plant Physiol. 125 2029–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uggla, C., Moritz, T., Sandberg, G., and Sundberg, B. (1996). Auxin as a positional signal in pattern formation in plants. Proc. Natl. Acad. Sci. USA 93 9282–9286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers, D., Schlereth, A., Ehrismann, J.S., Schwank, G., Kientz, M., and Jürgens, G. (2006). Auxin triggers transient local signaling for cell specification in Arabidopsis embryogenesis. Cell 10 265–270. [DOI] [PubMed] [Google Scholar]
- Yang, X., Lee, S., So, J.H., Dharmasiri, S., Dharmasiri, N., Ge, L., Jensen, C., Hangarter, R., Hobbie, L., and Estelle, M. (2004). The IAA1 protein is encoded by AXR5 and is a substrate of SCFTIR1. Plant J. 40 772–782. [DOI] [PubMed] [Google Scholar]
- Yang, X., Tuskan, G., and Cheng, M. (2006). Divergence of the Dof gene families in poplar, Arabidopsis, and rice suggests multiple modes of gene evolution after duplication. Plant Physiol. 142 820–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.