Abstract
Plastid genes are expressed at high levels in photosynthetically active chloroplasts but are generally believed to be drastically downregulated in nongreen plastids. The genome-wide changes in the expression patterns of plastid genes during the development of nongreen plastid types as well as the contributions of transcriptional versus translational regulation are largely unknown. We report here a systematic transcriptomics and translatomics analysis of the tomato (Solanum lycopersicum) plastid genome during fruit development and chloroplast-to-chromoplast conversion. At the level of RNA accumulation, most but not all plastid genes are strongly downregulated in fruits compared with leaves. By contrast, chloroplast-to-chromoplast differentiation during fruit ripening is surprisingly not accompanied by large changes in plastid RNA accumulation. However, most plastid genes are translationally downregulated during chromoplast development. Both transcriptional and translational downregulation are more pronounced for photosynthesis-related genes than for genes involved in gene expression, indicating that some low-level plastid gene expression must be sustained in chromoplasts. High-level expression during chromoplast development identifies accD, the only plastid-encoded gene involved in fatty acid biosynthesis, as the target gene for which gene expression activity in chromoplasts is maintained. In addition, we have determined the developmental patterns of plastid RNA polymerase activities, intron splicing, and RNA editing and report specific developmental changes in the splicing and editing patterns of plastid transcripts.
INTRODUCTION
Tomato (Solanum lycopersicum, formerly Lycopersicon esculentum) is one of the world's most important vegetable crops and has long been a classical model species for plant genetics. It belongs to the Solanaceae (nightshade) family and has its center of genetic diversity in South America. Recently, a tomato genome project was initiated by the international Solanaceae Genomics Network (SOL Genomics Network; http://www.sgn.cornell.edu/). The tomato nuclear genome has a haploid set of 12 chromosomes and comprises ∼950 Mb.
Among the three genomes of higher plants, the plastid genome (plastome) is the most gene-dense, with >100 genes in a genome of only 120 to 210 kb (for review, see Sugiura, 1992; Wakasugi et al., 2001). The genes encoded by the plastome fall into three major classes: (1) genetic system genes, including genes for a complete set of rRNAs and tRNAs, some ribosomal proteins, and the core subunits of a eubacterial-type of RNA polymerase; (2) photosynthesis-related genes; and (3) other genes and conserved open reading frames (Shimada and Sugiura, 1991). As a first contribution toward deciphering the complete genetic information of tomato, we recently determined the complete sequence of the tomato plastome (Kahlau et al., 2006). This 155,461-bp molecule maps as a circular genome and harbors 114 genes and conserved open reading frames (ycfs, for hypothetical chloroplast reading frames).
The genome organization and mechanisms of gene expression in plastids very much resemble those of their cyanobacterial ancestors. Clusters of genes are arranged in operons and cotranscribed as polycistronic mRNAs, translation occurs on 70S ribosomes, and most components of the translational apparatus have homologs in eubacteria (Zerges, 2000). Plastid primary transcripts undergo a complex series of processing and maturation steps, including cleavage of polycistronic mRNAs into monocistronic or oligocistronic units (also referred to as RNA cutting), 5′ and 3′ end processing, intron splicing, and RNA editing by C-to-U conversions (Barkan and Goldschmidt-Clermont, 2000; Bock, 2000).
In spite of the small size of the plastome, the transcriptional apparatus of plastids is much more complex than that of prokaryotes and has a dual evolutionary origin. A plastid-encoded RNA polymerase (PEP) is homologous with eubacterial RNA polymerases (Igloi and Kössel, 1992; Hess and Börner, 1999). A second RNA-synthesizing activity in the plastid is provided by bacteriophage-type enzymes that are encoded by nuclear genes (nucleus-encoded RNA polymerase [NEP]) (Hajdukiewicz et al., 1997; Hedtke et al., 1997; Hess and Börner, 1999; Filée and Forterre, 2005). While the four core subunits of the eubacterial-type, Escherichia coli-like, plastid RNA polymerase are encoded by the plastome, the σ factors, which are required for promoter recognition, are encoded by genes residing in the nuclear genome (Tanaka et al., 1996). The unicellular green alga Chlamydomonas reinhardtii has a single nuclear gene for a plastid σ factor (Bohne et al., 2006), which developed into a gene family in higher plants (Kanamaru et al., 1999; Lahiri et al., 1999). The members of this gene family confer promoter specificity of PEP transcription in that individual σ factors mediate mRNA synthesis from distinct (groups of) plastid genes (Kanamaru et al., 2001; Hanaoka et al., 2003; Tsunoyama et al., 2004; Favory et al., 2005). Adding to the complexity of the plastid's transcriptional machinery, PEP and NEP transcribe overlapping sets of genes. Some plastid genes are predominantly transcribed from either PEP or NEP, but a significant number of genes have promoters for both RNA polymerases (Allison et al., 1996; Hajdukiewicz et al., 1997; Lerbs-Mache, 2000; Legen et al., 2002).
Plastid gene expression is also extensively regulated at the posttranscriptional level, most importantly at the level of protein biosynthesis (Gillham et al., 1994; Bruick and Mayfield, 1999; Zerges, 2000). For most plastid genes in Chlamydomonas, translation initiation appears to be the most important step at which gene expression is regulated (Choquet and Wollman, 2002; Eberhard et al., 2002). The relative contributions of transcriptional and translational regulation to the control of plastid gene expression have been somewhat controversial. While some studies suggest a significant contribution of transcriptional regulation (Pfannschmidt et al., 1999; Tullberg et al., 2000), other studies conclude that translational regulation largely overrides fluctuations in mRNA levels and thus translation constitutes the rate-limiting step in the expression of nearly all plastid protein-coding genes (Eberhard et al., 2002).
Plastid gene expression and its regulation have been intensely studied in chloroplasts, the plastid type present in photosynthetically active tissues. By contrast, knowledge about gene expression in nongreen plastid types has remained scarce. Since a large fraction of plastid protein-coding genes is involved in photosynthesis, it is generally believed that plastid gene expression is drastically downregulated in nonphotosynthetic tissues (Piechulla et al., 1986, 1987). Ripe tomato fruits contain chromoplasts, carotenoid-accumulating plastids. Chloroplasts are present in green tomatoes and are then converted into chromoplasts during fruit ripening. The plastome in chromoplasts is still expressed. Ripe tomato fruits were shown to accumulate plastid rRNAs and at least some mRNAs and to carry out active protein biosynthesis, although protein accumulation levels are usually much lower than in photosynthetically active tissues (Bathgate et al., 1985; Piechulla et al., 1985, 1986). Likewise, chromoplasts in bell pepper (Capsicum annuum) fruits accumulate both plastid-encoded mRNAs and some proteins at low levels (Gounaris and Price, 1987; Kuntz et al., 1989) and are translationally active (Powell and Pryke, 1987).
The objective of this study was to systematically analyze the expression of the tomato plastid genome and to identify regulatory patterns of gene expression in different tissues and plastid types. To separate tissue-specific regulation from regulation triggered by plastid differentiation (during the chloroplast-to-chromoplast conversion), we have (1) compared gene expression in fruits with that in leaves and (2) analyzed a developmental series during tomato fruit ripening. In addition to a plastome-wide analysis of the transcriptome and the translatome, we have also determined developmental changes in two key RNA-processing steps: intron splicing and RNA editing. Our data reveal the contributions of transcriptional, posttranscriptional, and translational processes to the regulation of plastid gene expression. They also uncover a single plastid gene, accD, as probably the only protein-coding gene in the plastome for which plastid gene expression is maintained in chromoplasts. In addition, our data sets identify expression elements suitable for triggering high-level transgene expression from the plastid genome in fruit chromoplasts.
RESULTS
Design of an Oligonucleotide Microarray to Analyze Plastid Gene Expression in Solanaceae
Solanaceous plants have become important model species for studying plastid biology, not the least because their plastid genomes are amenable to genetic manipulation by chloroplast transformation (Svab and Maliga, 1993; Sidorov et al., 1999; Ruf et al., 2001). Knowledge about the regulation of plastid gene expression in response to developmental programs, environmental stimuli, and genetic perturbations is central to our understanding of photosynthesis and other plastid-localized metabolic pathways, plastid–nucleus communication by anterograde and retrograde signaling, and the application of plastid transformation in functional genomics and biotechnology. Therefore, we were interested in developing a plastome microarray for Solanaceae. As our primary interest was to provide a microarray that was applicable to the three most important solanaceous model species, tobacco (Nicotiana tabacum), tomato, and potato (Solanum tuberosum), we used the sequenced plastomes from those species to design an oligonucleotide microarray. We preferred an oligonucleotide array over a cDNA-based microarray for several reasons. First, an oligonucleotide microarray allows the strand-specific detection of transcripts. This is a significant advantage, because transcription from divergent promoters resulting in overlapping complementary transcripts is frequently observed in plastids (Kohchi et al., 1988; Haley and Bogorad, 1990; Meng et al., 1991; Vera et al., 1992). Second, an oligonucleotide microarray is much less prone to contamination and artifacts caused by nonspecific background amplification.
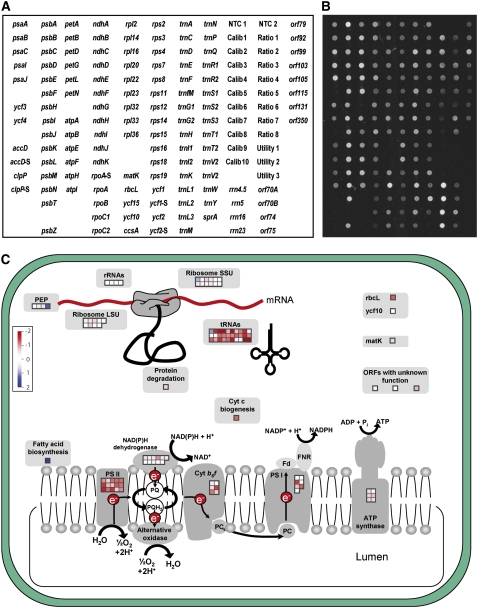
An oligonucleotide microarray containing all genes and conserved open reading frames present in solanaceous plastid genomes was designed using the complete sequences of the tobacco, tomato, and potato plastid genomes (Yukawa et al., 2005; Daniell et al., 2006; Kahlau et al., 2006). The lengths of the oligonucleotides were between 68 and 71 nucleotides, and a few additional species-specific oligonucleotides were added for poorly conserved genes (see Methods for details) (Figure 1A; see Supplemental Data Set 1 online). To facilitate the quantitation of hybridization signal intensities, a series of calibration and reference samples was spotted on each microarray (Figure 1A). All RNA samples analyzed were spiked with defined amounts of the corresponding reference RNAs, and extensive test hybridizations confirmed that this provided a reliable method for obtaining highly reproducible quantitative data (Figures 1A and 1B).
Figure 1.
Design of a Plastome Microarray for Solanaceous Plants.
(A) Spotting scheme of oligonucleotide probes on the array. The array contains all genes and conserved open reading frames (ycfs) contained in solanaceous plastid genomes and, in addition, a number of nonconserved open reading frames (orfs), which, however, are unlikely to represent functional genes (Kahlau et al., 2006). Oligonucleotides are 68 to 71 nucleotides long. Poorly conserved genes are covered by more than one oligonucleotide (indicated by the suffix -S for Solanum-specific oligonucleotide). NTC, negative control to assess nonspecific background hybridization; Calib, Ratio, and Utility, calibration and reference DNAs (artificial sequences corresponding to the Lucidea Universal ScoreCard; Amersham Biosciences). See Methods for details.
(B) Example of a microarray hybridization experiment with total leaf RNA showing that all genes and open reading frames are detected at sufficient sensitivity.
(C) Graphical visualization of data sets using a modified version of the MapMan software (Thimm et al., 2004). All processes in which plastome-encoded gene products participate are illustrated by pictograms containing a set of boxes corresponding to the genes involved in the respective process. The order of the boxes reflects the order of the genes on the array. For example, the first row of gene boxes for ribosomal proteins of the small ribosomal subunit represents the genes rps2, rps3, rps4, rps7, rps8, and rps11, and the second row represents rps12, rps14, rps15, rps16, rps18, and rps19. Red indicates downregulated, and blue indicates upregulated, expression. Expression changes are displayed on a log2 scale. The data set shown here as an example is a comparison of RNA accumulation in light red fruits and green fruits.
Two options exist for visualization of the data obtained with our microarrays. The TIGR (for The Institute for Genomic Research) MeV software (MultiExperiment Viewer; Saeed et al., 2003) can be used to display simple heat maps listing all plastome-encoded genes, providing a global overview of the expression changes (Figures 2 and 3). To be able to evaluate changes in plastid gene expression in a more pathway-dependent manner, we also adapted the MapMan software (Thimm et al., 2004) with a plastid genome output mask (downloadable at http://gabi.rzpd.de/database/java-bin/MappingDownloader). This provides a simple tool for visualizing the specific changes in different gene classes that reflect processes involving plastid-encoded genes (Figure 1C).
Figure 2.
Transcriptomics Analysis of Tomato Plastid Genome Expression.
(A) and (B) Transcriptomics of fruit development. Changes in RNA accumulation in the different stages of fruit ripening (green, turning, light red, and red, as schematically depicted above the displayed array data) are shown relative to the RNA accumulation levels in green leaves. Levels of downregulation (red) or upregulation (blue) are shown on a log2 scale from −8 to +8 (corresponding to a 256-fold change in expression from 0 to −8 and 0 to +8, respectively). The total data set is displayed in (A); the data set shown in (B) is limited to those changes that are statistically significant according to the tests described in Methods.
(C) and (D) Transcriptomics of chloroplast-to-chromoplast differentiation in fruit ripening. Changes in RNA accumulation during chromoplast development (turning, light red, and red ripening stages) are shown relative to the RNA accumulation levels in green, chloroplast-containing fruits. The total data set is displayed in (C); the data set shown in (D) is limited to those changes that are statistically significant according to the tests described in Methods. All data shown in (A) to (D) represent means of three independent experiments (RNA isolations).
(E) Confirmation of selected changes in transcript abundance by RNA gel blot analyses. Equal amounts of RNA extracted from green leaves and the four stages of fruit ripening were separated by denaturing agarose gel electrophoresis, blotted, and hybridized to radiolabeled probes specific for psbB, rpoB, matK, psbD, and accD. To control for equal loading, the ethidium bromide–stained agarose RNA gels are shown below each blot. The plant material used for these RNA extractions was generated independently from the materials used for the three microarray experiments.
Figure 3.
Translatomics Analysis of Tomato Plastid Genome Expression by Measuring the Levels of Polysome-Associated mRNAs.
(A) and (B) Translatomics of fruit development. Changes in the polysome association of all plastid protein-coding genes are shown for the different stages of fruit ripening (see Figure 2) relative to the translation levels in green leaves. Blue indicates upregulation and red indicates downregulation. Expression changes are displayed on a log2 scale from −8 to +8 (corresponding to a 256-fold change in expression from 0 to −8 and 0 to +8, respectively). The total data set is displayed in (A); the data set shown in (B) is limited to those changes that are statistically significant according to the tests described in Methods.
(C) and (D) Translatomics of chloroplast-to-chromoplast differentiation in fruit ripening. Changes in polysome association during chromoplast development (turning, light red, and red ripening stages) are shown relative to the translation rates in green, chloroplast-containing tomato fruits. The total data set is displayed in (C); the data set shown in (D) is limited to those changes that are statistically significant according to the tests described in Methods. All data shown in (A) to (D) represent means of two independent experiments (polysome isolations).
(E) Confirmation of selected changes in polysome association by RNA gel blot analyses. Polysome gradients were fractionated into six fractions, and equal aliquots of extracted RNAs were separated by denaturing agarose gel electrophoresis, blotted, and hybridized to radiolabeled probes specific for accD and psbD. The wedges above each blot indicate the gradient from low to high sucrose concentration. As a control, a sample was treated with puromycin to cause dissociation of ribosomes from the mRNAs. The ethidium bromide–stained agarose RNA gels are shown below each blot. The plant material used for these polysome preparations was generated independently from the materials used for the two microarray experiments.
Transcriptional Regulation
To assess the relative contributions of transcriptional and translation regulation to the changes in plastid gene expression upon fruit development and chloroplast-to-chromoplast conversion, we first determined the changes in RNA accumulation for all plastid genome–encoded genes and open reading frames. In addition to green leaves, we analyzed a developmental series of tomato fruits (green, turning, light red, and ripe red tomatoes; Figure 2). All developmental stages were analyzed in triplicate, each representing an independent RNA isolation.
We recorded the changes in plastid gene expression in two ways. First, we used the expression levels in leaves as a reference and, in this way, analyzed the changes in the expression of all genes in fruits compared with leaves (Figures 2A and 2B). This analysis determined the changes triggered by the fruit developmental program. In order to dissect the relative contributions of fruit development and plastid differentiation, we also used the expression levels in green, chloroplast-containing fruits as a reference (Figures 2C and 2D). This allowed us to identify those changes that are induced by chloroplast-to-chromoplast differentiation and thus are independent of the initiation of the fruit developmental program.
When gene expression in fruits was compared with that in leaves, it became apparent that most plastid genes are strongly downregulated in fruits. This is already the case in green fruits, suggesting that the downregulation is triggered by the developmental program rather than the chloroplast-to-chromoplast conversion. Interestingly, the photosynthesis-related genes were much more strongly downregulated than the genetic system genes, the latter including, for example, the ribosomal proteins and the subunits of the plastid-encoded E. coli–type RNA polymerase PEP (Figures 2A and 2B; the photosynthesis-related genes are at the top, ranging from psbA to ycf10, and the genetic system genes then follow from rpoA to clpP and may additionally include the conserved reading frames ycf1 and ycf2) (Drescher et al., 2000). Many tRNAs were also strongly downregulated, which would be consistent with a drastically reduced demand for charged tRNAs upon downregulation of the expression of the photosynthetic apparatus (Figures 2A and 2B). The fact that the transcripts of nearly all plastid genes are strongly downregulated in fruits compared with leaves (Figures 2A and 2B) suggests that the ratio of total plastid RNA to total cytosolic RNA may decrease in fruits, possibly reflecting a reduced demand for plastid gene expression capacity.
Interestingly, the chloroplast-to-chromoplast conversion was not accompanied by drastic changes in transcript abundance (Figures 2C and 2D). For most plastid genes, only minor (statistically insignificant) changes were seen, suggesting that, at the level of RNA accumulation, plastid differentiation has a much lower impact on the regulation of plastid gene expression than the developmental program. Notable exceptions include the trnA gene (encoding the tRNA-Ala) and the rpoC2 gene (encoding an RNA polymerase subunit), which tended to be upregulated. Remarkably, the protein-coding gene that displayed the strongest change in expression was accD, which encodes a subunit of the acetyl-CoA carboxylase and is the only known plastid-encoded gene involved in fatty acid biosynthesis. Its transcript abundance increased during the chloroplast-to-chromoplast conversion (Figure 2D), correlating with the high demand for lipid biosynthesis in fruit ripening to provide the storage matrix for the accumulating carotenoids.
To confirm that the microarray data faithfully represented the developmental changes occurring in mRNA accumulation, selected genes were analyzed by RNA gel blot experiments (using plant material generated independently from the material used in the microarray experiments; Figure 2E). We tested two photosynthesis genes (psbB and psbD), two genetic system genes (rpoB and matK), and accD, the gene displaying the strongest upregulation during fruit ripening. In all cases, the mRNA accumulation patterns seen in the RNA gel blots corresponded exactly to the data obtained by microarray analyses, confirming that our microarray reliably detected changes in plastid gene expression patterns.
Translational Regulation
Many, if not all, plastid genes are regulated at the translational level, and it has been suggested that this translational regulation may largely override changes in RNA accumulation (Eberhard et al., 2002). To assess the relevance of the RNA accumulation patterns determined by our microarray analyses with extracted RNA samples, we explored the translational regulation of plastid genes upon fruit development and chromoplast differentiation. We purified polysomes (mRNAs loaded with ribosomes), which can be separated from free mRNAs in sucrose density gradients and represent the fraction of transcripts that is actively translated. We analyzed the same developmental series as for the total RNA, analyzing all stages in duplicate, each experiment representing an independent polysome isolation (Figure 3). To separate the changes triggered by the initiation of the fruit developmental program from those induced by the chloroplast-to-chromoplast conversion, we evaluated the data using two different data sets as reference: (1) the polysome data set for leaves (Figures 3A and 3B) and (2) the polysome data set for green fruits (Figures 3C and 3D).
When the polysome association of plastid mRNAs in fruits was compared with that in leaves, a strong downregulation was seen for most plastid genes (Figures 3A and 3B). However, there were two notable exceptions. Three of the four plastid-encoded subunits of the E. coli–like RNA polymerase (PEP) were only weakly downregulated in fruits (rpoB, rpoC1, and rpoC2; Figures 3A and 3B). The only gene whose polysome association remained essentially unchanged during fruit development was accD, the acetyl-CoA carboxylase subunit gene that we had found to be the most strongly upregulated gene at the RNA level during the chloroplast-to-chromoplast conversion (Figure 2D).
While our analysis of plastid RNA accumulation revealed no significant overall changes during chromoplast development (Figure 2D), there was a pronounced general trend in polysome association (Figures 3C and 3D). Polysome association successively declined during fruit ripening, suggesting that, while the RNA levels remain largely constant, plastid translation is gradually downregulated during chloroplast-to-chromoplast differentiation. This trend was particularly pronounced in the photosynthesis gene group (genes psbA to ycf10; Figures 3C and 3D), correlating with the gradual loss of photosynthesis during the chloroplast-to-chromoplast conversion. A single exception was seen: accD, whose polysome association did not only stay high in fruits (Figures 3A and 3B) but even increased at the onset of ripening (Figures 3C and 3D).
In order to confirm that accD is actively translated in chromoplasts, we analyzed fractionated polysome gradients by RNA gel blotting (Figure 3E). As a control for an mRNA that is translationally downregulated during fruit ripening, we also determined ribosome association for psbD (Figure 3E). These experiments revealed that, while accD was still strongly loaded with ribosomes even in ripe red tomatoes, the ribosome association of the psbD mRNA declined gradually during ripening and was almost undetectable in red fruits. The translational upregulation of accD during the onset of the chloroplast-to-chromoplast conversion (transition from the green to the turning stage) was seen equally clearly in both the microarray analysis and the RNA gel blot experiments (cf. Figures 3C and 3D with Figure 3E).
It must be borne in mind that ribosome association does not necessarily correlate directly with translational regulation. This is because any change in mRNA accumulation can potentially translate into a corresponding change in polysome association. For example, in the extreme case that solely the amount of transcript limits gene expression, a transcriptional upregulation would be seen proportionally as an increase in polysome association, simply because any available mRNA molecule would become translated. Thus, the polysome association data reflect the net gene expression levels in that they directly correlate with the total amount of protein synthesized. In this sense, the data set presented in Figure 3 provides an overview of the changes in plastid gene expression during fruit development. Three main conclusions can be drawn. First, the expression of all photosynthesis-related genes is already drastically downregulated in green, chloroplast-containing fruits and declines further during fruit ripening and chloroplast-to-chromoplast differentiation. Second, the genetic system genes (i.e., genes involved in gene expression) show a similar trend, although the downregulation is somewhat less intense than for the photosynthesis genes (Figures 3A and 3B). Third, the only protein-coding gene that is expressed at constitutively high levels throughout fruit development is the fatty acid biosynthesis gene accD (Figures 3A and 3B). This raises the possibility that the main reason for maintaining some protein biosynthetic capacity in fruit plastids is to sustain fatty acid biosynthesis.
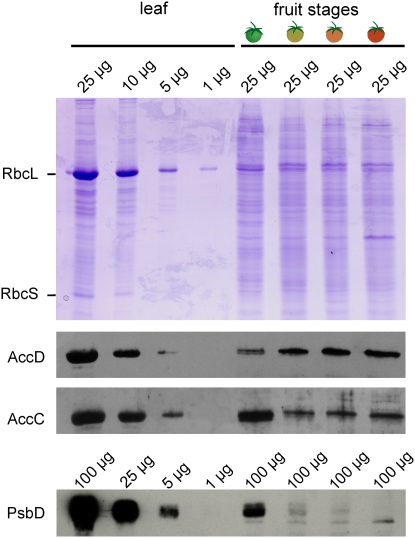
To confirm the significance of the sustained accD expression in fruit ripening at the protein level, protein gel blot analyses with antibodies against the AccD protein were conducted. These analyses revealed that the AccD protein accumulated in all fruit ripening stages and even increased slightly during the onset of chloroplast-to-chromoplast conversion (transition from the green to the turning stage; Figure 4). By contrast, PsbD, a photosystem II protein, was only detectable in green, chloroplast-containing tomato fruits (Figure 4). The acetyl-CoA carboxylase enzyme consists of four subunits, three of which are encoded in the nuclear genome (AccA, AccB, and AccC) and one of which is encoded in the plastid genome (AccD). To test whether the nucleus-encoded subunits also accumulate in chromoplasts, protein gel blot analysis with an anti-AccC antibody was performed. As expected, AccC protein accumulation was seen in all fruit ripening stages (Figure 4), suggesting that both the plastid-encoded and the nucleus-encoded components of the enzyme are present in chromoplasts. The accumulation of AccC did not strictly parallel the accumulation of AccD in all ripening stages (cf. green and turning fruits in Figure 4), indicating that some subunits can stably accumulate even if not immediately incorporated into the complex.
Figure 4.
Confirmation of AccD Expression in Chromoplasts at the Protein Level.
Protein gel blots for two subunits of the plastid acetyl-CoA carboxylase are shown: the plastid genome–encoded AccD protein and the nuclear genome–encoded AccC subunit (Sasaki et al., 1993, 1995; Madoka et al., 2002). As an example of a plastid-encoded photosynthesis protein, the photosystem II subunit PsbD was analyzed. For each protein, a dilution series of total leaf protein was compared with total proteins extracted from the four stages of fruit ripening. To control for correct protein quantitation, a Coomassie blue–stained polyacrylamide gel is also shown. The two subunits of ribulose-1,5-bis-phosphate carboxylase/oxygenase (RbcL and RbcS), representing the most abundant proteins in green leaves, are labeled. Note that while the PsbD protein is undetectable in chromoplast-containing tomato fruits, the AccD protein still accumulates in ripe red fruits.
RNA Polymerase Activity and Promoter Usage
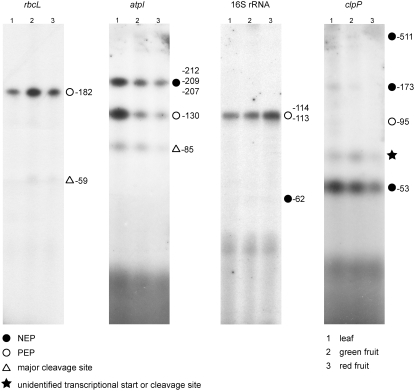
Having seen complex patterns of changes in RNA accumulation in fruits versus leaves (Figure 2), we were interested in determining whether these changes could be attributed to developmental changes in the relative activities of the two distinct transcription systems existing in plastids: the plastid-encoded E. coli–like RNA polymerase (PEP) and the nucleus-encoded bacteriophage-type RNA polymerase (NEP). To this end, we analyzed the transcription initiation sites of four plastid genes: rbcL, a gene predominantly transcribed by PEP; atpI, a gene transcribed by both PEP and NEP; clpP, a gene predominantly transcribed by NEP; and rrn16, the gene for the 16S rRNA, which is normally predominantly transcribed by PEP but is transcribed by NEP in the absence of PEP (as demonstrated by the analysis of PEP knockout plants) (Allison et al., 1996; Hajdukiewicz et al., 1997). RNA 5′ ends were mapped by primer extension analysis in leaves, green fruits, and ripe red fruits. No significant changes in the patterns of RNA 5′ ends were seen for rbcL, rrn16, and clpP (Figure 5). Analysis of atpI transcriptional start sites provides the most informative data on the relative activities of the two polymerases, because atpI possesses both a PEP and a NEP promoter that are about equally strong (Miyagi et al., 1998). Primer extension analysis of atpI transcripts revealed that both promoters are used in leaves and fruits, suggesting that both plastid RNA polymerases are active during fruit development. We observed, however, a change in the relative polymerase activities in that, in leaves, the PEP promoter was more intensively used than the NEP promoter, whereas in red fruits, transcription from the NEP promoter prevailed (Figure 5). This may suggest that the plastid-encoded RNA polymerase PEP is more strongly downregulated in fruit development than the nucleus-encoded RNA polymerase NEP. Together with the earlier finding that PEP transcribes mainly photosynthesis-related genes whereas NEP transcribes mainly genetic system genes (Hajdukiewicz et al., 1997; Maliga, 1998), this may provide a mechanistic explanation for our observation that photosynthesis genes are generally more strongly downregulated in fruits than genetic system genes (Figure 2).
Figure 5.
Developmental Analysis of Plastid RNA Polymerase Activities by Determining PEP and NEP Promoter Usage in Leaves and Green and Red Tomato Fruits.
Transcriptional start sites for the rbcL, atpI, rrn16, and clpP genes were determined by primer extension analysis. PEP promoters, NEP promoters, and RNA processing sites for the genes analyzed here were identified previously (Allison et al., 1996; Hajdukiewicz et al., 1997). Note that not all previously characterized 5′ ends were identified here, because some 5′ ends represent lowly abundant transcript species or are only detectable in a PEP knockout genetic background (Allison et al., 1996; Hajdukiewicz et al., 1997).
Intron Splicing
The expression of a number of genes in plastids is dependent upon the removal of intervening sequences (introns) that have to be removed by splicing to obtain a functional gene product. With a single exception, all introns in higher plant plastid genomes belong to the group II introns (Michel et al., 1989; Sugita and Sugiura, 1996; Bonen and Vogel, 2001). Group II introns are the likely evolutionary ancestors of the spliceosomal introns present in the nuclear genomes of eukaryotes (Copertino and Hallick, 1993; Hetzer et al., 1997). Compelling evidence has accumulated that most, if not all, introns in the plastomes of higher plants do not self-splice but instead require the assistance of proteinaceous splicing factors to be excised in vivo. While most splicing factors for plastid group II introns are encoded by the nuclear genome (Jenkins et al., 1997; Perron et al., 1999, 2004; Vogel et al., 1999; Jenkins and Barkan, 2001; Rivier et al., 2001; Till et al., 2001; Ostheimer et al., 2003), at least one appears to be encoded in the plastome. The matK reading frame present in the group II intron that interrupts the plastid trnK gene displays homology with intron maturases in fungal mitochondrial genomes (Neuhaus and Link, 1987; Mohr et al., 1993; Hess et al., 1994; Liere and Link, 1995).
As intron removal is an essential step in gene expression and a prerequisite for obtaining a functional gene product, it seems conceivable that splicing could be employed to regulate gene expression. The recent finding that some splicing factors act intron-specifically in that they promote the splicing of only a single intron in the plastid genome (Till et al., 2001; Ostersetzer et al., 2005) makes this possibility all the more reasonable. In fact, previous work in maize (Zea mays) has shown that the ratio of spliced to unspliced transcripts can be subject to tissue-dependent variation (Barkan, 1989). Therefore, we were interested in analyzing plastid intron splicing during fruit development and chloroplast-to-chromoplast conversion in tomato.
Splicing efficiency was compared in leaves and four different stages of fruit ripening by separating RNA samples on denaturing gels and hybridizing them to specific probes for intron-containing plastid genes. Five transcripts containing eight introns in total were investigated: petB/D (harboring one intron in petB and one in petD), rps12 (containing one intron spliced in cis and one spliced in trans), clpP (containing two introns), trnI-GAU, and ndhB. For most of these genes, there were no significant changes in the relative ratios of intron-containing precursor transcripts versus mature spliced RNAs (Figure 6). This was also the case for the only plastid mRNA that is processed by trans-splicing (rps12; Figure 6). By contrast, the intron in the ndhB transcript, encoding a subunit of the plastid NAD(P)H dehydrogenase, showed a strong decline in splicing during fruit development. In ripe red tomatoes, almost all ndhB transcripts remained unspliced, indicating that fruit development is accompanied by a gradual loss in ndhB intron splicing activity.
Figure 6.
Developmental Analysis of Plastid Intron Splicing and Processing of Polycistronic Transcripts by Intercistronic Cleavage.
RNA gel blots with probes specific for the transcripts indicated above each blot are shown. Introns are symbolized as open boxes, intron-free genes in polycistronic mRNAs are shown as closed boxes, and exons of intron-containing genes are depicted as shaded boxes. Lariats are indicated by circles. Asterisks denote mature (fully spliced) transcripts. Roman numerals above the shaded boxes indicate exon numbers. To control for equal loading, RNA gels are shown below each blot. Note that unspliced precursors are not detectable for petD and for the two introns of rps12 (the first of which is trans-spliced and the second cis-spliced). The processing patterns remain essentially unchanged during the four fruit ripening stages (lane 2, green fruit; lane 3, turning; lane 4, light red; lane 5, red) compared with green leaves (lane 1). Likewise, the splicing efficiency of the clpP and trnI-GAU introns does not change significantly during development, whereas the splicing efficiency of the ndhB intron declines during fruit development and the ndhB transcript remains largely unspliced in ripe red tomatoes.
In order to investigate this splicing deficiency in more detail, RNA gel blot experiments with an ndhB intron probe were conducted. These experiments revealed that the first intermediate of the group II intron splicing reaction, the lariat structure resulting from the first transesterification step, accumulated throughout fruit development and was still present in essentially unchanged amounts in the red fruit (Figure 6). Together with the nearly complete absence of the free intron and the ligated exons, this finding suggests that a splicing factor specifically mediating the second transesterification reaction of ndhB intron excision is absent from chromoplasts.
In addition to intron splicing, our RNA gel blot analyses also provided information about the processing patterns of plastid transcripts during fruit development. Even for transcripts displaying very complex RNA accumulation patterns in leaves (such as the transcripts from the rps12 and the pentacistroinc psbB/petBD operons), no significant changes were observed during fruit ripening, suggesting that the cleavage of polycistronic transcripts into monocistronic or oligocistronic mRNAs is probably not under developmental regulation.
mRNA Editing
mRNAs in plant cell organelles can be subject to RNA editing, a curious RNA-processing step altering the identities of single nucleotide residues. In higher plant chloroplasts, editing proceeds by C-to-U conversions at highly specific sites (Hoch et al., 1991; Kudla et al., 1992; reviewed in Bock, 2000). With very few exceptions (Hirose et al., 1996; Kudla and Bock, 1999), the vast majority of known plastid RNA editing events alter the coding properties of the affected transcripts, usually resulting in the restoration of phylogenetically conserved amino acid residues (Maier et al., 1992a, 1992b). Transgenic experiments creating plants with a noneditable version of a chloroplast transcript have provided direct evidence for the functional importance of plastid RNA editing (Bock et al., 1994). We recently determined the editing sites encoded in the plastid genome of tomato and compared the editing pattern of the tomato plastome with that of two other solanaceous plants, tobacco and deadly nightshade (Atropa bella-donna) (Kahlau et al., 2006). This analysis revealed that, while most editing sites are conserved between the three species, a few sites were species-specific or shared between only two species. Editing at most known plastid mRNA editing sites appears to be constitutive, in that virtually complete editing of the transcript population is seen in all tissues and developmental stages investigated. Very few editing sites have been shown to undergo changes in editing efficiency that are dependent on the developmental stage of the plant and/or the tissue type investigated (Bock et al., 1993; Karcher and Bock, 2002; Miyata and Sugita, 2004). To assess whether plastid RNA editing is regulated in plant development and/or even participates in the regulation of plastid gene expression, we performed a systematic analysis of mRNA editing in tomato plastids by comparing the editing in chloroplasts of green leaves with that in chloroplasts and chromoplasts of the different fruit ripening stages.
We analyzed 23 of the 36 RNA editing sites encoded in the plastid genome of tomato (Kahlau et al., 2006): the 9 editing sites in ndhB, the 6 sites in ndhD, the 2 sites in ndhA, 2 of the 5 sites in rpoB (sites 184 and 809), and the single sites in the ndhF, ndhG, atpF, and rps12 transcripts. Editing at each site was assessed by comparing the DNA sequence with that of the directly sequenced cDNA population amplified from green leaves and five different fruit ripening stages (Figure 7). For most sites analyzed, editing turned out to be equally efficient in all tissues and developmental stages (see Supplemental Table 1 online), indicating that editing at these sites is neither under developmental regulation nor affected by the plastid type (chloroplasts versus chromoplasts).
Figure 7.
Developmental Analysis of Plastid RNA Editing.
Editing at selected sites of the plastid ndhB, ndhF, and ndhD mRNAs is shown below the corresponding DNA sequences for a developmental series including green leaves and five different fruit stages. Editing sites are denoted by arrows pointing to the corresponding peak in the sequencing chromatogram. Note that C-to-U editing at ndhB site 279 and ndhD site 225 is complete in all developmental stages, whereas editing at ndhB sites 196 and 277 and ndhF site 97 is only partial in fruits. The ndhD sites 1 and 200 remain virtually completely unedited in fruits, with the exception of site 1, which is still partially edited in very young fruits. The relative sizes of the letters C and T above each editing site in the sequence chromatograms symbolize the editing efficiency as evidenced by relative peak intensities. Note that in some cases, the C and T peaks overlap exactly due to the presence of approximately equal amounts of edited and unedited mRNAs (ndhB-196 in red fruits, ndhB-277 in green and light red fruits, and ndhF-97 in green and red fruits).
However, editing in three transcripts was found to be altered in fruits compared with leaves. Interestingly, all three of these mRNAs encode subunits of the plastid NAD(P)H dehydrogenase, an enigmatic multiprotein complex of unclear physiological function. The complex is present in the thylakoid membranes of angiosperm plants (but not in gymnosperms and green algae) and has been implicated in cyclic electron flow and some abiotic stress responses (Burrows et al., 1998; Sazanov et al., 1998; Shikanai et al., 1998; Horváth et al., 2000; for a recent review, see Rumeau et al., 2007). The ndhB transcript contains nine RNA editing sites in tomato, tobacco, and deadly nightshade. While seven of these sites were constitutively fully edited in all analyzed tissues and developmental stages, two of the sites (sites 196 and 277) displayed strongly reduced editing efficiency in at least some fruit stages (Figure 7). Site 196 was nearly unedited in small green fruits. As fruit development proceeded, editing at site 196 first increased (in full-sized green tomatoes) and then declined during ripening, with ∼50% of the mRNA molecules remaining unedited in ripe tomatoes. By contrast, site 277 was still fully edited in small green fruits, but editing efficiency then gradually declined, reaching only 50% in ripe red tomatoes. RNA editing at the single editing site present in the ndhF transcript was already affected in small green fruits (with about half of the RNA molecules remaining unedited), and no pronounced further decline in editing efficiency was observed (Figure 7).
The most striking changes were seen in the ndhD transcript. Here, editing at two of six sites was affected. Editing at site 1 creates a canonical AUG translation initiation codon from a genomically encoded ACG codon, whereas editing at site 200 changes a genomically encoded TCA Ser codon to a UUA Leu codon at the RNA level. Editing at site 1 declined gradually during fruit development and was undetectable in ripe red fruits. An even more dramatic effect was seen at site 200. Editing was already completely lost in small green fruits, and no editing was seen at all later stages of fruit development (Figure 7). The early loss of editing at this site indicates that it is not an effect of chloroplast-to-chromoplast differentiation but rather the result of switched-off editing at the onset of fruit development. To further substantiate this conclusion, we also analyzed different tissues from small green fruits (fruit peel and pericarp). Site 200 remained unedited in the ndhD transcripts independent of the tissue type, indicating that the developmental switching off of editing is an organ-wide process affecting all fruit tissues (see Supplemental Table 2 online). The immediate consequence of this developmental regulation in RNA editing is that the NdhB, NdhF, and NdhD proteins in fruits differ from the proteins made in leaves. The NdhB and NdhF protein populations are probably heterogeneous in fruits due to partial editing in single amino acid positions. In the case of the NdhD protein, all protein molecules made in fruits are different from those in leaves. Whereas the NdhD protein in leaves carries a Leu at position 200, the protein in fruits has a Ser at that position.
No novel RNA editing sites were detected in fruits, possibly suggesting that the editing pattern seen in leaves represents the maximum RNA editing capacity of the plastid. However, as we have not sequenced fruit cDNAs for all plastid genes, there remains a slight possibility that some fruit-specific or chromoplast-specific RNA editing events could exist.
DISCUSSION
In the course of this work, we developed an oligonucleotide microarray suitable for obtaining complete transcriptomics and translatomics data sets for the plastid genome. We then used microarray-based analysis of RNA accumulation and polysome association to study the expression of the plastid genome during tomato fruit development and showed how transcriptomics and translatomics data sets can be integrated to assess the relative contributions of RNA metabolism and translation to the regulation of plastid gene expression. Our results demonstrate that both the regulation of RNA accumulation and translational regulation contribute to the regulation of gene expression in plastids. Interestingly, the global changes in gene expression in fruits compared with leaves are mainly brought about by changes in RNA accumulation, with translational regulation playing only a relatively modest role (cf. Figures 2A and 2B with 3A and 3B). By contrast, the global changes in gene expression during chloroplast-to-chromoplast conversion in fruit ripening are not accompanied by major changes in RNA abundance and, instead, are predominantly the result of translational regulation (cf. Figures 2C and 2D with 3C and 3D).
It should be borne in mind that the expression changes seen in transcriptomics-type microarray experiments reflect the sum of transcriptional regulation and the regulation of RNA stability and degradation. Thus, any change in RNA accumulation seen on the array could be due to either altered transcription or changes in RNA turnover (or both). Whether, for example, the strong global decline in RNA accumulation in fruit plastids (Figures 2A and 2B) is predominantly the result of downregulated plastid transcription or is, at least in part, also caused by an increase in RNA turnover remains to be established. From our data, however, it is clear that if the global downregulation of plastid RNA levels in fruits is transcriptional in nature, it cannot be specifically attributed to one of the two RNA-polymerizing activities in plastids, PEP and NEP. Although their relative activities change in fruit development, both are still active in ripe red tomatoes (Figure 5).
It is also important to note that most plastid genes are part of operons and transcribed as polycistronic mRNAs. Although most polycistronic precursor transcripts are posttranscriptionally processed into monocistronic or oligocistronic units (presumably by specific endonucleolytic cleavage) (Westhoff and Herrmann, 1988; Sugita and Sugiura, 1996; Herrin and Nickelsen, 2004), some mRNAs remain largely polycistronic. Examples of these are the psbE operon transcript comprising four small genes for polypeptides of photosystem II (psbE, psbF, psbL, and psbJ) (Carrillo et al., 1986; Willey and Gray, 1989) and the psaA/psaB transcript (Meng et al., 1988), which is also not regularly cleaved by intercistronic processing. Thus, the polysome association data do not allow us to distinguish between an altered translation of psaA or psaB or both. Although both genes encode reaction center proteins of photosystem I, it is theoretically possible that only one of the two is translationally regulated and is solely responsible for the change in ribosome association of the dicistronic psaA/psaB message.
Finally, it must be kept in mind that the translation rate is not the only determinant of protein accumulation. Posttranslational processes, including the differential stability of proteins, may additionally contribute to regulating the final protein accumulation levels. Protein degradation may become particularly relevant to all of those proteins that are components of multiprotein complexes and whose stability depends on the accumulation of the other subunits in stoichiometric amounts. It is well established, for example, that many photosystem subunits are condemned to rapid degradation if they are not properly incorporated into the complex (Goldschmidt-Clermont, 1998; Wostrikoff et al., 2004). The same may hold true for the ribosomal proteins, not all of which are strictly coregulated during fruit development (Figure 3) and thus could be additionally regulated at the posttranslational level. However, it has been suggested that in maize (Zea mays), the stoichiometry of plastid ribosomal protein subunits undergoes changes in response to environmental cues (Zhao et al., 1999). Thus, it remains to be established whether supernumerary ribosomal proteins are degraded or accumulate either freely or in ribosomes during tomato fruit development.
One of the most remarkable findings from our transcriptomics and translatomics analyses is that only a single protein-coding gene circumvented the global decline in gene expression during tomato fruit development: accD, the only gene on the plastome that is involved in fatty acid biosynthesis (Figures 3 and 4). This makes biological sense in that high fatty acid biosynthetic capacity must be sustained in fruit development to provide the membrane lipids that accommodate the storage carotenoids that are massively synthesized during fruit ripening. It is noteworthy in this respect that the accD gene is essential and cannot be deleted from the plastome (Kode et al., 2005).
The requirement for accD expression in fruit plastids makes it necessary that all plastid genes involved in gene expression (genetic system genes) also continue to be expressed. This is consistent with our observation that these genes are, on average, less strongly downregulated in fruits than are the photosynthesis genes (Figures 3A and 3B). Since, in green leaves, the photosynthesis genes are the most highly expressed plastid genes, it seems reasonable to assume that if no significant expression of the photosynthesis genes occurs, a low translational capacity is sufficient to (1) translate the genetic system gene mRNAs at a low level and (2) sustain the high expression of the accD gene in fruit plastids.
Although accD may be the only plastid-encoded protein needed in chromoplasts, at least one plastid-encoded RNA molecule should also be essential: the glutamyl-tRNA encoded by the trnE gene. In addition to its role in translation, the tRNA-Glu is also required in the first step of tetrapyrrole biosynthesis (Schön et al., 1986), presumably providing the precursors for both chlorophyll and heme syntheses. Expression of the trnE gene is downregulated in fruits compared with leaves (Figures 2A and 2B), presumably reflecting both the reduced need for tRNA-Glu in protein synthesis and the reduced demand by chlorophyll biosynthesis.
It is conceivable that, in addition to the regulation of RNA accumulation and translation, RNA processing also contributes to the regulation of plastid gene expression. Our analysis of the transcript patterns of several plastid operons revealed no evidence of a regulation at the level of intercistronic RNA processing and only a single example of developmentally downregulated splicing (Figure 6). The significance of our finding that the ndhB transcripts remain largely unspliced in chromoplasts is unclear, but the lack of splicing may contribute to the ndhB mRNA being the most strongly downregulated transcript (at the level of polysome association; Figures 3A to 3D) of all 11 plastid ndh genes. Moreover, the accumulation of the lariate intermediate suggests the existence of at least one splicing factor that is specifically involved in the second transesterification and/or exon ligation reaction in ndhB intron splicing.
Our comprehensive analysis of RNA editing in plastid transcripts during fruit development indicates that only three mRNAs undergo developmental changes in their editing patterns. Interestingly, all three mRNAs encode subunits of the plastid NAD(P)H dehydrogenase, a membrane protein complex of unclear physiological function (reviewed in Rumeau et al., 2007). The total loss of RNA editing at two sites in the ndhD transcript of red fruits (one of which, ndhD-200, is already fully unedited in small green fruits; Figure 7) is particularly interesting and raises the possibility that leaf and fruit plastids harbor different isoforms of the NdhD protein. One of the two RNA editing sites resides in the translation initiation codon and creates a canonical AUG start codon from a genomically encoded ACG codon. A lack of editing is unlikely to render the ndhD mRNA untranslatable, as previous work has established that ACG-to-AUG editing is not essential for ndhD translation in tobacco (Sasaki et al., 2003; Zandueta-Criado and Bock, 2004). However, in the absence of clearly defined functions of the NAD(P)H dehydrogenase in green and nongreen plastids, it remains difficult to assess the physiological consequences of the developmentally regulated RNA editing patterns in ndh transcripts.
Finally, our plastome-wide analysis of the transcriptional and translational regulation of gene expression in fruit plastids also provides a valuable data set for the design of expression cassettes for plastid transformation experiments. Plastid transformation is uniquely attractive for plant biotechnology because it offers high-level foreign protein accumulation, the absence of epigenetic effects, convenient transgene stacking in operons, and increased gene containment due to maternal plastid inheritance, which largely excludes transgenes from transmission via pollen (reviewed in Maliga, 2004; Bock, 2007). Efficient transgene expression in consumable plant organs (which frequently are nongreen) is central to the wide use of plastid transformation in biotechnology and is particularly critical to the production of pharmaceutical proteins (vaccines, antibodies, and antimicrobial agents), an area commonly referred to as molecular farming (Ma et al., 2005; Bock, 2007). Plastid transformation technology recently became available for tomato (Ruf et al., 2001; Wurbs et al., 2007). The identification of genes that are highly expressed at the transcriptional or translational level in tomato fruits will facilitate the design of novel expression cassettes that combine strong transcription signals (promoters) with strong translation signals (5′ untranslated regions) and, in this way, will help maximize foreign protein accumulation in tomato fruits.
METHODS
Plant Material
Seeds from Solanum lycopersicum cv IPA-6, a commercially grown South American tomato cultivar, were germinated and grown in a greenhouse with supplementary lighting of ∼200 μmol quanta·m−2·s−1. Fruits were harvested at the different stages of ripening defined by the California Tomato Commission (http://www.tomato.org/Member/Content.aspx?id=20).
RNA Isolation and RNA Gel Blot Analyses
Total RNA from tomato leaf and fruit samples was isolated using the peqGOLD TriFast reagent (Peqlab) and further purified with the NucleoSpin-RNA Clean Up kit (Macherey-Nagel). For RNA gel blot analyses, RNA samples were separated by denaturing gel electrophoresis in formaldehyde-containing 1% agarose gels, transferred onto Hybond nylon membranes (Amersham Biosciences) by capillary blotting, and hybridized to [32P]dCTP-radiolabeled probes. Hybridization probes were generated by labeling gene-specific PCR products with the Megaprime DNA labeling system (Amersham Biosciences) following the instructions of the manufacturer. Hybridizations were performed at 65°C in Church buffer (Church and Gilbert, 1984). mRNA-specific probes were prepared by PCR amplification from genomic DNA or cDNA, except for trnI, for which a synthetic oligonucleotide corresponding to the mature tRNA sequence was used for labeling (see Supplemental Data Set 1 online). PCR primers and conditions for their use are shown in Supplemental Table 3 online. For cDNA synthesis, RNA samples were treated with RNase-free DNase I (Roche) to remove residual contaminating DNA.
Isolation of Polysomes
Polysome isolation was performed as described (Barkan, 1998) with minor modifications. The gradients contained 4 × 0.9 mL of sucrose at the indicated concentrations, overlaid with 0.5 mL of sample in extraction buffer. Following centrifugation, each gradient was separated into 10 fractions for microarray analysis or 6 fractions for RNA gel blot analysis. Control gradients with puromycin were analyzed to identify fractions that contained only polysomes and were devoid of free mRNA and ribosomes. These fractions were pooled (typically, the five lowest fractions) and extracted with phenol:chloroform, and the RNA was precipitated with ethanol.
cDNA Synthesis
For analysis of RNA editing, samples of 2 μg of purified DNA-free RNA were used in 20-μL cDNA synthesis reactions. Reverse transcription was primed with random hexanucleotide primers (1 μg per reaction), and cDNA synthesis was performed with SuperScript III RNase H–free reverse transcriptase (Invitrogen) according to the manufacturer's instructions. For microarray analyses, labeled cDNA probes were generated by reverse transcription of 10 μg of total or polysomal RNA spiked with 2 μL of reference RNAs (Lucidea Universal ScoreCard; Amersham Biosciences) using the SuperScript Indirect cDNA Labeling kit (Invitrogen). The fluorescent dye Cy3 (Amersham Biosciences) was incorporated into the cDNA probes according to the manufacturer's instructions (Invitrogen).
Design and Production of a Plastome Oligonucleotide Microarray for Solanaceae
An oligonucleotide microarray containing all of the genes and conserved open reading frames present in solanaceous plastid genomes was designed using the published sequence of the tobacco (Nicotiana tabacum) plastid genome (accession number NC_001879). When the tomato (accession number NC_007898) and potato (Solanum tuberosum; accession number NC_008096) plastid genome sequences became available, five additional oligonucleotide probes were added to cover those genes with insufficient homology between the selected tobacco oligonucleotide and the corresponding tomato and potato sequences (accD, clpP, ycf1, ycf2, and rpoA; oligonucleotides designated –S for Solanum in Figure 1A). The lengths of the oligonucleotides were between 68 and 71 nucleotides. Whenever possible, the GC content was adjusted to ∼50%. A list of all oligonucleotides is presented in Supplemental Data Set 1 online. For printing onto SuperEpoxy 2 DNA substrates (ArrayIt), the concentrations of all HPLC-purified oligonucleotides (Metabion) were adjusted to 100 ng/μL (in Micro Spotting Solution Plus; ArrayIt). One hundred picograms of oligonucleotide was printed per spot using a Piezorray spotter (Perkin-Elmer). Each array pattern was printed in duplicate.
Microarray Hybridization Experiments
To reduce the volume of labeled cDNA probes, the samples were centrifuged for 10 min in YM-10 microcons (Millipore) and then dissolved in 130 μL of hybridization buffer (0.1% SDS, 25% formamide, 5× SSC [1× SSC is 0.15 M NaCl and 0.015 M sodium citrate], 10 mg/mL BSA, and 40 mM NaPP). The microarray slides were prehybridized according to the manufacturer's instructions (ArrayIt). Hybridization experiments were conducted in a HybArray 12 station (Perkin-Elmer). A standard hybridization protocol was 5 min of preheating of the slide at 75°C followed by cooling to 45°C and addition of the labeled and denatured (5 min at 95°C) cDNA. The slides were then incubated at 44, 43, and 42°C for 1 h per temperature step and then for 12 h at 41°C. Nonspecifically bound cDNAs were subsequently removed by three different washing steps conducted in the following order: (1) 2× SSC + 0.1% N-lauroylsarcosine sodium salt, (2) 0.2× SSC + 0.1% N-lauroylsarcosine sodium salt, and (3) 0.2× SSC. All washes were performed at 25°C in four cycles, with each cycle consisting of 20 s of wash buffer flowing through the hybridization chamber and a 40-s dwell time of the washing buffer inside the chamber. Afterward, the slides were dried by centrifugation at 500g for 10 min and scanned using a FLA-8000 scanner (Fujifilm). Hybridization signal intensities were determined using the GeneSpotter software (MicroDiscovery).
Microarray Data Evaluation
To facilitate signal quantitation, a series of 10 reference cDNAs (Lucidea Universal ScoreCard; Amersham Biosciences) was spotted onto all arrays (Calib 1 to 10 in Figure 1A). Prior to cDNA synthesis, all RNA samples were spiked with the corresponding 10 reference RNAs. The reference RNAs covered a quantitative range from 1 pg to 30 ng. The signal intensities resulting from cDNA hybridization to the calibration samples were plotted against the corresponding RNA amounts to record calibration curves, thus allowing the calculation of relative expression values and ensuring between-array comparability. Relative expression levels are displayed as log2 values of (1) the ratios of different stages of fruit ripening versus the green leaf sample or (2) later fruit ripening stages versus green fruits. The TIGR MeV software (Saeed et al., 2003) and the MapMan software (Thimm et al., 2004) adapted with a plastid genome output mask (downloadable at http://gabi.rzpd.de/database/java-bin/MappingDownloader) were used for data visualization.
The normalized microarray data were read into R (R Development Core Team, 2008), and, after taking the logarithm of the data, a linear model was fitted to them, using the limma package (Smyth, 2004). After extracting the contrasts of interest (red fruits versus leaves, light red fruits versus leaves, turning fruits versus leaves, and green fruits versus leaves as well as red fruits versus green fruits, light red fruits versus green fruits, and turning fruits versus green fruits) from the linear model, significant changes were estimated using the empirical Bayes procedure implemented in limma. Significance was tested controlling the false discovery rate at 5% and using the nestedF procedure, which adjusts genes down before classifying F tests. The false discovery rate was controlled as detailed by Benjamini and Hochberg (1995). The results are reproduced in Supplemental Data Sets 2 and 3 online. The same procedure was applied to the polysomal data sets.
PCR and DNA Sequencing
First-strand cDNAs were amplified in an Eppendorf thermal cycler using GoTaq Flexi DNA polymerase (Promega) and gene-specific primer pairs (see Supplemental Table 3 online). The standard PCR program was 30 to 40 cycles of 1 min at 94°C, 40 s at 58°C, and 40 s to 1.5 min at 72°C, with a 5-min extension of the first cycle at 94°C and a 5-min final extension at 72°C. PCR products were purified by electrophoretic separation on agarose gels followed by DNA extraction from excised gel slices using the NucleoSpin Extract II kit (Macherey-Nagel). To assess the RNA editing status of a given transcript, the amplified cDNA population was directly sequenced (MWG).
Primer Extension Analyses
The amount of RNA used for primer extension analysis was adjusted to the abundance of the target transcript in the RNA sample and thus varied greatly depending on the transcript and the source tissue. It ranged from 1 μg for the rbcL transcript in leaf tissue to 30 μg for the atpI transcript in fruits. Primer sequences were derived from published data (Allison et al., 1996; Hajdukiewicz et al., 1997) and are listed in Supplemental Table 3 online. The primers were radioactively labeled by incubation with [γ-32P]dATP and T4 polynucleotide kinase (New England Biolabs) according to the manufacturer's instructions. A total of 100 fmol of radiolabeled rbcL or 16S rRNA primers and 30 fmol of labeled atpI or clpP primers were used for gene-specific cDNA synthesis with the SuperScript III RNase H–free reverse transcriptase (Invitrogen) according to the manufacturer's instructions. The cDNAs were denatured for 5 min at 95°C and separated by electrophoresis on denaturing 8% polyacrylamide gels. To determine the size of the extension signals, a radiolabeled ΦX174 Hinf I DNA marker was run alongside the samples.
Protein Analysis
Total proteins were extracted according to published protocols (Kuroda and Maliga, 2001); extraction buffer was additionally supplemented with 0.02% N-lauroylsarcosine and 0.02% Triton X-100 (Cahoon et al., 1992). Twenty-five and 100 μg of total fruit protein and a dilution series of total leaf protein were separated by electrophoresis on 15% SDS-containing polyacrylamide gels. The gels were either stained with Coomassie Brilliant Blue (Serva) or blotted onto polyvinylidene difluoride membranes (Hybond-P; GE Healthcare). Membranes were treated with blocking buffer (20 mM Tris-HCl, pH 7.6, 137 mM NaCl, 0.1% Tween 20, and 0.5% BSA; BSA was replaced with 0.2% casein for the AccC antibody) overnight and subsequently incubated for 1 h with polyclonal antibodies against PsbD, AccD, and AccC (raised in rabbits) diluted in buffer (20 mM Tris-HCl, pH 7.6, 137 mM NaCl, and 0.1% Tween 20). Antibodies against the acetyl-CoA carboxylase subunits AccC and AccD were kindly provided by Yukio Nagano and Yukiko Sasaki (Madoka et al., 2002). An anti-PsbD antibody was purchased from Agrisera. Antibody dilutions were 1:2500 (AccC and AccD) and 1:1000 (PsbD). Detection was performed with the ECL Plus protein gel blotting detection system (GE Healthcare).
Accession Numbers
GenBank/EMBL database accession numbers for the sequences from which synthetic oligonucleotides were derived and hybridization probes were produced are listed in Supplemental Table 3 online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. Efficiency of Plastid RNA Editing in Tomato Leaves and Fruits.
Supplemental Table 2. RNA Editing at ndhD Sites 1, 200, and 225 in Different Fruit Tissues and Ripening Stages.
Supplemental Table 3. Primers Used in This Work.
Supplemental Data Set 1. Gene-Specific Oligonucleotides Used to Produce the Plastome Microarray.
Supplemental Data Set 2. Log2 Fold Change in Expression (AVG) of Total RNA Abundance and Significance Test at 5% False Discovery Rate.
Supplemental Data Set 3. Log2 Fold Change in Expression (AVG) of Polysomal RNA Abundance and Significance Test at 5% False Discovery Rate.
Supplementary Material
Acknowledgments
We thank the Max-Planck-Institut für Molekulare Pflanzenphysiologie Green Team for plant care and cultivation; Dirk Hincha, Susanne Freund, and Kerstin Petersen for help with microarray design and spotting; and Björn Usadel for help with statistical evaluation of the microarray data. We are grateful to Yukio Nagano and Yukiko Sasaki (Saga University and Nagoya University) for generously providing antibodies against the AccC and AccD proteins. This research was supported by a grant from the European Union (FP6 Plastomics Project LSHG-CT-2003-503238) and by the Max Planck Society.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Ralph Bock (rbock@mpimp-golm.mpg.de).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Allison, L.A., Simon, L.D., and Maliga, P. (1996). Deletion of rpoB reveals a second distinct transcription system in plastids of higher plants. EMBO J. 15 2802–2809. [PMC free article] [PubMed] [Google Scholar]
- Barkan, A. (1989). Tissue-dependent plastid RNA splicing in maize: Transcripts from four plastid genes are predominantly unspliced in leaf meristems and roots. Plant Cell 1 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan, A. (1998). Approaches to investigating nuclear genes that function in chloroplast biogenesis in land plants. Methods Enzymol. 297 38–57. [Google Scholar]
- Barkan, A., and Goldschmidt-Clermont, M. (2000). Participation of nuclear genes in chloroplast gene expression. Biochimie 82 559–572. [DOI] [PubMed] [Google Scholar]
- Bathgate, B., Purton, M.E., Grierson, D., and Goodenough, P.W. (1985). Plastic changes during the conversion of chloroplasts to chromoplasts in ripening tomatoes. Planta 165 197–204. [DOI] [PubMed] [Google Scholar]
- Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Statist. Soc. B 57 289–300. [Google Scholar]
- Bock, R. (2000). Sense from nonsense: How the genetic information of chloroplasts is altered by RNA editing. Biochimie 82 549–557. [DOI] [PubMed] [Google Scholar]
- Bock, R. (2007). Plastid biotechnology: Prospects for herbicide and insect resistance, metabolic engineering and molecular farming. Curr. Opin. Biotechnol. 18 100–106. [DOI] [PubMed] [Google Scholar]
- Bock, R., Hagemann, R., Kössel, H., and Kudla, J. (1993). Tissue- and stage-specific modulation of RNA editing of the psbF and psbL transcript from spinach plastids—A new regulatory mechanism? Mol. Gen. Genet. 240 238–244. [DOI] [PubMed] [Google Scholar]
- Bock, R., Kössel, H., and Maliga, P. (1994). Introduction of a heterologous editing site into the tobacco plastid genome: The lack of RNA editing leads to a mutant phenotype. EMBO J. 13 4623–4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohne, A.-V., Irihimovitch, V., Weihe, A., and Stern, D.B. (2006). Chlamydomonas reinhardtii encodes a single sigma70-like factor which likely functions in chloroplast transcription. Curr. Genet. 49 333–340. [DOI] [PubMed] [Google Scholar]
- Bonen, L., and Vogel, J. (2001). The ins and outs of group II introns. Trends Genet. 17 322–323. [DOI] [PubMed] [Google Scholar]
- Bruick, R.K., and Mayfield, S.P. (1999). Light-activated translation of chloroplast mRNA. Trends Plant Sci. 4 190–195. [DOI] [PubMed] [Google Scholar]
- Burrows, P.A., Sazanov, L.A., Svab, Z., Maliga, P., and Nixon, P.J. (1998). Identification of a functional respiratory complex in chloroplasts through analysis of tobacco mutants containing disrupted plastid ndh genes. EMBO J. 17 868–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoon, E.B., Shanklin, J., and Ohlrogge, J.B. (1992). Expression of a coriander desaturase results in petroselinic acid production in transgenic tobacco. Proc. Natl. Acad. Sci. USA 89 11184–11188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo, N., Seyer, P., Tyagi, A., and Herrmann, R.G. (1986). Cytochrome b-559 genes from Oenothera hookeri and Nicotiana tabacum show a remarkably high degree of conservation as compared to spinach. Curr. Genet. 10 619–624. [DOI] [PubMed] [Google Scholar]
- Choquet, Y., and Wollman, F.-A. (2002). Translational regulations as specific traits of chloroplast gene expression. FEBS Lett. 529 39–42. [DOI] [PubMed] [Google Scholar]
- Church, G.M., and Gilbert, W. (1984). Genomic sequencing. Proc. Natl. Acad. Sci. USA 81 1991–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copertino, D.W., and Hallick, R.B. (1993). Group II and group III introns of twintrons: Potential relationship with nuclear pre-mRNA introns. Trends Biochem. Sci. 18 467–471. [DOI] [PubMed] [Google Scholar]
- Daniell, H., Lee, S.-B., Grevich, J., Saski, C., Quesada-Vargas, T., Guda, C., Tomkins, J., and Jansen, R.K. (2006). Complete chloroplast genome sequences of Solanum bulbocastanum, Solanum lycopersicum and comparative analyses with other Solanaceae genomes. Theor. Appl. Genet. 112 1503–1518. [DOI] [PubMed] [Google Scholar]
- Drescher, A., Ruf, S., Calsa, T., Jr., Carrer, H., and Bock, R. (2000). The two largest chloroplast genome-encoded open reading frames of higher plants are essential genes. Plant J. 22 97–104. [DOI] [PubMed] [Google Scholar]
- Eberhard, S., Drapier, D., and Wollman, F.-A. (2002). Searching limiting steps in the expression of chloroplast-encoded proteins: Relations between gene copy number, transcription, transcript abundance and translation rate in the chloroplast of Chlamydomonas reinhardtii. Plant J. 31 149–160. [DOI] [PubMed] [Google Scholar]
- Favory, J.-J., Kobayshi, M., Tanaka, K., Peltier, G., Kreis, M., Valay, J.-G., and Lerbs-Mache, S. (2005). Specific function of a plastid sigma factor for ndhF gene transcription. Nucleic Acids Res. 33 5991–5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filée, J., and Forterre, P. (2005). Viral proteins functioning in organelles: A cryptic orgin? Trends Microbiol. 13 510–513. [DOI] [PubMed] [Google Scholar]
- Gillham, N.W., Boynton, J.E., and Hauser, C.R. (1994). Translational regulation of gene expression in chloroplasts and mitochondria. Annu. Rev. Genet. 28 71–93. [DOI] [PubMed] [Google Scholar]
- Goldschmidt-Clermont, M. (1998). Coordination of nuclear and chloroplast gene expression in plant cells. Int. Rev. Cytol. 177 115–180. [DOI] [PubMed] [Google Scholar]
- Gounaris, I., and Price, C.A. (1987). Plastid transcripts in chloroplasts and chromoplasts of Capsicum annuum. Curr. Genet. 12 219–224. [Google Scholar]
- Hajdukiewicz, P.T.J., Allison, L.A., and Maliga, P. (1997). The two RNA polymerases encoded by the nuclear and the plastid compartments transcribe distinct groups of genes in tobacco plastids. EMBO J. 16 4041–4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley, J., and Bogorad, L. (1990). Alternative promoters are used for genes within maize chloroplast polycistronic transcription units. Plant Cell 2 323–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanaoka, M., Kanamaru, K., Takahashi, H., and Tanaka, K. (2003). Molecular genetic analysis of chloroplast gene promoters dependent on SIG2, a nucleus-encoded sigma factor for the plastid-encoded RNA polymerase, in Arabidopsis thaliana. Nucleic Acids Res. 31 7090–7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedtke, B., Börner, T., and Weihe, A. (1997). Mitochondrial and chloroplast phage-type RNA polymerases in Arabidopsis. Science 277 809–811. [DOI] [PubMed] [Google Scholar]
- Herrin, D.L., and Nickelsen, J. (2004). Chloroplast RNA processing and stability. Photosynth. Res. 82 301–314. [DOI] [PubMed] [Google Scholar]
- Hess, W.R., and Börner, T. (1999). Organellar RNA polymerases of higher plants. Int. Rev. Cytol. 190 1–59. [DOI] [PubMed] [Google Scholar]
- Hess, W.R., Hoch, B., Zeltz, P., Hübschmann, T., Kössel, H., and Börner, T. (1994). Inefficient rpl2 splicing in barley mutants with ribosome-deficient plastids. Plant Cell 6 1455–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzer, M., Wurzer, G., Schweyen, R.J., and Mueller, M.W. (1997). Trans-activation of group II intron splicing by nuclear U5 snRNA. Nature 386 417–420. [DOI] [PubMed] [Google Scholar]
- Hirose, T., Fan, H., Suzuki, J.Y., Wakasugi, T., Tsudzuki, T., Kössel, H., and Sugiura, M. (1996). Occurrence of silent RNA editing in chloroplasts: Its species specificity and the influence of environmental and developmental conditions. Plant Mol. Biol. 30 667–672. [DOI] [PubMed] [Google Scholar]
- Hoch, B., Maier, R.M., Appel, K., Igloi, G.L., and Kössel, H. (1991). Editing of a chloroplast mRNA by creation of an initiation codon. Nature 353 178–180. [DOI] [PubMed] [Google Scholar]
- Horváth, E.M., Peter, S.O., Joet, T., Rumeau, D., Cournac, L., Horváth, G., Kavanagh, T.A., Schäfer, C., and Medgyesy, P. (2000). Targeted inactivation of the plastid ndhB gene in tobacco results in an enhanced sensitivity of photosynthesis to moderate stomatal closure. Plant Physiol. 123 1337–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igloi, G.L., and Kössel, H. (1992). The transcriptional apparatus of chloroplasts. Crit. Rev. Plant Sci. 10 525–558. [Google Scholar]
- Jenkins, B.D., and Barkan, A. (2001). Recruitment of a peptidyl-tRNA hydrolase as a facilitator of group II intron splicing in chloroplasts. EMBO J. 20 872–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins, B.D., Kulhanek, D.J., and Barkan, A. (1997). Nuclear mutations that block group II RNA splicing in maize chloroplasts reveal several intron classes with distinct requirements for splicing factors. Plant Cell 9 283–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahlau, S., Aspinall, S., Gray, J.C., and Bock, R. (2006). Sequence of the tomato chloroplast DNA and evolutionary comparison of solanaceous plastid genomes. J. Mol. Evol. 63 194–207. [DOI] [PubMed] [Google Scholar]
- Kanamaru, K., Fujiwara, M., Seki, M., Katagiri, T., Nakamura, M., Mochizuki, N., Nagatani, A., Shinozaki, K., Tanaka, K., and Takahashi, H. (1999). Plastidic RNA polymerase σ factors in Arabidopsis. Plant Cell Physiol. 40 832–842. [DOI] [PubMed] [Google Scholar]
- Kanamaru, K., Nagashima, A., Fujiwara, M., Shimada, H., Shirano, Y., Nakabayashi, K., Shibata, D., Tanaka, K., and Takahashi, H. (2001). An Arabidopsis sigma factor (SIG2)-dependent expression of plastid-encoded tRNAs in chloroplast. Plant Cell Physiol. 42 1034–1043. [DOI] [PubMed] [Google Scholar]
- Karcher, D., and Bock, R. (2002). The amino acid sequence of a plastid protein is developmentally regulated by RNA editing. J. Biol. Chem. 277 5570–5574. [DOI] [PubMed] [Google Scholar]
- Kode, V., Mudd, E.A., Iamtham, S., and Day, A. (2005). The tobacco plastid accD gene is essential and is required for leaf development. Plant J. 44 237–244. [DOI] [PubMed] [Google Scholar]
- Kohchi, T., Yoshida, T., Komano, T., and Ohyama, K. (1988). Divergent mRNA transcription in the chloroplast psbB operon. EMBO J. 7 885–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla, J., and Bock, R. (1999). RNA editing in an untranslated region of the Ginkgo chloroplast genome. Gene 234 81–86. [DOI] [PubMed] [Google Scholar]
- Kudla, J., Igloi, G.L., Metzlaff, M., Hagemann, R., and Kössel, H. (1992). RNA editing in tobacco chloroplasts leads to the formation of a translatable psbL mRNA by a C to U substitution within the initiation codon. EMBO J. 11 1099–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuntz, M., Evrard, J.-L., d'Harlingue, A., Weil, J.-H., and Camara, B. (1989). Expression of plastid and nuclear genes during chromoplast differentiation in bell pepper (Capsicum annuum) and sunflower (Helianthus annuus). Mol. Gen. Genet. 216 156–163. [Google Scholar]
- Kuroda, H., and Maliga, P. (2001). Complementarity of the 16S rRNA penultimate stem with sequences downstream of the AUG destabilizes the plastid mRNAs. Nucleic Acids Res. 29 970–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahiri, S.D., Yao, J., McCumbers, C., and Allison, L.A. (1999). Tissue-specific and light-dependent expression within a family of nuclear-encoded sigma-like factors from Zea mays. Mol. Cell Biol. Res. Commun. 1 14–20. [DOI] [PubMed] [Google Scholar]
- Legen, J., Kemp, S., Krause, K., Profanter, B., Herrmann, R.G., and Maier, R.M. (2002). Comparative analysis of plastid transcription profiles of entire plastid chromosomes from tobacco attributed to wild-type and PEP-deficient transcription machineries. Plant J. 31 171–188. [DOI] [PubMed] [Google Scholar]
- Lerbs-Mache, S. (2000). Regulation of rDNA transcription in plastids of higher plants. Biochimie 82 525–535. [DOI] [PubMed] [Google Scholar]
- Liere, K., and Link, G. (1995). RNA-binding activity of the matK protein encoded by the chloroplast trnK intron from mustard (Sinapis alba L.). Nucleic Acids Res. 23 917–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J.K.-C., et al. (2005). Molecular farming for new drugs and vaccines. EMBO Rep. 6 593–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madoka, Y., Tomizawa, K.-I., Mizoi, J., Nishida, I., Nagano, Y., and Sasaki, Y. (2002). Chloroplast transformation with modified accD operon increases acetyl-CoA carboxylase and causes extension of leaf longevity and increase in seed yield in tobacco. Plant Cell Physiol. 43 1518–1525. [DOI] [PubMed] [Google Scholar]
- Maier, R.M., Hoch, B., Zeltz, P., and Kössel, H. (1992. a). Internal editing of the maize chloroplast ndhA transcript restores codons for conserved amino acids. Plant Cell 4 609–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier, R.M., Neckermann, K., Hoch, B., Akhmedov, N.B., and Kössel, H. (1992. b). Identification of editing positions in the ndhB transcript from maize chloroplasts reveals sequence similarities between editing sites of chloroplasts and plant mitochondria. Nucleic Acids Res. 20 6189–6194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maliga, P. (1998). Two plastid RNA polymerases of higher plants: An evolving story. Trends Plant Sci. 3 4–6. [Google Scholar]
- Maliga, P. (2004). Plastid transformation in higher plants. Annu. Rev. Plant Biol. 55 289–313. [DOI] [PubMed] [Google Scholar]
- Meng, B.Y., Tanaka, M., Wakasugi, T., Ohme, M., Shinozaki, K., and Sugiura, M. (1988). Cotranscription of the genes encoding two P700 chlorophyll a apoproteins with the gene for ribosomal protein CS14: Determination of the transcriptional initiation site by in vitro capping. Curr. Genet. 14 395–400. [DOI] [PubMed] [Google Scholar]
- Meng, B.-Y., Wakasugi, T., and Sugiura, M. (1991). Two promoters within the psbK-psbI-trnG gene cluster in tobacco chloroplast DNA. Curr. Genet. 20 259–264. [DOI] [PubMed] [Google Scholar]
- Michel, F., Umesono, K., and Ozeki, H. (1989). Comparative and functional anatomy of group II catalytic introns—A review. Gene 82 5–30. [DOI] [PubMed] [Google Scholar]
- Miyagi, T., Kapoor, S., Sugita, M., and Sugiura, M. (1998). Transcript analysis of the tobacco plastid operon rps2/atpI/H/F/A reveals the existence of a non-consensus type II (NCII) promoter upstream of the atpI coding sequence. Mol. Gen. Genet. 257 299–307. [DOI] [PubMed] [Google Scholar]
- Miyata, Y., and Sugita, M. (2004). Tissue- and stage-specific RNA editing of rps14 transcripts in moss (Physcomitrella patens) chloroplasts. J. Plant Physiol. 161 113–115. [DOI] [PubMed] [Google Scholar]
- Mohr, G., Perlman, P.S., and Lambowitz, A.M. (1993). Evolutionary relationships among group II intron-encoded proteins and identification of a conserved domain that may be related to maturase function. Nucleic Acids Res. 21 4991–4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhaus, H., and Link, G. (1987). The chloroplast tRNALys(UUU) gene from mustard (Sinapis alba) contains a class II intron potentially coding for a maturase-related polypeptide. Curr. Genet. 11 251–257. [DOI] [PubMed] [Google Scholar]
- Ostersetzer, O., Cooke, A.M., Watkins, K.P., and Barkan, A. (2005). CRS1, a chloroplast group II intron splicing factor, promotes intron folding through specific interactions with two intron domains. Plant Cell 17 241–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostheimer, G.J., Williams-Carrier, R., Belcher, S., Osborne, E., Gierke, J., and Barkan, A. (2003). Group II intron splicing factors derived by diversification of an ancient RNA-binding domain. EMBO J. 22 3919–3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perron, K., Goldschmidt-Clermont, M., and Rochaix, J.-D. (1999). A factor related to pseudouridine synthases is required for chloroplast group II intron trans-splicing in Chlamydomonas reinhardtii. EMBO J. 18 6481–6490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perron, K., Goldschmidt-Clermont, M., and Rochaix, J.-D. (2004). A multiprotein complex involved in chloroplast group II intron splicing. RNA 10 704–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfannschmidt, T., Nilsson, A., and Allen, J.F. (1999). Photosynthetic control of chloroplast gene expression. Nature 397 625–628. [Google Scholar]
- Piechulla, B., Chonoles Imlay, K.R., and Gruissem, W. (1985). Plastid gene expression during fruit ripening in tomato. Plant Mol. Biol. 5 373–384. [DOI] [PubMed] [Google Scholar]
- Piechulla, B., Glick, R.E., Bahl, H., Melis, A., and Gruissem, W. (1987). Changes in photosynthetic capacity and photosynthetic protein pattern during tomato fruit ripening. Plant Physiol. 84 911–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piechulla, B., Pichersky, E., Cashmore, A.R., and Gruissem, W. (1986). Expression of nuclear and plastid genes for photosynthesis-specific proteins during tomato fruit development and ripening. Plant Mol. Biol. 7 367–376. [DOI] [PubMed] [Google Scholar]
- Powell, D.S., and Pryke, J.A. (1987). Gene expression in isolated plastids from fruits of Capsicum annuum. Plant Sci. 49 51–56. [Google Scholar]
- R Development Core Team (2008). R: A Language and Environment for Statistical Computing. (Vienna, Austria: R Foundation for Statistical Computing).
- Rivier, C., Goldschmidt-Clermont, M., and Rochaix, J.-D. (2001). Identification of an RNA-protein complex involved in chloroplast group II intron trans-splicing in Chlamydomonas reinhardtii. EMBO J. 20 1765–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruf, S., Hermann, M., Berger, I.J., Carrer, H., and Bock, R. (2001). Stable genetic transformation of tomato plastids and expression of a foreign protein in fruit. Nat. Biotechnol. 19 870–875. [DOI] [PubMed] [Google Scholar]
- Rumeau, D., Peltier, G., and Cournac, L. (2007). Chlororespiration and cyclic electron flow around PSI during photosynthesis and plant stress response. Plant Cell Environ. 30 1041–1051. [DOI] [PubMed] [Google Scholar]
- Saeed, A.I., et al. (2003). TM4: A free, open-source system for microarray data management and analysis. Biotechniques 34 374–378. [DOI] [PubMed] [Google Scholar]
- Sasaki, T., Yukawa, Y., Miyamoto, T., Obokata, J., and Sugiura, M. (2003). Identification of RNA editing sites in chloroplast transcripts from the maternal and paternal progenitors of tobacco (Nicotiana tabacum): Comparative analysis shows the involvement of distinct trans-factors for ndhB editing. Mol. Biol. Evol. 20 1028–1035. [DOI] [PubMed] [Google Scholar]
- Sasaki, Y., Hakamada, K., Suama, Y., Nagano, Y., Furusawa, I., and Matsuno, R. (1993). Chloroplast-encoded protein as a subunit of acetyl-CoA carboxylase in pea plant. J. Biol. Chem. 268 25118–25123. [PubMed] [Google Scholar]
- Sasaki, Y., Konishi, T., and Nagano, Y. (1995). The compartmentation of acetyl-coenzyme A carboxylase in plants. Plant Physiol. 108 445–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sazanov, L.A., Burrows, P.A., and Nixon, P.J. (1998). The chloroplast Ndh complex mediates the dark reduction of the plastoquinone pool in response to heat stress in tobacco leaves. FEBS Lett. 429 115–118. [DOI] [PubMed] [Google Scholar]
- Schön, A., Krupp, G., Gough, S., Berry-Lowe, S., Kannangara, C.G., and Söll, D. (1986). The RNA required in the first step of chlorophyll biosynthesis is a chloroplast glutamate tRNA. Nature 322 281–284. [DOI] [PubMed] [Google Scholar]
- Shikanai, T., Endo, T., Hashimoto, T., Yamada, Y., Asada, K., and Yokota, A. (1998). Directed disruption of the tobacco ndhB gene impairs cyclic electron flow around photosystem I. Proc. Natl. Acad. Sci. USA 95 9705–9709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada, H., and Sugiura, M. (1991). Fine structural features of the chloroplast genome: Comparison of the sequenced chloroplast genomes. Nucleic Acids Res. 19 983–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorov, V.A., Kasten, D., Pang, S.-Z., Hajdukiewicz, P.T.J., Staub, J.M., and Nehra, N.S. (1999). Stable chloroplast transformation in potato: Use of green fluorescent protein as a plastid marker. Plant J. 19 209–216. [DOI] [PubMed] [Google Scholar]
- Smyth, G.K. (2004). Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3 Article 3. [DOI] [PubMed] [Google Scholar]
- Sugita, M., and Sugiura, M. (1996). Regulation of gene expression in chloroplasts of higher plants. Plant Mol. Biol. 32 315–326. [DOI] [PubMed] [Google Scholar]
- Sugiura, M. (1992). The chloroplast genome. Plant Mol. Biol. 19 149–168. [DOI] [PubMed] [Google Scholar]
- Svab, Z., and Maliga, P. (1993). High-frequency plastid transformation in tobacco by selection for a chimeric aadA gene. Proc. Natl. Acad. Sci. USA 90 913–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, K., Oikawa, K., Ohta, N., Kuroiwa, H., Kuroiwa, T., and Takahashi, H. (1996). Nuclear encoding of a chloroplast RNA polymerase sigma subunit in a red alga. Science 272 1932–1935. [DOI] [PubMed] [Google Scholar]
- Thimm, O., Bläsing, O., Gibon, Y., Nagel, A., Meyer, S., Krüger, P., Selbig, J., Müller, L.A., Rhee, S.Y., and Stitt, M. (2004). MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37 914–939. [DOI] [PubMed] [Google Scholar]
- Till, B., Schmitz-Linneweber, C., Williams-Carrier, R., and Barkan, A. (2001). Crs1 is a novel group II intron splicing factor that was derived from a domain of ancient origin. RNA 7 1227–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunoyama, Y., Ishizaki, Y., Morikawa, K., Kobori, M., Nakahira, Y., Takeba, G., Toyoshima, Y., and Shiina, T. (2004). Blue light-induced transcription of plastid-encoded psbD gene is mediated by a nuclear-encoded transcription initiation factor, AtSig5. Proc. Natl. Acad. Sci. USA 101 3304–3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullberg, A., Alexciev, K., Pfannschmidt, T., and Allen, J.F. (2000). Photosynthetic electron flow regulates transcription of the psaB gene in pea (Pisum sativum L.) chloroplasts through the redox state of the pastoquinone pool. Plant Cell Physiol. 41 1045–1054. [DOI] [PubMed] [Google Scholar]
- Vera, A., Matsubayashi, T., and Sugiura, M. (1992). Active transcription from a promoter positioned within the coding region of a divergently oriented gene: The tobacco chloroplast rpl32 gene. Mol. Gen. Genet. 233 151–156. [DOI] [PubMed] [Google Scholar]
- Vogel, J., Börner, T., and Hess, W.R. (1999). Comparative analysis of splicing of the complete set of chloroplast group II introns in three higher plant mutants. Nucleic Acids Res. 27 3866–3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakasugi, T., Tsudzuki, T., and Sugiura, M. (2001). The genomics of land plant chloroplasts: Gene content and alteration of genomic information by RNA editing. Photosynth. Res. 70 107–118. [DOI] [PubMed] [Google Scholar]
- Westhoff, P., and Herrmann, R.G. (1988). Complex RNA maturation in chloroplasts. Eur. J. Biochem. 171 551–564. [DOI] [PubMed] [Google Scholar]
- Willey, D.L., and Gray, J.C. (1989). Two small open reading frames are co-transcribed with the pea chloroplast genes for the polypeptides of cytochrome b-559. Curr. Genet. 15 213–220. [DOI] [PubMed] [Google Scholar]
- Wostrikoff, K., Girard-Bascou, J., Wollman, F.-A., and Choquet, Y. (2004). Biogenesis of PSI involves a cascade of translational autoregulation in the chloroplast of Chlamydomonas. EMBO J. 23 2696–2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurbs, D., Ruf, S., and Bock, R. (2007). Contained metabolic engineering in tomatoes by expression of carotenoid biosynthesis genes from the plastid genome. Plant J. 49 276–288. [DOI] [PubMed] [Google Scholar]
- Yukawa, M., Tsudzuki, T., and Sugiura, M. (2005). The 2005 version of the chloroplast DNA sequence from tobacco (Nicotiana tabacum). Plant Mol. Biol. Rep. 23 359–365. [Google Scholar]
- Zandueta-Criado, A., and Bock, R. (2004). Surprising features of plastid ndhD transcripts: Addition of non-encoded nucleotides and polysome association of mRNAs with an unedited start codon. Nucleic Acids Res. 32 542–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerges, W. (2000). Translation in chloroplasts. Biochimie 82 583–601. [DOI] [PubMed] [Google Scholar]
- Zhao, Y.-Y., Xu, T., Zucchi, P., and Bogorad, L. (1999). Subpopulations of chloroplast ribosomes change during photoregulated development of Zea mays leaves: Ribosomal proteins L2, L21, L29. Proc. Natl. Acad. Sci. USA 96 8997–9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.