Abstract
The three amino acid loop extension (TALE) homeodomain superfamily, which comprises the KNOTTED-like and BEL1-like families, plays a critical role in regulating meristem activity. We previously demonstrated a function for KNAT6 (for KNOTTED-like from Arabidopsis thaliana 6) in shoot apical meristem and boundary maintenance during embryogenesis. KNAT2, the gene most closely related to KNAT6, does not play such a role. To investigate the contribution of KNAT6 and KNAT2 to inflorescence development, we examined their interactions with two TALE genes that regulate internode patterning, BREVIPEDICELLUS (BP) and PENNYWISE (PNY). Our data revealed distinct and overlapping interactions of KNAT6 and KNAT2 during inflorescence development. Removal of KNAT6 activity suppressed the pny phenotype and partially rescued the bp phenotype. Removal of KNAT2 activity had an effect only in the absence of both BP and KNAT6 or in the absence of both BP and PNY. Consistent with this, KNAT6 and KNAT2 expression patterns were enlarged in both bp and pny mutants. Thus, the defects seen in pny and bp are attributable mainly to the misexpression of KNAT6 and to a lesser extent of KNAT2. Hence, our data showed that BP and PNY restrict KNAT6 and KNAT2 expression to promote correct inflorescence development. This interaction was also revealed in the carpel.
INTRODUCTION
The aerial part of the plant results from the activity of the shoot apical meristem (SAM). The SAM has two main functions: the maintenance of a population of stem cells and the production of organs. In Arabidopsis thaliana, the three amino acid loop extension (TALE) homeodomain superfamily plays a critical role in determining meristem function (for review, see Ragni et al., 2007). This family comprises the KNAT (for KNOTTED-like from Arabidopsis thaliana) members and the BEL1-like (BELL) members that can form heterodimers to regulate plant development. KNAT class I contains four members: SHOOT MERISTEMLESS (STM), BREVIPEDICELLUS (BP)/KNAT1, KNAT2, and KNAT6. STM is required for the initiation of the meristem during embryogenesis and its maintenance during postembryonic development (Clark et al., 1996; Endrizzi et al., 1996; Long et al., 1996). BP contributes with STM to SAM maintenance since the loss of function of BP reduces the residual meristematic activity of the weak allele stm-2 (Byrne et al., 2002, 2003). Like BP, KNAT6 contributes redundantly with STM to SAM function, as the inactivation of KNAT6 abolishes the meristematic potential of the weak allele stm-2 (Belles-Boix et al., 2006). In addition, a specific role for KNAT6 in boundary maintenance is indicated by a severe defect in cotyledon separation observed in the stm-2 knat6 double mutant (Belles-Boix et al., 2006). KNAT2 also is expressed in the SAM, but its role is unclear as its inactivation does not aggravate stm-2, knat6 stm-2, or knat6 bp stm-2 phenotypes (Belles-Boix et al., 2006). PENNYWISE (PNY), a member of the BELL family, also known as BELLRINGER, REPLUMLESS (RPL), or VAAMANA, contributes to SAM maintenance redundantly with STM (Byrne et al., 2003).
In addition to their contributions to SAM function, BP and PNY regulate inflorescence development. BP plays a primary role in inflorescence growth, as indicated by bp mutants that exhibit reduced internode and pedicel lengths, bends at nodes, and downward-oriented siliques (Douglas et al., 2002; Venglat et al., 2002; Smith and Hake, 2003). PNY influences inflorescence architecture and fruit development (Byrne et al., 2003; Smith and Hake, 2003; Bhatt et al., 2004). PNY also interacts with BP to regulate inflorescence patterning and fruit development (Smith and Hake, 2003; Alonso-Cantabrana et al., 2007). A role for STM in inflorescence growth also is suggested as weak alleles of STM show aberrant organ position, fusion defects, and abnormal flowers (Endrizzi et al., 1996; Kanrar et al., 2006). However, these defects may be indirect effects resulting from the abnormal structure of the SAM. KNAT2 and KNAT6 are the most closely related members of class I; however, knat2, knat6, and knat2 knat6 mutants show wild-type development (Belles-Boix et al., 2006). Here, we investigated the genetic interaction of KNAT6 and KNAT2 with BP and PNY during inflorescence development. Our studies show that in wild-type plants, PNY and BP restrict KNAT6 and KNAT2 expression to promote correct inflorescence growth.
RESULTS
The Inactivation of KNAT6 Partially Rescues the bp Phenotype
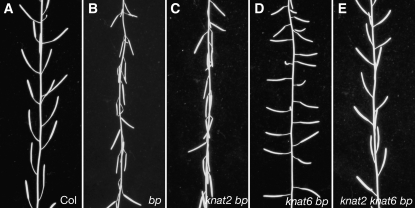
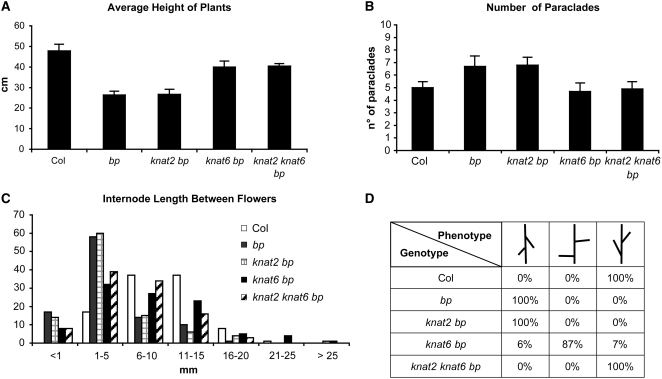
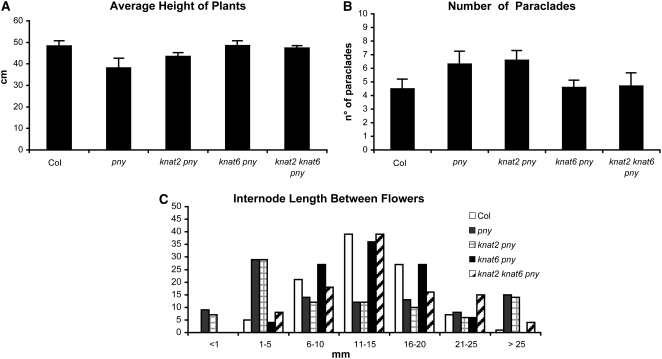
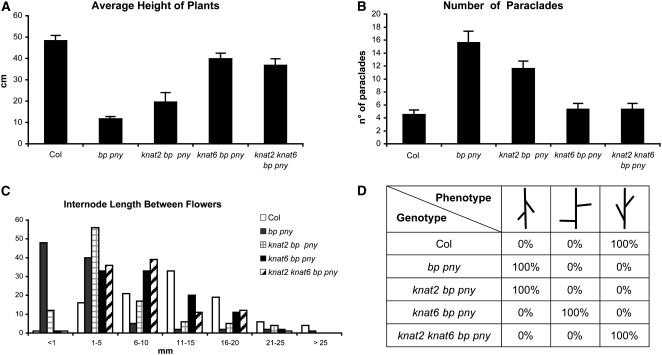
To analyze the interaction of KNAT6 and KNAT2 with BP, we generated the knat6 bp, knat2 bp, and knat2 knat6 bp mutants. bp mutants are characterized by reduced stature, partial loss of apical dominance, and downward-oriented siliques (Douglas et al., 2002; Venglat et al., 2002; Smith and Hake, 2003). Figure 1 shows the phenotypes of the main inflorescences of the bp, knat2 bp, knat6 bp, and knat2 knat6 bp mutants. The inactivation of KNAT2 alone did not alter the bp phenotype, whereas the inactivation of KNAT6 partially rescued it (Figures 1C and 1D). The downward orientation of siliques in bp mutants was partially rescued in knat6 bp double mutants and was completely rescued in the absence of both KNAT6 and KNAT2 (Figures 1D, 1E, and 2D). We performed quantitative phenotypic analyses on 10 plants of each genotype to further characterize the double and triple mutants. The height of the plants, the number of rosette paraclades, and the internode size between siliques along the main inflorescence were determined. These measurements first were performed on the knat2 and knat6 single mutants and on the knat2 knat6 double mutant and confirmed that these mutants exhibit essentially wild-type development with respect to these features (Belles-Boix et al., 2006; see Supplemental Figures 1 and 2 online). The same analyses were then performed on bp, knat2 bp, knat6 bp, and knat2 knat6 bp mutants. The average heights of the bp and knat2 bp mutants were 21 cm shorter, and those of knat6 bp and knat6 knat2 bp were 7 cm shorter than that of the wild type (Figure 2A). The bp mutants showed a partial loss of apical dominance as they produced six to eight rosette paraclades, whereas wild-type plants produced three to five paraclades on average (Figure 2B). This defect was rescued in the knat6 bp and knat6 knat2 bp mutants but not in the knat2 bp mutants (Figure 2B). We then examined the distribution of siliques along the main inflorescence. The bp mutants showed short internodes as 75% of bp internodes ranged in length from 0 to 5 mm, whereas 74% of wild-type internodes ranged in length from 6 to 15 mm (Figure 2C). The distribution of internode length along the main inflorescence of the knat2 bp double mutants was similar to that of bp (Figure 2C). By contrast, an increase of internode length was observed in the knat6 bp and knat6 knat2 bp mutants, since 50% of internodes ranged in length from 6 to 15 mm in both genotypes (Figure 2C). Thus, the inactivation of KNAT6 partially rescued the bp phenotypic features; by contrast, the inactivation of KNAT2 did not show such an effect. However, a redundant role of KNAT2 with KNAT6 in silique orientation was revealed (Figure 2D).
Figure 1.
Phenotype of the knat2 bp, knat6 bp, and knat2 knat6 bp Mutants.
(A) Wild-type inflorescence.
(B) bp inflorescence with downward oriented siliques.
(C) knat2 bp inflorescence. The inactivation of KNAT2 did not alter the bp phenotype.
(D) knat6 bp inflorescence. The inactivation of KNAT6 partially rescued the bp phenotype.
(E) knat2 knat6 bp inflorescence. The orientation of the siliques was wild type in the knat2 knat6 bp triple mutant.
Figure 2.
Quantitative Phenotypic Analyses of the knat2 bp, knat6 bp, and knat2 knat6 bp Mutants.
Ten plants for each genotype were analyzed.
(A) Average (+sd) height of wild-type, bp, knat2 bp, knat6 bp, and knat2 knat6 bp plants. The inactivation of KNAT6 partially rescued the defect in size of bp.
(B) Average (+sd) number of rosette paraclades of wild-type, bp, knat2 bp, knat6 bp, and knat2 knat6 bp plants.
(C) Distribution of the length of the internode between two successive siliques. Ten internodes between the 1st and 11th siliques (counting acropetally) were measured.
(D) Orientation of the pedicels in the wild type, bp, knat2 bp, knat6 bp, and knat2 knat6 bp.
KNAT6 and KNAT2 Expression Domains Are Enlarged in the bp Mutant
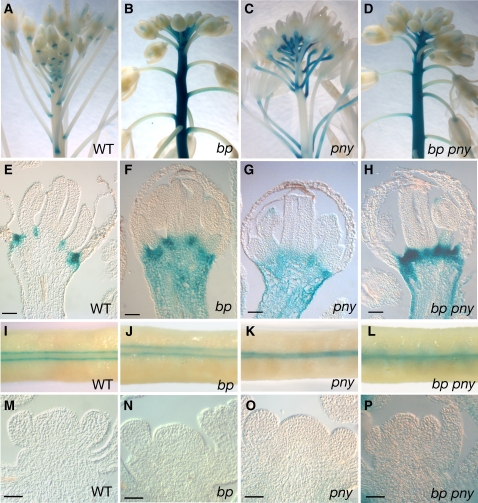
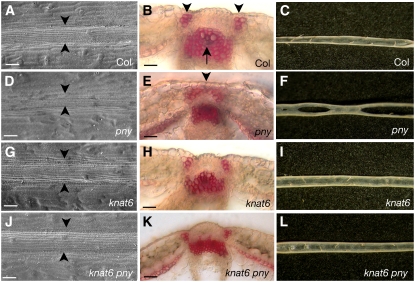
Since we observed that bp defects were partially rescued in the knat6 bp and knat2 knat6 bp mutants, we examined the expression patterns of KNAT2 and KNAT6 in bp mutants. The expression patterns of PKNAT6-GUS (for β-glucuronidase) and PKNAT2:GUS fusions were analyzed in the bp-9 background. In wild-type inflorescences, KNAT6 and KNAT2 expression overlapped partially. Both KNAT6-GUS and KNAT2-GUS activities were detected in the floral pedicel axils (Figures 3A and 4A). In floral buds, KNAT6-GUS was restricted to the boundaries between floral organs, whereas KNAT2-GUS was expressed at the base of the floral meristem (Figures 3E and 4E). By contrast, in the bp inflorescences, the domains of KNAT6 and KNAT2 expression were enlarged, although their patterns were distinct. In bp mutants, an ectopic KNAT6-GUS activity was present in the main stem and in young pedicels, whereas an ectopic KNAT2-GUS activity was detected only in the pedicels (Figures 3B, 3F, 4B, and 4F). Thus, in wild-type inflorescences, BP restricted KNAT6 expression in pedicels and stem and KNAT2 expression in pedicels.
Figure 3.
KNAT6-GUS Expression in bp, pny, and bp pny Mutants.
(A) Wild-type inflorescence showing GUS activity in the axil and in the distal part of the pedicel.
(B) bp inflorescence showing GUS activity in the main stem and in the pedicels.
(C) pny inflorescence showing GUS activity in the pedicels.
(D) bp pny inflorescence showing GUS activity in the main stem and in the pedicels.
(E) Median longitudinal section through a wild-type stage 12 flower showing GUS activity restricted to the boundaries between floral organs (bar = 40 μm).
(F) Median longitudinal section through a bp stage 12 flower showing GUS activity in the pedicel (bar = 40 μm).
(G) Median longitudinal section through a pny stage 12 flower showing GUS activity in the pedicel (bar = 40 μm).
(H) Median longitudinal section through a bp pny stage 12 flower showing GUS activity in the pedicel (bar = 40 μm).
(I) Wild-type carpel showing GUS activity in boundaries between the valves and the replum.
(J) bp carpel showing GUS activity in valves margins.
(K) pny carpel showing GUS activity between the valves. The pny carpel has a narrower replum.
(L) bp pny carpel showing GUS activity between the valves.
(M) Longitudinal median section through a wild-type inflorescence meristem showing no GUS activity (bar = 40 μm).
(N) bp inflorescence meristem with no GUS activity (bar = 40 μm).
(O) pny inflorescence meristem with no GUS activity (bar = 40 μm).
(P) bp pny inflorescence meristem with no GUS activity (bar = 40 μm).
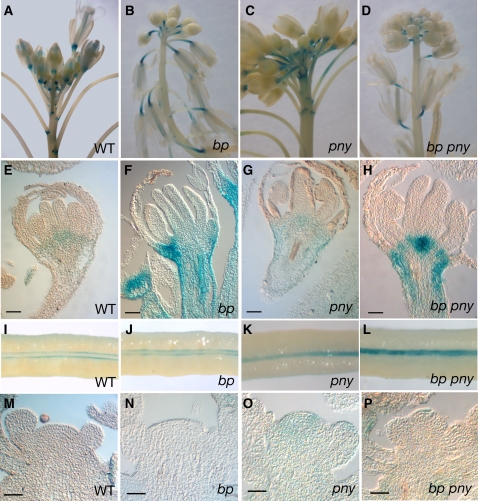
Figure 4.
KNAT2-GUS Expression in bp, pny, and bp pny Mutants.
(A) Wild-type inflorescence showing GUS activity in the axil and in the distal part of the pedicels.
(B) bp inflorescence showing GUS activity in the pedicels.
(C) pny inflorescence showing GUS activity in the pedicels.
(D) bp pny inflorescence showing GUS activity in the pedicels.
(E) Median longitudinal section through a wild-type stage 12 flower showing GUS activity restricted to the boundaries between floral organs (bar = 40 μm).
(F) Median longitudinal section through a bp stage 12 flower showing GUS activity in the pedicel (bar = 40 μm).
(G) Median longitudinal section through a pny stage 12 flower showing GUS activity in the pedicel (bar = 40 μm).
(H) Median longitudinal section through a bp pny stage 12 flower showing GUS activity in the pedicel (bar = 40 μm).
(I) Wild-type carpel showing GUS activity in boundaries between the valves and the replum.
(J) bp carpel showing GUS activity in valves margins.
(K) pny carpel showing GUS activity between the valves. The pny carpel has a narrower replum.
(L) bp pny carpel showing GUS activity between the valves.
(M) Longitudinal median section through a wild-type inflorescence meristem showing no GUS activity (bar = 40 μm).
(N) bp inflorescence meristem with no GUS activity (bar = 40 μm).
(O) Longitudinal section through a pny inflorescence meristem. GUS activity barely was detected (bar = 40 μm).
(P) bp pny inflorescence meristem with no GUS activity (bar = 40 μm).
The Inactivation of KNAT6 Rescues the pny Phenotype
Since BP and PNY cooperate to regulate inflorescence development (Smith and Hake, 2003), we examined the interaction of KNAT2 and KNAT6 with PNY. pny mutants are characterized by phyllotaxy defects, partial loss of apical dominance, and reduced stature (Byrne et al., 2003; Roeder et al., 2003; Smith and Hake, 2003; Bhatt et al., 2004). In addition, the pny mutant shows fruit alterations, such as defects in replum differentiation and in septum fusion (Byrne et al., 2003; Roeder et al., 2003). To investigate the interaction between KNAT2, KNAT6, and PNY, we generated the knat2 pny, knat6 pny, and knat2 knat6 pny mutants. The knat2 pny double mutants showed a pny phenotype (Figures 5 and 6; see Supplemental Figure 3 online) and confirmed previous data (Byrne et al., 2003). Surprisingly, the knat6 pny double mutant showed a wild-type phenotype (Figure 5D). The same phenotype was obtained using both alleles of KNAT6 (data not shown). The distribution of organs along inflorescences in the knat6 pny double mutant was the same as in the wild type. Moreover, the average height of the knat6 pny double mutant plants was the same as that of the wild type, whereas the pny mutant plants were 10 cm shorter on average than the wild type (Figure 6A). The partial loss of apical dominance seen in the pny mutants was rescued in the knat6 pny double mutant (Figure 6B). Whereas pny produced five to eight rosette paraclades, the knat6 pny double mutants produced four to five paraclades. In addition, the distribution of the internode lengths between flowers was comparable in the wild type and in the knat6 pny double mutant (Figure 6C). The pny mutants were characterized by internodes with irregular lengths, being shorter than 6 mm in 38% of the internodes, and longer than 25 mm in 15% of the internodes. By contrast, 63 and 60% of the internodes ranged in length from 6 to 15 mm in inflorescences from the knat6 pny double mutant and the wild type, respectively.
Figure 5.
Phenotypes of the knat2 pny, knat6 pny, and knat2 knat6 pny Mutants.
(A) Wild-type inflorescence.
(B) pny inflorescence with irregular internode lengths.
(C) knat2 pny inflorescence. The inactivation of KNAT2 did not alter the pny phenotype.
(D) knat6 pny inflorescence. The inactivation of KNAT6 rescued the pny phenotype.
(E) knat2 knat6 pny inflorescence. The inactivation of KNAT2 did not alter knat6 pny double mutant phenotype.
Figure 6.
Quantitative Phenotypic Analyses of the knat2 pny, knat6 pny, and knat2 knat6 pny Mutants.
Ten plants for each genotype were analyzed.
(A) Average (+sd) height of wild-type, pny, knat2 pny, knat6 pny, and knat2 knat6 pny plants. The inactivation of KNAT6 rescued the size defect of pny.
(B) Average (+sd) number of rosette paraclades of wild-type, pny, knat2 pny, knat6 pny, and knat2 knat6 pny plants. The partial loss of apical dominance of pny was rescued in the absence of KNAT6.
(C) Distribution of the length of the internode between two successive siliques. Ten internodes between the 1st and 11th siliques (counting acropetally) were measured. The distribution of siliques was wild type in knat6 pny and in knat2 knat6 pny mutants.
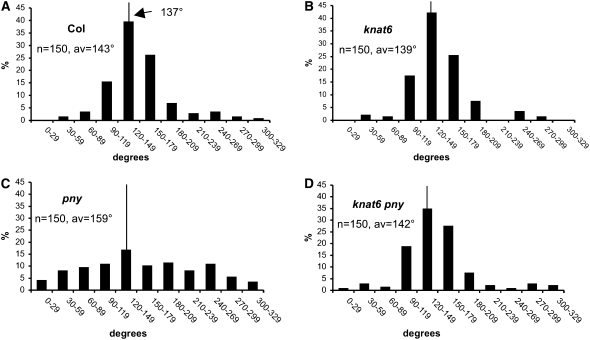
To further describe the internode patterning in the knat6 pny double mutant, we measured the phyllotactic pattern using the device and the method described previously (Peaucelle et al., 2007). The divergence angle between the insertion points of two successive floral pedicels along the main inflorescence was determined. Fifteen divergence angles between the 1st and 16th siliques (counting acropetally) of each inflorescence were measured according to the orientation of the generative spiral. In wild-type Columbia (Col) ecotype, the mean divergence angle was 143° (n = 150). Only 39% of the divergence angles fell within the range from 120 to 149, which contains the theoretical angle of 137.5° (Figure 7A). This proportion increased to 80% if the class was enlarged to encompass angles ranging from 90° to 179°. The value of the mean divergence angle was higher than the theoretical value and more variable. This was consistent with the recent observation showing that the phyllotactic pattern in Arabidopsis is less robust during reproductive development than in the inflorescence meristem and results from growth variation (Peaucelle et al., 2007). No differences in the phyllotactic pattern could be detected in the knat6 single mutant since 84% of the divergence angles fell into the 90° to 179° classes (Figure 7B). By contrast, a uniform distribution of divergence angles was observed in pny, confirming the disruption of the phyllotactic pattern in this mutant (Figure 7C). This defect was rescued in knat6 pny double mutants as 81% of the divergence angles fell into the 90° to 179° classes (Figure 7D). Together with the measurements of internode lengths between flowers, these analyses indicate that the inactivation of KNAT6 rescued the phyllotaxy defects of pny.
Figure 7.
Phyllotaxy Measurements.
The divergence angle between two successives siliques along the main inflorescence was determined. Fifteen angles between the 1st and 16th siliques counting acropetally were measured. Ten plants for each genotype were analyzed. On each panel, the average value (av) is labeled, and the class containing the theoretical value (137°) is marked with a line.
(A) The distribution of divergence angles in the wild-type ecotype is shown.
(B) The knat6 mutant phyllotactic pattern showed a wild-type distribution.
(C) The pny mutant had a uniform distribution of divergence angles.
(D) The knat6 pny double mutant phyllotactic pattern showed a wild-type distribution.
The pny mutant also exhibits defects in fruit development as the pny fruit shows a narrower replum, an abnormal pattern of lignification, and septum fusion defects (Roeder et al., 2003; Figures 8D to 8F compared with 8A to 8C). The replum and the septum of the knat6 mutant were wild type (Figures 8G to 8I). Characterization of the knat6 pny fruit showed that the fruit defects seen in pny were rescued (Figures 8J to 8L). The knat6 knat2 pny triple mutant exhibited a wild-type phenotype (Figures 5 and 6; see Supplemental Figure 3 online). All together, these results show that the inactivation of KNAT6 rescued the pny defects, whereas the inactivation of KNAT2 did not have such an effect.
Figure 8.
Phenotypes of the pny, knat6, and knat6 pny Siliques.
(A) to (C) Wild-type stage 17 fruit.
(A) Scanning electron micrograph showing the replum region (bar = 20 μm). The replum is the ridge between the two valves (arrowheads).
(B) Transverse section of the replum stained with phloroglucinol to detect lignification (pink). In the wild type, the lignification was detected at the valves margins (arrowheads) and in the inner replum (arrow) (bar = 40 μm).
(C) Septum.
(D) to (F) pny stage 17 fruit.
(D) Scanning electron micrograph showing a narrow replum region (arrowheads) (bar = 20 μm).
(E) Transverse section of pny fruit showing the lignified layer that extends across the replum.
(F) pny septum with fusion defects.
(G) to (I) knat6 stage 17 fruit.
(G) Scanning electron micrograph showing a wild-type replum surface (bar = 20 μm).
(H) Transverse section of knat6 fruit showing a wild-type lignification pattern (bar = 40 μm).
(I) The knat6 septum was wild type.
(J) to (L) knat6 pny stage 17 fruit. The inactivation of KNAT6 rescued pny fruit defects.
(J) Scanning electron micrograph showing a wild-type replum surface (bar = 20 μm).
(K) Transverse section of knat6 pny fruit showing a wild-type lignification pattern in the replum region (bar = 40 μm).
(L) The knat6 pny septum showing no fusion defects.
KNAT6 and KNAT2 Expression Domains Are Enlarged in the pny Mutant
We then examined the expression patterns of KNAT6 and KNAT2 in the pny mutants using PKNAT6-GUS and PKNAT2:GUS, respectively. As shown in Figures 3C, 3G, 4C, and 4G, domains of both KNAT6 and KNAT2 expression were enlarged in pny pedicels. In wild-type plants, KNAT2 and KNAT6 are expressed in the SAM (Pautot et al., 2001; Belles-Boix et al., 2006) and are downregulated during floral transition. Figures 3M and 4M show that KNAT6-GUS and KNAT2-GUS activities were not detected in wild-type inflorescence meristems. In pny inflorescence meristems, KNAT6-GUS activity was not detected, whereas a weak KNAT2-GUS activity was detected (Figures 3O and 4O). Thus, the downregulation of KNAT6 in the pny inflorescence meristem still was maintained, suggesting that the phyllotaxy defects seen in pny mutant results from the ectopic expression of KNAT6 in the pedicels. By contrast, the altered expression of KNAT2 observed in the pny mutant has no effect, as the knat2 pny double mutant exhibited a pny phenotype. Since the inactivation of KNAT6 rescued the replum and septum pny defects, we also examined the expression pattern of KNAT6 in pny carpels. The KNAT2 expression was analyzed in parallel. In wild-type carpels, KNAT2 and KNAT6 expressions were restricted to the boundaries between the replum and ovary valves (Figures 3I and 4I; see Supplemental Figures 4C and 4D online; Pautot et al., 2001). In pny siliques, KNAT6 and KNAT2 expression domains coalesced due to the absence of the replum (Figures 3K and 4K).
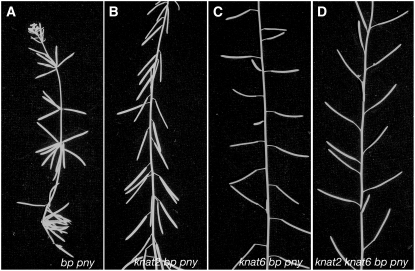
The Inactivation of KNAT6 Rescues Partially the bp pny Double Mutant Phenotype
To further examine the interaction with BP and PNY, we constructed the knat6 bp pny triple mutant. In parallel, we generated the knat2 bp pny and the knat2 knat6 bp pny mutants to test a potential redundancy of KNAT6 with KNAT2. The bp pny double mutant shows an additive phenotype, with extremely short internodes, downward-oriented organ clusters, increased branching, and more severe fruit defects (Figures 9A and 10; Byrne et al., 2003; Smith and Hake, 2003; Alonso-Cantabrana et al., 2007). The inactivation of KNAT2 and KNAT6 in the bp pny background had distinct effects. While the bp pny alterations were attenuated in the absence of KNAT2, they were greatly reduced in the absence of KNAT6 (Figures 9B, 9C, and 10). The average height of the bp pny mutant was 11.6 cm, whereas the average height of the wild type was 48.3 cm. The average heights of knat2 bp pny and knat6 bp pny were 20.7 and 39.8, respectively (Figure 10A). The bp pny mutant plants exhibited a strong loss of apical dominance as they produced 13 to 19 paraclades, whereas wild-type plants produced four to five paraclades (Figure 10B). This defect was partially rescued in the absence of KNAT2 as knat2 bp pny produced 10 to 13 paraclades and almost restored in the absence of KNAT6 as knat6 bp pny produced four to six paraclades (Figure 10B). One of the most striking features of the bp pny phenotype is the presence of extremely short internodes, with 88% of internodes ranging in length from 0 to 5 mm. This feature was slightly reversed in the absence of KNAT2, as 68% of the internodes ranged in length from 0 to 5 mm, and reversed to a greater extent in the absence of KNAT6, as 53% of the knat6 bp pny internodes were 6 to 15 mm long (Figure 10C). The knat2 knat6 bp pny quadruple mutant displayed the same phenotypic features as the knat6 bp pny triple mutant except for the orientation of the siliques. The average size of the quadruple mutant was 36.7 cm, with 50% of its internodes ranging in length from 6 to 15 mm, and the number of paraclades did not exceed 6 (Figure 10). The silique orientation defect was partially rescued in the absence of KNAT6 and was suppressed in the absence of both KNAT2 and KNAT6 (Figure 10D). Thus, the defects seen in the bp pny double mutant were reduced greatly in the absence of KNAT6, confirming the higher impact of the KNAT6 inactivation compared with KNAT2. Nevertheless, we found a slight effect of the KNAT2 inactivation in the absence of both BP and PNY. The KNAT2 effect on the orientation of the siliques was seen only when both KNAT6 and BP were absent, as previously seen in the knat2 knat6 bp triple mutant.
Figure 9.
Phenotype of the knat2 bp pny, knat6 bp pny, and knat2 knat6 bp pny Mutants.
(A) bp pny inflorescence with clusters of downward-oriented organs.
(B) knat2 bp pny inflorescence. The inactivation of KNAT2 partially rescued the bp pny phenotype.
(C) knat6 bp pny inflorescence. The inactivation of KNAT6 partially rescued the bp pny phenotype.
(D) knat2 knat6 bp pny inflorescence showing a stronger restoration than the knat2 bp pny or knat6 bp pny inflorescences.
Figure 10.
Quantitative Phenotypic Analyses of the bp pny, knat2 bp pny, knat6 bp pny, and knat2 knat6 bp pny Mutants.
Ten plants for each genotype were analyzed.
(A) Average (+sd) height of wild-type, bp pny, knat2 bp pny, knat6 bp pny, and knat2 knat6 bp pny plants. The shorter size of bp pny was partially rescued in the absence of KNAT6 and to a lesser extent in the absence of KNAT2.
(B) Average (+sd) number of rosette paraclades of wild-type, bp pny, knat2 bp pny, knat6 bp pny, and knat2 knat6 bp pny plants. The bp pny partial loss of apical dominance was partially rescued in the absence of KNAT6 and to a lesser extent in the absence of KNAT2.
(C) Distribution of the length of the internode between two successive siliques. Ten internodes between the 1st and 11th siliques (counting acropetally) were measured. The reduced size of internodes observed in bp pny was partially rescued in the absence of KNAT6 and to a lesser extent in the absence of KNAT2.
(D) Orientation of the pedicels in the wild type, bp pny, knat2 bp pny, knat6 bp pny, and knat2 knat6 bp pny.
We also checked whether KNAT6 inactivation restored the replum defect seen in bp pny. BP is expressed in the replum, and a role for BP in replum development has been suggested recently (Alonso-Cantabrana et al., 2007). The bp mutant shows wild-type replum phenotypes, but its inactivation enhances the replum defect of rpl mutant alleles (Alonso-Cantabrana et al., 2007). By contrast, we showed that the inactivation of KNAT6 rescued the bp pny replum defect (see Supplemental Figures 3I and 3K online). Thus, the inactivation of KNAT6 had the opposite effect when compared with that of BP on replum development. This was consistent with the expression pattern of KNAT6 that marked the boundaries between the replum and the valves (see Supplemental Figure 4D online). On the other hand, KNAT2 expression overlaps with that of KNAT6 in the carpel, but the inactivation of KNAT2 did not have such an effect (Figures 3I and 4I; see Supplemental Figures 3G, 4C, and 4D online; Alonso-Cantabrana et al., 2007). knat2 bp, knat6 bp, and knat2 knat6 bp showed wild-type replum phenotypes, confirming that KNAT6 and KNAT2 are not redundant with BP for replum specification (see Supplemental Figures 3D to 3F online). By contrast, both STM and BP are expressed in the replum (see Supplemental Figures 4A and 4B online; Long et al., 1996; Alonso-Cantabrana et al., 2007). Together, this suggests that STM plays a redundant role with BP in replum specification and that KNAT class I members play distinct roles during carpel development.
KNAT6 and KNAT2 Expression Domains Are Enlarged in the bp pny Mutant
The expressions of KNAT6 and KNAT2 were examined in parallel in the bp pny double mutant. KNAT6-GUS activity was detected in the stems and pedicels of the bp pny double mutant (Figures 3D and 3H) and was not detected in bp and bp pny inflorescence meristems (Figures 3N and 3P). KNAT2-GUS activity was present in pedicels and barely was detected in inflorescence meristems of bp pny double mutants and was absent from bp inflorescence meristems (Figures 4D, 4H, 4N, and 4P). In bp carpels, KNAT6-GUS and KNAT2-GUS activities were restricted to the boundaries of the replum and ovary valves as in the wild type (Figures 3J and 4J). The expression domains of KNAT6 and KNAT2 in bp pny carpels coalesced as in pny (Figures 3L and 4L).
DISCUSSION
In this article, we examined the interaction of KNAT6 and KNAT2 with BP and PNY during inflorescence development. We previously showed that KNAT6 contributes with STM to SAM function and boundary maintenance. KNAT2 does not play such a role (Belles-Boix et al., 2006). Here, we showed that the loss of KNAT6 partially rescues bp defects and suppresses pny alterations. The loss of KNAT2 activity also partially rescues bp defects but only in the absence of KNAT6 or in the absence of PNY.
BP and PNY Restrict KNAT6 and KNAT2 Expression to Promote Correct Inflorescence Development
Our genetic and expression analyses indicate that the defects seen in pny, and to a lesser extent in bp, are likely attributable to the misexpression of KNAT6 since the removal of KNAT6 activity completely suppressed the pny defects and partially suppressed the bp defects. KNAT2 also was misexpressed in pny and bp, but its inactivation had a weaker impact than that of KNAT6. In bp mutants, the effect of the KNAT2 inactivation was restricted to the orientation of the siliques. Further experiments should be performed to determine whether BP and PNY directly interact with KNAT6 and KNAT2. Motifs nearly identical to the BP/KNOX binding site were found in KNAT2 and KNAT6 genes (Mele et al., 2003). In wild-type plants, BP is expressed in the internodes and the pedicels and promotes their growth. The downward-pointing phenotype in bp is due to a defect in pedicel differentiation and fewer cell divisions (Venglat et al., 2002; Smith and Hake, 2003; Douglas and Riggs, 2005). Our data showed that this phenotypic feature was correlated with a misexpression of both KNAT6 and KNAT2. Consistent with these data, the inactivation of KNAT6 and KNAT2 restored the wild-type orientation of the pedicel. Given that the pedicel phenotype in bp is due to a shift toward differentiation, we found that KNAT class I proteins show distinct activities in the development of the pedicel, with BP counteracting the undifferentiated state maintained by KNAT6 and KNAT2.
The interactions of KNAT6 and KNAT2 with PNY further confirmed their distinct activities: the inactivation of KNAT6 suppressed the pny defects, while the inactivation of KNAT2 did not. One of the most obvious defects of the pny mutant is its aberrant organ positioning. The pny mutants had internodes with irregular sizes and clusters of organs. In wild-type plants, organs are initiated successively on the flanks of the SAM with a spiral arrangement. This pattern, referred to as phyllotaxy, depends on predictive auxin gradients in the meristem (Reinhardt, 2005). However, previous studies indicated that the majority of pny mutants have a wild-type meristem structure. The size of the inflorescence meristem was slightly smaller and only 2 out 16 pny mutants showed an aberrant initiation of organs (Byrne et al., 2003). This suggests that the phyllotaxy defects seen in the pny mutant probably occurred during inflorescence growth. Our analyses indicated that the knat6 pny double mutant inflorescences had wild-type phyllotaxy. However, our expression analyses showed that the downregulation of KNAT6 was maintained in pny inflorescence meristems. Therefore, the suppression of the aberrant organ positions in knat6 pny is likely attributable to the misexpression of KNAT6 in pny pedicels, leading to abnormal pedicel differentiation. Our data are in agreement with recent data showing that phyllotaxy results not only from the pattern initiated in the meristem, but also from the subsequent growth during stem development. This was illustrated with plants disrupted in the microRNA regulation of the CUP-SHAPED COTYLEDON2 (CUC2) gene: ectopic expression of CUC2 in internodes leads to aberrant phyllotaxy (Peaucelle et al., 2007; Sieber et al., 2007). Both CUC2 and KNAT6 genes are expressed in restricted domains in wild-type meristems to specify boundaries. When they are ectopically expressed in the pedicel or in the internodes, they prevent the proper differentiation of those tissues. This indicates a similar effect of these two genes in and out of the SAM. Thus, our data suggest that in wild-type inflorescences, PNY restricts KNAT6 expression to maintain the proper phyllotaxy during stem development. KNAT2 also was misexpressed in pny, but its inactivation had no effect, suggesting a redundancy with other TALE members or a different role.
The interactions of KNAT2 and KNAT6 with BP and PNY confirmed the greater impact of KNAT6 inactivation compared with KNAT2 inactivation. This genetic analysis revealed an effect of KNAT2 inactivation in the absence of both PNY and BP. This data confirms the redundancy between KNAT2 and KNAT6 in the absence of BP. Overlapping and distinct roles of transcription factors have been described for the homeodomain-leucine zipper family (Prigge et al., 2005). This type of interaction could provide flexibility in the regulation of distinct phases of development during plant growth.
The Loss of KNAT6 Also Suppresses the pny Fruit Defect
In carpels, KNAT6 and KNAT2 are expressed in the boundaries between the replum and the valves, while BP, STM, and PNY are expressed in the replum (this work; Long et al., 1996; Roeder et al., 2003; Alonso-Cantabrana et al., 2007). Both its layered structure and its role in generating ovule primordia make the replum a true meristem. Recently, a role for BP in replum development has been suggested since the inactivation of BP enhances the replum defect of the rpl allele (Alonso-Cantabrana et al., 2007). By contrast, we showed that the inactivation of KNAT6 suppresses the replum and septum defects seen in pny and in bp pny. Consistent with the absence of replum in pny siliques, KNAT6 expression domains coalesced. This suggests that PNY restricts KNAT6 in the wild type to promote replum identity. However, PNY is not required per se for replum specification, as double or triple mutant combinations involving pny or rpl alleles (asymmetric leaves1 rpl, shatterproof1 [shp1] shp2 rpl, jagged rpl, filamentous flower rpl, and knat6 pny) develop a normal replum (this work; Roeder et al., 2003; Dinneny et al., 2005; Alonso-Cantabrana et al., 2007). This suggests that other factors are redundant to specify replum identity. Since the knat2 knat6 bp triple mutants had a normal replum, STM could play such a role. This is in agreement with the expression of STM in the replum.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana plants were grown either on soil or in vitro as described by Hamant et al. (2002). All mutants used in this study are in the Col-0 background. Mutant alleles of knat6-1, knat6-2, and knat2-5 were obtained from the Nottingham Arabidopsis Stock Centre (http://nasc.nott.ac.uk) and were previously described (Belles-Boix et al., 2006). The bp-9 mutant allele was provided by Naomi Ori (Smith and Hake, 2003). The pny (pny-40126 or rpl-2, Col background) mutant allele was obtained from the Nottingham Arabidopsis Stock Centre (http://nasc.nott.ac.uk) and described by Smith and Hake (2003). The translational KNAT2-GUS (Dockx et al., 1995) and transcriptional KNAT6-GUS (Belles-Boix et al., 2006) fusions have been described previously.
Plant Genetics
To construct the knat6-1 pny double mutant, plants homozygous for knat6-1 were crossed to plants homozygous for pny (pny-40126), and knat6-1 pny double mutants were selected. Only two phenotypes segregated in the F2: the wild type and the pny phenotype. Genomic DNA was extracted and analyzed by PCR to select knat6-1 pny double mutants. To detect the knat6-1 allele, the T-DNA left border primer Lba 1 (5′-TGGTTCACGTAGTGGGCCATCG-3′) and the KNAT6-specific primer KNAT6-03 (5′-GAAGATAAACCCTAGCTACAAG-3′) were used. The KNAT6 wild-type allele was detected using the KNAT6-03 and KNAT6-04 (5′-AACCTGCATCGATCTATTTTC-3′) primers. The pny-40126 allele was genotyped as described (Smith and Hake, 2003). The knat6-1 pny double mutant showed the wild-type phenotype. The same phenotype was observed when using the knat6-2 allele. To detect the knat6-2 allele, the T-DNA left border primer Lba 1 and the KNAT6-specific primer KNAT6-15 (5′-AGATAAGTCGGTTCTGATGATG-3′) were used. The KNAT6 wild-type allele was detected using KNAT6-15 and KNAT6-41 (5′-GCTACCAATCATTTTTCAGAAACTGGTCA-3′) primers.
To generate the knat2-5 pny double mutant, plants homozygous for knat2-5 were crossed to plants homozygous for pny (pny-40126), and knat2-5 pny double mutants were selected. Only two phenotypes segregated in the F2: the wild type and the pny phenotype. Plants were analyzed by PCR to select the knat2-5 pny double mutant. To detect the knat2-5 mutant allele, the T-DNA left border primer Lba 1 and the KNAT2-specific primer 4K1 (5′-CGCTTCTCATCCTTTGTATC-3′) were used. The KNAT2 wild-type allele was detected using 4K1 and K11 (5′-TACCCATCAGTCTCTTAATG-3′) primers. The knat2-5 pny double mutant showed the pny phenotype.
To generate the knat6-1 knat2-5 pny triple mutant, plants homozygous for knat6-1 pny were crossed to plants homozygous for knat2-5 pny. Only two phenotypes segregated in the F2: the wild type and the pny phenotype. Plants were analyzed by PCR to select the knat6-1 knat2-5 pny triple mutant. The triple mutant exhibits the wild-type phenotype.
To generate the knat6-1 bp-9 double mutant, plants homozygous for knat6-1 were crossed to bp-9. Double mutants were selected by PCR analysis. To detect the bp-9 mutant allele, the dSpm1 (5′- CTTATTTCAGTAAGAGTGTGGGGTTTTGG-3′) and the BP-specific primer BP-14 were used. The BP wild-type allele was detected with BP-14 (5′-TGTTAAGGGTTAGAACACCATG-3′) and BP-3 (5′-GACAACAGCACCACTCCTCAAA-3′) primers. The bp phenotype was partially rescued in the knat6-1 bp-9 double mutant.
To generate the knat2-5 bp-9 double mutant, plants homozygous for knat2-5 were crossed to bp-9, and double mutants were selected by PCR analysis in the F2. The knat2-5 bp-9 double mutant exhibits a bp phenotype.
To generate the knat6-1 knat2-5 bp-9 triple mutant, plants homozygous for knat6-1 bp-9 were crossed to plants homozygous for knat2-5 bp-9. Plants homozygous for knat6-1 bp-9 and knat2-5 were selected by PCR analysis. The bp phenotype was partially rescued in the knat6-1 knat2-5 bp-9 triple mutant.
To generate the bp-9 pny double mutant, plants homozygous for pny were crossed to plants homozygous for bp-9. F2 plants showing the bp-9 pny phenotype were selected and checked by PCR analysis.
To generate the knat6-1 bp-9 pny triple mutant, the knat6-1 pny double mutant was crossed to the knat6-1 bp-9 double mutant. F2 plants homozygous for knat6-1 bp-9 pny were selected by PCR analysis. The bp pny phenotype was partially rescued in the knat6-1 bp-9 pny triple mutant.
To generate the knat2-5 bp-9 pny triple mutant, the knat2-5 pny double mutant was crossed to the knat2-5 bp-9 double mutant. F2 plants homozygous for knat2-5 bp-9 pny were selected by PCR analysis. The knat2-5 bp pny triple mutant showed a bp pny phenotype.
To generate the knat6-1 knat2-5 bp-9 pny quadruple mutant, the knat6-1 bp-9 pny triple mutant was crossed to the knat2-5 bp-9 pny triple mutant. F2 plants homozygous for knat6-1 knat2-5 bp-9 pny were selected by PCR analysis. The bp pny phenotype was partially rescued in the knat6-1 knat2-5 bp-9 pny quadruple mutant.
Phenotypic analyses were conducted on 10 plants for each genotype: knat2-5, knat6-1, knat6-2, knat2-5 knat6-1, knat2-5 knat6-2, knat6-1 bp-9, knat2-5 bp-9, knat6-1 knat2-5 bp-9, knat6-1 pny, knat2-5 pny, knat6-2 pny, knat6-1 knat2-5 pny, knat2-5 bp-9 pny, knat6-1 bp-9 pny, and knat6-1 knat2-5 bp-9 pny.
To examine the expression of KNAT6 in pny, bp, and bp pny, the PKNAT6-GUS homozygous line was crossed to pny, bp-9, and the bp-9 pny double mutant. The F2 plants showing the pny, the bp-9, or the bp-9 pny phenotype, respectively, and KNAT6-GUS activity were selected. The F3 progenies of these F2 plants were examined for GUS activity to select plants homozygous for the PKNAT6-GUS construct.
To examine the expression of KNAT2 in pny, bp, and bp pny, the PKNAT2:GUS homozygous line was crossed to the pny, the bp-9, or the bp-9 pny double mutant. F2 plants showing the pny, the bp-9, or the bp-9 pny phenotype, respectively, and KNAT2-GUS activity were selected. The F3 progenies of these F2 plants were examined for GUS activity to select plants homozygous for the PKNAT2:GUS construct.
Phenotypic Analysis
Quantitative analyses of the different genotypes were performed on 9-week-old plants as described (Smith and Hake, 2003). Ten plants for each genotype were used for phenotypic analyses. The phyllotaxy measurements were performed using the device described previously (Peaucelle et al., 2007). The divergence angle between the insertion points of two successive floral pedicels along the main inflorescence was measured. Fifteen divergence angles between the 1st and 16th siliques (counting acropetally) of each inflorescence were measured according to the orientation of the generative spiral.
In Situ Localization of GUS Activity
GUS staining was performed as described previously (Pautot et al., 2001). Tissue samples were fixed in 4% formaldehyde, dehydrated, and embedded in paraffin as described (Pautot et al., 2001). Paraffin sections (10 μm thick) were cut on a Leica-Jung (Leica Rueil-Malmaison) RM2055 rotary microtome carrying a disposable metal knife. Sections were attached to a precoated glass slide (Fisher) and photographed.
In Situ Hybridization
In situ hybridizations were performed as described previously (Belles-Boix et al., 2006). The BP antisense probe was generated using pDONOR201 that contains the BP cDNA as a template and using BP-03 (5′-GACAACAGCACCACTCCTCAAA-3′) and BP T7 (5′-TGTAATACGACTCACTATAGGGCTTATGGACCGAGACGATAAGG-3′), which incorporates a binding site for T7 polymerase, as primers. The STM antisense probe was generated using pDONOR201 that contains the STM cDNA as a template and using STM-16 (5′-GGTTGTGGCGAGGCTAGAGG-3′) and STM T7 (5′-TGTAATACGACTCACTATAGGGCTCAAAGCATGGTGGAGGA-3′) as primers.
Lignin Staining
Carpels were embedded in 6% agarose (Sigma-Aldrich). Sections (70 μm thick) were cut on a Leica-VT 1000S (Leica Rueil-Malmaison) vibratome and stained with phloroglucinol (VWR Prolabo). Sections were photographed using a Nikon Microphot FXA microscope and a Jenoptik ProgRes C10 plus digital camera (Clara Vision).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers At1g70510 (KNAT2), At1g23380 (KNAT6), At4g08150 (BP), At5g02030 (PNY), and At1g62360 (STM).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Phenotypes of the Wild Type knat2-5, knat6-1, knat6-2, knat2-5 knat6-1, and knat2-5 knat6-2 Mutants.
Supplemental Figure 2. Quantitative Phenotypic Analyses of the knat2-5, knat6-1, knat6-2, knat2-5 knat6-1, and knat2-5 knat6-2 Mutants.
Supplemental Figure 3. Lignin Pattern in knat2, knat2 knat6, bp, knat2 bp, knat6 bp, knat2 knat6 bp, knat2 pny, knat2 knat6 pny, bp pny, knat2 bp pny, knat6 bp pny, and knat2 knat6 bp pny Stage 17 Carpels.
Supplemental Figure 4. KNAT Class I Member Expression Domains Were Distinct in the Carpel.
Supplementary Material
Acknowledgments
We thank the Salk Institute Genomic Analysis Laboratory for providing the sequence-indexed Arabidopsis T-DNA insertion mutants. We thank Naomi Ori for providing the bp-9 mutant allele. We thank Bruno Letarnec for greenhouse management, Patrick Laufs for his hepful discussions, and Elisabeth Truernit and Olivier Hamant for critical reading of the manuscript. Enric Belles-Boix was funded by the Region Ile de France and the Institut National de la Recherche Agronomique. Laura Ragni and Markus Günl were supported by the European Marie-Curie (FP6) Program.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Véronique Pautot (pautot@versailles.inra.fr).
Online version contains Web-only data.
References
- Alonso-Cantabrana, H., Ripoll, J.J., Ochando, I., Vera, A., Ferrandiz, C., and Martinez-Laborda, A. (2007). Common regulatory networks in leaf and fruit patterning revealed by mutations in the Arabidopsis ASYMMETRIC LEAVES1 gene. Development 134 2663–2671. [DOI] [PubMed] [Google Scholar]
- Belles-Boix, E., Hamant, O., Witiak, S.M., Morin, H., Traas, J., and Pautot, V. (2006). KNAT6: An Arabidopsis homeobox gene involved in meristem activity and organ separation. Plant Cell 18 1900–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt, A.M., Etchells, J.P., Canales, C., Lagodienko, A., and Dickinson, H. (2004). VAAMANA – A BEL1-like homeodomain protein, interacts with KNOX proteins BP and STM and regulates inflorescence stem growth in Arabidopsis. Gene 328 103–111. [DOI] [PubMed] [Google Scholar]
- Byrne, M.E., Groover, A.T., Fontana, J.R., and Martienssen, R.A. (2003). Phyllotactic pattern and stem cell fate are determined by the Arabidopsis homeobox gene BELLRINGER. Development 130 3941–3950. [DOI] [PubMed] [Google Scholar]
- Byrne, M.E., Simorowski, J., and Martienssen, R.A. (2002). ASYMMETRIC LEAVES1 reveals knox gene redundancy in Arabidopsis. Development 129 1957–1965. [DOI] [PubMed] [Google Scholar]
- Clark, S.E., Jacobsen, S.E., Levin, J.Z., and Meyerowitz, E.M. (1996). The CLAVATA and SHOOT MERISTEMLESS loci competitively regulate meristem activity in Arabidopsis. Development 122 1567–1575. [DOI] [PubMed] [Google Scholar]
- Dinneny, J.R., Weigel, D., and Yanofsky, M.F. (2005). A genetic framework for fruit patterning in Arabidopsis thaliana. Development 132 4687–4696. [DOI] [PubMed] [Google Scholar]
- Dockx, J., Quaedvlieg, N., Keultjes, G., Kock, P., Weisbeek, P., and Smeekens, S. (1995). The homeobox gene ATK1 of Arabidopsis thaliana is expressed in the shoot apex of the seedling and in flowers and inflorescence stems of mature plants. Plant Mol. Biol. 28 723–737. [DOI] [PubMed] [Google Scholar]
- Douglas, S.J., Chuck, G., Dengler, R.E., Pelecanda, L., and Riggs, C.D. (2002). KNAT1 and ERECTA regulate inflorescence architecture in Arabidopsis. Plant Cell 14 547–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas, S.J., and Riggs, C.D. (2005). Pedicel development in Arabidopsis thaliana: Contribution of vascular positioning and the role of the BREVIPEDICELLUS and ERECTA genes. Dev. Biol. 284 451–463. [DOI] [PubMed] [Google Scholar]
- Endrizzi, K., Moussian, B., Haecker, A., Levin, J.Z., and Laux, T. (1996). The SHOOT MERISTEMLESS gene is required for maintenance of undifferentiated cells in Arabidopsis shoot and floral meristems and acts at a different regulatory level than the meristem genes WUSCHEL and ZWILLE. Plant J. 10 967–979. [DOI] [PubMed] [Google Scholar]
- Hamant, O., Nogue, F., Belles-Boix, E., Jublot, D., Grandjean, O., Traas, J., and Pautot, V. (2002). The KNAT2 homeodomain protein interacts with ethylene and cytokinin signaling. Plant Physiol. 130 657–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanrar, S., Onguka, O., and Smith, H.M. (2006). Arabidopsis inflorescence architecture requires the activities of KNOX-BELL homeodomain heterodimers. Planta 224 1163–1173. [DOI] [PubMed] [Google Scholar]
- Long, J.A., Moan, E.I., Medford, J.I., and Barton, M.K. (1996). A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379 66–69. [DOI] [PubMed] [Google Scholar]
- Mele, G., Ori, N., Sato, Y., and Hake, S. (2003). The knotted1-like homeobox gene BREVIPEDICELLUS regulates cell differentiation by modulating metabolic pathways. Genes Dev. 17 2088–2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pautot, V., Dockx, J., Hamant, O., Kronenberger, J., Grandjean, O., Jublot, D., and Traas, J. (2001). KNAT2: Evidence for a link between knotted-like genes and carpel development. Plant Cell 13 1719–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peaucelle, A., Morin, H., Traas, J., and Laufs, P. (2007). Plants expressing a miR164-resistant CUC2 gene reveal the importance of post-meristematic maintenance of phyllotaxy in Arabidopsis. Development 134 1045–1050. [DOI] [PubMed] [Google Scholar]
- Prigge, M.J., Otsuga, D., Alonso, J.M., Ecker, J.R., Drews, G.N., and Clark, S.E. (2005). Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell 17 61–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragni, L., Truernit, E., and Pautot, V. (2007). KNOXing on the BELL:TALE homeobox genes and meristem activity. Int. J. Plant Dev. Biol. 1 42–44. [Google Scholar]
- Reinhardt, D. (2005). Regulation of phyllotaxis. Int. J. Dev. Biol. 49 539–546. [DOI] [PubMed] [Google Scholar]
- Roeder, A.H., Ferrandiz, C., and Yanofsky, M.F. (2003). The role of the REPLUMLESS homeodomain protein in patterning the Arabidopsis fruit. Curr. Biol. 13 1630–1635. [DOI] [PubMed] [Google Scholar]
- Sieber, P., Wellmer, F., Gheyselinck, J., Riechmann, J.L., and Meyerowitz, E.M. (2007). Redundancy and specialization among plant microRNAs: Role of the MIR164 family in developmental robustness. Development 134 1051–1060. [DOI] [PubMed] [Google Scholar]
- Smith, H.M., and Hake, S. (2003). The interaction of two homeobox genes, BREVIPEDICELLUS and PENNYWISE, regulates internode patterning in the Arabidopsis inflorescence. Plant Cell 15 1717–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venglat, S.P., Dumonceaux, T., Rozwadowski, K., Parnell, L., Babic, V., Keller, W., Martienssen, R., Selvaraj, G., and Datla, R. (2002). The homeobox gene BREVIPEDICELLUS is a key regulator of inflorescence architecture in Arabidopsis. Proc. Natl. Acad. Sci. USA 99 4730–4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.