Abstract
Nucleoside analog reverse transcriptase inhibitors (NRTI) are known to directly inhibit mitochondrial complex I activity as well as various mitochondrial kinases. Recent observations that complex I activity and superoxide production are modulated through cAMP-dependent phosphorylation suggests a mechanism through which NRTIs may affect mitochondrial respiration via kinase-dependent protein phosphorylation. In the current study we examine the potential for NRTIs to inhibit the cAMP-dependent phosphorylation of complex I and the associated NADH:CoQ oxidoreductase activities and rates of superoxide production using HepG2 cells. Phosphoprotein staining of immunocaptured complex I revealed that 3′-azido-3′-deoxythymidine (AZT; 10 and 50 μM), AZT monophosphate (150 μM), and 2′,3′-dideoxycytidine (ddC; 1μM) prevented the phosphorylation of the NDUFB11 subunit of complex I. This was associated with a decrease in complex I activity with AZT and AZT monophosphate only. In the presence of succinate, superoxide production was increased with 2′,3′-dideoxyinosine (ddI; 10 μM) and ddC (1 μM). In the presence of succinate + cAMP AZT showed an inverse dose-dependent effect on superoxide production. None of the NRTIs examined inhibit PKA activity suggesting that the observed effects are due to a direct interaction with complex I. These data demonstrate a direct effect of NRTIs on cAMP-dependent regulation of mitochondrial bioenergetics independent of DNA polymerase-γ activity; in the case of AZT these observations may provide a mechanism for the observed long-term toxicity with this drug.
Introduction
Nucleoside analog reverse transcriptase inhibitors (NRTIs) represent the primary therapy for HIV infection, forming the cornerstone of highly active antiretroviral therapy (HAART), which consists of combination therapies including NRTIs, protease inhibitors, and non-nucleoside reverse transcriptase inhibitors. NRTIs act by mimicking endogenous deoxynucleosides and competing for incorporation into DNA. Once incorporated, NRTIs halt DNA chain elongation because of the missing or substituted 3′-hydroxyl moiety. By terminating viral reverse transcription NRTIs are effective at preventing HIV replication, diminishing viral burden, and improving patient immune function. However, therapies including NRTIs are unfortunately associated with a host of adverse pathologies including: lactic acidosis with hepatic steatosis, cardiac and skeletal mypopathy, neuropathy, and pancreatitis. It is generally held that the primary mechanism for NRTI toxicity is through the inhibition of mitochondrial DNA polymerase-γ (pol-γ) and depletion of mitochondrial DNA (mtDNA) leading to mitochondrial dysfunction. While this mechanism may be significant for dideoxy NRTIs (ddC, ddI, and d4T), pol-γ has a low affinity for AZT, which is unlikely to significantly inhibit pol-γ at relevant therapeutic doses (Martin et al., 1994; Lim and Copeland, 2001). Regardless, AZT is associated with a significant level of toxicity both clinically (Moyle, 2000; White, 2001) and experimentally (Pan-Zhou et al., 2000; Walker et al., 2002; Lund et al., 2007).
There is a substantial body of evidence which suggests NRTIs also have direct, pol-γ independent effects on mitochondria; however, this evidence is sometimes problematic and contradictory. This is further confounded by the observations that there are drug- and organ-specific effects; it is therefore difficult to draw broad conclusions regarding NRTI toxicity. Various investigators have shown that AZT and other NRTIs affect each of the electron transport complexes (ETC) along with other metabolically critical enzymes (reviewed in (Lund and Wallace, 2004)). Several of these direct targets share a common feature in that they bind endogenous nucleotides/nucleosides. This feature opens up a vast array of potential macromolecular targets by NRTIs considering the large number of biochemical reactions that involve nucleosides, and structurally-related analogs.
Several studies have shown a direct effect on complex I by NRTIs, for example we have demonstrated an apparent decoupling of complex I with AZT in vitro (Lund and Wallace, 2004), and others have suggested an alteration of electron flow through complex I (Pereira et al., 1998; Valenti et al., 2002) as well as alterations in NADH linked respiration with AZT and ddC (Modica-Napolitano, 1993; Skuta et al., 1999; Szabados et al., 1999). Although complex I does have a nucleotide binding site (NADH), there is no evidence to suggest AZT or other NRTIs interfere with NADH binding. It is the hypothesis of this study that the mechanism through which AZT and other NRTIs affect complex I (and possibly other ETC complexes) is instead by interfering with its regulation by protein kinases which do contain susceptible ATP/ADP nucleotide binding domains.
Beyond substrate availability, the regulatory pathways of complex I are not well understood. A slow active/inactive transition has been demonstrated for complex I which is sensitive to the presence of divalent cations and sulfhydryl reagents (Grivennikova et al., 2003). Previous studies have shown a decrease in complex I activity in the presence of ATP though the mechanism behind this effect is unknown (Rouslin, 1991; Raha et al., 2002). Additionally, it is becoming increasingly apparent that complex I is modulated by cAMP-dependant phosphorylation of critical protein targets (Technikova-Dobrova et al., 2001).
Within the inner mitochondrial membrane there are relatively few phosphoproteins; studies involving complex I (primarily from bovine heart) have shown upwards of five phosphoproteins with apparent molecular weights of 42, 39, 29, 18, and 6.5 kDa. The 42 kDa protein has been identified as both the E1α subunit of pyruvate dehydrogenase loosely associated with complex I (Chen et al., 2004) and the NDUFA10 (bovine/human) subunit of mammalian complex I (Schilling et al., 2005). The physiological importance of the latter is unknown since the phosphoserine residue in the 42 kDa subunit is absent in mouse and human (Brandt, 2006). The 39 kDa protein has been identified as the α–subunit of succinyl CoA ligase (Chen et al., 2004). The identity of the 29 kDa protein is unknown, though it is suspected to be another loosely associated matrix protein (Papa et al., 1999). The 18 kDa subunit was initially identified as the AQDQ/NDUSF4 subunit (Papa et al., 1996), but was later shown to be the transmembrane ESSS/NDUFB11 subunit in the hydrophobic arm of complex I (Chen et al., 2004). Finally, the 6.5 kDa protein has been identified as the MWFE/NDUFA1 subunit, also a transmembrane protein in the hydrophobic portion of complex I (Chen et al., 2004; Scheffler et al., 2004). The ESSS and MWFE subunits are of particular interest because their phosphorylation is dependent, or greatly enhanced, in the presence of cAMP (Sardanelli et al., 1995; Chen et al., 2004).
The cAMP-dependent phosphorylation of the ESSS/NDUFB11 and MWFE/NDUFA1 subunits has been shown in various mammalian models: bovine (Technikova-Dobrova et al., 2001; Chen et al., 2004; Schulenberg et al., 2004), rat (Raha et al., 2002; Pasdois et al., 2003), mouse (Technikova-Dobrova et al., 2001) and human (Papa et al., 2001). The specific function of these phosphorylations is unknown and is unlikely to become clear until the coupling/proton-translocation mechanism of complex I is better characterized. It is clear, however, that complex I activity is enhanced in the presence of cAMP. Using various models, NADH:Q oxidoreductase, NADH: cytochrome c reductase, and glutamate + malate dependent respiration rates have all been shown to increase in the presence of cAMP analogs (N6,2′-O-dibutyryladenosine 3′,5′-cyclic monophosphate, dbcAMP; 8-bromoadenosine 3′,5′-cyclic monophosphate, 8BrcAMP) (Scacco et al., 2000; Technikova-Dobrova et al., 2001; Raha et al., 2002; Pasdois et al., 2003). These observations have been associated with an increase in ESSS and MWFE phosphorylation as well as a decrease in superoxide produced at complex I (Raha et al., 2002).
The phosphoregulation of the ESSS and MWFE subunits is suspected to be governed by the activity of a mitochondrial localized cAMP-dependent protein kinase A (PKA) (Thomson, 2001; Horbinski and Chu, 2005) and a Mg2+-dependent and Ca2+ inhibitable PP2C-type phosphatase (Signorile et al., 2002). Mitochondrially localized protein kinases such as PKA -which regulate complex I activity – may provide a mechanistic link between NRTI related disruptions in complex I activity/respiration and evidence of endogenous kinase inhibition by NRTIs. NRTIs (AZT in particular) have been shown to inhibit various nucleotide kinases (Munch-Petersen et al., 1991; McKee et al., 2004; Rylova et al., 2005) as well as metabolic and regulatory kinases (Barile et al., 1994; Pereira et al., 1998; Carnicelli et al., 2006). However, to date, no studies involving PKA and NRTIs have been published.
Another important feature of NRTI induced toxicity is oxidative stress. The high probability of long-term exposure to NRTIs means that any alteration in redox homeostasis caused by these compounds could be detrimental to cell/tissue functioning. Although it is a common feature, especially with AZT, it is not always clear whether oxidative stress is a direct effect of the NRTI or a result of malfunctioning complexes due to mtDNA depletion/mutation. Regardless, various studies have demonstrated evidence of oxidative stress in the absence of mtDNA depletion suggesting a direct stimulation of oxidative stress (Skuta et al., 1999; Szabados et al., 1999; Valenti et al., 2002; de la Asuncion et al., 2004).
Complex I is a well established source of superoxide in the mitochondria (Jezek and Hlavata, 2005); though the mechanisms of superoxide generation at complex I have been elusive (due to its abundant electron carriers) (Brandt, 2006). The location of maximal superoxide production is believed to be the site of quinone reduction (Q-site), based on the stimulation of superoxide by specific Q-site inhibitors (rotenone, pericidin) (St-Pierre et al., 2002; Lambert and Brand, 2004) and also by reverse electron flow from succinate (which is blocked by Q-site inhibitors) (Lambert and Brand, 2004). The latter is the most likely physiological source of superoxide from this complex (Liu et al., 2002); alternately, a low steady-state production of superoxide may occur under high metabolic flux simply by mass action. Interestingly, superoxide production at complex I also appears to be modulated by PKA (Raha et al., 2002).
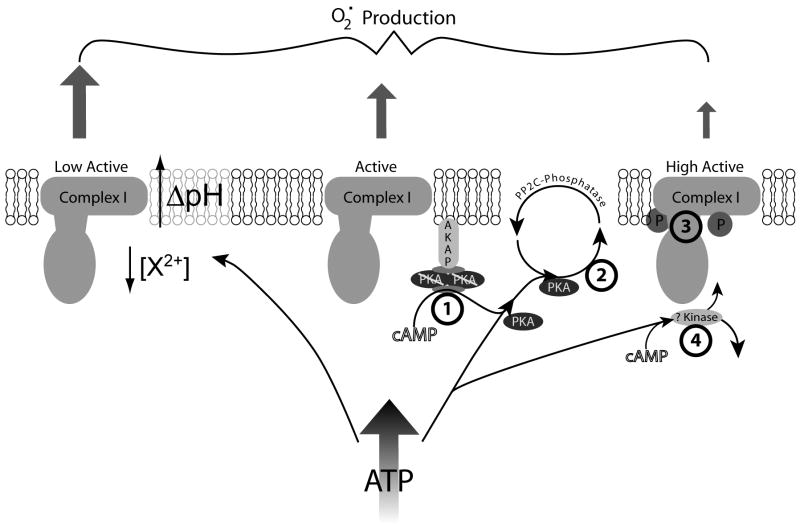
With this last piece of evidence a provocative picture regarding NRTIs effects on complex I can be put forth, centered around the phosphoregulation of this complex (figure 1). Complex I activity and ROS generation appear to be modulated by phosphorylation events resulting from mitochondrial cAMP-dependent PKA activation. Additionally, NRTIs have been shown to affect complex I activity and increase oxidative stress, therefore we hypothesize that these two phenomena are causally linked. The nucleotide-analog structures of NRTIs provide several potential loci of inhibition: 1) cAMP activation of PKA, 2) ATP/ADP binding site of PKA (i.e. PKA activity), and/or 3) interaction at nucleotide binding sites of complex I (at the phosphorylation site or elsewhere). There is also the possibility that 4) phosphoregulation of complex I is accomplished by an unknown cAMP-dependent protein kinase which could be inhibited by NRTIs.
Figure 1.
Theory of ATP and cAMP mediated regulation of Complex I and potential sites of NRTI inhibition. An increase in mitochondrial ATP can lead to two states of complex I, depending on the availability of cAMP. In the absence of cAMP, ATP causes a decrease in activity and an increase in superoxide generation by an unknown process, potentially by allosteric inhibition, alteration in divalent cation concentration, or increasing ΔpH across the inner membrane. In the presence of cAMP, AKAP localized PKA is activated/released from its regulatory subunits and phosphorylates complex I at the ESSS and MWFE phosphorylation sites leading to an increase in activity and a decrease in superoxide production. The hypothesized points of NRTI inhibition of this process occur at the four indicated points: 1) cAMP binding to the tetrameric PKA complex, thus preventing PKA activation, or 2) PKA’s phosphorylation of complex I (by interfering with the ATP/ADP binding site of PKA, 3) some combination of ATP, PKA, and complex I binding, or 4) inhibition of an unknown cAMP-dependent kinase which phosphorylates complex I.
ADP, adenosine 5′-diphosphate, AKAP, A kinase anchor protein; ATP, adenosine 5′-triphosphate, cAMP, adenosine 3′,5′-cyclic monophosphate; NRTI, nucleoside reverse transcriptase inhibitor;PKA, protein kinase A
In the current study we investigate the potential for NRTIs to interfere with the phosphorylation of complex I and affect its resultant activity and potential for superoxide generation. Using HepG2 human hepatoma cells and their isolated mitochondria we explore the ability of AZT (1, 10, 50 μM) and its phosphates, ddI (10 μM), and ddC (1 μM) to interfere with phosphorylation of complex I subunits, inhibit resulting complex I activity, and affect production of superoxide in whole cells. Additionally the effects of NRTIs on PKAc and cAMP-dependent PKA activity are also investigated.
Materials and Methods
Reagents
All reagents were of research or cell culture quality. AZT was purchased from Toronto Research Chemicals (North York, On., Canada); ddI, and ddC were purchased from Sigma (St. Louis, MO, USA); mono-, di-, and triphosphates of AZT were purchased from Moravek Biochemicals (Brea, CA, USA). Tissue culture reagents were purchased from Invitrogen (Carlsbad, CA, USA). Pro-Q® Diamond phosphoprotein gel stain, MitoSOX Red™ and MitoTracker™ Green were obtained from Molecular Probes (Eugene, OR, USA). All other reagents were from Sigma unless specified otherwise.
NRTIs were dissolved in PBS and stocks frozen at −20° C when not in use. New stocks were prepared each month or after four successive freeze-thaws. Digitonin was purified before use by cold ethanol precipitation as described by Sigma. Protein kinase A, purified catalytic subunit (PKAc), from bovine heart (Sigma) and crude cAMP dependent protein kinase A from bovine heart (Sigma) were prepared immediately before use in PKA dilution buffer (350 mM K3PO4, pH 6.8, 0.1 mM dithiothreitol).
Cell Culture
HepG2/C3A human hepatoma cells obtained from American Type Culture Collection (ATCC, Rockville, MD, USA) were cultured in Minimum Essential Medium (MEM) supplemented with 1.5 g/L sodium bicarbonate, 110 mg of sodium pyruvate, and 10% FBS. Cells were plated on 75 cm2 tissue culture flasks and grown in a 5% CO2 incubator at 37 °C with saturating humidity with media changes every 2 days. Cells were passaged by detachment with 0.05% trypsin and 0.5 mM EDTA at 80% confluency. Each experiment represents a separate culture of cells started from a separate vial of master stock.
HepG2 Mitochondrial Isolation
Mitochondria were isolated from digitonin permeabilized HepG2 cells by differential centrifugation. Briefly, eight to ten highly confluent T75 flasks were harvested by trypsinization, rinsed with PBS, and resuspended in PBS with protease inhibitors (1 μL/mL protease inhibitor cocktail). Cells were frozen and stored at −80° C until isolation. Total cell protein was determined by the Bradford method (Bradford, 1976) and the cells were resuspended in isolation buffer (0.25 M sucrose, 0.2 mM EDTA, 10 mM Tris-HCl (pH 7.8), 10 mM triethanolamine, 0.01% digitonin) at 5 mg total cell protein/mL. Cell suspension was incubated on ice for 10 minutes before homogenizing in a glass-Teflon® (Potter-Elvehjem) homogenizer at 500 rpm with 15 up and down strokes. The cell homogenate was then centrifuged at 1000 × g for 10 minutes at 4° C. The first supernatant was saved and the pellet resuspended in the same volume of isolation buffer, homogenized, and centrifuged as before. Supernatants were then combined and centrifuged at 12,000 × g for 15 minutes at 4° C. Final supernatant was discarded and the mitochondria pellet resuspended in 500 μL PBS with protease inhibitor cocktail (Sigma-Aldrich). Mitochondrial protein was determined by the Bradford method and ~1 mg aliquots were stored at −80° C until use. Approximate yield was 0.15 mg mitochondria/mg whole cell protein.
In Vitro Phosphorylation of Complex I
In vitro phosphorylation of complex I was accomplished as described previously (Schulenberg et al., 2004) with some modification. Briefly, ~1 mg aliquots of frozen mitochondria were thawed and centrifuged at 12,000 × g for 15 minutes at 4°C. The supernatant was discarded and the mitochondria resuspended in 200 μL hypotonic buffer (5 mM MgCl2, 40 mM potassium phosphate buffer, pH 7.4) then disrupted by rapid freeze-thaw in liquid nitrogen and 30° C water. The mitochondrial fragments were then pelleted by centrifugation at 12,000 × g at 4° C and resuspended in 1 mL phosphorylation buffer (5 mM ATP, 5 mM MgCl2, 1 mM EDTA, 100 mM Tris-HCl, pH 7.5, 20 μg/mL oligomycin, 60 μM dbcAMP, 1 μL/mL protease inhibitor cocktail, 10 μL/mL phosphatase inhibitor cocktails 1 and 2 (Sigma-Aldrich)). NRTIs were added individually to aliquots at the following concentrations AZT (1, 10, 50 μM), AZT-MP (50, 150 μM), AZT-DP (3 μM), AZT-TP (1.5 μM), ddI (10 μM), ddC (1 μM), control was PBS. To facilitate phosphorylation, 20 U of protein kinase A, catalytic subunit, were added before incubating at 30° C for 30 minutes; a control without exogenous PKA was also included. Following the incubation a 100 μL aliquot was taken from each sample for enzyme activity and the remainder centrifuged at 12,000 × g for 15 minutes at 4° C. The resulting pellet was resuspended in 180 μL PBS with 1 μL/mL protease inhibitor cocktail and 10 μL/mL phosphatase inhibitor cocktails 1 and 2. Mitochondrial membranes were solubilized by the addition of 20 μL of 10% lauryl maltoside (n-dodecyl-β-D-maltopyranoside) in PBS followed by incubation on ice for 10 minutes with frequent mixing. Using this detergent effectively disrupts the mitochondrial membranes while keeping the embedded multi-subunit complexes intact. Insoluble material was removed by centrifuging at 20,000 × g for 30 minutes at 4° C. The supernatant was then used for immunocapture.
NADH:CoQ1 Oxidoreductase Activity
Rotenone-sensitive complex I activity was measured spectrophotometrically on a DU® 7400 (Beckman-Coulter, USA) by monitoring the oxidation of NADH at 340 nm as previously described (Birch-Machin et al., 1993). In order to measure the effects of NRTIs on several regulatory states of complex I, activity was measured under three separate conditions: naïve (phosphorylation buffer without ATP, dbcAMP, or PKAc), phosphorylation buffer with 5 mM ATP, and phosphorylation buffer with 5 mM ATP, 60 μM dbcAMP, and 20 U PKAc. Mitochondrial prepartions were incubated in the respective conditions at 30° C for 30 minutes, as described above. Following the incubation, 100 μL of mitochondrial suspensions (~100 μg mitochondrial protein) were mixed with 900 μL assay buffer (final concentrations: 40 mM potassium phosphate buffer, pH 7.4, 5 mM MgCl2, 3 mM KCN, 60 μM CoQ1, 2.5 mg/mL bovine serum albumin (BSA), and 2 μg/mL antimycin A) and read at 340 nm (425 nm background) at 2 second intervals for 15 seconds before the addition of NADH (50 μM final concentration). NADH oxidation was monitored for 45 seconds before the addition of 5 μg/mL rotenone (~13 μM) and followed for an additional 30 seconds. Rotenone sensitive complex I activity is reported as nmol NADH oxidized/min/mg mitochondrial protein (ε340 NADH = 6.22 mM−1 cm−1).
Immunocapture and Visualization of Complex I
Immunocapture of complex I was accomplished using a mouse monoclonal antibody (mAb) anti-complex I which has been covalently cross linked to Protein G-Agarose beads (Mitosciences, Eugene, OR). Following in vitro phosphorylation, the solubilized mitochondria were combined with a slurry containing 5 μL (solid bead volume) of the complex I immunocapture beads (expressing 50 μg capture antibody) in 40 μL PBS (1 % lauryl maltoside) and incubated 18 hours at 4° C on a microtube rotator (20 rpm). Following incubation the beads were washed and complex I eluted using 36 μL of an acidic glycine buffer(0.05% w/v lauryl maltoside, 0.2 M glycine-HCl, pH 2.5) according to the manufacture’s instructions. Use of this buffer allowed the beads to be reused for further immunocapture once the complex has been removed. Cleaned beads were resuspended in PBS with 0.02% azide and stored at 4° C for future use; immunocapture beads performed well through 5–7 uses with this procedure.
The first elution of purified complex I was combined with 8 μL 6x sample buffer (30% glycerol, 10% sodium dodecyl sulfate (SDS), 300 mM Tris-HCl, pH 6.8, 0.012% bromophenol blue, and 0.6 M dithiothreitol) to produce a yellow solution (due to the acidic glycine buffer). To neutralize the solution, 1 M Tris.HCl, pH 7.5 was added until the bromophenol blue dye returned to blue (6–8 μL). To further denature the proteins, samples were heated to 95° C for 5 minutes in a block heater before loading onto a gel. To each well, 40 μL of sample were loaded into PAGEr Gold pre-cast 10–20% tris-glycine gels (Cambrex, ME, USA) in running buffer (25 mM Tris, 192 mM glycine, 0.1% SDS). In order to accurately establish protein size and phosphoprotein content 1 μL Molecular Probes’ protein molecular weight standards (Sigma) and PeppermintStick™ phosphoprotein molecular weight standard (Molecular Probes) were each combined with 40 μL 1x sample buffer and loaded into separate wells. Proteins were separated using a Bio-Rad Protean 3 cell gel box at 120 V for 90 minutes or until the dye-front reached the bottom of the gel.
Proteins were initially visualized using Pro-Q® Diamond phosphoprotein gel stain (Molecular Probes) which selectively stains phosphate groups attached to tyrosine, serine, or threonine. Gels were fixed overnight in ~100 mL of 50% methanol and 10% acetic acid and subsequently stained/destained according to the manufacture’s instructions. Gels were photographed using a FluorChem Imaging Station (Alpha Innotech, CA, USA) with 302 nm UV transilluminator and a digital camera equipped with a with a 595/40 nm bandpass filter.
Following Pro-Q Diamond staining the gels were stained for total protein using SYPRO® Ruby protein gel stain (Sigma) according to the manufacture’s instructions. Destaining of Pro-Q® Diamond stain and fixing the gel are not required after following the Pro-Q® Diamond protocol. Following the final rinse the gels were imaged as described above.
Lane analysis and band densitometry was performed using Kodak Molecular Imaging Software version 4.04 (Eastman Kodak Company, USA). Relative phosphorlyation was determined by calculating the ratio of the background subtracted, net intensity for Pro-Q® Diamond stained bands to their respective SYPRO® Ruby band intensity.
Protein Kinase A Activity
The qualitative activity of PKA was determined using the PepTag® non-radioactive PKA assay kit (Promega, WI, USA). This kit utilizes a fluorescently labeled peptide fragment (Kemptide; L-R-R-A-S-L-G) which is specifically phosphorylated at the serine residue by PKA thus changing the peptide’s charge from +1 to −1; therefore allowing phosphorylated and non-phosphorylated peptide to be separated and quantified on an agarose gel. Reactions were prepared according to the manufacture’s instructions; to minimize variation a master mix containing the appropriate volumes of 5x reaction buffer (100 mM Tris-HCl, pH 7.4, 50 mM MgCl2, 5 mM ATP), A1 peptide (0.4 μg/μL Kemptide), peptide protection solution, and PKA activator solution (5 μM cAMP) was prepared for each experiment. To 16 μL of the master mix was added 5 μL PBS (control) or NRTI at final concentrations of AZT (1, 10, 50 μM), AZT-MP (50, 150 μM), AZT-DP (3 μM), AZT-TP (1.5 μM), ddI (10 μM), ddC (1 μM) and 4 μL of either purified catalytic subunit of PKA (2 μg/mL in PKA dilution buffer; see reagents) or crude cAMP dependent PKA (50 μg/mL in PKA dilution buffer). For the latter a second control substituting water for PKA activator solution (cAMP) was included to measure cAMP independent activity. Reactions were mixed well (25 μL final volume) and incubated in a Perkin Elmer GeneAmp® PCR system 2400 thermocycler at 22° C for 30 minutes followed by 10 minutes at 95° C to stop the reaction. Reactions were then mixed with 80% glycerol (1 μL) and 20 μL (containing approximately 1.23 nmol Kemptide) of the reaction were loaded onto an agarose gel (0.8% agarose in 50 mM Tris-HCl, pH 8.0). The gel was run at 115 V for 35 minutes in 50 mM Tris-HCL, pH 8.0 before visualizing on a FluorChem Imaging Station (Alpha Innotech, CA, USA) with 302 nm UV transilluminator and a digital camera equipped with a 595/40 nm bandpass filter.
Region of interest densitometry on gel images was performed using Kodak Molecular Imaging Software version 4.04 (Eastman Kodak Company, USA). Qualitative assessment of PKA activity was calculated using the background subtracted net intensity of phosphorylated and non-phosphorylated regions. Activity is reported as the percent of total intensity represented by phosphorylated Kemptide.
Mitochondrial Superoxide Production
The rate of mitochondrial superoxide production was determined using the cell permeable, mitochondrial localizing, dihydroethidine analog MitoSOX® Red (Invitrogen). Localization of MitoSOX® to the mitochondria in HepG2 cells was confirmed by fluorescence microscopy which showed that MitoSOX fluorescence had strong concordance with the fluorescence of MitoTracker™ Green (data not shown). The oxidation of MitoSOX by superoxide produces a hydroxylated ethidium product which has a characteristic excitation at 396 nm, which is not present for other dihydroethidine oxidation products (Zhao et al., 2005; Robinson et al., 2006). Excitation of the hydroxylated product at 396 nm results in a much enhanced emission at 579 nm, facilitating the specificity of this probe to superoxide determination. To take advantage of these properties the MitoSOX fluorescence was measured in HepG2 cells using a Fluoroskan Ascent® fluorometer (Thermo Scientific/ Labsystems), at 390/590 nm (ex/em).
HepG2 cells were harvested by trypsinization (2–3 75 cm2 flasks) and resuspended in PBS. Cells were counted on an improved Neubauer hemacytometer (Hausser Scientific, PA, USA) and diluted to 106 cells/mL in PBS. Cells were loaded into a 96-well plate at 105 cells/well (100 μL) and mixed with 100 μL PBS with 8 μM MitoSOX® Red (final concentration 4 μM); a cell-free well was included as a blank. Fluorescence was measured every minute for 60 minutes at 32° C. Superoxide production was monitored under six separate conditions: control (PBS), 5 μM rotenone, 60 μM dbcAMP/3-isobutyl-1-methylxanthine (IBMX), 60 μM dbcAMP/IBMX + 5 μM rotenone, 10 mM succinate, and 10 mM succinate + 60 μM dbcAMP/IBMX. NRTIs were added to each of these conditions at the following concentrations: AZT (1, 10, 50 μM), ddI (10 μM), ddC (1 μM). The rate of superoxide production was calculated following the first 20 readings and is reported as the blank subtracted increase in relative fluorescence units (RFU)/minute; each experiment was performed in duplicate.
Statistics
Data were analyzed using GraphPad Prism® 4 software (San Diego, CA, USA). Protein phosphorylation and complex I activity were analyzed by one-way ANOVA. Protein kinase A activity was analyzed by one-way ANOVA paired by enzyme preparation. Mitochondrial superoxide production was analyzed by two-way ANOVA paired by cell culture. Data is expressed as the mean of four separate experiments + mean standard error; statistical difference from control was assumed at p ≤ 0.05.
Results
HepG2 human hepatoma cells were chosen for this study due to their relatively rich mitochondrial content and demonstrated use in the study of NRTI toxicity (Pinti et al., 2003; Hoschele, 2006); additionally, these features allowed for the investigation of NRTIs effects on the phosphoregulation of complex I to be performed on a ready source of human mitochondria. As reported earlier, the average yield of mitochondria was 0.15 mg/mg whole cell protein; ten highly confluent flasks generated 4–6 mg of mitochondria. Separate isolations were performed on individual cultures to ensure enough mitochondria to perform all of the required assays.
The concentrations of AZT and ddI were chosen to reflect maximum serum concentrations of patients receiving NRTI therapy (Moyer et al., 1999; Estrela et al., 2004). A high dose for AZT was included to examine dose dependent effects, though 50 μM AZT concentrations have been observed clinically (Blum et al., 1988). The concentrations of AZT-phosphates were determined using previously published data which showed that AZT-MP is capable of accumulating to 2–20 times that of the parent drug intracellularly (Furman et al., 1986; Slusher et al., 1992); the di- and triphosphate concentrations represent maximal conceivable concentrations based on the same studies. A supratherapeutic dose of ddC (10x) was chosen to compare to previously published work by this laboratory at the same concentration (Lund and Wallace, 2004; Lund et al., 2007).
Immunocapture and Phosphorylation of Complex I
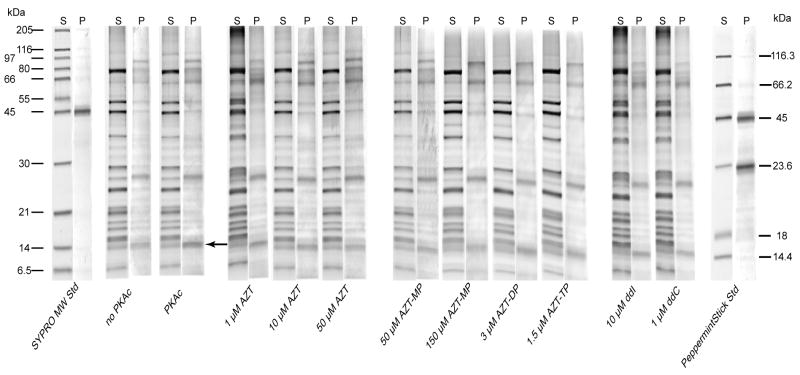
SYPRO staining of immunocaptured complex I resolved 21 bands in a pattern consistent with previously published purified complex I from human heart (Murray et al., 2003) (figure 2, S lanes). Under the reported phosphorylation conditions, Pro-Q® Diamond staining of immunocapture-purified complex I consistently showed four bands which were sufficiently darker than the respective SYPRO stained bands (ratio ≥ 3.5 at 595 nm; figure 2, P lanes) at the following apparent sizes: 95 kDa, 64 kDa, 23.9 kDa, and 14.5 kDa. The identity of the 95 kDa protein is unknown (the largest complex I subunit is 75–77 kDa) and may represent non-denatured proteins containing phosphoproteins of lower molecular weight or contaminating IgG (Murray et al., 2003). The 64 kDa protein may represent contaminating complex II core protein 2 (a 70 kDa phosphoprotein) observed in previous immunocaptured complex I (Murray et al., 2003). The 23.9 kDa protein most likely represents the unknown 29 kDa matrix protein observed previously in rat, mouse, and bovine (Technikova-Dobrova et al., 2001; Pasdois et al., 2003; Chen et al., 2004). The only band which consistently showed dbcAMP/PKAc dependence was the 14.5 kDa phosphoprotein (figure 2, arrow). The inclusion of 60 μM dbcAMP and 20 U PKAc essentially doubled the Pro-Q® stain intensity of this band (figure 3). This band most likely contains the ESSS/NDUFB11 subunit of complex 1; depending on the species and gel conditions this protein runs at 14–19 kDa (Sardanelli et al., 1995; Technikova-Dobrova et al., 2001; Pasdois et al., 2003; Chen et al., 2004; Schulenberg et al., 2004).
Figure 2.
Total and phosphoprotein staining of immunocaptured complex I. Representative, paired-gel lanes of immunocaptured complex I from HepG2 cells run by PAGE and stained first with Pro-Q® Diamond phosphoprotein gel stain (P lanes) followed by SYPRO® Ruby protein stain (S lanes) as described in materials and methods. HepG2 isolated mitochondria were incubated in phosphorylation buffer (see methods) with the indicated NRTI for 30 minutes before complex I was immunocaptured as described. Pro-Q® stained phosphoproteins of apparent molecular weight of 95, 64, 23.9, and 14.5 kDa were resolved. The 14.5 kDa band (indicated by arrow) showed cAMP/PKAc dependent phosphorylation.
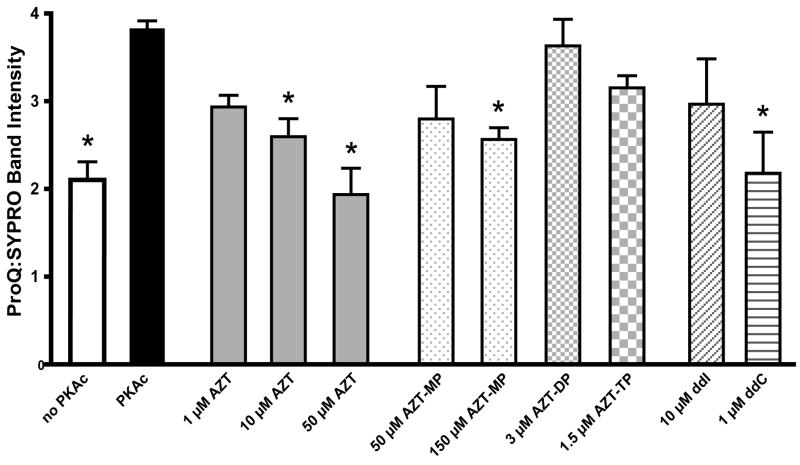
Figure 3.
Phosphorylation of 14.5 kDa subunit of complex I. Relative phosphorylation of the 14.5 kDa band determined by digital densitometry as described in the materials and methods. Data represents the ratio of net intensities for Pro-Q® Diamond and SYPRO® Ruby stain of the 14.5 kDa band of the same gel. Data is the mean + SEM of four separate gels.
* Statistically different from PKAc (p < 0.05).
NRTI-related effects on complex I phosphorylation were observed at the 14.5 kDa protein, none of the other phosphoproteins showed drug related changes in phosphorylation. Densitometric analysis revealed that AZT (10 and 50 μM) inhibited PKAc related phosphorylation (figure 3). The monophosphate of AZT also prevented phosphorylation at 150 μM, but the di- and triphosphate forms had no effect. The effects of ddI and ddC were mixed with 1 μM ddC inhibiting phosphorylation and 10 μM ddI having no effect (figure 3).
Complex I activity
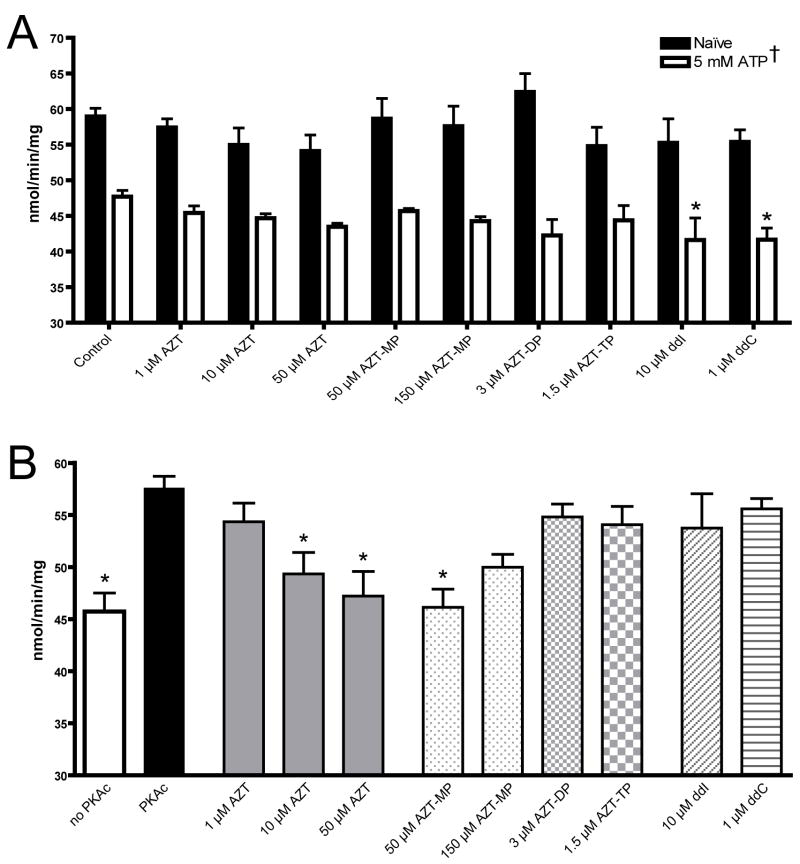
Rotenone sensitive NADH:CoQ1 oxidoreductase activity of complex I revealed several trends. HepG2 isolated mitochondria incubated in naïve conditions for 30 minutes demonstrated an activity of 59.97 ± 1.14 nmol/min/mg; 5 mM ATP resulted in a significantly lower rate (47.71 ± 0.86) (figure 4a). NRTIs had no effect on naïve complex I activity, but in the presence of ATP both ddI (10 μM) and ddC (1 μM) decreased the rate of NADH oxidation (figure 4a). The addition of dbcAMP and PKAc (to promote phosphorylation of complex I) prevented the decrease in complex I activity caused by ATP (57.55 ± 1.16); AZT (10 and 50 μM) as well as AZT-MP (50 μM) inhibited this rate (figure 4b). None of the other NRTIs significantly affected the cAMP/PKAc-dependent stimulation of complex I activity.
Figure 4.
Complex I activity. Rotenone sensitive NADH:CoQ1 oxidoreductase activity of HepG2 isolated mitochondria incubated for 30 minutes in phosphorylation buffer (see materials and methods) A) without dbcAMP/PKAc or B) with dbcAMP/PKAc. Data is the mean + SEM of four separate experiments.
† 5 mM ATP was statistically different from naïve under all treatments (p < 0.05)
* Statistically different from A) respective control or B) PKAc (p < 0.05).
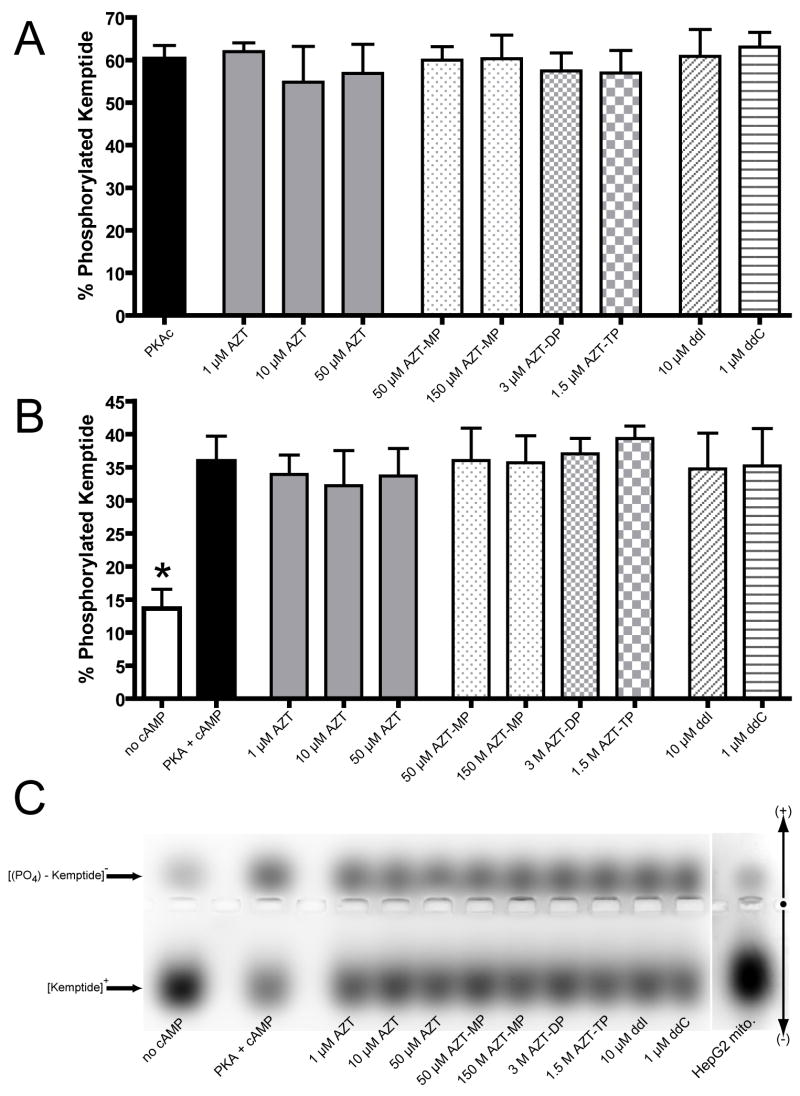
Qualitative activity of Protein Kinase A
Commercially purified PKAc effectively phosphorylated 60.3 ± 3.15% of the added Kemptide over the 30 minute incubation (24.5 ± 1.3 pmol/min) (figure 5a). In contrast, cAMP-dependent PKA phosphorylated 35.9 ± 3.8% and 13.6 ± 2.9% (14.6 ± 1.5 and 5.5 ± 1.9 pmol/min) in the presence and absence of cAMP respectively (figure 5b). None of the NRTIs examined had a significant effect on either PKAc or cAMP-dependent PKA phosphorylation of Kemptide (figure 5a and b). A representative gel demonstrating the separation of phosphorylated and non-phosphorylated Kemptide is shown (figure 5c). A qualitative assessment of cAMP-dependent PKA activity in isolated HepG2 mitochondria is also shown (figure 5c); based on the phosphorylation of 11.4% of the Kemptide a rough estimate of PKA activity was calculated to be 130 pmol/min/mg mitochondrial protein. The component of this activity which is solely mitochondrial is unknown, as the mitochondrial preparations were not tested for purity.
Figure 5.
Protein kinase A activity. Qualitative determination of A) PKAc and B) cAMP-dependent PKA activity as determined by the PepTag® PKA assay kit. The extent of Kemptide phosphorylation was determined by area of interest densitometry on gels run as described in the materials and methods. Values represent the percent net intensity of negatively charged (phosphorylated) Kemptide of the total net intensity of Kemptide from each well; displayed as the mean + SEM of four separate experiments. C) Representative composite gel of cAMP-dependent phosphorylation of Kemptide; the far right well represents the PKA activity of 35 μg of isolated HepG2 mitochondrial protein.
* Statistically different from PKA + cAMP (p < 0.05)
Mitochondrial Superoxide Production
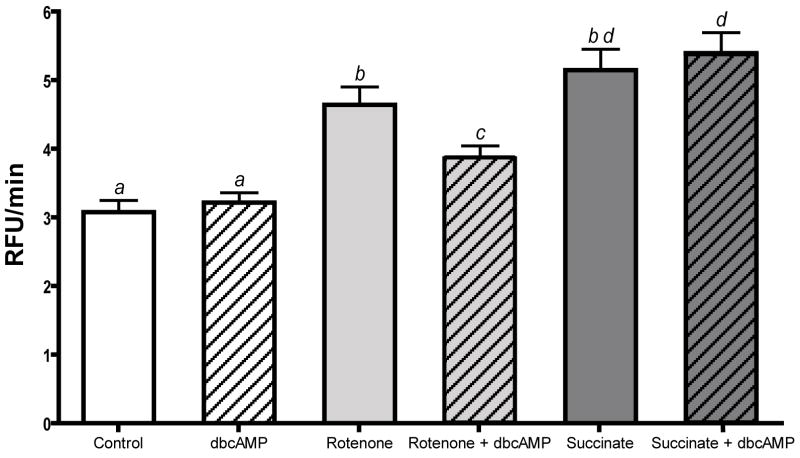
The rate of mitochondrial superoxide production in HepG2 cells as determined by MitoSOX® Red fluorescence generally followed the expected trends under the respective conditions. Superoxide formation from forward-electron flow showed a significant rate increase in the presence of rotenone (4.75 ± 0.22 versus 3.43 ± 0.37 RFU/min) and reverse-electron flow from succinate also generated a significantly elevated rate of superoxide production (5.34 ± 0.29 RFU/min) though this was not greater than with rotenone alone (figure 6). Dibutyryl-cAMP alone did not affect the rate of superoxide production (3.36 ± 0.18 RFU/min), but did significantly (but not completely) prevent the increase caused by rotenone (4.14 ± 0.18 RFU/min) (figure 6). Somewhat surprisingly, the maximum rate of superoxide production was observed in the presence of dbcAMP + succinate (5.59 ± 0.30 RFU/min) (figure 6).
Figure 6.
Mitochondrial superoxide production in whole cells. Rate of mitochondrial superoxide production in HepG2 cells as determined by MitoSOX® Red fluorescence (390/590 nm ex/em). 105 cells were incubated in 4 μM MitoSOX® Red in PBS (control) with the addition of the following conditions: 60μM dbcAMP/IBMX, 5 μM Rotenone, 5 μM Rotenone + 60 μM dbcAMP/IBMX, 10 mM succinate, or 10 mM succinate + 60 μM dbcAMP. Fluorescence was monitored for 60 minutes; data is the cell-free blank subtracted rate (relative fluorescence units, RFU/min) of the final 40 minutes. Values are the mean of four separate experiments + SEM.
a–d Different letters are significantly different from each other (p < 0.05).
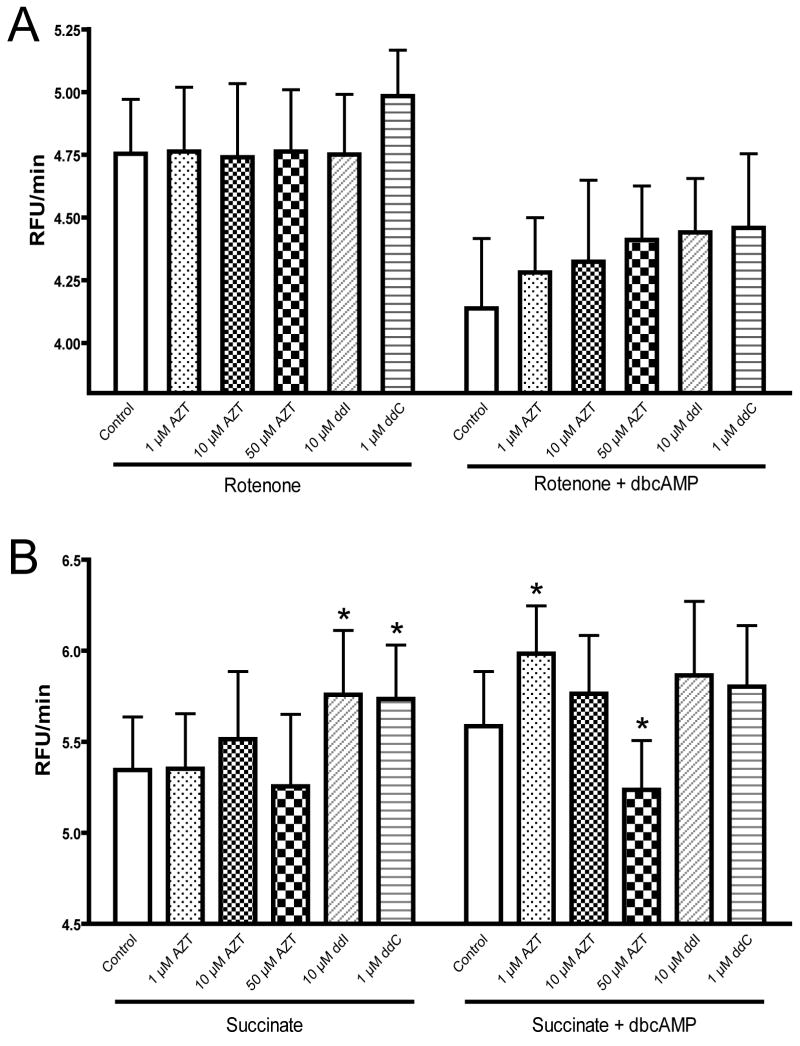
The addition of NRTIs produced assorted changes in superoxide production depending on the condition; the phosphates of AZT were not included in this experiment because they have limited ability to cross the plasma membrane of whole cells (Wagner et al., 2000). Under the control and dbcAMP alone conditions, no drug related effects were shown (data not shown). In the presence of rotenone and rotenone + dbcAMP no significant drug effects were observed (figure 7a). In the presence of succinate, both ddI (10 μM) and ddC (1 μM) caused an increase in superoxide production, though this difference was not observed in the presence of dbcAMP (figure 7b). The most peculiar results were shown with AZT in the presence of succinate + dbcAMP, which showed an inverse dose-dependence on the rate of superoxide production. AZT (1 μM) caused a significant increase in the rate of superoxide generation with succinate + dbcAMP, but at 50 μM caused a significant decrease; 10 μM was not different from the respective control (figure 7b).
Figure 7.
Effect of NRTIs on maximal superoxide production. The effect of NRTIs on mitochondrial superoxide production in whole HepG2 cells in the presence of A) rotenone with or without dbcAMP and B) succinate with or without dbcAMP. Conditions are as described in materials and methods with treatments as indicated. Fluorescence was monitored for 60 minutes; data is the cell-free blank subtracted rate (relative fluorescence units, RFU/min) of the final 40 minutes. Values are the mean of four separate experiments + SEM.
* Significantly different from respective control (p < 0.05)
Discussion
Previous studies on the inhibition of mitochondrial complex I activity by NRTIs provided mixed results (reviewed in (Lund and Wallace, 2004)). The role this complex provides in oxidative phosphorylation as well as NAD+/NADH homeostasis (and upstream lactate/pyruvate homeostasis) makes it a critical center for metabolic control. Prior work by this laboratory has consistently demonstrated the function of this complex is altered by some NRTIs, especially AZT (Lund and Wallace, 2004; Lund et al., 2007); however, the mechanism of this direct effect was unknown. In the current study we have demonstrated that, to varying degrees, NRTIs are capable of inhibiting the cAMP-dependent phosphorylation of the ESSS/NDUFB11 subunit – an important element of complex I regulation. In the case of AZT (and its monophosphate) this inhibition was associated with a decrease in cAMP-stimulated complex I activity. Additionally, we have shown that cAMP alters mitochondrial superoxide production at complex I and that NRTIs also interfere with this process.
In the original hypothesis for this study it was assumed that the most probable points of NRTI interference are the nucleotide binding sites of PKA, involving either cAMP binding/activation or the ATP consuming protein kinase reaction. This assumption was made based on the structural similarity of NRTIs to these substrates as well as prior evidence suggesting NRTIs inhibit other endogenous kinases (Barile et al., 1994; Valenti et al., 1999; McKee et al., 2004; Rylova et al., 2005; Carnicelli et al., 2006). However, as demonstrated by the PepTag® assays (figure 5), neither PKAc nor cAMP-dependent PKA activity were directly affected by any of the NRTIs examined here. While there is the possibility of an unknown mitochondrial cAMP-dependent kinase (sensitive to NRTIs), given the evidence for mitochondrial PKA and the fact that the complex I phosphorylations have been identified at PKA consensus sequences, this possibility is quite remote. Instead, it seems more probable that the inhibition of complex I phoshorylation and its associated activity result from a direct interaction between the NRTIs and complex I (figure 1, point 3). Considering the uncertainty surrounding this complex and the many suspected nucleotide binding sites it may harbor (Hegde, 1998; Zakharova et al., 1999; Brandt, 2006), this observation may not be surprising.
As mentioned previously, the protein and phosphoprotein banding pattern of isolated complex I observed here was generally consistent with previous reports (Sardanelli et al., 1995; Murray et al., 2003; Chen et al., 2004). While the apparent molecular weight of the cAMP-dependent ESSS phosphoprotein was on the low end of similar studies (Technikova-Dobrova et al., 2001; Pasdois et al., 2003; Chen et al., 2004; Schulenberg et al., 2004), our observed molecular weight of 14.5 kDa agrees well with the expected mature mass of this subunit (14,453.1 Da) (Carrol et al., 2003). Variability in the extent of subunit dissociation and co-migration should be expected with variations in the denaturing conditions or ionic environment of the electrophoresis. With the limited number of cAMP-dependent phosphoproteins in the mitochondria there is a very high probability that this band represents the ESSS/NDUFB11 subunit of complex I.
In the current study we did not observe any evidence of phosphorylation at the smaller MWFE/NDUFA1 subunit, but this may be an artifact of the method of detection rather than a lack of phosphorylation at this subunit. SYPRO® Ruby staining revealed a band at 6.5 kDa, but no staining by Pro-Q® Diamond was observed (Figure 2). Using similar phosphorylation conditions and Pro-Q® Diamond stain Schulenberg et al. (Schulenberg et al., 2004) also failed to show MWFE phosphorylation. Studies using phosphoserine antibodies have also shown inconsistent MWFE phosphorylation (Technikova-Dobrova et al., 2001; Pasdois et al., 2003), while the use of [γ-32P]-ATP radiolabeling and mass spectroscopy seems to be the most reliable methods to measure phosphorylation at this subunit (Sardanelli et al., 1995; Raha et al., 2002; Chen et al., 2004). These latter observations may suggest secondary structure of this protein within the gel which prevents antibody/stain interaction with the phosphoserine.
Both the ESSS and MWFE subunits are transmembrane proteins in the hydrophobic, proton transduction regions of complex I (Scheffler et al., 2004; Brandt, 2006); therefore, one would expect that they would be involved in proton translocation. However, the observation that phosphorylation of these subunits may affect NADH:CoQ oxidoreductase activity (Technikova-Dobrova et al., 2001; Raha et al., 2002; Pasdois et al., 2003) suggests that they are somehow integrated into the electron-transport process. It is reasonable to speculate that they may be involved in the coupling of the two processes and affect the hypothesized conformational change linked to the reduction of quinone (Brandt et al., 2003) – for example, phosphorylations of the ESSS/MWFE subunits could affect the free energy required for the conformational change leading to the reduction of quinone. In the case of AZT there was strong agreement between the level of phosphorylation of the ESSS subunit and complex I activity, which was also observed with AZT-monophosphate (figures 4 and 5b). This may suggest that the location of AZT’s interaction with complex I is near the coupling site of this process. In contrast, ddC showed an inhibitory effect on phosphorylation, but did not affect complex I activity. The location of the MWFE subunit near the Q-site (Scheffler et al., 2004) is suggestive of its potential to affect the coupling of electron transport to proton translocation; it is possible that AZT interferes at both the MWFE and ESSS sites and therefore affects activity while ddC only interferes with ESSS phosphorylation and does not affect activity. Alternative methods which would enable the visualization of MWFE phosphorylation (as described above) are required to test this hypothesis.
The effects of ATP on complex I activity observed in this study (figure 4a) are in agreement with previous studies which showed a concentration and/or time dependent decrease in complex I activity in the presence of ATP (Rouslin, 1991; Raha et al., 2002; Maj et al., 2004). However, the mechanism of this effect is unknown and may depend heavily on the assay conditions/preparation. The idea that ATP may allosterically inhibit complex I in a feedback mechanism makes conceptual sense as this complex is the primary entry point of electrons for oxidative phosphorylation. It is possible that the common methods for determining complex I activity (i.e. in the absence of ATP) (Birch-Machin et al., 1993) actually represent a maximal rate which would not occur in the presence of ATP, in which case the depressed rate observed in this study would represent physiological activity and the PKAc-dependent recovery would represent a stimulation of the physiological rate. The concentrations of ATP used in this study (5 mM) are certainly within the physiological intracellular range (1–10 mM), depending on cell-type and metabolic state (Allue et al., 1996; Kennedy et al., 1999). Alternatively, the decrease in complex I activity observed with ATP may have been caused by an alteration in availability of divalent cations (i.e. Mg2+) which have been shown to affect a slow active/inactive transition of complex I (Kotlyar et al., 1992; Grivennikova et al., 2003). Under the phosphorylation incubation conditions used in the current study (30° C for 30 minutes) the described transition is thermodynamically possible (Kotlyar et al., 1992). Under this latter scheme, the presence of ATP favored the transition to the inactive state during the incubation, but this was prevented by PKAc/cAMP. The inactive form has been shown to be sensitive to sulfhydryl reagents suggesting that the inactivation is due to a conformational change (Grivennikova et al., 2001); the observed inhibition of this low active state by ddI and ddC may suggest that this conformation change exposes a binding site for ddI and ddC which further inhibit its activity. Alternately, ddI and ddC may additionally affect divalent cation concentrations and further promote the inactive state.
The observation in the current study that cAMP affects superoxide production at complex I, depending on the direction of electron flow, is interesting on its own accord and may provide insights to the location(s) of superoxide production. In the current study, rotenone stimulated superoxide production was inhibited by dbcAMP/IBMX, but succinate stimulated superoxide production was not (figure 6). This suggests that whatever the effect on electron-transport the ESSS/MWFE subunits have, it occurs prior to the site of reverse-electron-transfer (RET) induced superoxide generation.
Under long-term exposure to these drugs (as is expected under current HIV management strategies) a minimal but persistent elevation of superoxide may prove detrimental to ROS homestasis (Jezek and Hlavata, 2005) and could explain the delayed toxicities and oxidative damage observed with AZT and other NRTIs. While the dbcAMP protection against rotenone stimulated ROS appeared to be influenced by NRTIs, none of these effects were statistically significant (figure 7a). As mentioned previously, the most likely physiological source of superoxide from complex I is from RET from succinate (Lambert and Brand, 2004). Under this condition both ddI and ddC stimulated superoxide production, but this elevation was not observed with dbcAMP present (figure 7b). The peculiar results shown by AZT with succinate + dbcAMP may provide further evidence that AZT has multiple binding sites at complex I. A primary binding site, occupied at low AZT concentrations, appears to increase RET superoxide with dbcAMP, but occupation of a second site at higher concentrations prevents superoxide production in this same state. The qualitative rates of ddC and ddI superoxide production with succinate were the same with or without cAMP suggesting their interaction with complex I is separate from this process. AZT stimulated superoxide, however, is dependent on the presence of cAMP which further suggests it interferes with both phosphorylation sites at complex I.
The ultimate picture which is forming is one in which complex I has several different states, depending on various potential regulatory mechanisms (i.e. concentration of ATP/divalent cations, cAMP, ΔpH, substrate availability, etc.) with which the NRTIs may interact to differing degrees (figure 1). This may offer one explanation of differences observed between studies and also between tissues (i.e. depending on the preparation or metabolic state). The study of complex I activity is certainly not exact and any invasive methodology (i.e. isolation/disruption of mitochondria) is likely to disturb the actual physiological state(s) of the enzyme. Given the differential effects caused by NRTIs in the current study it is reasonable to assume that under various physiological conditions NRTIs are capable of interfering with the proper function of this complex. In vitro studies at both therapeutic and supratherapeutic levels provide evidence of a rapid and direct effect of the NRTIs (Modica-Napolitano, 1993; Pereira et al., 1998; Lund and Wallace, 2004), but within the varied physiological states of complex I these effects could be subtle and may not affect at the tissue or system level until after prolonged use.
In this study we demonstrate that NRTIs are indeed capable of affecting complex I activity in a non-polymerase-γ/mtDNA mediated pathway. The direct effects observed here could alter NAD+/NADH homeostasis and lead to elevated lactate as observed experimentally and clinically (Lamperth et al., 1991; Brinkman, 2000; Lund et al., 2007). Additionally the elevations in superoxide produced at complex I caused by NRTIs could provide a mechanism for the oxidative stress observed with these drugs. In the case of AZT, which has limited activity against DNA polymerase-γ (Cheng et al., 1990; Kakuda, 2000), elevations in superoxide could represent the dominant mechanism for the observed long-term mitochondrial DNA damage observed both experimentally and clinically (Casademont et al., 1996; Yamaguchi et al., 2002).
Acknowledgments
Support: NIH grant number HL072715
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allue I, Gandelman O, Dementieva E, Ugarova N, Cobbold P. Evidence for rapid consumption of milimolar concentrations of cytoplasmic ATP during rigor-contracture of metabolically compromised single cardiomyocytes. Biochem J. 1996;319:463–469. doi: 10.1042/bj3190463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barile M, Valenti D, Hobbs GA, Abruzzese MF, Keilbaugh SA, Quadgliariello E, Passarella S, Simpson MV. Mechanisms of toxicity of 3′-azido-3′-deoxythymidine: its interaction with adenylate kinase. Biochem Pharmacol. 1994;48:1405–1412. doi: 10.1016/0006-2952(94)90564-9. [DOI] [PubMed] [Google Scholar]
- Birch-Machin M, Jackson S, Kler RS, Turnbul DM. Study of skeletal muscle mitochondrial dysfunction. In: Lash LH, Jones DP, editors. Mitochondrial Dysfunction. Academic Press Inc.; San Diego, CA: 1993. pp. 51–69. [Google Scholar]
- Blum MR, Liao SH, Good SS, de Miranda P. Pharmacokinetics and bioavailability of zidovudine in humans. Am J Med. 1988;85:189–194. [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brandt U. Energy converting NADH:quinone oxidoreductase (complex I) Annu Rev Biochem. 2006;75:69–92. doi: 10.1146/annurev.biochem.75.103004.142539. [DOI] [PubMed] [Google Scholar]
- Brandt U, Kerscher S, Drose S, Zwicker K, Zickermann V. Proton pumping by NADH:ubiquinone oxidoreductase. A redox driven conformational change mechanism? FEBS Lett. 2003;545:9–17. doi: 10.1016/s0014-5793(03)00387-9. [DOI] [PubMed] [Google Scholar]
- Brinkman K. Hyperlactatemia and hepatic steatosis as features of mitochondrial toxicity of nucleoside analogue reverse transcriptase inhibitors. Clin Infect Dis. 2000;31:167–169. doi: 10.1086/313921. [DOI] [PubMed] [Google Scholar]
- Carnicelli V, Giulio AD, Bozzi A, Strom R, Oratore A. Zidovudine inhibitis protein kinase C activity in human chronic myeloid (K562) cells. Pharmacol Toxicol. 2006;99:317–322. doi: 10.1111/j.1742-7843.2006.pto_486.x. [DOI] [PubMed] [Google Scholar]
- Carrol J, Fearnley IM, Shannon RJ, Hirst J, Walker JE. Analysis of the subunit composition of complex I from bovine heart mitochondria. Molecular and Cellular Proteomics. 2003;2:117–126. doi: 10.1074/mcp.M300014-MCP200. [DOI] [PubMed] [Google Scholar]
- Casademont J, Barrientos A, Grau JM, Pedrol E, Estivill X, Urbano-Marquez A, Nunes V. The effect of zidovudine on skeletal muscle mtDNA in HIV-1 infected patients with mild or no muscle dysfunction. Brain. 1996;119:1357–1364. doi: 10.1093/brain/119.4.1357. [DOI] [PubMed] [Google Scholar]
- Chen R, Fearnley IM, Peak-Chew SY, Walker JE. The phosphorylation of subunits of complex I from bovine heart mitochondria. J Biol Chem. 2004;279:26036–26045. doi: 10.1074/jbc.M402710200. [DOI] [PubMed] [Google Scholar]
- Cheng YC, Gao WY, Chen CH, Vazquez-Padua M, Starnes MC. DNA polymerases versus HIV reverse transcriptase in AIDS therapy. Ann N Y Acad Sci. 1990;616:217–223. doi: 10.1111/j.1749-6632.1990.tb17842.x. [DOI] [PubMed] [Google Scholar]
- de la Asuncion JG, del Olmo ML, Gomez-Cambronero LG, Sastre J, Pallardo FV, Vina J. AZT induces oxidative damage to cardiac mitochondria: protective effect of vitamins C and E. Life Sci. 2004;76:47–56. doi: 10.1016/j.lfs.2004.06.020. [DOI] [PubMed] [Google Scholar]
- Estrela RdCE, Salvadori MC, Suarez-Kurtz G. A rapid and sensitive method for simultaneous determination of lamivudine and zidovudine in human serum by on-line solid-phase extraction coupled to liquid chromatography/tandem mass spectrometry detection. Rapid Commun Mass Spectrom. 2004;18:1147–1155. doi: 10.1002/rcm.1446. [DOI] [PubMed] [Google Scholar]
- Furman PA, Fyfe JA, St Clair MH, Weinhold K, Rideout JL, Freeman GA, Lehrman SN, Bolognesi DP, Broder S, Mitsuya H, Barry DW. Phosphorylation of 3′-azido-3′-deoxythymidine and selective interaction of the 5′-triphosphate with human immunodeficiency virus reverse transcriptase. Proc Natl Acad Sci. 1986;83:8333–8337. doi: 10.1073/pnas.83.21.8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikova VG, Kapustin AN, Vinogradov AD. Catalytic activity of NADH-ubiquinone oxidoreductase (complex I) in intact mitochondria: evidence for the slow active/inactive transition. J Biol Chem. 2001;276:9038–9044. doi: 10.1074/jbc.M009661200. [DOI] [PubMed] [Google Scholar]
- Grivennikova VG, Serebryanaya DV, Isakova EP, Belozerskaya TA, Vinogradov AD. The transition between active and de-activated forms of NADH:ubiquinone oxidoreductase (complex I) in the mitochondrial membrane of Neurospora crassa. Biochem J. 2003;369:619–626. doi: 10.1042/BJ20021165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde R. The 24-kDa subunit of the bovine mitochondrial NADH:ubiqinone oxidoreductase is a G protein. Biochem Biophys Res Commun. 1998;244:620–629. doi: 10.1006/bbrc.1998.8304. [DOI] [PubMed] [Google Scholar]
- Horbinski C, Chu CT. Kinase signaling cascades in the mitochondrion: a matter of life or death. Free Radic Biol Med. 2005;38:2–11. doi: 10.1016/j.freeradbiomed.2004.09.030. [DOI] [PubMed] [Google Scholar]
- Hoschele D. Cell culture models for the investigation of NRTI-induced mitochondrial toxicity. Relevance for the prediction of clinical toxicity. Toxicol in Vitro. 2006;20:535–546. doi: 10.1016/j.tiv.2005.11.007. [DOI] [PubMed] [Google Scholar]
- Jezek P, Hlavata L. Mitochondria in homeostasis of reactive oxygen species in cell, tissues, and organism. Int J Biochem Cell Biol. 2005;37:2478–2503. doi: 10.1016/j.biocel.2005.05.013. [DOI] [PubMed] [Google Scholar]
- Kakuda TN. Pharmacology of nucleoside and nucleotide reverse transcriptase inhibitor-induced mitochondrial toxicity. Clin Ther. 2000;22:685–708. doi: 10.1016/S0149-2918(00)90004-3. [DOI] [PubMed] [Google Scholar]
- Kennedy HJ, Pouli AE, Ainscrow EK, Jouaville LS, Rizzuto R, Rutter GA. Glucose generates sub-plasma membrane ATP microdomains in single islet β-cells. J Biol Chem. 1999;274:13281–13291. doi: 10.1074/jbc.274.19.13281. [DOI] [PubMed] [Google Scholar]
- Kotlyar AB, Sled VD, Vinogradov AD. Effect of Ca2+ ions on the slow active/inactive transition of the mitochondrial NADH-ubiquinone reductase. Biochim Biophys Acta. 1992;1098:144–150. doi: 10.1016/s0005-2728(05)80329-9. [DOI] [PubMed] [Google Scholar]
- Lambert AJ, Brand MD. Inhibitors of the quinone-binding site allow rapid superoxide production from mitochondrial NADH:ubiquinone oxidoreductase (complex I) J Biol Chem. 2004;279:39414–39420. doi: 10.1074/jbc.M406576200. [DOI] [PubMed] [Google Scholar]
- Lamperth L, Dalakas MC, Dagani F, Anderson J, Ferrari R. Abnormal skeletal and cardiac muscle mitochondria induced by zidovudine (AZT) in human muscle in vitro and in an animal model. Lab Invest. 1991;65:742–751. [PubMed] [Google Scholar]
- Lim SE, Copeland WC. Differential incorporation and removal of antiviral deoxynucletides by human DNA polymeraseγ. J Biol Chem. 2001;276:23616–23623. doi: 10.1074/jbc.M101114200. [DOI] [PubMed] [Google Scholar]
- Liu Y, Fiskum G, Schubert D. Generation of reactive oxygen species by the mitochondrial electron transport chain. J Neurochem. 2002;80:780–787. doi: 10.1046/j.0022-3042.2002.00744.x. [DOI] [PubMed] [Google Scholar]
- Lund KC, Peterson LL, Wallace KB. Absence of a universal mechanism of mitochondrial toxicity by nucleoside analogs. Antimicrob Agents Chemother. 2007;51 doi: 10.1128/AAC.00039-07. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund KC, Wallace KB. Direct effects of nucleoside reverse transcriptase inhibitors on rat cardiac mitochondrial bioenergetics. Mitochondrion. 2004;4:193–202. doi: 10.1016/j.mito.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Lund KC, Wallace KB. Direct, DNA pol-γ-independent effects of nucleoside reverse transcriptase inhibitors on mitochondrial bioenergetics. Cardiovascular Toxicology. 2004;4:217–228. doi: 10.1385/ct:4:3:217. [DOI] [PubMed] [Google Scholar]
- Maj MC, Raha S, Myint T, Robinson BH. Regulation of NADH/CoQ oxidoreductase: do phosphorylation events affect activity? Protein Journal. 2004;23:25–32. doi: 10.1023/b:jopc.0000016255.17077.2c. [DOI] [PubMed] [Google Scholar]
- Martin JL, Brown CE, Matthews-Davis N, Reardon JE. Effects of antiviral nucleoside analogs on human DNA polymerases and mitochondrial DNA synthesis. Antimicrob Agents Chemother. 1994;38:2743–2749. doi: 10.1128/aac.38.12.2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee EE, Bentley AT, Hatch M, Gingerich J, Susan-Resiga D. Phosphorylation of thymidine and AZT in heart mitochondria: elucidation of a novel mechanism of AZT cardiotoxicity. Cardiovascular Toxicology. 2004;4:155–168. doi: 10.1385/ct:4:2:155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modica-Napolitano JS. AZT causes tissue-specific inhibiton of mitochondrial bioenergetic function. Biochem Biophys Res Commun. 1993;194:170–177. doi: 10.1006/bbrc.1993.1800. [DOI] [PubMed] [Google Scholar]
- Moyer TP, Temesgen Z, Enger R, Estes L, Charlson J, Oliver L, Wright A. Drug monitoring of antiretroviral therapy for HIV-1 infection: method validation and results of a pilot study. Clinical Chemistry. 1999;45:1465–1476. [PubMed] [Google Scholar]
- Moyle G. Clinical manifestations and management of antiretroviral nucleoside analog-related mitochondrial toxicity. Clin Ther. 2000;22:911–936. doi: 10.1016/S0149-2918(00)80064-8. [DOI] [PubMed] [Google Scholar]
- Munch-Petersen B, Cloos L, Tyrsted G, Erksson S. Diverging substrate specificity of pure human thymidine kinases 1 and 2 against antiviral dideoxynuclesides. J Biol Chem. 1991;266:9032–9038. [PubMed] [Google Scholar]
- Murray J, Zhang B, Taylor SW, Oglesbee D, Fahy E, Marusich MF, Ghosh SS, Capaldi RA. The subunit composition of the human NADH dehydrogenase obtained by rapid one-step immunopurification. J Biol Chem. 2003;278:13619–13622. doi: 10.1074/jbc.C300064200. [DOI] [PubMed] [Google Scholar]
- Pan-Zhou XR, Cui L, Zhou XJ, Sommadossi JP, Darley-Usmar VM. Differential effects of antiretroviral nucleoside analogs on mitochondrial function in HepG2 cells. Antimicrob Agents Chemother. 2000;44:496–503. doi: 10.1128/aac.44.3.496-503.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa S, Sardanelli AM, Cocco T, Speranza F, Scacco SC, Technikova-Dobrova Z. The nuclear-encoded 18 kDa (IP) AQDQ subunit of bovine heart complex I is phosphorylated by the mitochondrial cAMP-dependent protein kinase. FEBS Lett. 1996;379:299–301. doi: 10.1016/0014-5793(95)01532-9. [DOI] [PubMed] [Google Scholar]
- Papa S, Sardanelli AM, Scacco S, Technikova-Dobrova Z. cAMP-dependent protein kinase and the phosphoproteins in mammalian mitochondria. An extension of the cAMP-mediated intracellular signal transduction. FEBS Lett. 1999;444:245–249. doi: 10.1016/s0014-5793(99)00070-8. [DOI] [PubMed] [Google Scholar]
- Papa S, Scacco S, Sardanelli AM, Vergari R, Papa F, Budde S, Heuvel Lvd, Smeitink J. Mutation in the NDUFS4 gene of complex I abolishes cAMP-dependent activation of the complex in a child with fatal neurological syndrome. FEBS Lett. 2001;489:259–262. doi: 10.1016/s0014-5793(00)02334-6. [DOI] [PubMed] [Google Scholar]
- Pasdois P, Deveaud C, Voisin P, Bouchaud V, Rigoulet M, Beauvoit B. Contribution of the phosphorylable complex I in the growth phase-dependent respiration of C6 glioma cells in vitro. J Bioenerg Biomembr. 2003;35:439–450. doi: 10.1023/a:1027391831382. [DOI] [PubMed] [Google Scholar]
- Pereira LF, Oliveira MBM, Carnieri EGS. Mitochondrial sensitivity to AZT. Cell Biochem Funct. 1998;16:173–181. doi: 10.1002/(SICI)1099-0844(199809)16:3<173::AID-CBF783>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Pinti M, Troiano L, Nasi M, Ferraresi R, Dobrucki J, Cossarizza A. Hepatoma HepG2 cells as a model for in vitro studies on mitochondrial toxicity of antiretroviral drugs: which correlation with the patient? J Biol Regul Homeost Agents. 2003;17:166–171. [PubMed] [Google Scholar]
- Raha S, Myint AT, Johnstone L, Robinson BH. Control of oxygen free radical formation from mitochondrial complex I: roles for protein kinase A and pyruvate dehydrogenase kinase. Free Radic Biol Med. 2002;32:421–430. doi: 10.1016/s0891-5849(01)00816-4. [DOI] [PubMed] [Google Scholar]
- Robinson KM, Janes MS, Pehar M, Monette JS, Ross MF, Hagen TM, Murphy MP, Beckman JS. Selective fluorescent imaging of superoxide in vivo using ethidium-based probes. Proc Natl Acad Sci. 2006;103:15038–15043. doi: 10.1073/pnas.0601945103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouslin W. Effects of acidosos and ATP depletion on cardiac muscle electron transfer complex I. J Mol Cell Cardiol. 1991;23:1127–1135. doi: 10.1016/0022-2828(91)90202-w. [DOI] [PubMed] [Google Scholar]
- Rylova SN, Albertioni F, Flygh G, Eriksson S. Activity profiles of deoxynucleside kinases and 5′-nucleotidases in cultured adipocytes and myoblastic cells: insights into mitochondrial toxicity of nucleside analogs. Biochem Pharmacol. 2005;69:951–960. doi: 10.1016/j.bcp.2004.12.010. [DOI] [PubMed] [Google Scholar]
- Sardanelli AM, Technikova-Dobrova Z, Scacco SC, Speranza F, Papa S. Characterization of proteins phosphorylated by the cAMP-dependent protein kinase of bovine heart mitochondria. FEBS Lett. 1995;377:470–474. doi: 10.1016/0014-5793(95)01407-1. [DOI] [PubMed] [Google Scholar]
- Scacco S, Vergari R, Scarpulla RC, Technikova-Dobrova Z, Sardanelli A, Lambo R, Lorusso V, Papa S. cAMP-dependent phosphorylation of the nuclear encoded 18-kDa (IP) subunit of respiratory complex I and activation of the complex in serum-starved mouse fibroblast cultures. J Biol Chem. 2000;275:17578–17582. doi: 10.1074/jbc.M001174200. [DOI] [PubMed] [Google Scholar]
- Scheffler IE, Yadava N, Potluri P. Molecular genetics of complex I-deficient Chinese hamster cell lines. Biochim Biophys Acta. 2004;1659:160–171. doi: 10.1016/j.bbabio.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Schilling B, Aggeler R, Schulenberg B, Murray J, Row RH, Capaldi RA, Gibson BW. Mass spectrometric identification of a novel phosphorylation site in subunit NDUFA10 of bovine mitochondrial complex I. FEBS Lett. 2005;579:2485–2590. doi: 10.1016/j.febslet.2005.03.061. [DOI] [PubMed] [Google Scholar]
- Schulenberg B, Goodman TN, Aggeler R, Capaldi RA, Patton WF. Characterization of dynamic and steady-state protein phosphorylation using a fluorescent phosphoprotein gel stain and mass spectrometry. Electrophoresis. 2004;25:2526–2532. doi: 10.1002/elps.200406007. [DOI] [PubMed] [Google Scholar]
- Signorile A, Sardanelli AM, Nuzzi R, Papa S. Serine(threonine) phosphatase(s) acting on cAMP-dependent phosphoproteins in mammalian mitochondria. FEBS Lett. 2002;512:91–94. doi: 10.1016/s0014-5793(02)02226-3. [DOI] [PubMed] [Google Scholar]
- Skuta G, Fischer GM, Janaky T, Kele Z, Szabo P, Tozser J, Sumegi B. Molecular mechanism of the short-term cardiotoxicity caused by 2′,3′-dideoxycytidine (ddC): modulation of reactive oxygen species levels and ADP-ribosylation reactions. Biochem Pharmacol. 1999;58:1915–1925. doi: 10.1016/s0006-2952(99)00282-8. [DOI] [PubMed] [Google Scholar]
- Slusher JT, Kuwahara SK, Hamzeh FM, Lewis LD, Kornhauser DM, Lietman PS. Intracellular zidovudine (ZDV) and ZDV phosphates as measured by a validated combined high-pressure liquid chromatography-radioimmunoassay procedure. Antimicrob Agents Chemother. 1992;36:2473–2477. doi: 10.1128/aac.36.11.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Pierre J, Buckingham JA, Roebuck SJ, Brand MD. Topology of superoxide production from different sites in the mitochondrial electron transport chain. J Biol Chem. 2002;277:44784–44790. doi: 10.1074/jbc.M207217200. [DOI] [PubMed] [Google Scholar]
- Szabados E, Fischer GM, Toth K, Csete B, Nemeti B, Trombitas K, Habon T, Endrei D, Sumegi B. Role of reactive oxygen species and poly-ADP-ribose polymerase in the development of AZT-induced cardiomyopathy in rat. Free Radic Biol Med. 1999;26:309–317. doi: 10.1016/s0891-5849(98)00199-3. [DOI] [PubMed] [Google Scholar]
- Technikova-Dobrova Z, Sardanelli AM, Speranza F, Scacco S, Signorile A, Lorusso V, Papa S. Cyclic adenosine monophosphate-dependent phosphorylation of mammalian mitochondrial proteins: enzyme and substrate characterization and functional role. Biochemistry. 2001;40:13941–13947. doi: 10.1021/bi011066p. [DOI] [PubMed] [Google Scholar]
- Thomson M. Evidence of undiscovered cell regulatory mechanisms: phosphoproteins and protein kinases in mitochondria. Cell Mol Life Sci. 2001;59:213–219. doi: 10.1007/s00018-002-8417-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenti D, Atlante A, Barile M, Passarella S. Inhibition of phosphate transport in rat heart mitochondria by 3′-azido-3′-deoxythymidine due to stimulation of superoixide anion mitochondrial production. Biochem Pharmacol. 2002;64:201–206. doi: 10.1016/s0006-2952(02)01065-1. [DOI] [PubMed] [Google Scholar]
- Valenti D, Barile M, Quagliariello E, Passarella S. Inhibition of nucleoside diphosphate kinase in rat liver mitochondria by added 3′-azido-3′-deoxythymidine. FEBS Lett. 1999;444:291–295. doi: 10.1016/s0014-5793(99)00071-x. [DOI] [PubMed] [Google Scholar]
- Wagner CR, Iyer VV, McIntee EJ. Pronucleotides: towards the in vivo delivery of antiviral and anticancer nucleotides. Med Res Rev. 2000;20:417–451. doi: 10.1002/1098-1128(200011)20:6<417::aid-med1>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Walker UA, Setzer B, Venhoff N. Increased long-term mitochondrial toxicity in combinations of nucleoside analog reverse-transcriptase inhibitors. AIDS. 2002;16:2165–2173. doi: 10.1097/00002030-200211080-00009. [DOI] [PubMed] [Google Scholar]
- White AJ. Mitochondrial toxicity and HIV therapy. Sex Transm Infect. 2001;77:158–173. doi: 10.1136/sti.77.3.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Katoh I, Kurata S-i. Azidothymidine causes functional and structural destruction of mitochondria, glutathione deficiency and HIV-1 promoter sensitization. Eur J Biochem. 2002;269:2782–2788. doi: 10.1046/j.1432-1033.2002.02954.x. [DOI] [PubMed] [Google Scholar]
- Zakharova NV, Zharova TV, Vinogradov AD. Kinetics of transhydrogenase reaction catalyzed by the mitochondrial NADH-ubiquinone oxidoreductase (complex I) imply more than one catalytic nucleotide-binding sites. FEBS Lett. 1999;444:211–216. doi: 10.1016/s0014-5793(99)00062-9. [DOI] [PubMed] [Google Scholar]
- Zhao H, Joseph J, Fales HM, Sokoloski EA, Levine RL, Vasquez-Vivar J, Kalyanaraman B. Detection and characterization of the products of hydroethdine and intracellular superoxide by HPLC and limitations of fluorescence. Proc Natl Acad Sci. 2005;102:5727–5732. doi: 10.1073/pnas.0501719102. [DOI] [PMC free article] [PubMed] [Google Scholar]