Abstract
The interaction between the fungal pathogen Cladosporium fulvum and its host tomato (Solanum lycopersicum) is an ideal model to study suppression of extracellular host defenses by pathogens. Secretion of protease inhibitor AVR2 by C. fulvum during infection suggests that tomato papain-like cysteine proteases (PLCPs) are part of the tomato defense response. We show that the tomato apoplast contains a remarkable diversity of PLCP activities with seven PLCPs that fall into four different subfamilies. Of these PLCPs, transcription of only PIP1 and RCR3 is induced by treatment with benzothiadiazole, which triggers the salicylic acid–regulated defense pathway. Sequencing of PLCP alleles of tomato relatives revealed that only PIP1 and RCR3 are under strong diversifying selection, resulting in variant residues around the substrate binding groove. The doubled number of variant residues in RCR3 suggests that RCR3 is under additional adaptive selection, probably to prevent autoimmune responses. AVR2 selectively inhibits only PIP1 and RCR3, and one of the naturally occurring variant residues in RCR3 affects AVR2 inhibition. The higher accumulation of PIP1 protein levels compared with RCR3 indicates that PIP1 might be the real virulence target of AVR2 and that RCR3 acts as a decoy for AVR2 perception in plants carrying the Cf-2 resistance gene.
INTRODUCTION
Considering that invasion of the plant apoplast is a critical phase of the infection cycle of numerous pathogens, it can be postulated that this compartment serves as a molecular battlefield that contributes to the success of pathogen infection and plant resistance. This battlefield is likely to be ancient, predating the evolution of translocation mechanisms for effector proteins by which pathogens manipulate the host cytoplasm and suppress host defense responses (Jones and Dangl, 2006). Therefore, understanding the nature of plant defenses in the apoplast and the counter defense mechanisms that pathogens evolved to overcome these defenses is essential for a comprehensive understanding of host–pathogen interactions and should complement the body of knowledge that has emerged on cytoplasmic effectors and defense mechanisms.
The apoplast of tomato (Solanum lycopersicum) is easily accessible for biochemical experiments and ideal for studying apoplastic molecular plant–pathogen interactions. Tomato is the only host for the well-studied fungal pathogen Cladosporium fulvum and an important host for the devastating solanaceous oomycete pathogen Phytophthora infestans (Kamoun and Smart, 2005; Rivas and Thomas, 2005; Thomma et al., 2005). Infection of tomato with these pathogens triggers the accumulation of a large amount of pathogenesis-related (PR) proteins in the apoplast (Kombrink et al., 1988; Joosten and De Wit, 1989). For example, PR proteins such as β-1,3-glucanase (PR2), chitinase (PR3), and subtilase-like serine protease (P69B) directly target these pathogens by degrading their cell wall components (Van Loon et al., 2006; Ferreira et al., 2007). Pathogens, on the other hand, secrete different effector proteins during infection to prevent this degradation. P. infestans secretes glucanase and subtilase inhibitors, whereas C. fulvum secretes chitin binding Avr4 proteins to protect its cell wall (Rose et al., 2002; Tian et al., 2004, 2005; Van den Burg et al., 2006; Van Esse et al., 2007). Selection pressure on host enzymes to evade inhibition and pathogen inhibitors to adapt to new host enzymes cause variant residues at the interaction surface for many of these enzyme–inhibitor interactions (Misas-Villamil and Van der Hoorn, 2008).
Although apoplast-localized plant proteases can play a role in defense, they might also act in signaling or perception upon pathogen infection (reviewed in Van der Hoorn and Jones, 2004). A role in signaling has been indicated for secreted proteases CDR1 and CathB. CDR1 is an aspartic protease that probably releases systemic signaling molecules that trigger defense responses in Arabidopsis thaliana (Xia et al., 2004). CathB is a secreted papain-like cysteine protease (PLCP) required for the development of the hypersensitive response (HR) in Nicotiana benthamiana (Gilroy et al., 2007). HR is an effective defense response at the site of pathogen infection that includes programmed cell death.
A very interesting role has been documented for RCR3, a secreted PLCP of tomato. RCR3 is essential for the function of the tomato resistance gene Cf-2, which mediates recognition of the pathogenic fungus C. fulvum carrying the avirulence gene Avr2 (Krüger et al., 2002). This gene-for-gene recognition event triggers an effective defense response that includes HR, making RCR3 an essential component of a pathogen perception system. However, RCR3 is transcriptionally regulated as a PR protein, and the secreted AVR2 protein binds and inhibits RCR3 (Krüger et al., 2002; Rooney et al., 2005). This mechanism of perception suggests that RCR3 is rather a virulence target of AVR2 that became guarded by the Cf-2 resistance protein to monitor pathogen entry (Van der Hoorn et al., 2002; Rooney et al., 2005; Jones and Dangl, 2006).
The hypothesis that PLCPs can be virulence targets is also supported by the discovery that P. infestans secretes PLCP inhibitors during infection of tomato (Tian et al., 2007). However, the cystatin-like proteins EPIC1 and EPIC2B are not homologous to AVR2 and bind and inhibit PIP1, a PLCP that is closely related to RCR3 and accumulates in the apoplast as a PR protein (Tian et al., 2007). These data indicate that tomato plants secrete PLCPs during defense to create a proteolytic apoplast that is harmful to pathogens. However, it is unknown what the full content of apoplastic PLCP activities is and to what extent these secreted PLCPs are inhibited by AVR2 and other pathogen-derived inhibitors. Description of these defensive proteases and their inhibition would therefore uncover a new layer of molecular host–pathogen interactions that would allow us to study coevolution of proteases, substrates, and inhibitors as well as resistance proteins that monitor the inhibition of these proteases.
In this study, we used a biochemical approach to investigate which PLCPs are active in the tomato apoplast during the benzothiadiazole (BTH)-triggered defense response and to determine the extent to which these PLCPs are inhibited by AVR2. Our results suggest a remarkable diversity of PLCP activities in the apoplast. Tomato secretes seven different active PLCPs during defense that belong to four phylogenetic subfamilies. Among these, PIP1 and RCR3 are transcriptionally upregulated during BTH-induced defense, with PIP1 dominating apoplastic PLCP activities. Sequencing PLCP alleles of wild tomato species showed that PIP1 and RCR3 are under diversifying selection, suggesting their participation in coevolutionary arms races with plant pathogens. Finally, by expressing the proteases separately through transient agroinfiltration assays, we demonstrated that AVR2 inhibits PIP1 and RCR3 and that a naturally occurring variant residue in RCR3 alters its susceptibility to being inhibited.
RESULTS
BTH Treatment Results in Increased PLCP Activity in the Tomato Apoplast
To investigate secreted PLCP activities during the defense response of tomato, we isolated apoplastic fluids (AFs) 5 d after treating tomato plants with water or with the salicylic acid (SA) analog BTH. The AF of BTH-treated tomato contains the expected PR proteins, demonstrating that the BTH treatment was successful (Figure 1A). We employed protease activity profiling as a tool to display activities of PLCPs in the AFs (Van der Hoorn et al., 2004). Protease activity profiling is based on the use of a biotinylated, broad-range PLCP inhibitor E-64, which reacts with the catalytic Cys residue of the protease in an activity-dependent manner (Greenbaum et al., 2002). E-64 lacks specificity-determining binding groups and is often used for diagnostic purposes since it is reactive to the entire range of PLCPs. Although the readout does not contain information on substrate specificity and conversion rates, signals represent the availability and abundance of active sites of PLCPs. We previously used protease activity profiling with biotinylated E-64 (called DCG-04) to show that AVR2 inhibits RCR3 and that EPIC2B inhibits PIP1 (Rooney et al., 2005; Tian et al., 2007).
Figure 1.
BTH-Induced Protease Activities in the Tomato Apoplast.
(A) Apoplastic PR accumulation in BTH-treated tomato plants. Five-week-old tomato plants were treated with water or BTH. AFs were isolated after 5 d, and equal volumes were separated on a 17% protein gel to visualize PR protein accumulation in the AF.
(B) Protease activity profiling on equal volumes of total extract and AF of combined leaves from water- or BTH-treated tomato plants. Extracts were labeled at pH 5.0 with DCG-04 in the absence or presence of an excess of E-64. Proteins were separated on 12% protein gels, and biotinylated proteins were detected on a protein blot with streptavidin-HRP. Total extracts corresponds to ∼0.17 cm2 leaf area/lane; AF corresponds to ∼7.4 cm2 leaf area/lane. A representative of seven independent experiments is shown. An enlarged Coomassie blue–stained gel (cbb) shows equal loading and the accumulation of PR3 proteins.
(C) Protease activity profiling of AFs from young leaves of water- and BTH-treated tomato plants. A Coomassie blue–stained gel (cbb) shows equal loading and the accumulation of PR3 proteins.
(D) Protease activity profiling on AFs isolated from leaves of different age of the same water-treated plant (indicated in illustration on the right). E-64 competition was incomplete, resulting in some remaining 25-kD signal.
In this work, protease activity profiling with DCG-04 revealed three biotinylated signals in AF of water-treated tomato plants: a strong signal at 25 kD, a weaker signal at 30 kD, and a weak signal at 35 kD (Figure 1B, lane 5). All three of these signals can be competed by adding an excess of E-64 during labeling (Figure 1B, lane 6). These three signals are upregulated in AF of BTH-treated tomato plants (Figure 1B, lane 7). Quantification of these signals from films showed that the overall upregulation of signal intensity was 1.93-fold (±0.38) (n = 7). The differential activity is less pronounced in the total extracts (TE), suggesting that most of the differential PLCP activities upon BTH treatment reside in the apoplast and not inside the cells (Figure 1B, lanes 1 and 3). The difference in PLCP activities upon BTH treatment is stronger in AFs isolated from younger leaves of BTH-treated plants (Figure 1C) when compared with AFs isolated from all the leaves (Figure 1B). This is explained by a lower PLCP activity in AFs in young leaves when compared with old leaves from the same nontreated tomato plant (Figure 1D). Thus, tomato plants create a proteolytic apoplast by secreting active PLCPs upon BTH treatment, especially in young leaves.
Tomato Apoplast Contains Activities of Different PLCPs
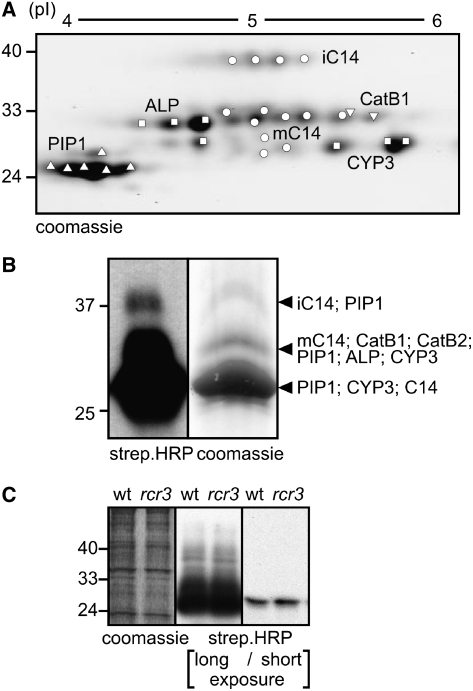
To identify the PLCPs in the tomato apoplast, AFs were labeled with DCG-04 and biotinylated proteins were purified, separated on two-dimensional (2D) gels, and identified by tandem mass spectrometry (MS). The 28 protein spots on the 2D gel represent five different PLCPs called C14, PIP1, CYP3, ALP, and CatB1 (Figure 2A) (discussed below). The experimental pI and molecular weight of the identified proteases roughly coincide with that predicted for the mature proteins (Figures 2A and 3A). The horizontal rows of spots for most of the proteases probably come from different isoforms that exist in planta or are generated during extraction. Peptides were found only for the protease domains of these proteins, consistent with the prediction that the autoinhibitory prodomain should be cleaved off to activate the protease (Figure 3A). Peptides containing the active site Cys were missing from the spectra since these peptides are modified by DCG-04.
Figure 2.
Identification of BTH-Induced Tomato Apoplastic Proteases.
(A) Identification of in vitro–labeled tomato PLCPs from AFs. AF of BTH-treated leaves was labeled with DCG-04. After labeling, biotinylated proteins were purified and separated on a 2D gel. Proteins were isolated from Coomassie blue–stained spots/bands and analyzed by tandem MS.
(B) Identification of in vivo–labeled tomato PLCPs from AFs. Tomato leaflets (183) were vacuum infiltrated with 2 μM DCG-04 and labeled in situ. AF was isolated, and biotinylated proteins were purified and separated on protein gel. A small portion of the sample was analyzed on protein blots with streptavidin-HRP to visualize the biotinylated proteins (left panel). Proteins were isolated from Coomassie blue–stained spots/bands and analyzed by tandem MS.
(C) RCR3 is not a major protease in AFs of BTH-treated tomato plants. Cf2/Rcr3 and Cf2/rcr3-3 plants were treated with BTH for 5 d, and AF was isolated. AF was labeled with DCG-04, and biotinylated proteins were displayed on a protein blot using streptavidin-HRP. A representative of three independent experiments is shown.
Figure 3.
In Silico Analysis of Identified Secreted Tomato Proteases.
(A) Domains encoded by the open reading frames (ORFs) of the identified secreted tomato proteases. The peptides that are reproducibly found in MS analysis are indicated with black bars. sp, signal peptide; pro-, autoinhibitory prodomain; Cys, catalytic Cys residue; protease, mature protease domain; p, Pro-rich domain; granulin, granulin-like domain; *, vacuolar targeting signal NPIR; pI/MW, isoelectric point and molecular weight of the protease after removal of the signal peptide and prodomain. The color coding represents the similarity-colored subfamilies in (B).
(B) Unrooted phylogenetic tree of 145 plant papain-like Cys proteases showing the diversity of the identified tomato proteases (colored names). The classes are subdivided into eight subfamilies (Beers et al., 2004). Accession codes of other plant PLCPs (numbered) are in Supplemental Data Set 1 online. S24988 is a tomato protease of group 6 that was not identified in AFs in this study. Bootstrap values for critical nodes are indicated in gray bold italics.
(C) Conservation of proteases and control proteins within solanaceous species. tBLASTp searches of the tomato proteins to translated EST libraries of tobacco, pepper, N. benthamiana, potato, and petunia resulted in the identification of close homologs from each of these solanaceous species. The amino acid identity over the protease domain was scored and combined for the different species. Resistance proteins, PR proteins, and various household proteins were used as controls (gray; see Methods). Asterisk indicates mature proteins used for tBLASTp analysis. Error bars represent sd.
To confirm the identity of these apoplastic PLCPs independently, we infiltrated tomato plants with DCG-04 and isolated AF after in situ labeling. Since DCG-04 is not membrane permeable (Lennon-Dumenil et al., 2002), this approach ensures that only apoplastic proteases are labeled and not proteases that might leak from cells during AF isolation. Biotinylated proteins were purified and analyzed on a one-dimensional (1D) protein gel (Figure 2B, left). Analysis of the proteins by tandem MS revealed the same five proteases as were found by 2D gel analysis (Figure 2B, right). An additional cathepsin B–like protease (CatB2) was identified from the 30-kD region. This protease has a calculated pI of 7.2 and was therefore missed in the 2D analysis. PIP1 was identified in every band, probably because this protein is highly abundant and contaminated the other signals during excision of the bands.
Importantly, RCR3 was missing from both 1D and 2D analyses. The pI/molecular weight of RCR3 is identical to that of PIP1, which indicates that the abundant PIP1 probably hides the presence of RCR3 on 1D and 2D gels and therefore interferes with its detection by MS. To investigate the contribution of RCR3 to the overall activity profile, we labeled AF from wild-type and rcr3-3 tomato plants. The rcr3-3 mutation results in a premature stop codon that prevents RCR3 protein accumulation (Krüger et al., 2002). The apoplastic PLCP activity profile of the rcr3-3 mutant plants is identical to that of wild-type plants, indicating that the amount of active RCR3 is minor when compared with PIP1 and other apoplastic PLCPs (Figure 2C).
Secreted Tomato PLCPs Belong to Four Different Classes
A phylogenetic tree constructed with 145 different plant PLCPs (Beers et al., 2004), including the seven identified tomato PLCPs, shows that the seven tomato PLCPs (Figure 3A) belong to four different subfamilies (Figure 3B). C14/TDI-65/CYP1/SENU2 belongs to subfamily 1, which includes PLCPs that are encoded by stress-inducible (heat, cold, or senescence) genes (Schaffer and Fischer, 1988, 1990; Drake et al., 1996; Harrak et al., 2001). Characteristic for many PLCPs of subfamily 1 is that they carry a C-terminal granulin domain (Bateman and Bennett, 1998; Yamada et al., 2001). PIP1 and RCR3 are closely related and belong to subfamily 5 (Figure 3B). These two proteases were described in the introduction. CYP3 and ALP are two aleurain-like proteases of subfamily 7 that both carry the vacuolar targeting signal NPIR in their prodomain (Figure 3A) (Holwerda et al., 1992). CatB1 and CatB2 are two cathepsin-B like proteases of subfamily 8 (Figure 3B). The Cathepsin B–like protease of N. benthamiana (CathB) is secreted as an active protease, and gene silencing of CathB in N. benthamiana prevents HR induced by nonhost bacteria and gene-for-gene interactions (Gilroy et al., 2007). In summary, the seven identified apoplastic PLCPs represent four subfamilies that probably fulfill different biological functions.
Only PIP1 and RCR3 Are Induced by BTH Treatment
We used fluorescent protease activity profiling using TMR-DCG-04 (Greenbaum et al., 2002) to investigate which of the PLCPs is differentially active in tomato AFs upon BTH treatment. Fluorescent protease activity profiling facilitates a direct, quantitative readout and reduces background caused, for example, by binding of streptavidin-horseradish peroxidase (HRP) to other proteins. The signals detected by fluorescent protease activity profiling on 1D gels are similar to those observed using streptavidin-HRP (Figure 4A, top). However, in contrast with streptavidin-HRP detection, BTH treatment results in an upregulation of only the 25-kD signal (Figure 4A, top). The fluorescent signals were further analyzed on 2D gels. Comparison of the fluorescent 2D map (Figure 4A, bottom) with the Coomassie blue–stained 2D gel containing purified PLCPs (Figure 2A) indicates that we can detect the activities of PIP1, C14, CatB1, CYP3, and perhaps even ALP simultaneously using fluorescent protease activity profiling. Comparison of the fluorescent 2D maps of water- and BTH-treated tomato plants indicates that only the signal corresponding to PIP1 is upregulated upon BTH treatment (Figure 4A, bottom). The signals corresponding to the other PLCPs remain the same when compared with the standard. These data indicate that PIP1 dominates the increased PLCP activity during BTH treatment.
Figure 4.
Induction of PIP1 and RCR3 upon BTH Treatment by Fluorescent Protease Activity Profiling and Real-Time RT-PCR.
(A) Fluorescent protease activity profiling of AFs. Plants were treated with BTH for 5 d, and AF was isolated. Equal volumes of AF were used for protease activity profiling with TMR-DCG-04. Proteins were separated by 1D (top) and 2D (bottom) gel electrophoresis, and fluorescent proteins were detected by fluorescent protease activity profiling. Please note that only signals coinciding with PIP1 are significantly stronger from BTH-treated plants. A representative of three independent experiments is shown.
(B) Induction of transcript levels by BTH treatment. Tomato leaves were harvested at 5 d after water (H) or BTH (B) treatment. Quantitative real-time RT-PCR was performed using gene-specific primers. The difference in threshold cycles (dCt) between the protease transcript and ubiquitin transcripts was calculated from three independent samples. Error bars represent sd. A representative of five independent biological experiments is shown.
To investigate the transcriptional regulation of the protease-encoding genes during BTH-induced defense responses, we performed quantitative real-time RT-PCR on BTH- and water-treated tomato plants. This analysis shows that PIP1 and RCR3 transcripts are eightfold upregulated upon BTH treatment, whereas the other PLCP transcript levels are not significantly induced by BTH treatment (Figure 4B). The induction by BTH treatment is similar to those of PR genes (Sanz-Alferez et al., 2008; see Supplemental Figure 1 online), indicating that PIP1 and RCR3 gene expression is regulated through the SA pathway and that they can be considered as being PR genes.
PIP1 and RCR3 Are Less Conserved Than the Other Proteases
To investigate the conservation of PLCP sequences in other solanaceous species, we searched for homologous genes in The Institute for Genomic Research (TIGR) database. tBLASTp searches of the seven tomato proteases to translated EST libraries of tobacco, pepper (Capsicum annuum), N. benthamiana, potato (Solanum tuberosum), and petunia (Petunia hybrida) resulted in the identification of close homologs from each of these solanaceous species. The amino acid identity over the protease domain was scored and combined for the different species. This in silico analysis indicates that the proteases fall into two groups: a conserved group, consisting of C14, CYP3, ALP, CatB1, and CatB2 proteins; and a diverse group, containing PIP1 and RCR3 (Figure 3C). The degree of sequence identity of the conserved proteases among the examined species is similar to housekeeping proteins with known, conserved functions (Figure 3C, right columns). By contrast, the degree of identity of diverse proteases PIP1 and RCR3 is as low as that of resistance proteins (Cf9, Mi, and Prf) or PR proteins (PR1, PR2, PR3, or P69A) (Figure 3C, middle columns). Although this analysis is limited by the absence of complete genome sequences for the examined species and by the size of the EST libraries, the relative expression levels of each gene, and the tissues from which these EST libraries were generated, these data suggest that the protease domains of PIP1 and RCR3 are more diverse and rapidly evolving within solanaceous species than those of the other PLCPs.
PIP1 and RCR3 Are Subject to Diversifying Selection
The observation that PIP1 and RCR3 are probably more diverse inspired us to investigate if these genes are subject to diversifying selection. We therefore sequenced the region encoding the protease domain of eight wild tomato relatives: S. cheesmanniae, S. pimpinellifolium, S. chilense, S. pennellii, S. habrochates(hirsutum), S. peruvianum, S. schiewlskii, and S. parviflorum. Sequences of theses alleles were validated (see Methods) and found to be 98% identical to the reported tomato sequences. Amino acids encoded by the polymorphic codons of all the protease domains are shown in Figure 5A. The protease-coding part of each gene contains ∼20 variant nucleotides, except for RCR3, which has 41 variant nucleotides (Figure 5B). Some of the variant nucleotides are shared among different species, indicating that part of the variation predates speciation (Figure 5A; see Supplemental Alignment 3 online). Most of the polymorphic nucleotides, however, are allele specific. The consequence of these variant residues at amino acid level is striking. Variant codons hardly change the encoded amino acids in C14, CYP3, ALP, CatB1, and CatB2 (Figure 5A, bottom, white and gray residues). By contrast, nearly all variant codons of PIP1 and RCR3 cause nonsimilar amino acid substitutions (Figure 5A, bottom, red residues). The ratio between nonsimilar and similar amino acids indicates that C14, CYP3, ALP, CatB1, and CatB2 are under conservative selection, whereas PIP1 and RCR3 are under diversifying selection (Figure 5C). These data are consistent with the indication from tBLASTp searches that PIP1 and RCR3 proteases are less conserved in solanaceous species than the other PLCPs (Figure 3C). Taken together, these observations demonstrate that PIP1 and RCR3 are under diversifying selection, possibly to adapt to diversifying substrates or inhibitors, whereas the other proteases are under conservative selection.
Figure 5.
Sequence Analyses of Proteases from Tomato Relatives.
(A) Summary of amino acids encoded by variant codons in the protease domains of C14, PIP1, RCR3, CYP3, ALP, CatB1, and CatB2 alleles sequenced from various wild tomato relatives (indicated top right). Amino acids encoded by the variant codons are summarized by leaving out the amino acids of nonvariant codons from the protein alignment. Amino acids encoded by codons different from the S. lycopersicum (lyc) allele are indicated with gray, blue, and red residues if they are identical, similar, or nonsimilar, respectively, compared with the lyc sequence. Dashes indicate missing sequence information. RCR3 of S. cheesmanniae is not shown since it contained a premature stop codon and could be amplified from genomic DNA and not from cDNA (see Supplemental Alignment 2 online).
(B) Number of single nucleotide (nt) polymorphisms per protease.
(C) Ratio of nonsimilar/similar amino acid (aa) substitutions calculated from (A). PIP1 and RCR3 are under diversifying selection; the other proteases are under conservative selection.
(D) Position of variant residues in structural models of PIP1 and RCR3. Positions with nonsimilar variance and similar variance are indicated in red and blue, respectively.
Diversified Residues in PIP1 and RCR3 Reside around the Substrate Binding Groove
To investigate where within the protease structure the variation of PIP1 and RCR3 resides, we generated structural models for PIP1 and RCR3 to indicate the position of the variant amino acids (Figure 5D). Like with all PLCPs, the PIP1 and RCR3 models consists of two lobes with a substrate binding groove in between (Kim et al., 1992). The active site Cys is in the middle of this groove. All of the nonsimilar residue variance of PIP1 and RCR3 are on the surface of the protein (red in Figure 5D). By contrast, similar residue variance is mostly located inside the protein structure (blue in Figure 5D). Five of the variant amino acids of PIP1 and RCR3 coincide at a similar location, but the other variant residues are unique to either PIP1 or RCR3. These residues are nearly all on the front side, around the substrate binding groove, but not in the groove itself. The location of the variant residues could coincide with a binding region for substrates or inhibitors.
AVR2 Targets Specific PLCPs in the Tomato Apoplast
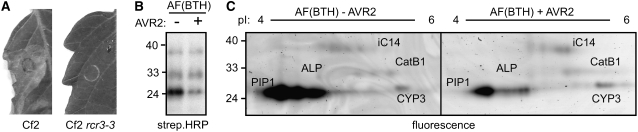
Having identified the PLCPs that pathogens encounter when they invade the tomato apoplast, we next investigated which PLCPs are inhibited by AVR2 of the fungus C. fulvum (Rooney et al., 2005). AVR2 was produced as an N-terminally FLAG-tagged protein in Escherichia coli. This FLAG-AVR2 triggers hypersensitive cell death when injected into tomato plants carrying the Cf-2 resistance gene, demonstrating that AVR2 is folded properly (Figure 6A).
Figure 6.
Production of AVR2 and Inhibition in AF.
(A) Recombinant AVR2 triggers hypersensitive cell death in tomato leaves of MM-Cf2 plants. Recombinant AVR2 was injected into leaves of MM-Cf2 and MM-Cf2/rcr3-3 plants. Picture was taken 4 d after injection.
(B) Inhibition of PLCPs in AFs by AVR2. AF of BTH-treated tomato plants were preincubated with 66 nM AVR2 before adding DCG-04 to label the remaining noninhibited proteases. Biotinylated proteins were detected using streptavidin-HRP. A representative of three independent experiments is shown.
(C) Fluorescent protease activity profiling of PLCPs in AFs in presence or absence of AVR2. AFs of BTH-treated tomato plants were preincubated with 83 nM AVR2 before adding TMR-DCG-04 to label the remaining noninhibited proteases. Proteins were separated by 2D gel electrophoresis, and fluorescent proteins were visualized. A representative of three independent experiments is shown.
To investigate the inhibition of PLCPs by AVR2 in crude AFs of BTH-treated tomato plants, we preincubated these AFs for 30 min with AVR2 and then added DCG-04 to label the remaining, noninhibited proteases. These assays revealed that AVR2 prevents the labeling of most of the 25-kD proteases, whereas the other signals remain unaltered (Figure 6B). To determine the selectivity of AVR2 inhibition in more detail, we performed fluorescent protease activity profiling using TMR-DCG-04 on AF of BTH-treated tomato plants with and without preincubation with AVR2. The 2D images were normalized using the standard and compared. Comparing the 2D fluorescent maps in the absence or presence of AVR2 shows that fluorescent labeling of C14, CYP3, and CatB1 is unchanged, whereas labeling of PIP1 is significantly reduced in the presence of AVR2 (Figure 6C). These data indicate that AVR2 selectively targets PIP1 and does not inhibit other C14, CYP3, or CatB1 in the apoplast.
AVR2 Inhibits PIP1 and RCR3
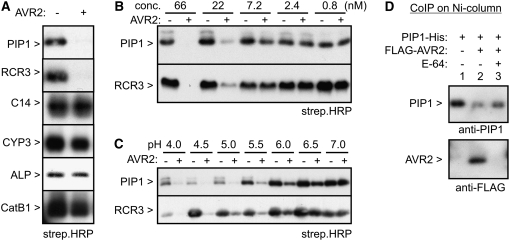
To investigate the inhibition of each protease separately, we overexpressed each of the seven tomato proteases in planta through agroinfiltration of N. benthamiana (Van der Hoorn et al., 2000; Voinnet et al., 2003). Protease activity profiling with DCG-04 on total extracts or AFs from these agroinfiltrated leaves revealed strong additional biotinylated signals in addition to weaker signals of the endogenous proteases of N. benthamiana (see Supplemental Figure 2 online). The intensities of these signals were used to dilute the extracts to contain comparable concentrations of active proteases. The sizes of the signals are consistent with the sizes of these proteases observed in tomato AFs and the sizes predicted for the mature protease domains (Figures 2A and 2B). This transient expression system allowed us to monitor activities of in planta–produced tomato proteases separately.
To monitor the inhibition by pathogen effector proteins, we preincubated the protease-containing extracts with AVR2 and then added DCG-04 to label the noninhibited proteases. This revealed that AVR2 inhibits labeling of PIP1 and RCR3 but not C14, CYP3, ALP, or CatB1 (Figure 7A). These inhibition data are consistent with the data observed for the tomato apoplastic PLCPs (Figure 6C).
Figure 7.
Inhibition of Agroinfiltrated Tomato Proteases by AVR2.
(A) Extracts from agroinfiltrated N. benthamiana leaves overexpressing different tomato proteases (indicated on the left) were preincubated for 30 min with 66 nM AVR2. DCG-04 was added after preincubation to label the noninhibited proteases. Biotinylated proteins were visualized on protein blots using streptavidin-HRP. Representatives of at least three independent experiments are shown.
(B) Concentration dependency of inhibition by AVR2. Protease-containing extracts were incubated with different AVR2 concentrations for 30 min. DCG-04 was added after preincubation to label the noninhibited proteases. Biotinylated proteins were visualized on protein blots using streptavidin-HRP. A representative of three independent experiments is shown.
(C) pH dependency of inhibition by AVR2. Extracts from agroinfiltrated leaves overexpressing the proteases were preincubated for 30 min at different pH with or without 66 nM AVR2. DCG-04 was added after preincubation to label the noninhibited proteases. Biotinylated proteins were visualized on protein blots using streptavidin-HRP. A representative of three independent experiments is shown.
(D) AVR2 physically interacts with PIP1. Extracts from agroinfiltrated leaves expressing His-tagged PIP1 were preincubated with or without an excess E-64 and incubated with FLAG-AVR2 at pH 5. Protein complexes containing PIP1-His were purified using Ni-NTA columns and analyzed using anti-PIP and anti-FLAG antibodies. A representative of three independent experiments is shown.
The inhibition assays described above were done at 66 nM inhibitor concentrations. To investigate inhibition at lower inhibitor concentrations, PIP1 and RCR3 were incubated with or without various AVR2 concentrations and then incubated with DCG-04. This revealed that AVR2 inhibits both PIP1 and RCR3 at concentrations above 7.2 nM (Figure 7B), indicating that AVR2 inhibits both RCR3 and PIP1 to a similar level. However, precise comparisons of inhibition strengths were not possible with these assays.
The inhibition experiments were so far performed at the pH of the apoplast (pH 5). To investigate if the inhibition could also take place under a wider pH range, inhibition assays were performed at pH 4 to 7. Although PIP1 and RCR3 are similarly active at all tested pH, inhibition by AVR2 only occurs at pH 4 to 5.5 (Figure 7C). This shows that inhibition of both PIP1 and RCR3 by AVR2 is a pH-dependent process that occurs at apoplastic pH.
Coimmunoprecipitation experiments were performed to investigate if inhibition is caused by a physical interaction between AVR2 and PIP1. C-terminally His-tagged PIP1 (Tian et al., 2007) was produced by agroinfiltration and immobilized on nickel-nitrilotriacetic acid agarose (Ni-NTA) columns. These PIP1-coated beads were incubated with FLAG-AVR2, washed, and eluted, and the eluate was used for protein blots using anti-PIP1 and anti-FLAG antibodies. This revealed that AVR2 is in the eluate of immobilized PIP1 columns (Figure 7D, lane 2). The interaction is prevented by preincubation of PIP1-His with an excess E-64 before immobilization (Figure 7D, lane 3), indicating that the interaction is at the active site. Taken together, these data show that AVR2 physically interacts with PIP1, similar to what was demonstrated previously for RCR3 (Rooney et al., 2005).
A Naturally Occurring Variant Residue in RCR3 Affects Inhibition by AVR2
To investigate if variant residues in RCR3 can affect its inhibition by AVR2, we selected variant residues in RCR3 that are close to the active site. Four of the variant residues in RCR3 (orange in Figure 8A) were not tested since these variant residues are present in RCR3pim, and this protein is inhibited by AVR2 to a similar extent as RCR3lyc (Rooney et al., 2005). Four other variant residues (H148N, R151Q, N194D, and Q267H; red in Figure 8A) were selected and introduced into RCR3 using site-directed mutagenesis. In addition, one double mutant was generated (H148N-R151Q).
Figure 8.
The Naturally Occurring N194D Mutation in RCR3 Affects Inhibition by AVR2.
(A) Position of variant residues in a structural model of RCR3. Differences between the RCR3lyc and RCR3pim alleles (orange) were not affecting AVR2 inhibition (Rooney et al., 2005). Four variant residues (red) were selected for targeted mutagenesis.
(B) The RCR3N194D mutant is less sensitive for AVR2 inhibition. Extracts from agroinfiltrated leaves expressing (mutant) RCR3 proteins were preincubated at pH 5.0 with or without 65 nM AVR2 (A) or 20 μM E-64 (E). DCG-04 was added to label the remaining noninhibited proteases, and biotinylated proteins were detected on protein blots using streptavidin-HRP. The experimental differences in accumulation of RCR3 is shown using RCR3 antibodies. A representative of four independent assays is shown.
Mutant RCR3 proteins were produced by agroinfiltration and subjected to protein blot analysis with anti-RCR3 antibodies and protease activity profiling with DCG-04. All mutant RCR3 proteins accumulate to similar levels and react with DCG-04, indicating that these proteins are stable, active proteases (Figure 8B). Preincubation of the (mutant) RCR3 proteins with AVR2 showed that mutants H148N, R151Q, and N194D and the double mutant H148N-R151Q can be inhibited by AVR2 to a similar extent as wild-type RCR3 (Figure 8B). By contrast, the N194D mutant RCR3 protein is less sensitive to AVR2 inhibition, even though it can be inhibited by E-64 (Figure 8B). These data demonstrate that the variance of a single amino acid can affect the inhibition by AVR2. However, in addition to N194D variance, the S. chilense RCR3 isoform carries another five variant residues. The effect of these residues on AVR2 inhibition will be investigated in future studies.
DISCUSSION
This work demonstrates that the apoplast of BTH-treated tomato contains at least seven active PLCPs that fall into four different subfamilies based on their homology, structural features, and putative cellular functions. PIP1 and RCR3 are the only proteases that are strongly transcriptionally upregulated upon BTH treatment. In tomato species, these proteases are rapidly evolving and are under strong diversifying selection, resulting in variant residues around the substrate binding groove of the protease. The proteases of other subfamilies are not induced by BTH treatment and are under conservative selection. The Cladosporium AVR2 effector protein specifically targets the defense-related proteases PIP1 and RCR3, and one of the naturally occurring variant residues in RCR3 affects AVR2 inhibition.
The described proteases are probably only a subset of the proteases that are present in the apoplast of BTH-treated tomato plants. P69B, for example, is another predominant PR protein and an active subtilisin-like Ser protease (Tornero et al., 1997; Figure 1A). P69B is inhibited by Kazal-like EPI1 and EPI10 proteins of P. infestans (Tian et al., 2004, 2005). We focused on papain-like proteases by applying protease activity profiling using the broad-range probe DCG-04, which reacts with PLCPs with limited selectively (Van der Hoorn et al., 2004). The seven PLCPs that we identified are probably not the full set of secreted PLCPs but certainly comprise the majority of active PLCPs in the tomato apoplast. It is likely that extracellular pathogens will encounter these secreted PLCPs during infection.
We identified proteases in the apoplast by careful analysis that excludes leakage from internal sources. However, we did find aleurain-like proteases that carry vacuolar targeting signals. In fact, many of the apoplastic proteases we identified have Arabidopsis orthologs that were identified in the vegetative vacuole proteome (Carter et al., 2004). These Arabidopsis vacuolar PLCPs contain representatives of group 1 (RD21A and XBCP3), group 6 (number 132), group 7 (aleurain-like, AALP), and group 8 (CatB-like, numbers 142 and 143) (indicated in Figure 3B) (Carter et al., 2004). In addition, AALP is used as a vacuolar marker protein, and RD21 was found in stress-induced protease storage vesicles (Ahmed et al., 2000; Hayashi et al., 2001; Watanabe et al., 2004). The data presented here indicate that pools of some of these proteases are also partly secreted. This partial secretion probably occurs with RD21-like (group 1), aleurain-like (group 7), and cathepsin B-like (group 8) proteases and may result from a default secretion pathway that is followed if sorting by the subcellular targeting machinery is reduced. However, it remains to be investigated how these proteases are secreted and why.
Transcript analysis showed that only PIP1 and RCR3 are induced by BTH treatment. This indicates that these genes are under control of the SA signaling pathway and that PIP1 and RCR3 belong to the class of PR proteins that accumulate during the basal immune response. The other PLCP genes are not induced by BTH treatment. Consistent with BTH induction, RCR3 is also induced during infection with C. fulvum (Krüger et al., 2002), and PIP1 is upregulated during infection with Pseudomonas syringae and P. infestans (Zhao et al., 2003; Tian et al., 2007). However, since SA signaling is not the only pathway that is regulated during infection, the final expression levels of PLCPs will strongly depend on the time point, cell type, and pathogen. Infection of tomato with P. syringae, for example, induces the expression of C14 but not CatB2 (Zhao et al., 2003). Furthermore, infection of potato plants with avirulent P. infestans results in upregulation of both C14 and CatB2 (Avrova et al., 1999, 2004). Thus, although the other PLCP genes are not induced by the SA analog BTH, their expression can be induced during infection, probably through other pathways. However, a distinction based on BTH induction appears to be relevant given the fact that only BTH-induced proteases are under diversifying selection and targeted by AVR2.
Allele sequencing revealed that PIP1 and RCR3 are under diversifying selection, whereas the other PLCPs are under conservative selection. This indicates that BTH-induced proteases are under diversifying selection to coevolve with a changing environment imposed by pathogens. The variant residues within the PIP1 and RCR3 proteins are at the surface, around the substrate binding groove, but not within. This region could coincide with a binding region for pathogen-derived substrates and inhibitors. Diversifying selection at interaction surfaces between enzyme and their inhibitors are common in plant–pathogen interactions (reviewed in Misas-Villamil and Van der Hoorn, 2008). Similar diversifying residues at the interaction surfaces have been reported for xylanase, polygalacturonase, and glucanase inhibiting proteins and their target enzymes (Stotz et al., 2000; Bishop et al., 2005; Beliën et al., 2007; Raiola et al., 2008).
That AVR2 selectively targets PIP1 and RCR3 among the apoplastic PLCPs was demonstrated by fluorescent protease activity profiling and by studying PLCPs separately through agroinfiltration. The fact that only BTH-induced PLCPs are inhibited is an interesting observation, although it cannot be excluded that C. fulvum secretes other proteins that target other PLCPs. Although these data suggest that RCR3 and PIP1 play a role in defense in the absence of Cf-2, this remains to be demonstrated. Conversely, a contribution of AVR2 to virulence remains to be shown.
The hypothesis that some of the variant residues in PIP1 and RCR3 might affect inhibition by pathogen-derived inhibitors was tested with AVR2 on a series of RCR3 mutants. Although weak inhibition cannot be measured with these assays, we showed that one mutant (RCR3N194D) is less sensitive to inhibition by AVR2. This variant N194D residue is close to the active site of RCR3, which indicates that this surface is important for binding AVR2. The other tested residues and those present in RCR3pim (orange and red in Figure 8A) do not alter AVR2 inhibition. These experiments indicate that single variant residues can affect inhibition by AVR2 and other pathogen-derived inhibitors, such as the cystatin-like inhibitors EPIC1 and EPIC2B of P. infestans, which also inhibit PIP1 and RCR3 (Tian et al., 2007; S. Kamoun, personal communication). The contribution of single variant residues in the context of other variance in the different RCR3 alleles remains to be investigated since some variant residues may compensate others. Diversifying selection on PLCPs and their inhibitors might be common to plant–pathogen/pest interactions since PLCPs are frequently employed on the battlefield in these interactions, for example, in the apoplast, plant cytoplasm, and herbivore gut (reviewed in Shindo and Van der Hoorn, 2008).
RCR3 has accumulated twice as many variant residues in comparison to PIP1. This is an interesting observation since it probably results from the adaptation of RCR3 to Cf resistance proteins. The RCR3lyc allele triggers necrosis in the presence of the Cf-2 resistance gene, explaining why the Cf2 resistance locus was introgressed from S. pimpinellifolium together with the RCR3pim allele (Krüger et al., 2002). The RCR3esc and RCR3pim proteases differ in four nonsimilar residues in the protease domain, located around the active site (Figure 8A, orange residues). The adaptation of RCR3 to Cf proteins is probably essential to prevent self-recognition. A similar accidental self-recognition is probably also the basis of HR triggered by expression of recombinant tomato Cf proteins and Cf proteins from S. peruvianum in tobacco (Wulff et al., 2004). The responses of these auto-activators differed between different tobacco species, suggesting that an endogenous polymorphic factor contributes to the degree of autoimmune responses. These data are consistent with the model that non-self-recognition in plants causes necrosis that is often observed in hybrids (Bomblies and Weigel, 2007). In Arabidopsis, for example, an NB-LRR resistance gene was found to trigger immune responses in hybrids, when combined with a specific allele at a second locus (Bomblies et al., 2007). These observations suggest that some of the variation in RCR3 is to adjust its interaction to Cf resistance proteins preventing accidental necrosis induced by Cf-2 in the absence of the pathogen.
These genetic data indicate that non-self RCR3 proteins could trigger accidental immune responses by an inadvertent physical interaction with Cf-2 proteins. Cf proteins consist predominantly of Leu-rich repeats that are folded as a horseshoe, having the inner concave surface with solvent-exposed residues available for specific protein–protein interactions. A direct interaction between Cf-2 and RCR3 remains to be shown, but docking studies of RCR3 with Cf proteins (Van der Hoorn et al., 2005) suggest that RCR3 could fit inside the Leu-rich repeat curvature of Cf-2 with the variant amino acids of RCR3 interacting with the solvent-exposed residues of Cf-2. In this model, AVR2 could act as a molecular glue to enhance interactions of RCR3 with Cf-2.
PIP1 is the most dominant PLCP in the apoplast of BTH-treated tomato. RCR3 accumulates to a much lower level and is even hard to detect, partly because its presence is overshadowed by PIP1. This observation was further substantiated with the fact that there is no significant difference in overall protease activity profiles between BTH-treated wild-type plants and rcr3-3 mutants (Figure 3D). The additional adaptation of RCR3, and its low abundance compared with PIP1, suggests that RCR3 could be a decoy to trap the fungus into a recognition event, rather than a defensive protease itself. The real target of AVR2 would then be the abundant PIP1 protease.
The approach taken in this study to identify (defense-related) secreted proteases and analyze their inhibition by pathogen-derived proteins has revealed interesting aspects of an apoplastic molecular battlefield between plants and pathogens. Given these observations, it seems likely that many pathogens secrete PLCP inhibitors during infection and that these inhibitors, the proteases, and their substrates are involved in a continuous coevolutionary battle. How coevolution shaped the apoplastic defense, how these defense-related proteases discriminate between self and non-self, and how RCR3 became part of a pathogen perception mechanism involving the Cf-2 resistance gene remain exciting questions to resolve.
METHODS
Plant Materials
Nicotiana benthamiana and tomato (Solanum lycopersicum Money Maker [MM] Cf0, Cf2, and Cf2/rcr3-3) were grown in a climate chamber at a 14-h light regime at 18°C (night) and 22°C (day). Four- to six-week old plants were used for experiments. BTH treatment was done by watering 5-week-old tomato plants with 25 μg/mL BTH (Actigard; Syngenta) or water every second day. Samples were taken at 5 d after starting the BTH treatment, unless otherwise indicated.
Production and Purification of AVR2 Protein
All DNA manipulations were performed using standard protocols (Sambrook et al., 1989). The ORF of Avr2 (Luderer et al., 2002) was amplified from Cladosporium fulvum race 5 by RT-PCR using primers 5′-TCCCCGCGGGCCAAAAAACTACCTGGCTGCGA-3′ and 5′-CGGGGTACCTCAACCGCAAAGACCAAAACAGCA-3′ and cloned into pFLAG-ATS (Sigma-Aldrich) using SacII and KpnI restriction enzymes, resulting in pFLAG-AVR2 (pFK3). FLAG-AVR2 was expressed and purified on an anti-FLAG matrix as described previously (Tian et al., 2004). Protein purities were checked on Coomassie Brilliant Blue–stained 17% protein gels and quantified using the Bradford assay (Bio-Rad).
Protein Work
AFs were isolated by vacuum infiltration of tomato leaves with ice-cold water. Infiltrated leaves were dried on the surface and centrifuged (10 min, 1600g) in a tube with holes in the bottom. The AF was collected below in a larger collection tube. Equal volumes (200 to 450 μL) were used for protease activity profiling, protein gel blot analysis, or Coomassie blue staining. In each case, samples were 10-fold concentrated by precipitating with 67% ice-cold acetone and dissolving the protein pellet in protein gel-loading buffer. TEs were generated by grinding six randomly taken 28-mm2 leaf discs into 1 mL of water. TEs were centrifuged (1 min, 16,100g), and the supernatant was analyzed as described for AFs. Protease activity profiling was performed as described previously (Van der Hoorn et al., 2004). Unless otherwise indicated, 200 to 450 μL of TE or AF was labeled for 5 h at room temperature with 2 μM DCG-04 (Van der Hoorn et al., 2004) in the presence of 50 mM sodium acetate, pH 5.0, and 1 mM l-cysteine. Competition with E-64 was done by adding 20 μM E-64 (Sigma-Aldrich) 30 min before adding DCG-04. Proteins were precipitated by adding 1 mL ice-cold acetone and centrifugation (1 min, 16,100g). Pellets were dissolved in SDS gel loading buffer. Detection of biotinylated proteins was done as previously described using streptavidin-HRP polymer (Sigma-Aldrich) (Van der Hoorn et al., 2004). Protein gel blot analysis was performed as described for detection of biotinylated proteins, using RCR3 and PIP1 antibodies (Rooney et al., 2005; Tian et al., 2007), followed by detection with HRP-conjugated anti-rabbit antibodies (Amersham).
For large-scale labeling of AF, 100 mL of AF was isolated as described above from 6-week-old tomato plants, 7 d after BTH treatment, and labeled with 2 μM DCG-04 at pH 5.0 (25 mM sodium acetate and 1 mM l-cysteine). Large-scale in situ labeling of tomato leaves by DCG-04 infiltration was done by vacuum infiltrating ∼183 tomato leaflets with water containing 2 μM DCG-04. Leaves were incubated for 5 h at room temperature. Ice-cold water was vacuum infiltrated, and AF was quickly isolated as explained above. AF proteins were precipitated using 67% ice-cold acetone, dissolved in TBS (50 mM Tris, pH 7.5, and 150 mM NaCl) containing a protease inhibitor cocktail (Complete tablets; Roche), and incubated for 1 h at room temperature. Undissolved proteins were removed by centrifugation (10 min, 3000g), and 500 μL of washed magnetic streptavidin beads (Promega) were added. Biotinylated proteins were captured by rotating overnight at room temperature. Beads were collected in a magnetic holder (Promega) and washed three times with washing buffer (1% Triton X-100, 1 M NaCl, and 50 mM Tris, pH 7.5), three times with TBS, and three times with water. Beads were boiled in gel-loading buffer for 1D analysis or eluted with 50% formic acid for 2D analysis.
For 2D analysis of purified biotinylated PLCPs, the formic acid eluate was dried and dissolved 160 μL of IEF buffer (7 M urea, 2 M thiourea, 2% CHAPS, 0.5% IPG buffer [pI 3 to 10; Amersham], 0.002% bromephenol blue, and 20 mM DTT). The sample was impregnated overnight into an IPG ready strip (7 cm, 3-6L; Bio-Rad). Isoelectric focusing was done in ZOOM IPG runner cassettes (Invitrogen) (15 min, 175 V; 45 min, 175 to 2000 V ramp; 25 min, 2000 V). After IEF, strips were incubated for 15 min in 4.5 mL LDS sample buffer (Invitrogen) supplemented with 0.5 mL sample reducing agent (Invitrogen) and then incubated for 15 min in 5 mL of LDS containing 116 mg iodoacetamide (Sigma-Aldrich). Strips were loaded on precasted NuPage 4 to 12% ZOOM gradient gels (Invitrogen) for 50 min at 200 V, and the gel was stained with SYPRO Ruby (Invitrogen).
After 2D electrophoresis, spots of various intensities were automatically excised and tryptically digested using the DP Chemical 96 kits for fully automated in-gel digestion (Proteineer spII/dp; Bruker). Aliquots of the digests were automatically prepared (Proteineer dp; Bruker) for subsequent matrix-assisted laser-desorption ionization time of flight (MALDI-TOF) MS analysis on AnchorChip targets (Bruker) according to Gobom et al. (2001). Mass spectra of tryptic peptides were taken with a Bruker Reflex IV MALDI-TOF MS. The obtained peptide mass fingerprints were processed in Xmass 5.1.16 (Bruker) and used to identify the corresponding proteins in the ProteinScape 1.3 database system (Protagen), which triggered MASCOT (Matrix Science) and Profound (Genomic Solutions) searches against the National Center for Biotechnology Information (NCBI) and TIGR genome databases.
For fluorescent protease activity profiling, 565 μL of AF isolated from water- or BTH-treated tomato plants was preincubated for 30 min with and without 83 nM AVR2 in 50 mM NaAc, pH 5. The remaining noninhibited proteases were labeled for 5 h with 3 μM DCG-04-TMR (Greenbaum et al., 2002). Proteins were precipitated with trichloroacetic acid and acetone, washed, dissolved in IEF buffer, and separated on 2D gels as described above. Fluorescent proteins were detected using the Typhoon fluorescent imager (Molecular Dynamics). Gels were scanned simultaneously, and the standards had equal intensities.
For coimmunoprecipitation experiments, PIP1-His (Tian et al., 2007) was expressed by agroinfiltration of N. benthamiana (see below), and AF was isolated at 5 dpi. Forty microliters of PIP1-His–containing AFs were incubated with 50 μL of Ni-NTA agarose beads (Qiagen) for 2 h in 50 mM NaPO4, pH 8, 150 mM NaCl, and 1 mM imidazole. Beads were washed with 50 mM NaAc, pH 6.0, and incubated overnight in a cold room in the presence of absence of 10 mM E-64. Beads were washed with 50 mM NaAc, pH 6.0, and incubated for 20 min with or without 50 μL 260 nM AVR2. Beads were quickly washed with ice-cold 50 mM NaAc, pH 6, and eluted with 50 μL of 50 mM NaPO4, pH 8.0 150 mM NaCl, and 500 mM imidazole. Eluted proteins were analyzed on protein blots using anti-PIP1 and anti-FLAG antibodies.
Agroinfiltration of Tomato Proteases
The pFK26 cloning vector was constructed as follows: the multiple cloning site (mcs) of pBluescript KS+ (Stratagene) was modified by digesting with KpnI and HindIII, treating with Klenow polymerase, and self-ligation, resulting in pFK18. The promoter-terminator (35S-TPI) from pRH80 (Van der Hoorn et al., 2000) was amplified by PCR using primers r014 and r015 (see Supplemental Table 1 online). Digesting the PCR product with PstI-XhoI resulted in a 300- and 500-bp fragment. The 500-bp fragment (containing the 35S promoter of pRH80) was cloned into pFK18 using PstI and XbaI restriction sites, resulting in pFK19. The terminator region (TPI) was amplified from pRH80 using primers r013 and r014 (see Supplemental Table 1 online) and cloned into pGEM-T (Promega), resulting in pFK22. The TPI fragment was cloned from pFK22 into pFK19 using NcoI and EcoRI restriction sites, resulting in pFK26. The pTP5 binary vector was generated by cloning the 35S-mcs-TPI cassette of pFK26 into pRH385 (Van der Hoorn et al., 2005) using XbaI and EcoRI restriction enzymes.
The tomato protease ORFs were amplified by RT-PCR from RNA extracted from tomato leaves, cloned into pFK26, and sequenced. Expression cassettes containing correct ORF sequences were shuttled into binary vector pTP5 as follows. Details of these cloning procedures are summarized in Supplemental Table 2 online.
RCR3 mutant proteins were generated by introducing mutations into pTP28 (35S:RCR3:term) using the Quickchange site-directed mutagenesis kit (Stratagene) and primers (see Supplemental Table 1 online). Mutations were confirmed by sequencing, and correct expression cassettes were shuttled into pTP5 using XbaI-SalI, resulting in pMS34(RCR3H148N), pMS35(RCR3R151Q), pMS36(RCR3H148N,R151Q), pMS37(RCR3N194D), and pMS38(RCR3Q267H).
Agrobacterium tumefaciens strain GV3101 was transformed with binary vectors and used for agroinfiltration as described previously (Van der Hoorn et al., 2000). Agrobacterium was grown overnight at 28°C in 10 mL Luria-Bertani medium containing 50 μg/mL kanamycin and 50 μg/mL rifampicin. The culture was centrifuged (10 min, 3000g) and the bacterial pellet resuspended in 10 mM MES, pH 5, 10 mM MgCl2, and 1 mM acetosyringone to a final OD of 2. Agrobacterium cultures containing binary protease expression vectors were mixed with Agrobacterium cultures containing binary expression vector for silencing inhibitor p19 (Voinnet et al., 2003). Agrobacterium cultures were infiltrated into 5-week-old N. benthamiana plants using a syringe without a needle. AF was isolated and the remaining tissue was ground in a mortar with water. This TE was centrifuged (10 min, 16,100g) and the supernatant stored in aliquots until further use. Both AF and TE were stored in aliquots at −20°C.
Inhibition assays with agroinfiltrated proteases were done as follows. AFs or total extracts from agroinfiltrated N. benthamiana were isolated at 5 dpi. In a total volume of 400 μL, 40 μL extract was preincubated in 50 mM NaAc, pH 5.0, 1 mM l-cysteine with 65 nM (or less) AVR2, or 10 μM E-64 for 30 min at room temperature. DCG-04 was added to a final concentration of 2 μM, and the labeling was performed for another 5 h at room temperature. Proteins were analyzed on protein blots using streptavidin-HRP or anti-RCR3 antibodies.
Phylogenetic Analysis
Sequences of 143 PLCPs (accession numbers are provided in Supplemental List 1 online) were aligned using ClustalW 1.83 (Higgins et al., 1994). The alignment was optimized manually, resulting in a file with a minimum number of gaps. Protein distances were calculated using the program Protdist from the PHYLIP suite of programs (Felsenstein, 1989) with the JTT matrix on 1000 bootstraps. This data set (see Supplemental Data Set 1 and Supplemental Alignment 1 online) was then used to generate phylogenetic trees based on neighbor joining. The resulting trees were then joined to a consensus tree using the Consense program and plotted with Drawtree (Felsenstein, 1989). The output from Drawtree was further edited in Adobe Illustrator.
Bioinformatic Analysis
For comparative tBLAST searches through the solanaceae TIGR database, the TIGR database of each of the solanaceous species was searched with queries (below), and the percentage of identity from the highest tBLASTp score was taken and averaged for all solanaceous species. The TIGR database contained 26,918 N. benthamiana, 29,894 pepper (Capsicum annuum), 8336 petunia (Petunia hybrida), 219,407 potato (Solanum tuberosum), 72,850 tobacco, and 213,947 tomato EST sequences (August, 2007). The accession codes used for the queries are in Supplemental List 2 online. If known, only the domain of the mature protein was taken for tBLASTp searches.
Structural models of RCR3 and PIP1 were created using SWISS-MODEL (Schwede et al., 2003), using papain (PDB code 9papA) as a template. This template was chosen since it was one of the top hits given by the template search by SWISS-MODEL and because it is close to RCR3 and PIP1 in phylogenetic tree (Beers et al., 2004). The models were evaluated using Ramachandran plots and other outlier-detecting software of the MOE package (Molecular Operating Environment; Chemical Computing Group), and amino acid environment was evaluated using VERIFY-3D (Lüthy et al., 1992). All parameters were within the acceptable ranges, and no disturbing outliers were detected.
Sequencing, Analysis of Variant Nucleotides and Residues, and Quantitative RT-PCR
For sequencing protease alleles from wild tomato species, RNA was isolated from (old) leaves (BTH-treated) using the Qiagen RNeasy mini kit, and cDNA was synthesized using Superscript II reverse transcriptase (Invitrogen) using oligo(dT) 20 primer (Invitrogen) according to the instructions of the manufacturer. cDNA was used as a template for PCR using primers at the start and stop codons (see Supplemental Table 1 online). PCR products were sequenced using forward sequencing primers (see Supplemental Table 1 online). Sequences were aligned using Multialign (Corpet, 1988), and variant nucleotides were verified in the trace data. Except for one nucleotide in CatB1 and one in CatB2, all generated sequences from S. lycopersicum MM-Cf0 and S. lycopersicum var cerasiforme were identical to those at the NCBI database. The new sequence of the RCR3pim allele differs at one position compared with the sequence at the NCBI database. The RCR3pen allele was identical to the previously published sequence at the NCBI database. In addition, except for one nucleotide in PIP1, all sequences from the two cheesmaniae species were identical. The polymorphism causing the N194D mutation in RCR3chi was confirmed by sequencing genomic DNA. Taken together, these data underline the confidence of the generated sequences and indicate the presence of some polymorphic sites within some tomato species. Variant codons were analyzed in translation frames for their effect on the encoded amino acid. The ratio of nonsimilar/similar residues was calculated from these data. To superimpose the variant residues onto the papain structure, an alignment between PIP1 and papain and between RCR3 and papain was used to select residues that are variant in PIP1 and RCR3. These residues were colored in the papain structure (1PPP) using PYMOL (DeLano, 2002; http://pymol.sourceforge.net/) (Kim et al., 1992).
For real-time RT-PCR, cDNA was synthesized as described above. Gene-specific primers were designed using Pearl Primer software and validated for specificity. Reaction mixtures for SYBR green (Roche)-based real-time RT-PCR was made as described previously (Karsai et al., 2002). DNA synthesis was recorded with the IQ5 Multicolor Real Time PCR detection system (Bio-Rad). Threshold cycles (Ct) were recorded in triplicate over five independent biological samples, corrected for the Ct of ubiquitin (Rotenberg et al., 2006), and subjected to statistical analysis following the manufacturer's guidelines (Bio-Rad).
Accession Numbers
Accession numbers for the sequences used to construct the phylogenetic tree shown in Figure 3B can be found in Supplemental List 1 online and those used for Figure 3C are in Supplemental List 2 online. Accession numbers for tomato relatives used as a template for sequencing are as follows: S. lycopersicum MM-Cf0; LA0927 (S. cheesmaniae), LA1407 (S. cheesmaniae), LA0442 (S. pimpinellifolium); LA1930 (S. chilense); LA0716 (S. pennellii); LA1777 (S. habrochates/hirsutum); (S. peruvianum); LA1028 (S. schiewlskii); LA1322 (S. parviflorum); and LA1204 (S. lycopersicum var cerasiforme).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. mRNA Accumulation upon BTH Treatment.
Supplemental Figure 2. Activities of Tomato PLCPs upon Agroinfiltration.
Supplemental Table 1. Primers Used in This Study.
Supplemental Table 2. Cloning Procedure of Tomato Proteases into Expression Vectors.
Supplemental List 1. Plant PLCP Accession Numbers Used for Phylogenetic Analysis.
Supplemental List 2. Tomato Accession Numbers Used for pBLASTp Searches.
Supplemental Alignment 1. Alignment of Plant PLCPs Used for Phylogenetic Analysis.
Supplemental Alignment 2. Alignment of Protein Sequences Encoded by PLCP Alleles of Tomato Relatives.
Supplemental Alignment 3. Alignment of Nucleotide Sequences Encoded by PLCP Alleles of Tomato Relatives.
Supplemental Data Set 1. Text File of Alignments Used to Create the Phylogenetic Tree in Figure 3B.
Supplementary Material
Acknowledgments
We thank Jürgen Schmidt, Ursula Wieneke, Florian Kaffarnik, and Alex Jones for their MS support; Klaus Theres and Ali Ahmed Naz for providing seeds and leaf material of wild tomato species; Matt Bogyo and Steven Verhelst for providing TMR-DCG-04; Jonathan Jones for providing Cladosporium isolates and RCR3 antibodies; George Coupland, Paul Schulze-Lefert, and members of the departments of plant-microbe interactions and developmental biology for useful suggestions; and Brande Wulff for critically reading the manuscript. This work was financially supported by the Max Planck Society and the Deutsche Forschungsgemeinschaft (F.K.; DFG project HO3983/3-1). S.K. was supported by the Gatsby Charitable Foundation.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Renier A.L. van der Hoorn (hoorn@mpiz-koeln.mpg.de).
Online version contains Web-only data.
References
- Ahmed, S.U., Rojo, E., Kovaleva, V., Venkataraman, S., Dombrowski, J.E., Matsuoka, K., and Raikhel, N.V. (2000). The plant vacuolar sorting receptor AtELP is involved in transport of NH(2)-terminal propeptide-containing vacuolar proteins in Arabidopsis thaliana. J. Cell Biol. 149 1335–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avrova, A.O., Stewart, H.E., De Jong, W.D., Heilbronn, J., Lyon, G.D., and Birch, P.R. (1999). A cysteine protease is expressed early in resistant potato interactions with Phytophthora infestans. Mol. Plant Microbe Interact. 12 1114–1119. [DOI] [PubMed] [Google Scholar]
- Avrova, A.O., et al. (2004). Potato oxysterol binding protein and cathepsin B are rapidly up-regulated in independent defence pathways that distinguish R gene-mediated and field resistances to Phytophthora infestans. Mol. Plant Pathol. 5 45–56. [DOI] [PubMed] [Google Scholar]
- Bateman, A., and Bennett, H.P.J. (1998). Granulins: The structure and function of an emerging family of growth factors. J. Endocrinol. 158 145–151. [DOI] [PubMed] [Google Scholar]
- Beers, E.P., Jones, A.M., and Dickerman, A.W. (2004). The S8 serine, C1A cysteine and A1 aspartic protease families in Arabidopsis. Phytochemistry 65 43–58. [DOI] [PubMed] [Google Scholar]
- Beliën, T., Van Campenhout, S., Van Acker, M., Robben, J., Courtin, C.M., Delcour, J.A., and Volckaert, G. (2007). Mutational analysis of endoxylanases XylA and XylB from the phytopathogen Fusarium graminearum reveals comprehensive insights into their inhibitor insensitivity. Appl. Environ. Microbiol. 73 4602–4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop, J.G., Ripoll, D.R., Bashir, S., Damasceno, C.M.B., Seeds, J.D., and Rose, J.K.C. (2005). Selection on Glycine β-1,3-endoglucanase genes differentially inhibited by a Phytophthora glucanase inhibitor protein. Genetics 169 1009–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomblies, K., Lempe, J., Epple, P., Warthmann, N., Lanz, C., Dangl, J.L., and Weigel, D. (2007). Autoimmune response as a mechanism for a Dobzhansky-Muller-type incompatibility syndrome in plants. PLoS Biol. 5 1962–1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomblies, K., and Weigel, D. (2007). Hybrid necrosis: Autoimmunity as a potential gene-flow barrier in plant species. Nat. Rev. Genet. 8 382–393. [DOI] [PubMed] [Google Scholar]
- Carter, C., Pan, S., Zouhar, J., Avila, E.L., Girke, T., and Raikhel, N.V. (2004). The vegetative vacuole proteome of Arabidopsis thaliana reveals predicted and unexpected proteins. Plant Cell 16 3285–3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet, F. (1988). Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16 10881–10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano, W.L. (2002). The PyMOL User's Manual. (Palo Alto, CA: DeLano Scientific).
- Drake, R., John, I., Farrell, A., Cooper, W., Schuch, W., and Grierson, D. (1996). Isolation and analysis of cDNAs encoding tomato cysteine proteases expressed during leaf senescence. Plant Mol. Biol. 30 755–767. [DOI] [PubMed] [Google Scholar]
- Felsenstein, J. (1989). PHYLIP - Phylogeny Inference Package (Version 3.2). Cladistics 5 164–166. [Google Scholar]
- Ferreira, R.B., Monteiro, S., Freitas, R., Santos, C.N., Chen, Z., Batista, L.M., Duarte, J., Borges, A., and Teixeira, A.R. (2007). The role of plant defence proteins in fungal pathogenesis. Mol. Plant Pathol. 8 677–700. [DOI] [PubMed] [Google Scholar]
- Gilroy, E.M., et al. (2007). Involvement of cathepsin B in the plant disease resistance hypersensitive response. Plant J. 52 1–13. [DOI] [PubMed] [Google Scholar]
- Gobom, J., Schuerenberg, M., Mueller, M., Theiss, D., Lehrach, H., and Nordhoff, E. (2001). Alpha-cyano-4-hydroxycinnamic acid affinity sample preparation. A protocol for MALD-MS peptide analysis in proteomics. Anal. Chem. 73 434–438. [DOI] [PubMed] [Google Scholar]
- Greenbaum, D., Baruch, A., Hayrapetian, L., Darula, Z., Burlingame, A., Medzihradszky, K.F., and Bogyo, M. (2002). Chemical approaches for functionally probing the proteome. Mol. Cell. Proteomics 1 60–68. [DOI] [PubMed] [Google Scholar]
- Harrak, H., Azelmat, S., Baker, E.N., and Tabaeizadeh, Z. (2001). Isolation and characterisation of a gene encodiong a drought-induced cysteine protease in tomato (Lycopersicon esculentum). Genome 44 368–374. [DOI] [PubMed] [Google Scholar]
- Hayashi, Y., Yamada, K., Shimada, T., Matsushima, R., Nishizawa, N.K., Nishimura, M., and Hara-Nishimura, I. (2001). A proteinase-storing body that prepares for cell death or stresses in the epidermal cells of Arabidopsis. Plant Cell Physiol. 42 894–899. [DOI] [PubMed] [Google Scholar]
- Higgins, D., Thompson, J., and Gibson, T. (1994). CLUSTRAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holwerda, B.C., Padgett, H.S., and Rogers, J.C. (1992). Proaleurain vacuolar targeting is mediated by short contiguous peptide interactions. Plant Cell 4 307–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.D.G., and Dangl, J.L. (2006). The plant immune system. Nature 444 323–329. [DOI] [PubMed] [Google Scholar]
- Joosten, M.H.A.J., and De Wit, P.J.G.M. (1989). Identification of several pathogenesis-related proteins in tomato leaves inoculated with Cladosporium fulvum (syn. Fulvia fulva) as 1,3-β-glucanases and chitinases. Plant Physiol. 89 945–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamoun, S., and Smart, C.D. (2005). Late blight of potato and tomato in the genomics era. Plant Dis. 89 692–699. [DOI] [PubMed] [Google Scholar]
- Karsai, A., Müller, S., Platz, S., and Hauser, M.T. (2002). Evaluation of a home-made SYBR Green I reaction mixture for real-time PCR quantification of gene expression. Biotechniques 32 790–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M.-J., Yamamoto, D., Matsumoto, K., Inoue, M., Ishida, T., Mizuno, H., Sumiya, S., and Katamura, K. (1992). Crystal structure of papain-E64-c complex. Biochem. J. 287 797–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kombrink, E., Schröder, M., and Halbrock, K. (1988). Several ‘pathogenesis-related’ proteins in potato are 1,3-β-glucanases and chitinases. Proc. Natl. Acad. Sci. USA 85 782–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger, J., Thomas, C.M., Golstein, C., Dixon, M.S., Smoker, M., Tang, S., Mulder, L., and Jones, J.D.G. (2002). A tomato cysteine protease required for Cf-2-dependent disease resistance and suppression of autonecrosis. Science 296 744–747. [DOI] [PubMed] [Google Scholar]
- Lennon-Dumenil, A.M., Bakker, A.H., Maer, R., Fiebiger, E., Overkleeft, H.S., Rosemblatt, M., Ploegh, H.L., and Lagaudriere-Gesbert, C. (2002). Analysis of protease activity in live antigen-presenting cells shows regulation of the phagosomal proteolytic contents during dendritic cell activation. J. Exp. Med. 196 529–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luderer, R., Takken, F.L.W., De Wit, P.J.G.M., and Joosten, M.H.A.J. (2002). Cladosporium fulvum overcomes Cf-2-mediated resistance by producing truncated AVR2 elicitor proteins. Mol. Microbiol. 45 875–884. [DOI] [PubMed] [Google Scholar]
- Lüthy, R., Bowie, J.U., and Eisenberg, D. (1992). Assessment of protein models with three-dimensional profiles. Nature 356 83–85. [DOI] [PubMed] [Google Scholar]
- Misas-Villamil, J.C., and Van der Hoorn, R.A.L. (2008). Enzyme-inhibitor interactions at the plant-pathogen interface. Curr. Opin. Plant Biol. 11 in press. [DOI] [PubMed]
- Raiola, A., Sella, L., Castiglioni, C., Balmas, V., and Favaron, F. (2008). A single amino acid substitution in highly similar endo-PGs from Fusarium verticillioides and related Fusarium species affects PGIP inhibition. Fungal Genet. Biol. 45 776–789. [DOI] [PubMed] [Google Scholar]
- Rivas, S., and Thomas, C.M. (2005). Molecular interactions between tomato and the leaf mold pathogen Cladosporium fulvum. Annu. Rev. Phytopathol. 43 395–436. [DOI] [PubMed] [Google Scholar]
- Rooney, H.C.E., Van't Klooster, J.W., van der Hoorn, R.A.L., Joosten, M.H.A.J., Jones, J.D., and De Wit, P.J.G.M. (2005). Cladosporium Avr2 inhibits tomato Rcr3 protease required for Cf-2-dependent disease resistance. Science 308 1783–1786. [DOI] [PubMed] [Google Scholar]
- Rose, J.K., Ham, K.S., Darvill, A.G., and Albersheim, P. (2002). Molecular cloning and characterisation of glucanase inhibitor proteins: coevolution of a counterdefense mechanism by plant pathogens. Plant Cell 14 1329–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotenberg, D., Thompson, T.S., German, T.L., and Willis, D.K. (2006). Methods for effective real-time RT-PCR analysis of virus-induced gene silencing. J. Virol. Methods 138 49–59. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T.A. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Sanz-Alferez, S., Mateos, B., Alvarado, R., and Sanches, M. (2008). SAR induction in tomato plants is not effective against root-knot nematode infection. Eur. J. Plant Pathol. 120 417–425. [Google Scholar]
- Schaffer, M.A., and Fischer, R.L. (1988). Analysis of mRNAs that accumulate in response to low temperature identifies a thiol protease gene in tomato. Plant Physiol. 87 431–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer, M.A., and Fischer, R.L. (1990). Transcriptional activation by heat and cold of a thiol protease gene in tomato. Plant Physiol. 93 1486–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwede, T., Kopp, J., Guex, N., and Peitsch, M.C. (2003). SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 31 3381–3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo, T., and Van der Hoorn, R.A.L. (2008). Papain-like cysteine proteases: Key players at molecular battlefields employed by both plants and their invaders. Mol. Plant Pathol. 9 119–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stotz, H.U., Bishop, J.G., Bergmann, C.W., Koch, M., Albersheim, P., Darvill, A.G., and Labavitch, J.M. (2000). Identification of target amino acids that affect interactions of fungal polygalacturonases and their plant inhibitors. Physiol. Mol. Plant Pathol. 56 117–130. [Google Scholar]
- Thomma, B.P.H.J., Van Esse, H.P., Vrous, P.W., and De Wit, P.J.G.M. (2005). Cladosporium fulvum (syn. Passalora fulva), a highly specialized plant pathogen as a model for functional studies on plant pathogenic Mycosphaerellaceae. Mol. Plant Pathol. 6 379–393. [DOI] [PubMed] [Google Scholar]
- Tian, M., Benetti, B., and Kamoun, S. (2005). A second Kazal-like protease inhibitor from Phytophthora infestans inhibits and interacts with the apoplastic pathogenesis-related protease P69B of tomato. Plant Physiol. 138 1785–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, M., Huitema, E., Da Cunha, L., Torto-Alalibo, T., and Kamoun, S. (2004). A Kazal-like extracellular serine protease inhibitor from Phytophthora infestans targets the tomato pathogenesis-related protease P69B. J. Biol. Chem. 279 26370–26377. [DOI] [PubMed] [Google Scholar]
- Tian, M., Win, J., Van der Hoorn, R., Van der Knaap, E., and Kamoun, S. (2007). A Phytophthora infestans cystatin-like protein targets a novel tomato papain-like apoplastic protease. Plant Physiol. 143 364–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornero, P., Conejero, V., and Vera, P. (1997). Identification of a new pathogen-induced member of the subtilisin-like processing protease family from plants. J. Biol. Chem. 272 14412–14419. [DOI] [PubMed] [Google Scholar]
- Van den Burg, H.A., Harrison, S.J., Joosten, M.H.A.J., Vervoort, J., and De Wit, P.J.G.M. (2006). Cladosporium fulvum Avr4 protects fungal cell walls against hydrolysis by plant chitinases accumulating during infection. Mol. Plant Microbe Interact. 19 1420–1430. [DOI] [PubMed] [Google Scholar]
- Van der Hoorn, R.A.L., De Wit, P.J.G.M., and Joosten, M.H.A.J. (2002). Balancing selection favors guarding resistance proteins. Trends Plant Sci. 6 67–71. [DOI] [PubMed] [Google Scholar]
- Van der Hoorn, R.A.L., and Jones, J.D.G. (2004). The plant proteolytic machinery and its role in defence. Curr. Opin. Plant Biol. 7 400–407. [DOI] [PubMed] [Google Scholar]
- Van der Hoorn, R.A.L., Laurent, F., Roth, R., and De Wit, P.J.G.M. (2000). Agroinfiltration is a versatile tool that facilitates comparative analysis of Avr9/Cf-9-induced and Avr4/Cf-4-induced necrosis. Mol. Plant Microbe Interact. 13 439–446. [DOI] [PubMed] [Google Scholar]
- Van der Hoorn, R.A.L., Leeuwenburgh, M.A., Bogyo, M., Joosten, M.H.A.J., and Peck, S.C. (2004). Activity profiling of papain-like cysteine proteases in plants. Plant Physiol. 135 1170–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Hoorn, R.A.L., Wulff, B., Rivas, S., Durrant, M.C., Van der Ploeg, A., De Wit, P.J.G.M., and Jones, J.D.G. (2005). Structure-function analysis of Cf-9, a receptor-like protein with extracytoplasmic leucine-rich repeats. Plant Cell 17 1000–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Esse, H.P., Bolton, M.D., Stergiopoulos, I., De Wit, P.J.G.M., and Thomma, B.P.H.J. (2007). The chitin-binding Cladosporium fulvum effector protein Avr4 is a virulence factor. Mol. Plant Microbe Interact. 20 1092–1101. [DOI] [PubMed] [Google Scholar]
- Van Loon, L.C., Rep, M., and Pieterse, C.M.J. (2006). Significance of inducible defence-related proteins in infected plants. Annu. Rev. Phytopathol. 44 135–162. [DOI] [PubMed] [Google Scholar]
- Voinnet, O., Rivas, S., Mestre, P., and Baulcombe, D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33 949–956. [DOI] [PubMed] [Google Scholar]
- Watanabe, E., Shimada, T., Tamura, K., Matsushima, R., Koumoto, Y., Nishimura, M., and Hara-Nishimura, I. (2004). An ER-localized form of PV72, a seed-specific vacuolar sorting receptor, interferes the transport of an NPIR-containing proteinase in Arabidopsis leaves. Plant Cell Physiol. 45 9–17. [DOI] [PubMed] [Google Scholar]
- Wulff, B.B.H., Kruijt, M., Collins, P.L., Thomas, C.M., Ludwig, A.A., De Wit, P.J.G.M., and Jones, J.D.G. (2004). Gene shuffling-generated and natural variants of the tomato resistance gene Cf-9 exhibit different auto-necrosis-inducing activities in Nicotiana species. Plant J. 40 942–956. [DOI] [PubMed] [Google Scholar]
- Xia, Y., Suzuki, H., Borevitz, J., Blount, J., Guo, Z., Patel, K., Dixon, R.A., and Lamb, C. (2004). An extracellular aspartic protease functions in Arabidopsis disease resistance signaling. EMBO J. 23 980–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada, K., Matsushima, R., Nishimura, M., and Hara-Nishimura, I. (2001). A slow maturation of a cysteine protease with a granulin domain in the vacuoles of senescing Arabidopsis leaves. Plant Physiol. 127 1626–1634. [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y., Thilmony, R., Bender, C.L., Schaller, A., He, S.Y., and Howe, G.A. (2003). Virulence systems of Pseudomonas syringae pv. tomato promote bacterial speck disease in tomato by targeting the jasmonate signaling pathway. Plant J. 36 485–499. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.