Abstract
Stem cells in shoot and floral meristems of Arabidopsis thaliana secrete the signaling peptide CLAVATA3 (CLV3) that restricts stem cell proliferation and promotes differentiation. The CLV3 signaling pathway is proposed to comprise the receptor kinase CLV1 and the receptor-like protein CLV2. We show here that the novel receptor kinase CORYNE (CRN) and CLV2 act together, and in parallel with CLV1, to perceive the CLV3 signal. Mutations in CRN cause stem cell proliferation, similar to clv1, clv2, and clv3 mutants. CRN has additional functions during plant development, including floral organ development, that are shared with CLV2. The CRN protein lacks a distinct extracellular domain, and we propose that CRN and CLV2 interact via their transmembrane domains to establish a functional receptor.
INTRODUCTION
Plant aerial organs originate from the shoot apical meristem (SAM), a self-maintaining structure harboring stem cells (Stahl and Simon, 2005). Stem cells reside at the meristem center, and their descendants that are displaced to more lateral positions are destined for differentiation. In Arabidopsis thaliana, stem cell fate at the SAM is promoted by a group of underlying cells that express the homeodomain protein WUSCHEL (WUS) (Mayer et al., 1998). A mobile signaling peptide, CLAVATA3 (CLV3), that is secreted from the stem cells and restricts WUS expression acts as a negative feedback regulator (Fletcher et al., 1999; Brand et al., 2000; Schoof et al., 2000). Loss of the promotive factor WUS induces untimely cellular differentiation and the cessation of organ formation from the meristem. Loss of the restrictive CLV3 signal causes stem cell overproliferation and the enlargement of shoot and floral meristems, resulting in the formation of extra floral organs and club-shaped siliques (Clark et al., 1995).
Mutant screens and molecular genetic analysis identified the receptor-like kinase CLV1 and the receptor-like protein CLV2 as components of the CLV3 signaling pathway (Clark et al., 1993, 1997; Kayes and Clark, 1998; Jeong et al., 1999). CLV1 carries extracellular Leu-rich repeats, a transmembrane domain (TMD), and a Ser/Thr kinase domain on its cytoplasmic site; CLV2 resembles CLV1 but lacks the kinase. WUS expression expands in clv1, clv2, and clv3 mutants, while CLV3 overexpression arrests meristem function and downregulates WUS if CLV2 and CLV1 are functional (Brand et al., 2000; Schoof et al., 2000). Together, these data indicate that the CLV1, -2, and -3 proteins act together in a stem cell–repressing pathway that acts upon WUS as its major target. The prevalent model suggests that CLV3 encodes the ligand that binds to and activates a receptor complex consisting of CLV1 and CLV2, and a CLV3 peptide binding affinity of the extracellular domain of CLV1 was recently shown (Ogawa et al., 2008). Mutagenesis experiments have uncovered a range of mutant alleles for CLV1, among them several gain-of-function alleles that act as dominant negative repressors of CLV3 signaling (Dievart et al., 2003). One explanation for this is that the dominant clv1 mutants interfere with a second receptor system that acts independently of CLV1 to transmit the CLV3 signal.
Several other essential components of the pathway have been identified, mostly by genetic screens. Mutations in the POLTERGEIST (POL) gene, encoding a protein phosphatase 2C, suppress weak clv1 and clv2 mutants, suggesting that POL is negatively regulated by CLV3 signaling (Yu et al., 2000). Further analysis revealed that POL expression can activate WUS expression in the meristem (Yu et al., 2000, 2003; Song and Clark, 2005; Song et al., 2006). Mutations in the SHEPHERD (SHD) gene, encoding a HSP90-like protein, cause clv-like phenotypes. The SHD protein is possibly involved in the folding of CLV3 signaling components or the assembly of a functional receptor complex (Ishiguro et al., 2002).
The CLV3 peptide is processed from a larger precursor protein (Kondo et al., 2006). The active peptide consists of a 12–amino acid sequence that is conserved among members of the CLE protein family (Sharma et al., 2003). Misexpression of several different CLE peptides, including CLV3, can cause a rapid arrest of root meristem activity (Casamitjana-Martinez et al., 2003; Hobe et al., 2003). The main target in the root for CLE signaling pathways has not been identified, but in contrast to the situation in the shoot system, developmental inhibition is not due to the differentiation of stem cells. Instead, the proliferation rate of stem cell daughters is reduced and cell identities in the pericycle and endodermis layers are misspecified (Fiers et al., 2005). CLE-dependent developmental arrest of root meristem growth has been employed in suppressor screens that revealed the identity of additional pathway components (Casamitjana-Martinez et al., 2003). Mutations in SOL-1 (for Suppressor of Overexpression of LLP1-1), encoding a Zn2+-carboxypeptidase, rendered plants resistant to high-level expression of CLE19. Although sol-1 mutants were aphenotypic, SOL-1 function could be involved in the processing of CLE peptides.
We have identified additional components of the CLV3 signaling pathway using a similar strategy. Plants overexpressing CLV3 were mutagenized and screened for suppressor mutants that permit meristem maintenance. We report here on the cloning and analysis of CORYNE (CRN), encoding a membrane-associated receptor kinase that is required for CLV3 signaling. crn mutants exhibit clv-like phenotypes, indicating that CRN is an essential component of the CLV3 pathway. CRN shares many functions with CLV2, and we propose that CRN and CLV2 act closely together to transmit the CLV3 signal independently of CLV1.
RESULTS
A Suppressor Screen to Identify Components of the CLV3 Signaling Pathway
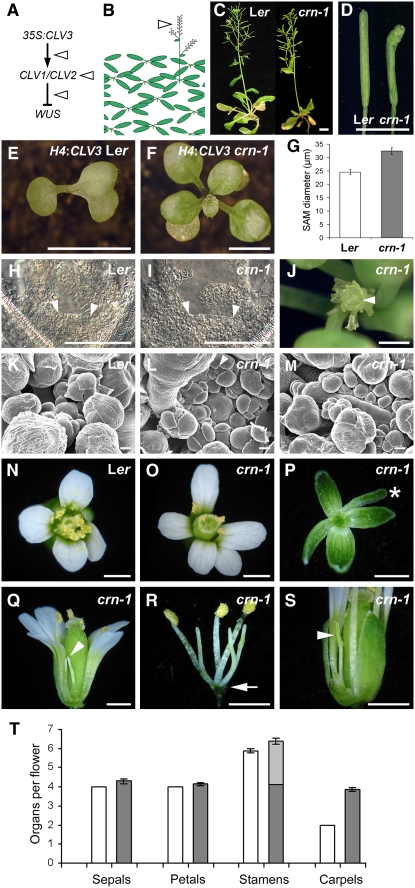
Because increased CLV3 signaling arrests meristem function due to the repression of WUS, we reasoned that essential components of the CLV3 signaling pathway can be identified in a screen for suppressor mutants (Figures 1A and 1B). We used a CLV3-overexpressing Arabidopsis line (Brand et al., 2000) that exhibits a milder meristem arrest phenotype and produces seeds within 12 weeks of germination. After ethyl methanesulfonate (EMS) mutagenesis, M2 seedlings were screened for earlier bolting, indicating that meristems were functional and that CLV3 signaling was impeded. Progeny from 40,000 M1 plants were analyzed, and eight mutants were recovered. We expected also to isolate new alleles of clv1 and clv2 in this screen, which we confirmed by allelism tests for five new clv1 mutants and two new clv2 mutants. One new mutant was not allelic to clv1 or clv2 and was named coryne, the Greek word for club (allele crn-1), due to its aberrant silique shape that resembled those of clv mutants (see below) (Figures 1C and 1D).
Figure 1.
Phenotype of crn-1 Mutants.
(A) Concept of the mutant screen. The arrowheads indicate the possible positions of suppressors in the CLV3 signaling pathway.
(B) Cartoon illustrating that suppressor mutants form inflorescence shoots with flowers.
(C) Wild-type (Ler) and crn-1 plants, 5 weeks after germination.
(D) crn-1 siliques are club-shaped and carry more carpels than wild-type siliques.
(E) and (F) An H4:CLV3 transgene arrests SAM development in wild-type (E) but not in crn-1 (F) seedlings.
(G) to (I) Increased meristem size in crn-1. SAM diameter was measured across the surface (arrowheads in [H] and [I]) (n = 40; ±se).
(J) crn-1 inflorescence at 29°C. The enlarged SAM arrests (arrowhead) with the formation of carpelloid organs.
(K) to (M) Wild-type (K) and crn-1 ([L] and [M]) meristems at 22°C. The arrowhead in (L) indicates the extra sepal. In (M), the inflorescence meristem is strongly fasciated.
(N) Wild-type flower, with four petals and six stamens.
(O) to (S) crn-1 flowers grown under continuous light.
(O) and (P) The gynoecium is enlarged, and some flowers carry five petals or additional sepals (asterisk in [P]).
(Q) Lateral view of the same flower shown in (O). Two stamens are shorter and lack anthers (arrowhead).
(R) Third whorl organs; three stamens lack anthers. The arrow indicates the flower basis.
(S) crn-1 stamens may remain immature (arrowhead).
In (P) to (S), organs or complete whorls were removed.
(T) Organ number in crn-1 mutant flowers (gray bars; n = 20) compared with wild-type flowers (white bars; n = 10) grown under continuous light. The light gray bars indicate the number of stamens lacking anthers. se is indicated.
Bars = 10 mm in (C), (E), and (F), 1 mm in (D), (J), and (N) to (S), and 50 μm in (H), (I), and (K) to (M).
The F1 progeny of crn-1 crossed to Landsberg erecta (Ler) showed a wus-like phenotype, indicating that the cauliflower mosaic virus (CaMV)35S:CLV3 transgene was still active and that the crn-1 phenotype was not due to cosuppression of the CLV3 transgene. F2 seedlings segregated for wild-type and wus-like plants and plants with club-shaped (clv-like) siliques in an ∼3:1 ratio (n = 92, segregating 70:22), indicating that the crn-1 mutation is fully recessive. Of 22 plants with clv-like siliques, 14 carried the CaMV35S:CLV3 transgene, confirming that the crn-1 mutant was responsible for the suppression of CLV3 overexpression.
For all further analyses, crn-1 mutant lines lacking the CaMV35S:CLV3 transgene were obtained by repeated outcrossing against Ler. We had initially used a CaMV35S:CLV3 transgenic line that permitted a limited recovery of meristem activity, probably due to reduced transgenic CLV3 expression or intermittent silencing of the CaMV35S promoter. Therefore, we reassayed the suppression capacity of crn-1 mutants by transformation of crn-1 mutants with a T-DNA that caused high-level CLV3 expression, either from the CaMV35S promoter or from the Histone4 (H4) promoter. The H4 promoter was chosen because it controls the expression of an essential gene in Arabidopsis and is unlikely to be inactivated by gene silencing. In a wild-type (Ler) background, all transgenic seedlings showed the expected wus-like growth behavior (n = 13), while all crn-1 mutant plants expressing the CLV3 transgene were indistinguishable from their untransformed siblings (n = 13) (Figures 1E and 1F). This complete suppression of CLV3 overexpression phenotypes in a crn-1 mutant led us to conclude that CRN is an essential component of the CLV signaling pathway that restricts WUS expression in Arabidopsis meristems.
CRN Controls Shoot Apical and Floral Meristem Size
Already at 10 d after germination, the surface diameter of crn-1 mutant meristems was increased by ∼30% compared with that in the wild type (Figures 1G to 1I). After bolting, crn-1 inflorescence meristems were enlarged and occasionally fasciated (Figures 1K to 1M), resulting in increased stem width. We noted that the stem fasciation of crn-1 mutant plants was more severe when the growth temperature was increased from 22 to 29°C. At elevated temperature, most crn-1 inflorescence meristems terminated with the production of carpelloid structures (22 of 29, at 56 d after germination) (Figure 1J). This could indicate either that crn-1 is a hypomorph with residual CRN activity, which is lost at higher temperatures, or that the development of the SAM becomes temperature-sensitive when CRN activity is lost.
Increased meristem size is mostly reflected by the production of additional organs during flower development. The crn-1 floral meristems appeared larger, and sepal primordia were broad with indentations at their tip that could mark the formation of supernumerary sepals. The floral organ number of crn-1 mutants was occasionally increased in all four whorls, compared with the 4-4-6-2 pattern of the wild type (Figures 1N to 1P). However, in most crn-1 flowers, only four stamens developed to maturity, carrying fertile anthers at their tips (Figures 1Q to 1T). The other two or three stamens either remained immature and did not produce fertile pollen or consisted of only short filaments that lacked anthers entirely. All crn-1 flowers carried an increased number of carpels in the fourth whorl that fused to form a club-shaped silique (Figure 1T). Similar to clv1, clv2, and clv3 mutants, a fifth whorl of organs was observed (see Supplemental Figure 1 online.)
The crn-1 mutant was identified in the Ler background, which carries a mutation in the ERECTA (ER) gene that causes a strong reduction in pedicel length (Torii et al., 1996). This effect of the er mutation is partially suppressed by clv2 and clv3 mutants but not by clv1 (Figures 2A and 2B) (Kayes and Clark, 1998). Pedicels subtending crn-1 mutant flowers were consistently longer than those of the corresponding Ler wild type. Together, the phenotype of crn-1 mutants suggested that CRN acts like the CLV genes in the control of shoot and floral meristem size. CRN exerts additional functions during stamen and pedicel development and shows extensive functional overlap with CLV2 in these organs (Kayes and Clark, 1998).
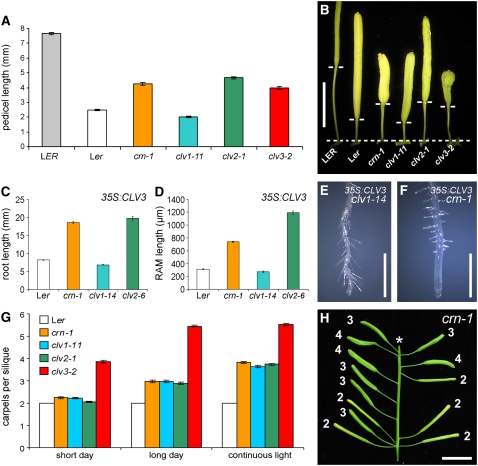
Figure 2.
Similarities and Differences between crn and clv Mutants.
(A) and (B) Pedicel lengths of crn and clv mutants (n = 120; ±se). Pedicel lengths are reduced in Ler due to a mutation in the ER gene. crn-1, clv2-1, and clv3-2, but not clv1-11, partially suppress the er phenotype.
(C) to (F) Overexpression of CLV3 restricts root growth by reducing root apical meristem (RAM) size (n = 12; ±se). crn-1 and clv2-6, but not clv1-14, restore normal root growth.
(G) Carpel number of crn-1, clv1-11, and clv2-1 mutants increases from two to four with increased daylengths (n = 200; ±se). clv3-2 mutants respond similarly, but carpel number is higher under all conditions.
(H) Carpel number of crn-1 increases when the plant is shifted from short days to continuous light during flowering. The asterisk indicates that young flowers at the shoot tip were removed.
Bars = 5 mm in (B) and (H) and 500 μm in (E) and (F).
Role of CRN in Root Meristem Development
The overall root development of crn-1 mutants was unaffected by the mutation, which indicates that CRN is not required for root meristem activity per se or for cellular differentiation. However, increased expression of CLV3 or related CLE proteins was shown to strongly restrict root meristem maintenance, suggesting that a CLV-like and CLE-controlled signaling pathway acts in the root (Casamitjana-Martinez et al., 2003; Hobe et al., 2003; Fiers et al., 2004). Therefore, we assayed root length and meristem size to compare the response of the wild type and clv1-14, clv2-6, and crn-1 mutants to increased CLV3 expression (Figures 2C to 2F). We found that wild-type and clv1-14 plants showed a strong reduction in root meristem activity upon CLV3 expression, while crn-1 and clv2-6 mutant plants were unaffected by increased CLV3 signaling. Because the expression levels of the CaMV35S:CLV3 transgene employed in these assays could vary between individual transgenic lines, we also assayed the responses of wild-type and crn-1 roots to incubation with CLE peptides. Plants were germinated on standard growth agar and transferred to Gamborg's medium agar with 1 μM CLE40 peptide or a mutant peptide carrying an amino acid exchange (G to A) at position 6, which should render the peptide inactive (mCLE40) (Fletcher et al., 1999; Fiers et al., 2005). Within 7 d after transfer, only the presence of the active peptide induced a reduction to 50% in overall root length and root meristem size of wild-type roots. The development of crn-1 mutants was unaffected by the peptide treatments (see Supplemental Figure 2 online). Mutations in clv2 also rendered plants insensitive to CLE peptides (Fiers et al., 2005), thus extending the functional commonalities between CRN and CLV2. Here, we showed that although CRN function is not required for root development under normal growth conditions, CRN is involved in transmitting CLE signals that can restrict root meristem activity.
Extended Expression of CLV3 and WUS in crn-1 Mutants
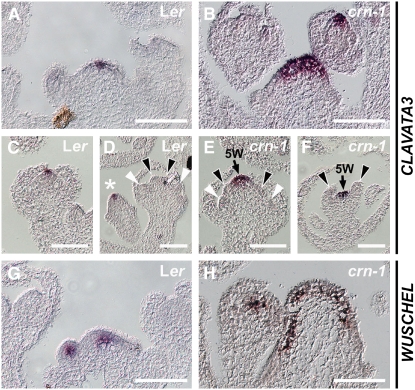
We analyzed the expression patterns of CLV3 and WUS in crn-1 mutant meristems by RNA in situ hybridization (Figure 3). In the wild type, CLV3 was expressed in stem cells at the meristem tip. The CLV3 expression domain appeared triangular and three to five cells wide in longitudinal sections. crn-1 shoot and floral meristems were drastically enlarged and expressed CLV3 in an expanded domain of >10 cells, indicating that crn-1 mutants accumulate more stem cells in the meristem center. Furthermore, CLV3 expression was also temporally extended and maintained beyond stage 6 of flower development, consistent with the increased and prolonged stem cell activity that induces the formation of a fifth floral whorl (Figures 3E and 3F). Expression of the known target gene of CLV signaling, WUS, was similarly expanded in both inflorescence and floral meristems of crn-1 mutants (Figures 3G and 3H). These overall changes in gene expression patterns are reminiscent of those seen in clv1, -2, and -3 mutants (Brand et al., 2000; Schoof et al., 2000), indicating that CRN and the CLV genes act closely together to restrict the stem cell domain in shoot and floral meristems.
Figure 3.
CLV3 and WUS Expression in crn-1 Mutants.
RNA in situ hybridization was performed on wild-type and crn-1 meristem sections. Bars = 100 μm.
(A) to (F) CLV3 expression.
(A) and (B) CLV3 is expressed in an expanded domain in crn-1 mutant SAMs.
(C) to (F) In wild-type flowers at stage 6, CLV3 expression in the stem cell domain is lost when carpels are formed (asterisk in [D]: stage 3 flower; black arrowheads, carpel primordia; white arrowheads, stamen primordia). Expression is maintained in crn-1 flowers, and a fifth whorl (5W) is formed ([E] and [F]).
(G) and (H) WUS expression. Like CLV3 expression, WUS expression expands in a crn-1 SAM.
Integrating CRN into the CLV Signaling Pathway
We used genetics, supported by physiological analysis, to position CRN relative to the known components of the CLV3 signaling pathway. First, we tested the relationship between CRN and WUS. In clv mutants, WUS expression expands, and wus was shown to be epistatic to clv (Brand et al., 2000; Schoof et al., 2000). In the homozygous state, the strong wus-1 mutant was found to be also fully epistatic to crn-1 (Figures 4A to 4C, Table 1), suggesting that WUS acts downstream of CRN.
Figure 4.
Genetic Interactions of CRN.
(A) to (C) wus-1 mutants are epistatic to crn-1. wus-1 (A), crn-1 wus-1 (B), and crn-1 (C) plants at 30 d after germination. Arrowheads mark the arrested SAMs.
(D) Siliques of crn-1 and pol-1 crn-1.
(E) to (H) Top view of inflorescences at 4 weeks after germination. Shoot meristems (arrowheads) of crn clv1-11 double mutants are larger than those of crn-1 and clv1-11 single mutants and comparable to the SAM of clv3-2. In the insets, the SAM is outlined.
(I) Stem diameter of crn-1 clv1-11 double mutants is increased compared with that in the single mutants.
(J) The wild-type phenotype is restored in crn-1 mutants carrying a transgenic copy of the At5g13290 region.
(K) and (L) Overexpression of a CRN cDNA does not affect wild-type development (K) but rescues the crn-1 mutant (L).
Arrows in (J) to (L) indicate siliques with two carpels. Bars = 10 mm in (A) to (D), 1 mm in (E) to (I), and 5 mm in (J) to (L).
Table 1.
Analysis of CLV3 Signaling Activity
| CRN and CLV2 Act Together and Independently of CLV1
| ||||
|---|---|---|---|---|
| Genotype | Carpels per Flower | se | n | Groupa |
| Ler | 2.0 | 0.0 | 100 | A |
| crn-1 | 3.9 | 0.1 | 100 | B |
| pol-1 | 2.0 | 0.0 | 40 | A |
| clv1-11 | 3.9 | 0.1 | 40 | B |
| clv1-1 | 4.2 | 0.1 | 40 | |
| clv2-1 | 3.9 | 0.1 | 40 | B |
| clv3-2 | 6.1 | 0.2 | 40 | C |
| crn-1 pol-1 | 2.1 | 0.1 | 200 | A |
| crn-1 clv1-11 | 5.3 | 0.1 | 200 | |
| crn-1 clv1-1 | 4.5 | 0.1 | 100 | |
| crn-1 clv2-1 | 3.8 | 0.1 | 100 | B |
| crn-1 clv3-2 | 6.0 | 0.1 | 100 | C |
| Recessive wus-1 and Dominant pol-1 Mutants Suppress crn-1
| |||||
| F2 Phenotype
|
χ2 Test (3:1) | ||||
| F1 Genotype | Wild Type | crn-1 | wus-1 | n | |
| crn-1/crn-1 wus-1/+ | – | 94 | 31 | 125 | P = 0.96 |
| crn-1/crn-1 pol-1/+ | 76 | 20 | – | 96 | P = 0.41 |
Groups without significant differences (P > 0.05) based on Kruskal-Wallis H test and LSD test.
The protein phosphatases POL and PLL1 are downstream intermediates of CLV3 signaling and are required for the maintenance of WUS expression. Semidominant pol-1 mutants can suppress the stem cell accumulation of clv mutants, suggesting that POL1 is a target for repression by CLV3 signaling (Song et al., 2006). crn-1 mutants produced on average 3.9 carpels per silique, but pol-1 mutants dominantly suppressed the floral meristem defects in pol-1 crn-1 double mutant plants (Figure 4D, Table 1). This indicated that CRN shares functions with the CLV genes to repress WUS expression by controlling POL activity.
The severity of floral meristem defects in clv mutants decreases with shorter daylength (Kayes and Clark, 1998; Jeong and Clark, 2005), indicating that loss of CLV3 signaling activity can be compensated by another pathway that may be active exclusively under short-day conditions. Of note, this pathway appears to act only in floral meristems, because the main shoot meristem remains less affected by daylength variations. We used carpel number as a precise indicator to measure the activity of the CLV3 signaling pathway under different photoperiods and compared it to those of the null mutants clv1-11, clv2-1, clv3-2, and crn-1 (Figure 2G). For all mutant genotypes, carpel number was smallest under short days (8 h of light/16 h of dark) and increased with extended daylength from long days (16 h of light/8 h of dark) to continuous light (24 h of light) (Figure 2G). Under short days, carpel number was close to wild-type values for clv1-11, clv2-1, and crn-1 (between 2.0 and 2.2, se = 0.1; n = 200) but still increased to 3.9 for clv3-2. The strict correlation of carpel number with daylength was further tested by shifting crn-1 mutants from short days to continuous light. Siliques formed before the transfer carried two carpels, but carpel number increased to three or four on siliques that initiated after the transfer (Figure 2H). This indicates that an additional, CLV3-dependent stem cell–restricting pathway is activated with decreasing daylength, which can fully substitute for CLV1, CLV2, and CRN under short-day conditions. Importantly, the common response of clv1, clv2, and crn-1 mutants to daylength variation suggested that their gene products act closely together in the control of stem cell number.
CLV3 Signals via Two Separable Pathways
To further extend the analysis of commonalities between crn-1 and clv mutants, we created double mutant combinations and analyzed their effects on the size of the stem cell domain. For CLV1, the null allele clv1-11 and the dominant negative allele clv1-1 were used (Dievart et al., 2003). Compared with the single mutants, the inflorescence meristems of crn-1 clv1-11 double mutants were increasingly fasciated and wider in diameter, resembling clv3-2 mutants, which resulted in thick stems (Figures 4E to 4I). Carpel number was increased to 5.3 in crn-1 clv1-11, compared with 3.9 in crn-1 and clv1-11 single mutants (Table 1). Thus, crn-1 mutants strongly enhanced clv1-11 loss-of-function mutants, indicating that CRN and CLV1 act independently of each other to transmit the stem cell–restricting CLV3 signal.
The meristem phenotypes of loss-of-function mutants of CLV2, such as clv2-1, were similar to those of clv1-11 and crn-1. Double mutants of crn-1 clv2-1 were indistinguishable from their respective single mutants, suggesting that CRN and CLV2 act in the same pathway (Table 1). Furthermore, clv1-4 mutants have been shown to enhance the meristem phenotype of clv2-1 in double mutant combinations (Kayes and Clark, 1998), which also indicates that CLV1 and CLV2 can act in separate pathways. From this genetic and phenotypic analysis, we concluded that the CLV3 signal can be transmitted via two independent pathways, one that comprises CLV1 and a second involving CLV2 and CRN. Is there crosstalk between the two proposed pathways? We used the dominant negative allele clv1-1 to address this question. All dominant negative mutants of CLV1 are phenotypically stronger than clv1 null mutants, such as clv1-11 (Dievart et al., 2003). One explanation for this observation is that the mutant clv1-1 protein interferes with the activity of another signaling pathway (Dievart et al., 2003). We found that the phenotype of clv1-1 was further enhanced by the crn-1 mutant, suggesting that CRN remains active in the clv1-1 mutant background (Table 1). Surprisingly, crn-1 clv1-1 double mutants were phenotypically weaker than crn-1 clv1-11 double mutants. This observation could be explained if we assume that the clv1-1 protein maintains some signaling activity and that the dominant negative effect of clv1-1 is exerted by inhibition of the proposed CRN/CLV2 pathway.
Molecular Nature of CRN
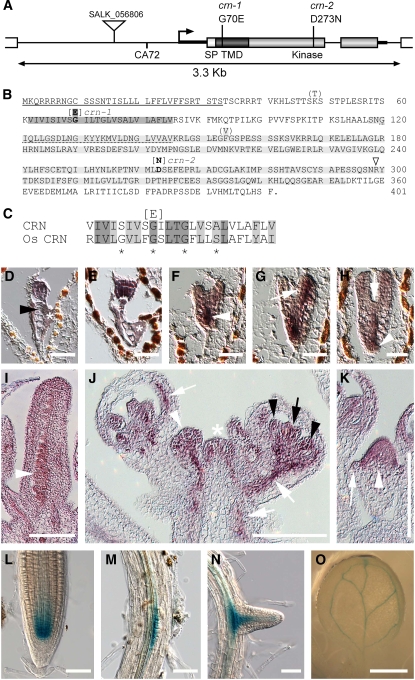
The CRN gene was isolated via a map-based cloning approach. Briefly, an F2 population of crn-1 (Ler) × CRN (Columbia [Col]) was generated, and plants showing the crn-1 mutant phenotype were genotyped using simple sequence length polymorphism (SSLP) and cleaved-amplified polymorphic sequence (CAPS) markers. The SSLP marker CA72 was strictly cosegregating with crn-1 among 554 analyzed plants. CA72 maps to the 5′ untranslated region of the gene At5g13290, which is predicted to encode a protein kinase (Figure 5A). DNA sequence determination re2vealed a nucleotide difference between crn-1 and CRN plants that would cause an amino acid exchange of the predicted protein.
Figure 5.
Gene Structure and Expression Pattern of CRN.
(A) Genomic region of CRN (At5g13290). The 3.3-kb genomic DNA fragment was used for the complementation test. The open reading frames of CRN (boxes, divided into three regions: SP, signal peptide; TMD; and kinase domain) and of the neighboring genes (open boxes) are shown. The mutation in crn-1 causes an exchange of the amino acid at position 70 from G to E. The crn-2 mutant results in an exchange of the amino acid at position 273 from D to N. The T-DNA insertion in the SALK line 056806, the SSLP marker CA72, and the 5′ and 3′ untranslated regions (thick black lines) are indicated.
(B) Amino acid sequence of CRN (Col). Amino acids predicted from the Ler sequence and exchanges (boldface) found in the mutant alleles are in brackets. The signal sequence is underlined. The dark-shaded area indicates the TMD, and the light-shaded area indicates the kinase domain. The dotted line indicates amino acids deleted by facultative splicing of the first intron. The triangle shows the position of the second intron.
(C) Alignment between the TMDs of CRN and the putative rice ortholog Os CRN. Regions of identity (dark shaded), similarity (light shaded), and difference (unshaded) are indicated. The asterisks mark the four (sm)xxx(sm) motifs.
(D) to (H) CRN expression in wild-type embryos. The asterisks indicate weak staining of the embryo-surrounding region.
(D) Sixteen-cell stage. The embryo but not the suspensor (black arrowhead) is stained.
(E) Late globular stage.
(F) to (H) From heart to torpedo stage, CRN expression focuses to the vascular initials of the hypocotyl (arrowhead), the inner cells of the cotyledons (arrow in [G]), and the primary root (arrowhead in [H]). Only weak expression is detected at the position of the developing SAM (arrow in [H]).
(I) CRN is expressed in ovules (arrowhead).
(J) In the inflorescence, CRN is expressed in the inflorescence meristem (asterisk), young flower primordia (white arrowhead), vasculature of sepals, pedicels, and stem (white arrows), stamen (black arrowheads), and carpel primordia (black arrow).
(K) Flower primordium, stage 3. CRN is expressed in the whole flower meristem (white arrowhead) but not in the sepal primordia (arrow).
(L) to (O) CRN:GUS reporter line.
(L) Expression in the root meristem comprises the quiescent center and surrounding initials and extends apically into the root meristem.
(M) Expression in the pericycle at sites of lateral root initiation.
(N) Lateral root meristem.
(O) CRN is expressed in the cotyledon vasculature.
Bars = 25 μm in (D) to (H), 100 μm in (I) to (K), 50 μm in (L) to (N), and 1 mm in (O).
A 3.3-kb genomic DNA fragment containing the promoter and coding sequences of At5g13290 was sufficient to complement crn-1 mutant plants and fully restore the wild-type phenotype (Figure 4J). Therefore, we concluded that At5g13290 is the CRN gene.
Complementation with a genomic copy of CRN had shown that the crn-1 mutant is recessive. To test for any dominant effects of the mutant allele, we expressed crn-1 (mutant) or CRN (wild type) cDNA clones from the strong constitutive CaMV35S promoter in Arabidopsis (Figures 4K and 4L). crn-1 mutants (n = 14) were rescued by expression of a CRN cDNA but not the crn-1 cDNA (n = 6). Overexpression of either the wild-type CRN (n = 17) or the mutant crn-1 allele (n = 13) in a wild-type background did not affect normal development. We conclude that the crn-1 mutant represents a true hypomorphic or amorphic allele of CRN and that CRN function does not require the tight control of CRN transcript levels.
CRN is predicted to encode a membrane-associated Ser/Thr kinase that falls into the non-RD group of kinases (Figure 5B). Most predicted kinases can be phylogenetically grouped according to their amino acid sequence, but CRN maps to an isolated branch with no close relative (Shiu and Bleecker, 2001, 2003). DNA sequence determination of a CRN cDNA clone and comparison with the genomic sequence identified a 1203-bp open reading frame that is interrupted by an intron of 86 bp and that encodes a predicted protein of 401 amino acids (The Arabidopsis Information Resource [TAIR] gene model At5g13290.2). A second intron located upstream of the other intron is recognized and spliced in <3% of all CRN mRNAs (TAIR gene model At5g13290.1). This shortened transcript would encode a protein that lacks 26 amino acids (Figure 5B).
The N terminus of the predicted longer CRN protein carries a signal peptide of 33 to 34 amino acids (SignalP; http://www.cbs.dtu.dk/services/SignalP/), a 24–amino acid hydrophobic TMD (http://www.ch.embnet.org/software/TMPRED_form.html), and a Ser/Thr kinase domain. The mutation in crn-1 causes an exchange of the Gly (G) residue at position 70, which is conserved between the Arabidopsis CRN protein and the putative ortholog from rice (Oryza sativa), with Glu (E) (Figure 5C). The introduction of the charged amino acid would shorten the predicted TMD to only 19 amino acids, which could affect intracellular localization or protein stability (Watson and Pessin, 2001; Gallagher et al., 2005). Interestingly, Gly-70 belongs to one of four (sm)xxx(sm) motifs within this region. Such motifs, consisting of any three amino acids flanked by two small amino acids (Ser, Gly, or Ala), mediate helix-to-helix interactions within TMDs for protein homodimerization or heterodimerization (Curran and Engelman, 2003; Bennasroune et al., 2004). A recent phylogenomic analysis identified this motif in all functionally characterized receptor-like proteins or receptor-like kinases, including CLV2 (Fritz-Laylin et al., 2005). Exchange of Gly-70 with a charged and bulky Glu could hinder or eliminate interactions with partner proteins via the TMD or could contribute to protein misfolding. In agreement with this, we noted that the floral meristem phenotype of crn-1 is significantly enhanced by growth at increased temperatures (Figure 1J).
To isolate additional mutant alleles of CRN, we analyzed five Arabidopsis stocks that carried T-DNA insertions in or around the At5g13290 region, which were available from public collections. All insertions mapped to the predicted promoter of CRN, and all transgenic lines homozygous for the T-DNA insertions were aphenotypic. Analysis by RT-PCR showed that a T-DNA insertion (Salk_056806) within ∼650 bp of the predicted CRN transcription start site (TAIR gene model At5g13290.2) did not cause a significant reduction in CRN mRNA levels. An EMS-mutagenized Arabidopsis population was screened via Targeting-Induced Local Lesions in Genomes (TILLING), and a new allele, crn-2, causing a D273N exchange within the kinase domain, was recovered. Flowers of plants homozygous for crn-2 carried a normal set of outer floral organs, but carpel number was increased significantly, from 2.0 carpels per flower in the Col wild type (se = 0.0; n = 36) to 3.2 carpels per flower (se = 0.1; n = 57). Thus, the crn-2 mutant is phenotypically weaker than crn-1, which produced on average 3.9 carpels per flower (Table 1).
The CRN Expression Pattern Suggests Multiple Roles in Shoot and Root Development
A tissue survey using RT-PCR revealed significant CRN expression predominantly in shoot tips and to a lesser extent in young organs and roots (see Supplemental Figure 3 online). We analyzed CRN expression using RNA in situ hybridization and a CRN promoter:GUS (for β-glucuronidase) transgene (Figures 5D to 5N). CRN mRNA was first detected in embryos and in the region surrounding embryos at the 16-cell stage but not in suspensor cells (Figures 5D to 5H). From early heart to early torpedo stage, CRN expression was focused on the developing vasculature of the hypocotyl and cotyledons and the developing root. The emerging SAM showed only weak staining at the torpedo stage. After germination, CRN RNA was detected in shoot and inflorescence meristems and, slightly stronger, throughout young flower primordia and in ovules (Figures 5I to 5K). Expression was maintained in stamen and carpel primordia, consistent with the floral organ defects observed in the crn-1 mutant. We also noted expression in young provascular tissue of floral organs and stem tissue, which receded with progressing tissue differentiation. The expression pattern of CRN in meristems is consistent with a role in the CLV3 pathway, while the expression in organ primordia and vascular cells suggests a separate role in other signaling cascades. RT-PCR analysis had shown that CRN is also expressed during root development. Using a CRN:GUS transgene, we detected expression in the basal end of the root meristem, comprising the quiescent center, surrounding initials, and vascular precursors. CRN was also expressed in the vasculature of primary and lateral roots, in the pericycle at sites of lateral root initiation, in lateral root meristems, and in the vasculature of the leaves (Figures 5L to 5O). Notably, we were unable to detect any phenotypic alteration in crn mutant roots or vasculature tissues, suggesting that CRN is not required in these tissues for normal development.
DISCUSSION
The Role of CRN in CLV3 Signaling
Previous studies proposed that the CLV3 signaling peptide is secreted from stem cells and perceived by a receptor complex, consisting of CLV1 and CLV2. The role for CLV2, which lacks a kinase domain, was seen as stabilizing the CLV1 protein (Jeong et al., 1999). In clv2 loss-of-function mutants, CLV1 protein levels were reported to be reduced by >90%. Overexpression of CLV3 would cause increased signaling through a CLV1/CLV2 complex, complete downregulation of WUS, and the consequential loss of stem cells from the meristem. Because mutations in CLV2 or CLV1 are epistatic to CLV3 overexpression, both genes have been regarded as essential for CLV3-dependent signaling (Brand et al., 2000). Loss-of-function mutants in clv3 are phenotypically stronger than clv1 or clv2 null mutants, indicating that CLV3 can signal in the absence of a CLV1/CLV2 complex (Dievart et al., 2003). Together with the observation that all strong mutant alleles of clv1 act as dominant negatives, this led to the suggestion that the defective CLV1 proteins interfere with signaling via other receptor-like kinases that also restrict stem cell number. The closest relatives of CLV1, the BAM1, -2, and -3 proteins, were unlikely candidates, because they are expressed in the meristem periphery (where CLV1 is not expressed) and their function in the meristem is opposite to that of CLV1 (DeYoung et al., 2006).
However, some observations do not easily match with this model. For example, clv1 mutants drastically enhance the carpel number phenotype of clv2, indicating that CLV1 is significantly active even in the absence of CLV2 (Kayes and Clark, 1998). This is inconsistent with the proposed requirement of CLV2 to maintain CLV1 in a stable state but indicates that CLV1 and CLV2 can act independently of each other.
The isolation and characterization of CRN and genetic analysis allow us to modify the current model for CLV3 signaling. Our screen was designed to identify components of CLV3 signaling as suppressors of CLV3 overexpression, and as expected from this setup, we identified several new alleles of clv1 and clv2 and the novel crn-1 mutant. Like clv mutants, crn mutants accumulate stem cells in shoot and floral meristems. Complementation tests and misexpression experiments with crn-1 mutant or wild-type cDNAs confirmed that the identified crn-1 allele is fully recessive. The second mutant allele, crn-2, is phenotypically weaker and carries a point mutation causing an amino acid exchange in the predicted kinase domain, which could reduce kinase function or protein–protein interactions.
The phenotype of crn-1 mutants most closely resembles that of clv2 mutants. Both mutants show defects in lateral organ formation, such as stamen and anther development, and suppress the reduced pedicel length of er mutants. When comparing the phenotypes of clv1 and clv2 null mutants with the crn-1 mutant, it is evident that all three genes contribute equally to the control of stem cell number in meristems. The finding that crn-1 and clv2 null mutants are fully epistatic to each other but additive with clv1 mutants suggests that the CLV3 signal is perceived by two separable pathways: one involving CLV1 as a receptor and the second consisting of CRN and CLV2 (Figure 6). Taking carpel number as an accurate readout for CLV3 signaling activity, CLV3 signaling is almost fully abolished in clv1 crn and clv1 clv2 double mutants but not in crn clv2 or in a clv1 single mutant. This is consistent with a model of two parallel-acting pathways; however, a function for individual components, such as CLV2, in both pathways cannot be ruled out prior to any biochemical characterization. Furthermore, crosstalk between the two pathways appears likely. The dominant negative character of strong clv1 mutants, such as clv1-1, could then be explained by interference with the proposed CRN/CLV2 pathway. Interestingly, combinations of crn-1 with the dominant mutant clv1-1 display a weaker mutant phenotype than crn-1 clv1-11 (a null allele) double mutants, which could indicate that the mutant clv1-1 receptor has residual signaling potential.
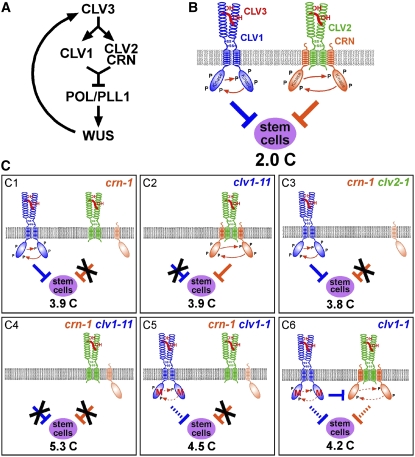
Figure 6.
Models of CLV3 Signaling via Two Independent Receptor Systems.
(A) Integration of CRN into the CLV/WUS feedback loop.
(B) Speculative model of CLV3 receptor interactions. P, Phosphorylation. CLV3 is depicted as a hydroxylated peptide. Both receptor systems act to restrict stem cell fate in shoot and floral meristems, and their joint activities permit the formation of only two carpels (2.0 C) in the wild type.
(C) Model of CLV3 receptor complexes in crn and clv mutants. Recessive mutations in CRN (crn-1; [C1]), CLV1 (clv1-11; [C2]), or CLV2 (data not shown) cause stem cell accumulation to a similar degree, as indicated by the increased carpel number. (C3) crn and clv2 mutants are epistatic to each other, indicating that they act together and independently of CLV1. (C4) CLV1 acts separately from CRN and CLV2; therefore, the double mutant phenotypes are additive. (C5) Double mutants of crn-1 with the dominant allele clv1-1 are phenotypically weaker than the combination of null alleles, suggesting that the clv1-1 receptor has residual signaling activity. (C6) The CRN/CLV2 pathway is subject to inhibition by the clv1-1 protein and therefore is only partially active.
The comparable severity of the mutant phenotypes indicates that the two proposed pathways contribute to CLV3 signaling at a similar level. Both pathways should be inactivated in a crn-1 clv1 double mutant, and these indeed show only a small difference in carpel number from clv3 null mutants. It is still possible, therefore, that a third signaling route of minor importance exists; alternatively, crn-1 could be a hypomorphic allele. However, crn-1 mutants display a strong phenotype like clv2 null alleles, suggesting that crn-1 also represents a loss-of-function allele.
A similar scenario, in which two receptor systems contribute equally to signal perception and transmission, was described for the control of vulva development in Caenorhabditis elegans. Here, the Wnt-type ligands signal via the two receptors LIN-17 and LIN-18, which act in parallel and do not depend on each other. LIN-17 and LIN-18 are not redundant, because null mutations synergize, and only the lin-17 lin-18 double mutants show a fully penetrant bivulva phenotype (Inoue et al., 2004). Both receptors can be activated by at least three Wnt ligands, which differ in their preferences for LIN-17 or LIN-18. For CLE peptides, it was shown that both CLV3 and CLE40 can activate CLV signaling if they are expressed from the stem cell domain (Hobe et al., 2003), but receptor affinities have not been analyzed. Such parallel receptor systems that are activated by several ligands could permit fine-tuning of responses to a given signal by acting differentially through downstream intermediates. Signaling via CLV1 and CRN converges ultimately on the repression of WUS, and because mutations in either CLV or CRN are suppressed by the dominant pol-1 mutants (Song et al., 2006), POL1 and related phosphatases could act as upstream nodes that integrate the output from both pathways. Meristem homeostasis has been shown to tolerate large fluctuations in CLV3 levels (Müller et al., 2006). The integration of signals from two parallel receptor systems would provide a mechanism for achieving such increased system stability.
The CRN protein kinase carries only a short presumably extracellular domain, while CLV2 lacks a larger intracellular domain. It is tempting to speculate that CRN and CLV2 interact via their TMDs to form a functional receptor heterodimer. For many receptor Tyr kinases, such as epidermal growth factor receptor, receptor dimerization mediated by their TMDs forms the signaling-competent structure, which is stabilized upon ligand binding (Li and Hristova, 2006). Mutations in the TMD, as we found for crn-1, could disrupt dimer formation and thereby cause receptor inactivation. A range of dominant mutations in animal receptor kinases has been described, in which single amino acid substitutions within the TMD induce unregulated signaling, such as the G380R mutation in the TMD of human FGFR3, which causes achondroplasia (Shiang et al., 1994). However, we can exclude a dominant activity for the crn-1 protein, because the Crn− phenotype was fully rescued by expression of the wild-type gene and overexpression of the mutant cDNA in plants was aphenotypic. We propose that the G70E exchange in the TMD destabilizes a CRN/CLV2 heterodimer, thereby reducing or even abolishing signaling through this pathway. The latter is entirely consistent with our observation that clv2 null mutants do not further enhance crn-1.
In contrast to the restricted expression domain of CLV1, both CLV2 and CRN are expressed in many plant tissues. CRN RNA was detected from embryogenesis onward in the vasculature and in shoot, flower, and root meristems. CRN shares a wide expression pattern with CLV2, and it is likely that both proteins act together in diverse signaling pathways throughout plant development. In addition to the common function in shoot and flower meristems, these include stamen development, pedicel elongation, and possibly root meristem growth. Both CRN and CLV2 (Fiers et al., 2005) mediate CLE signaling in the root, and it is intriguing that CRN is expressed in root meristems and vasculature, suggesting an auxin-dependent regulation of CRN in these tissues. However, neither crn nor clv2 mutants affect root growth under normal conditions, and the role of CLE pathways in the root remains to be elucidated. In shoot and flower meristems, CRN and CLV2 act in parallel with CLV1 to restrict stem cell activity. In contrast, CRN and CLV2 antagonize CLV1 function in ER-dependent pedicel elongation, because CRN and CLV2 repress the pedicel elongation of er mutants, while CLV1 can substitute for a loss of ER function in the pedicel (Dievart et al., 2003).
METHODS
Growth Conditions
Arabidopsis thaliana plants were grown on soil at 22°C under short-day (8 h day/16 h night), long-day (16 h day/8 h night), or continuous light conditions. Some plants were kept in short days for 6 weeks after germination before flowering was induced by transfer to continuous light.
Plant Material and Genetics
Arabidopsis wild-type and mutant lines were obtained from the Nottingham Arabidopsis Stock Centre (for exceptions, see Supplemental Table 1 online). crn-1, clv1-14, and clv2-6 are EMS-induced suppressor mutants of CLV3 overexpression isolated in a CaMV35S:CLV3 line (ecotype Ler) (Brand et al., 2000). clv1-4, clv1-11, clv2-1, and clv3-2 are in a Ler background and were described previously (Clark et al., 1993, 1995, 1997; Kayes and Clark, 1998; Dievart et al., 2003). To establish nontransgenic crn-1 plants, crn-1 was backcrossed to Ler. The crn-1 clv, crn-1 pol-1, and crn-1 wus-1 double mutants were identified in the F2 or F3 population by PCR, CAPS markers, or phenotype (see Supplemental Table 1 online). The crn-2 allele was identified in an EMS-mutagenized Arabidopsis population, provided by Thomas Altmann and Georg Strompen (University of Potsdam, Germany), and was generated in the context of the GABI-FUTURE project Establishment of a Central Platform for Testing Gene Function on TILLING.
Construction of Transgenic Plants
For the complementation test, the genomic CRN region was amplified by PCR from BAC T31B5 with the primers RM_C_1B (5′-CCCCGGATCCTAAAGATGCATAGGCTTGCGGACA-3′), introducing a BamHI site, and RM_C_2S (5′-CCCCACTAGTCTCGACAAGCTTATTGCTTGATCTGTG-3′), which introduces a SacI site. The 3.3-kb fragment was cloned into the pGreen vector (Hellens et al., 2000) using the BamHI and SacI sites to give pCRN.
To express the CRN cDNA from the constitutive CaMV35S promoter, we isolated the Ler, Col, and mutant versions of CRN by RT-PCR amplification from total RNA using the primers RM_C_GT_5 (5′-CACCTATGTAAGCTTTGAACAGCTTCA-3′), which introduces the CACC-TOPO cloning site, and RM_C_3 (5′-ATATATTGATGCAACTGCAGATGT-3′). The amplicons were cloned in pENTR/D-TOPO (Invitrogen) to give pRM_LC, pRM_CC, and pRM_mC. These entry vectors were used in the LR-Gateway reaction (Invitrogen) with pGWB2 (T. Nakagawa and T. Kimura; http://bio2.ipc.shimane-u.ac.jp/pgwbs/pgwb2.htm) to give p35S:L_CRN, p35S:C_CRN, and p35S:m_CRN. To analyze the expression pattern of CRN, genomic DNA was amplified using the primers AB_CRN-P_5 (5′-AAAAAGCAGGCTAAAGATGCATAGGCTTGC-3′) and AB_CRN-P_3 (5′-AAGAAAGCTGGGTGCTGCTTCTACGAATAA-3′) and then with AttB1v2 (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCT-3′) and AttB2v2 (5′-GGGGACCACTTTGTACAAGAAAGCTGGGT-3′) and cloned via pDONR201 into pGWB3 (T. Nakagawa and T. Kimura; http://bio2.ipc.shimane-u.ac.jp/pgwbs/pgwb3.htm) to create pAB42 (CRN:GUS).
The CaMV35S:CLV3 transgene was described previously (Brand et al., 2000), and the H4:CLV3 (pBU5) vector was created by replacing the CaMV35S promoter with the H4 promoter. The transgenes were transformed into Arabidopsis plants via the floral dip method (Bechtold and Pelletier, 1998). All transgenic plants were selected for resistance to the herbicide BASTA, except for transgenic plants carrying pGWB, which were selected on hygromycin.
Root Growth Assays
For peptide treatments, crn-1 and Ler seedlings were transferred at 3 d after germination to Gamborg's medium plates with 1 μM CLE40 peptide (RQVPTGSDPLHH) or with 1 μM inactive mCLE40 peptide carrying an exchange of the Gly at position 6 (underlined) to Ala. Seven days after transfer, plates were scanned and root lengths were analyzed using ImageJ software.
RNA in Situ Hybridization and Quantitative Real-Time RT-PCR
Protocols for RNA in situ hybridization, including probe preparation for CLV3 and WUS, have been described previously (Müller et al., 2006). The CRN antisense probe was obtained after PCR amplification from pRM_LC with M13 primer and RNA synthesis using T7 RNA polymerase. For RT-PCR, the primers RTP-CRN_5 (5′-GAGTCCTCTCAAAGTAACAGATACACAGA-3′) and RTP-CRN_3 (5′-TCCGGTTAAAAGAACACCCAAT-3′) were used. For expression quantification via real-time RT-PCR, total RNA was isolated from different tissues of flowering wild-type plants, and 5 μg of RNA was converted to cDNA using SuperScript II reverse transcriptase (Invitrogen) and oligo(dT) primer. cDNAs were quantified with the GeneAmp 5700 sequence detection system (Applied Biosystems) using the Platinum SYBR Green qPCR SuperMix UDG with 0.5 μL of ROX dye (Invitrogen). To amplify CRN-derived cDNAs, the following primers were used: RM-RTP-CRN-f (5′-GAGTCCTCTCAAAGTAACAGATACACAGA-3′) and RM-RTP-CRN-R (5′-TCCGGTTAAAAGAACACCCAAT-3′). For all samples, cDNAs were normalized using cytochrome b5 isoform 1 (At5g53560) and primers RM-RTP-CYTb5-f (5′-CGACACTGCAAGGGACATGA-3′) and RM-RTP-CYTb5-r (5′-ACGTATGTCCTAGTTGCTGGAACA-3′).
Phenotypic Analysis
Carpel counts were used to determine CLV3 signaling activity. Mature siliques were individually observed with a dissection microscope. Both ends of the silique were examined to determine the carpel number, and a partially formed carpel was counted as one.
Photographs were taken with a Canon Powershot G2 digital camera mounted onto a Zeiss dissecting microscope or with an Axiocam HR camera attached to a Zeiss Axioscope II microscope. Scanning electron microscopy images using replicas taken from the surface of shoot apices were made as described previously (Müller et al., 2006). Digital photographs were collated with Adobe Photoshop. Analysis of variance was followed by multiple comparison of means using the Kruskal-Wallis H test and the LSD test. Segregation ratios were determined with the χ2 test. Statistical software (Winstat) and Excel were used for this analysis.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: NM_121332/At5g13290.1 (CRN), NM_180481/At5g13290.2 (CRN), NP_200168.1 (cytochrome b5), NP_565429.1/At2g17950 (WUS), and NP_973541/At2g27250 (CLV3).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Formation of a Fifth Floral Whorl in crn-1, clv1-11, and clv2-1.
Supplemental Figure 2. Roots of crn-1 Mutants Are Resistant to CLE Peptides.
Supplemental Figure 3. Quantification of CRN Expression.
Supplemental Table 1. Molecular Markers and Origin of Mutants.
Supplementary Material
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft through Grant SFB590. We thank Ulrike Brand for the pBU5 vector, Carin Theres for excellent technical assistance, Steve Clark for clv1-11 and pol-1 seeds and information on CAPS markers, Hiroo Fukuda for CAPS markers on chromosome 5, Martin Hobe for wus alleles, Georg Strompen and Thomas Altmann for TILLING, Tsuyoshi Nakagawa for pGWB vectors, and the Nottingham Arabidopsis Stock Centre for seeds and DNA stocks.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Rüdiger Simon (ruediger.simon@uni-duesseldorf.de).
Online version contains Web-only data.
References
- Bechtold, N., and Pelletier, G. (1998). In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 82 259–266. [DOI] [PubMed] [Google Scholar]
- Bennasroune, A., Fickova, M., Gardin, A., Dirrig-Grosch, S., Aunis, D., Cremel, G., and Hubert, P. (2004). Transmembrane peptides as inhibitors of ErbB receptor signaling. Mol. Biol. Cell 15 3464–3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand, U., Fletcher, J.C., Hobe, M., Meyerowitz, E.M., and Simon, R. (2000). Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289 617–619. [DOI] [PubMed] [Google Scholar]
- Casamitjana-Martinez, E., Hofhuis, H.F., Xu, J., Liu, C.M., Heidstra, R., and Scheres, B. (2003). Root-specific CLE19 overexpression and the sol1/2 suppressors implicate a CLV-like pathway in the control of Arabidopsis root meristem maintenance. Curr. Biol. 13 1435–1441. [DOI] [PubMed] [Google Scholar]
- Clark, S.E., Running, M.P., and Meyerowitz, E.M. (1993). CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development 119 397–418. [DOI] [PubMed] [Google Scholar]
- Clark, S.E., Running, M.P., and Meyerowitz, E.M. (1995). CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1. Development 121 2057–2067. [Google Scholar]
- Clark, S.E., Williams, R.W., and Meyerowitz, E.M. (1997). The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89 575–585. [DOI] [PubMed] [Google Scholar]
- Curran, A.R., and Engelman, D.M. (2003). Sequence motifs, polar interactions and conformational changes in helical membrane proteins. Curr. Opin. Struct. Biol. 13 412–417. [DOI] [PubMed] [Google Scholar]
- DeYoung, B.J., Bickle, K.L., Schrage, K.J., Muskett, P., Patel, K., and Clark, S.E. (2006). The CLAVATA1-related BAM1, BAM2 and BAM3 receptor kinase-like proteins are required for meristem function in Arabidopsis. Plant J. 45 1–16. [DOI] [PubMed] [Google Scholar]
- Dievart, A., Dalal, M., Tax, F.E., Lacey, A.D., Huttly, A., Li, J., and Clark, S.E. (2003). CLAVATA1 dominant-negative alleles reveal functional overlap between multiple receptor kinases that regulate meristem and organ development. Plant Cell 15 1198–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiers, M., Golemiec, E., Xu, J., van der Geest, L., Heidstra, R., Stiekema, W., and Liu, C.M. (2005). The 14-amino acid CLV3, CLE19, and CLE40 peptides trigger consumption of the root meristem in Arabidopsis through a CLAVATA2-dependent pathway. Plant Cell 17 2542–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiers, M., Hause, G., Boutilier, K., Casamitjana-Martinez, E., Weijers, D., Offringa, R., van der Geest, L., van Lookeren Campagne, M., and Liu, C.M. (2004). Mis-expression of the CLV3/ESR-like gene CLE19 in Arabidopsis leads to a consumption of root meristem. Gene 327 37–49. [DOI] [PubMed] [Google Scholar]
- Fletcher, J.C., Brand, U., Running, M.P., Simon, R., and Meyerowitz, E.M. (1999). Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283 1911–1914. [DOI] [PubMed] [Google Scholar]
- Fritz-Laylin, L.K., Krishnamurthy, N., Tor, M., Sjolander, K.V., and Jones, J.D. (2005). Phylogenomic analysis of the receptor-like proteins of rice and Arabidopsis. Plant Physiol. 138 611–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher, M.J., Shen, W., Song, L., and Macdonald, R.L. (2005). Endoplasmic reticulum retention and associated degradation of a GABAA receptor epilepsy mutation that inserts an aspartate in the M3 transmembrane segment of the alpha1 subunit. J. Biol. Chem. 280 37995–38004. [DOI] [PubMed] [Google Scholar]
- Hellens, R.P., Edwards, E.A., Leyland, N.R., Bean, S., and Mullineaux, P.M. (2000). pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42 819–832. [DOI] [PubMed] [Google Scholar]
- Hobe, M., Müller, R., Grünewald, M., Brand, U., and Simon, R. (2003). Loss of CLE40, a protein functionally equivalent to the stem cell restricting signal CLV3, enhances root waving in Arabidopsis. Dev. Genes Evol. 213 371–381. [DOI] [PubMed] [Google Scholar]
- Inoue, T., Oz, H.S., Wiland, D., Gharib, S., Deshpande, R., Hill, R.J., Katz, W.S., and Sternberg, P.W. (2004). C. elegans LIN-18 is a Ryk ortholog and functions in parallel to LIN-17/Frizzled in Wnt signaling. Cell 118 795–806. [DOI] [PubMed] [Google Scholar]
- Ishiguro, S., Watanabe, Y., Ito, N., Nonaka, H., Takeda, N., Sakai, T., Kanaya, H., and Okada, K. (2002). SHEPHERD is the Arabidopsis GRP94 responsible for the formation of functional CLAVATA proteins. EMBO J. 21 898–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong, S., and Clark, S.E. (2005). Photoperiod regulates flower meristem development in Arabidopsis thaliana. Genetics 169 907–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong, S., Trotochaud, A.E., and Clark, S.E. (1999). The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. Plant Cell 11 1925–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayes, J.M., and Clark, S.E. (1998). CLAVATA2, a regulator of meristem and organ development in Arabidopsis. Development 125 3843–3851. [DOI] [PubMed] [Google Scholar]
- Kondo, T., Sawa, S., Kinoshita, A., Mizuno, S., Kakimoto, T., Fukuda, H., and Sakagami, Y. (2006). A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science 313 845–848. [DOI] [PubMed] [Google Scholar]
- Li, E., and Hristova, K. (2006). Role of receptor tyrosine kinase transmembrane domains in cell signaling and human pathologies. Biochemistry 45 6241–6251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, K.F., Schoof, H., Haecker, A., Lenhard, M., Jurgens, G., and Laux, T. (1998). Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95 805–815. [DOI] [PubMed] [Google Scholar]
- Müller, R., Borghi, L., Kwiatkowska, D., Laufs, P., and Simon, R. (2006). Dynamic and compensatory responses of Arabidopsis shoot and floral meristems to CLV3 signaling. Plant Cell 18 1188–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa, M., Shinohara, H., Sakagami, Y., and Matsubayashi, Y. (2008). Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science 319 294. [DOI] [PubMed] [Google Scholar]
- Schoof, H., Lenhard, M., Haecker, A., Mayer, K.F., Jurgens, G., and Laux, T. (2000). The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100 635–644. [DOI] [PubMed] [Google Scholar]
- Sharma, V.K., Ramirez, J., and Fletcher, J.C. (2003). The Arabidopsis CLV3-like (CLE) genes are expressed in diverse tissues and encode secreted proteins. Plant Mol. Biol. 51 415–425. [DOI] [PubMed] [Google Scholar]
- Shiang, R., Thompson, L.M., Zhu, Y.Z., Church, D.M., Fielder, T.J., Bocian, M., Winokur, S.T., and Wasmuth, J.J. (1994). Mutations in the transmembrane domain of FGFR3 cause the most common genetic form of dwarfism, achondroplasia. Cell 78 335–342. [DOI] [PubMed] [Google Scholar]
- Shiu, S.H., and Bleecker, A.B. (2001). Plant receptor-like kinase gene family: Diversity, function, and signaling. Sci. STKE 2001 RE22. [DOI] [PubMed] [Google Scholar]
- Shiu, S.H., and Bleecker, A.B. (2003). Expansion of the receptor-like kinase/Pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol. 132 530–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, S.K., and Clark, S.E. (2005). POL and related phosphatases are dosage-sensitive regulators of meristem and organ development in Arabidopsis. Dev. Biol. 285 272–284. [DOI] [PubMed] [Google Scholar]
- Song, S.K., Lee, M.M., and Clark, S.E. (2006). POL and PLL1 phosphatases are CLAVATA1 signaling intermediates required for Arabidopsis shoot and floral stem cells. Development 133 4691–4698. [DOI] [PubMed] [Google Scholar]
- Stahl, Y., and Simon, R. (2005). Plant stem cell niches. Int. J. Dev. Biol. 49 479–489. [DOI] [PubMed] [Google Scholar]
- Torii, K.U., Mitsukawa, N., Oosumi, T., Matsuura, Y., Yokoyama, R., Whittier, R.F., and Komeda, Y. (1996). The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell 8 735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson, R.T., and Pessin, J.E. (2001). Transmembrane domain length determines intracellular membrane compartment localization of syntaxins 3, 4, and 5. Am. J. Physiol. Cell Physiol. 281 C215–C223. [DOI] [PubMed] [Google Scholar]
- Yu, L.P., Miller, A.K., and Clark, S.E. (2003). POLTERGEIST encodes a protein phosphatase 2C that regulates CLAVATA pathways controlling stem cell identity at Arabidopsis shoot and flower meristems. Curr. Biol. 13 179–188. [DOI] [PubMed] [Google Scholar]
- Yu, L.P., Simon, E.J., Trotochaud, A.E., and Clark, S.E. (2000). POLTERGEIST functions to regulate meristem development downstream of the CLAVATA loci. Development 127 1661–1670. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.