Abstract
Binding of APOBEC3G to the nucleocapsid (NC) domain of the human immunodeficiency virus (HIV) Gag polyprotein may represent a critical early step in the selective packaging of this antiretroviral factor into HIV virions. Previously, we and others have reported that this interaction is mediated by RNA. Here, we demonstrate that RNA binding by APOBEC3G is key for initiation of APOBEC3G:NC complex formation in vitro. By adding back nucleic acids to purified, RNase-treated APOBEC3G and NC protein preparations in vitro, we demonstrate that complex formation is rescued by short (≥10 nucleotides) single-stranded RNAs (ssRNAs) containing G residues. In contrast, complex formation is not induced by add-back of short ssRNAs lacking G, by dsRNAs, by ssDNAs, by dsDNAs or by DNA:RNA hybrid molecules. While some highly structured RNA molecules, i.e., tRNAs and rRNAs, failed to rescue APOBEC3G:NC complex formation, other structured RNAs, i.e., human Y RNAs and 7SL RNA, did promote NC binding by APOBEC3G. Together, these results indicate that ternary complex formation requires ssRNA, but suggest this can be presented in the context of an otherwise highly structured RNA molecule. Given previous data arguing that APOBEC3G binds, and edits, ssDNA effectively in vitro, these data may also suggest that APOBEC3G can exist in two different conformational states, with different activities, depending on whether it is bound to ssRNA or ssDNA.
Keywords: HIV-1, APOBEC3G, intrinsic immunity, RNA binding, retrovirus
INTRODUCTION
The human APOBEC3G (A3G) protein is the prototype of a family of cytidine deaminases that function as antiretroviral defense factors (Sheehy et al. 2002; for review, see Cullen 2006). APOBEC3 proteins exert their inhibitory activity by being selectively packaged into retroviral virion particles (Alce and Popik 2004; Cen et al. 2004; Schafer et al. 2004; Svarovskaia et al. 2004; Wiegand et al. 2004; Zennou et al. 2004) and then interfering with reverse transcription of the retroviral RNA genome during the next infection cycle, at least in part by editing dC residues present in the nascent proviral DNA minus strand to dU (Harris et al. 2003; Mangeat et al. 2003; Shindo et al. 2003; Zhang et al. 2003; Yu et al. 2004; Miyagi et al. 2007; Schumacher et al. 2008). While there has been considerable work on the mechanism(s) by which the packaged A3G protein interferes with retroviral virion infectivity, the first step in the process, i.e., selective virion packaging, has remained poorly understood. A3G and/or other APOBEC3 proteins are able to inhibit a very wide range of retroviruses, including lentiviruses such as HIV-1 as well as members of the foamy virus family (e.g., primate foamy virus; Lochelt et al. 2005; Russell et al. 2005), α-retroviruses (e.g., Rous sarcoma virus; Wiegand and Cullen 2007), β-retroviruses (e.g., Mason–Pfizer monkey virus) (Doehle et al. 2006), γ-retroviruses (e.g., murine leukemia virus) (Kobayashi et al. 2004; Doehle et al. 2005), and human T-cell leukemia virus (Derse et al. 2007). Moreover, several APOBEC3 proteins have been shown to also function as inhibitors of endogenous retroviruses and LTR-retrotransposons, including the yeast retrotransposon Ty1 (Dutko et al. 2005; Esnault et al. 2005; Schumacher et al. 2005; Bogerd et al. 2006; Chen et al. 2006). In all cases where this has been examined, inhibition has been shown to correlate with the specific packaging of the inhibitory APOBEC3 protein into retroviral virions or retrotransposon virus-like particles (VLPs) (Kao et al. 2003; Doehle et al. 2005, 2006; Dutko et al. 2005; Derse et al. 2007; Goila-Gaur et al. 2007; Huthoff and Malim 2007; Opi et al. 2007; Aguiar et al. 2008). However, given the lack of any obvious protein sequence similarity across these very diverse retroelement families, the identity of the common target that is recognized by the APOBEC3 proteins, leading to virion or VLP incorporation, has remained unclear.
The majority of the published work focusing on A3G packaging into retroviral virions has used HIV-1 as a model system. This work has demonstrated that the HIV-1 Gag protein is sufficient for packaging of A3G into budding virion particles (Alce and Popik 2004; Cen et al. 2004; Schafer et al. 2004; Zennou et al. 2004). Within Gag, the key determinant is the nucleocapsid (NC) protein, and NC is both necessary for packaging of A3G into released virions and sufficient for specific binding to A3G in vitro (Alce and Popik 2004; Cen et al. 2004; Luo et al. 2004; Schafer et al. 2004; Zennou et al. 2004; Burnett and Spearman 2007).
An interesting, and poorly understood, aspect of A3G:NC complex formation is that this in vitro interaction is blocked by treatment with RNase A, i.e., complex formation is RNA dependent (Schafer et al. 2004; Svarovskaia et al. 2004; Zennou et al. 2004; Khan et al. 2005; Burnett and Spearman 2007). Here, we have analyzed this requirement in detail. We show that single-stranded RNA (ssRNA) renders A3G capable of forming a complex with HIV-1 NC, while RNA binding by NC, at least prior to complex formation, is dispensable. Fully double-stranded RNA (dsRNA), dsDNA, ssDNA, or DNA:RNA hybrids are unable to promote A3G:NC complex formation. This is intriguing, given the fact that A3G is known to bind ssDNA in vitro (Iwatani et al. 2006) and that it edits ssDNA substrates using its cytidine deaminase activity (Harris et al. 2003; Yu et al. 2004). Together, these data suggest that RNA binding by A3G serves to render A3G competent for incorporation into retroviral virion particles, while ssDNA binding may be selectively involved in the execution phase of A3G's biological activity, once reverse transcription of virion incorporated RNAs has initiated.
RESULTS
As a first step toward defining the role played by nucleic acids in mediating recruitment of A3G to the HIV-1 NC protein, we wished to confirm the previously reported observation (Alce and Popik 2004; Cen et al. 2004; Schafer et al. 2004; Zennou et al. 2004) that recombinant A3G binds NC specifically in vitro. For this purpose, we purified recombinant glutathione-S-transferase (GST) fused to full-length HIV-1 NC from bacteria. This fusion protein also contained the flanking amino-terminal (p2) and carboxy-terminal (p1) linker regions present in HIV-1 Gag. Nonfused GST was also prepared as a control. Full-length human A3G was expressed in transfected 293T cells as an A3G-Myc-His fusion protein (Khan et al. 2005). Similar Myc-His fusion proteins containing green fluorescent protein (GFP-Myc-His), firefly luciferase (Luc-Myc-His), or full-length human APOBEC3A (A3A-Myc-His) were also expressed in transfected 293T cells.
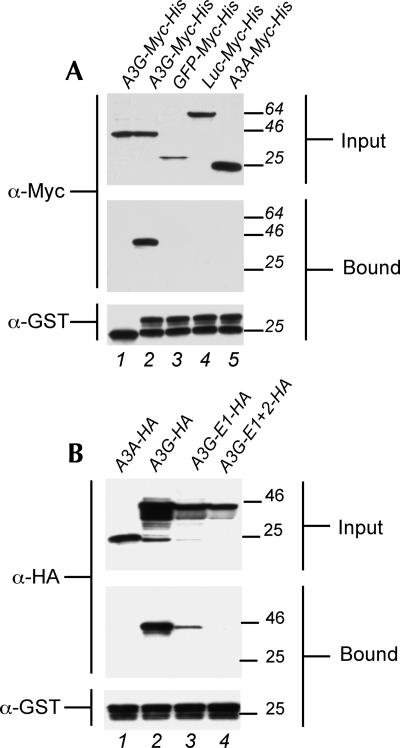
At 44 h after transfection, cells expressing the above fusion proteins were lysed, and the lysates were clarified by centrifugation and then used directly for Western analysis or binding assays. As shown in Figure 1A, the A3G-Myc-His fusion protein bound to recombinant bacterial GST-NC but not to GST itself (lanes 1 and 2). In contrast, GFP-Myc-His, Luc-Myc-His, and A3A-Myc-His all failed to detectably interact with GST-NC (Fig. 1A, lanes 3–5), even though they were expressed in the input lysate at comparable levels. The inability of A3A to interact with HIV-1 NC correlates with the inability of A3A to package into virion cores and inhibit HIV-1 infectivity (Goila-Gaur et al. 2007; Aguiar et al. 2008). We conclude that A3G indeed binds the HIV-1 NC protein specifically in vitro, as previously reported (Alce and Popik 2004; Cen et al. 2004; Schafer et al. 2004; Zennou et al. 2004).
FIGURE 1.
Formation of a specific complex between A3G and HIV-1 NC. (A) 293T cells were transfected with plasmids expressing A3G-Myc-His, GFP-Myc-His, Luc-Myc-His, or A3A-Myc-His. At 44 h after transfection, the cells were lysed and the lysates incubated with recombinant purified GST (lane 1) or GST-NC (lanes 2–5). Bound complexes were then collected using glutathione agarose beads. Input lysates (upper panel, 5% of total) and bound proteins (middle panel, 25% of total) were then visualized by Western analysis using an anti-Myc polyclonal antiserum. GST fusion proteins and non-fused GST were visualized by Western analysis using an anti-GST polyclonal (lower panel). (B) Similar to panel A, except that the 293T cells were transfected with plasmids expressing HA-tagged forms of A3A, A3G, A3G-E1, or A3G-E1+2. GST-NC bound proteins were collected using glutathione agarose beads and input lysates (5% of total) and bound proteins (25% of total) visualized by Western analysis using an HA-specific mouse monoclonal antibody. GST-NC (lower panel) was visualized as described in panel A.
The A3G protein contains two cytidine deaminase active (CDA) site consensus sequences, although only the more carboxy-terminal active site is enzymatically active (Hache et al. 2005; Navarro et al. 2005; Newman et al. 2005; Bogerd et al. 2007). The CDA sites also mediate nucleic acid biding by A3G (Iwatani et al. 2006), and both CDAs, but particularly the amino-terminal CDA, have been reported to play an important role in A3G virion incorporation (Navarro et al. 2005; Newman et al. 2005; Bogerd et al. 2007).
To address the physiological relevance of the observed in vitro A3G:Gag interaction, we asked whether HA-tagged A3G proteins bearing mutant CDAs would be able to bind Gag. The previously described (Bogerd et al. 2007) A3G-E1 mutant bears a glutamic acid to glutamine mutation at position 67 (E67Q), which disrupts the function of the amino-terminal CDA, while the A3G-E1+2 mutant contains not only the E67Q mutation but also a second mutation, E259Q, which mutates the analogous glutamic acid in the carboxy-terminal CDA to glutamine. The A3G-E1+2 mutant therefore lacks both an intact CDA and enzymatic activity (Bogerd et al. 2007). Previous work has shown that the A3G-E1 mutation is poorly packaged into HIV-1 virions, while A3G-E1+2 is not detectably packaged (Bogerd et al. 2007). Similarly, while the wild-type HA-tagged A3G protein bound the HIV-1 NC capsid effectively (Fig. 1B, lane 2), recombinant HA-tagged A3G-E1 showed weak but detectable binding and A3G-E1+2 failed to bind HIV-1 NC detectably (Fig. 1B, lanes 3,4). As expected, A3A-HA failed to detectably bind the HIV-1 NC protein (Fig. 1B, lane 1). These data are consistent with the hypothesis that the ability of A3G mutants to bind to HIV-1 Gag correlates with the efficiency of packaging into HIV-1 virion particles.
RNA binding renders A3G competent to bind HIV-1 NC
Both A3G and HIV-1 NC are known to bind RNA (Damgaard et al. 1998; De Guzman et al. 1998; Navarro et al. 2005; Iwatani et al. 2006), and several groups have previously reported that formation of the NC:A3G complex is RNA dependent (Schafer et al. 2004; Svarovskaia et al. 2004; Zennou et al. 2004; Khan et al. 2005; Burnett and Spearman 2007). We wished to confirm this result and to ask whether NC:A3G binding requires RNA binding by NC, by A3G or by both proteins. For this purpose, we isolated A3G-Myc-His protein from transfected 293T cells and measured its ability to bind to purified bacterial GST-NC in the presence or absence of copurifying RNA.
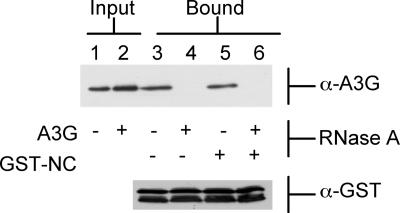
As shown in Figure 2 (lane 3), the purified A3G-Myc-His protein again bound the GST-NC fusion protein effectively in vitro. This interaction was, however, blocked if both the A3G-Myc-His and the GST-NC proteins, or the A3G-Myc-His protein alone, had been previously incubated with RNase A (lanes 4 and 6). This treatment had no apparent effect on the electrophoretic mobility of either protein (Fig. 2). In contrast, extensive incubation of only the recombinant GST-NC with RNase A, prior to elution from the glutathione-agarose beads, had no effect on complex formation (Fig. 2, lane 5). Therefore, these data indicate that it is RNA binding by A3G, not RNA binding by NC, that is critical for initiation of A3G:NC complex formation.
FIGURE 2.
RNase A treatment of A3G blocks formation of an A3G:NC complex. A3G-Myc-His was purified from transfected 293T cells and either left untreated or incubated with RNase A, as indicated. Similarly, GST-NC was purified from bacteria and left untreated or incubated with RNase A. Treated or untreated protein samples were then mixed and incubated together, and any A3G:NC complexes collected using glutathione-agarose beads. The input A3G proteins are shown in the upper panel, lanes 1 and 2, while the bound A3G proteins are shown in lanes 3–6. The GST-NC protein used is shown in the lower panel. Proteins were visualized by Western analysis using a rabbit polyclonal anti-A3G antiserum (upper panel) or an anti-GST polyclonal antibody (lower panel).
Rescue of A3G:NC complex formation by ssRNAs
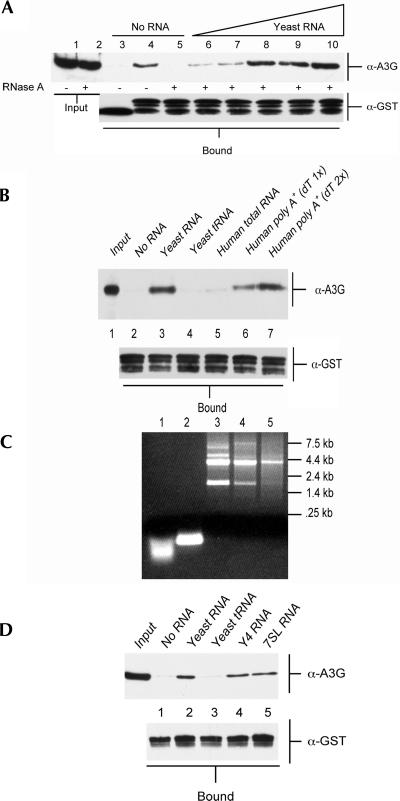
We next asked if we could rescue formation of the A3G:NC complex by adding back RNA to the RNase A-treated recombinant proteins. As shown in Figure 3A (lanes 5–10), we indeed observed a dose-dependent reconstitution of A3G:NC complex formation when we added increasing amounts of a commercial preparation of fragmented Torulla yeast RNA. Although this preparation was claimed to consist of random 300–500-nucleotide (nt) RNA fragments, gel analysis of this preparation showed that it, in fact, consisted of small RNA oligonucleotides that migrated more rapidly than a preparation of purified yeast tRNAs, which are expected to be only 70–80 nt in length (Fig. 3C, cf. lanes 1 and 2). Nevertheless, this result clearly demonstrated that added RNA could reconstitute NC binding by RNase A-treated A3G. To extend these data, we next asked if A3G:NC complex formation could be rescued by the yeast tRNA preparation analyzed in Figure 3C (lane 2) or by a preparation of total human cell RNA (Fig. 3C, lane 3). As shown in Figure 3B (lane 4), yeast tRNA was not able to rescue complex formation, while rescue by total human RNA was very weak (Fig. 3B, lane 5).
FIGURE 3.
Formation of the A3G:NC complex is mediated by unstructured RNAs. (A) The purified A3G-Myc-His was untreated (lanes 3,4) or had been incubated with RNase A (all other lanes). Input A3G-Myc-His protein (5% of total) is shown in lanes 1 and 2. A3G-Myc-His protein was incubated with purified GST (lane 3) or GST-NC (lanes 4–10). In lanes 6–10, yeast RNA was added to the incubation: lane 6, 1.25 μg; lane 7, 2.5 μg; lane 8, 5 μg; lane 9, 10 μg; lane 10, 20 μg. A3G:NC complexes were recovered using glutathione-agarose beads and bound A3G-Myc-His protein visualized by Western analysis using a rabbit polyclonal anti-A3G antiserum (upper panel). Input GST or GST-NC protein was also analyzed by Western blot (lower panel). (B) Similar to panel A, except that the added RNAs (10 μg in each case) represent yeast total RNA (lane 3); purified yeast tRNA (lane 4); total human cell RNA (lane 5), or total human RNA that had been subjected to one round (lane 6) or two rounds (lane 7) of purification on an oligo-dT column. This experiment used exclusively RNase A treated A3G-Myc-His protein. (C) Ethidium bromide-stained agarose gel visualizing 5 μg of each of the RNA samples used in panel B. As may be seen, the yeast RNA sample (Ambion) in lane 1 is degraded to the point where it is smaller than the 70–80-nt-long yeast tRNAs shown in lane 2. Lanes 3–5 reveal the removal of human rRNA as the total human RNA sample (lane 3) was subjected to one (lane 4) or two (lane 5) rounds of oligo-dT purification. (D) Similar to panels A and B, except that this experiment also analyzed in vitro transcribed human Y4 RNA and 7SL RNA.
It has previously been reported that, while A3G binds to ssRNA with high affinity, A3G does not bind dsRNA (Iwatani et al. 2006). We therefore considered the possibility that the poor rescue seen with yeast tRNA and total human RNA (Fig. 3B) was due to the fact that tRNAs are highly structured while total human RNA consists largely of rRNA (Fig. 3C, lane 3), which is also highly structured. To address this issue for total human RNA, we subjected this RNA preparation to either one round (Fig. 3C, lane 4) or two rounds (Fig. 3C, lane 5) of oligo dT purification, which greatly decreased the level of rRNA in these preparations, while increasing the relative level of less-structured mRNA molecules (Fig. 3C, lanes 3–5). As shown in Figure 3B, the level of rescue of A3G:NC complex formation observed after addition of the same amount of (primarily) poly(A)+ human RNA was significantly enhanced when compared to the level of complex formation seen with unfractionated total human RNA (Fig. 3B, cf. lane 5 and lanes 6,7). Therefore, these data suggest that A3G:NC complex formation is effectively rescued by total human poly(A)+ RNA but is not rescued by either yeast tRNA or by human rRNA, i.e., by highly structured RNA species. In contrast, previous work has reported that A3G is found complexed with Y RNAs and 7SL RNA in human cells (Chiu et al. 2006; Wang et al. 2007) and has suggested that 7SL might actually be critical for A3G packaging into HIV-1 virions. Importantly, both Y RNAs and 7SL are short, structured noncoding RNAs.
To address this apparent contradiction, we generated recombinant human Y4 and 7SL RNA and asked whether these RNAs could rescue HIV-1 NC binding by A3G in vitro. As shown in Figure 3D, Y4 RNA and 7SL RNA clearly differed from yeast tRNA in that they would indeed support this protein:protein interaction. A similar rescue was also seen with recombinant human Y1 and Y3 RNA (data not shown). One potential caveat with this result is that we cannot be sure that the recombinant Y4 or 7SL RNAs are fully and/or appropriately folded. However, gel analysis shows that both RNAs are of the expected size and readily stained by the intercalating dye ethidium bromide, which only binds to dsRNA (data not shown).
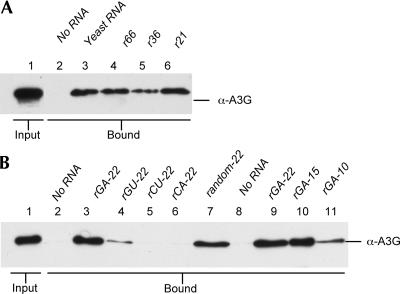
We wished to ask whether A3G:NC complex formation required a specific RNA sequence and whether there was a length requirement for the RNA. As shown in Figure 4A, complex formation was efficiently rescued by recombinant, random ssRNAs of 66 nt, 36 nt, or 21 nt. This result therefore implies that any ssRNA sequence requirement for complex formation must be fairly limited, i.e., NC binding by A3G is unlikely to require a unique RNA sequence present in only one or a few specific RNAs present in the human cell.
FIGURE 4.
Formation of A3G:NC complexes is rescued by short, G-containing ssRNAs. (A) Similar to Figure 3B, except that the added RNAs (10 μg) are derived from the fragmented yeast RNA preparation shown in Figure 3C (lane 3) or represent 66 nt (lane 4), 36 nt (lane 5), or 21 nt (lane 6) long ssRNA transcripts generated in vitro using T7 RNA polymerase. (B) Similar to panel A, except that we are here analyzing short, synthetic ssRNAs (0.5 nM). With the exception of random-22 (lane 7), these were all designed to contain only two out the four possible bases and to lack the ability to form duplex molecules. Rescue of A3G:NC complex formation required G residues in the ssRNA (compare lanes 3,4,7 and lanes 5,6) and was more efficient if the oligonucleotide was over 10 nt in length (compare lanes 9,10 and lane 11). Control Western blots analyzing the input level of GST-NC are shown in Supplemental Figure 1.
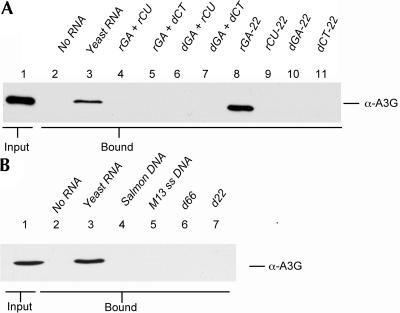
We next asked if 22-nt-long synthetic ssRNAs consisting entirely of G and A (rGA-22), G and U (rGU-22), C and U (rCU-22), or C and A (rCA-22) would rescue A3G:NC complex formation. As shown in Figure 4B, A3G:NC complex formation was efficiently rescued by rGA-22 (Fig. 4B, lane 3) and by a 22-nt-long random ssRNA (Fig. 4B, lane 7), but was not rescued by the similar rCU-22 or rCA-22 (Fig. 4B, lanes 5,6). The rGU-22 oligonucleotide gave a weak, intermediate level of rescue (Fig. 4B, lane 4). While truncation of the rGA-22 oligomer to 15 nt did not inhibit complex formation (Fig. 4B, lane 10), further truncation to 10 nt did significantly reduce complex formation (Fig. 4B, lane 11). Together, these data argue that formation of the A3G:NC complex can be rescued by single-stranded RNAs of ≥10 nt in length that contain G residues.
We also examined whether complex formation could be enhanced by addition of ssDNA, dsDNA, or DNA:RNA hybrid molecules. As shown in Figure 5A, the ability of the synthetic rGA-22 to rescue A3G:NC complex formation (Fig. 5A, lane 8) was blocked when this oligonucleotide was annealed to a fully complementary RNA molecule (Fig. 5A, rCU, lane 6) or DNA molecule (Fig. 5A, dCT, lane 5). Moreover, complex formation was not rescued by ssDNA oligonucleotides of 22 nt in length (Fig. 5A, lanes 10,11) or by a dsDNA of 22 base pairs (Fig. 5A, lane 7). This is of interest, given previous data showing that A3G readily binds ssDNA molecules of ≥16 nt in vitro (Iwatani et al. 2006).
FIGURE 5.
Rescue of A3G:NC complex formation by ssRNA. (A) Similar to Figure 4B, except that in this experiment the A3G and GST-NC proteins were incubated in the presence of ssRNA oligos (lanes 8,9), ssDNA oligos (lanes 10,11), two complementary ssRNA oligos that had been annealed to form a dsRNA (lane 4), ssRNA oligos that had been annealed to a complementary ssDNA oligo (lanes 5,6) or two complementary ssDNA oligos that had been annealed to form a dsDNA (lane 7) (0.5 nM in each case). Only the G-containing ssRNA oligo (lane 8) and the fragmented yeast RNA positive control (lane 3) rescued complex formation. (B) Similar to Figure 4A, except that this experiment analyzed the ability of 10 μg of denatured salmon testes DNA (lane 4), bacteriophage M13 genomic ssDNA (lane 5), or ssDNA oligonucleotides (lanes 6,7) to rescue formation of the A3G:NC complex. Control Western blots analyzing the input level of GST-NC are shown in Supplemental Figure 1.
To confirm that the inability of ssDNA to rescue A3G:NC complex formation is not due to the specific sequence or short length of the 22-nt-long ssDNAs tested in Figure 5A, we also analyzed the effect of denatured salmon testes DNA, bacteriophage M13 genomic ssDNA, or 63-nt and 22-nt random ssDNA oligonucleotides. In none of these cases was complex formation observed when RNase A-treated A3G and recombinant GST-NC were incubated together (Fig. 5B). We conclude that formation of the A3G:NC complex can be rescued by the addition of G-containing ssRNA molecules with a minimum length of ≥10 nt. In contrast, complex formation is not induced by the addition of dsRNA, ssDNA, dsDNA, or DNA:RNA hybrid molecules.
DISCUSSION
Several lines of evidence have previously demonstrated that the human A3G protein binds RNA both in vitro and in vivo. First, recombinant A3G has been previously shown to directly bind ssRNA, as well as ssDNA, but not either dsRNA or dsDNA in vitro (Yu et al. 2004; Navarro et al. 2005; Iwatani et al. 2006). Second, binding of A3G by NC in vitro requires RNA and a ternary complex consisting of A3G, NC, and RNA has been reported (Iwatani et al. 2006). Finally, in dividing cells, A3G has been found to form a “high molecular mass” (HMM) ribonucleoprotein complex that consists of A3G, a wide range of cellular RNAs, and almost 100 different cellular RNA binding proteins (Chiu et al. 2006; Kozak et al. 2006; Wichroski et al. 2006; Gallois-Montbrun et al. 2007). The HMM complex falls apart when treated with RNase A, thus suggesting that almost all of these cellular proteins are bound to A3G indirectly, via bridging RNA molecules.
In this article, we have focused on the nucleic acid requirements for formation of the A3G:NC complex that is thought to mediate packaging of A3G into retroviral particles. Our data argue that short stretches of ssRNA are necessary and sufficient to render A3G competent to bind NC in vitro (Fig. 2). Perhaps surprisingly, given the known affinity of NC for RNA (Damgaard et al. 1998; De Guzman et al. 1998; Iwatani et al. 2006), exhaustive incubation of the recombinant GST-NC fusion protein used in these experiments with RNase A did not affect the ability of GST-NC to bind A3G in vitro (Fig. 2). The hypothesis that it is A3G binding to RNA, rather than NC binding to RNA, which is critical for at least the initial step in A3G:NC complex formation is also supported by our observation that complex formation by RNase A-treated A3G can be rescued by add-back of 15-nt-long, or even 10-nt-long, ssRNA oligonucleotides (Fig. 4B), as it has previously been reported that ssRNAs of <18 nt in length bind HIV-1 NC very inefficiently (Iwatani et al. 2006).
The observation that formation of a complex between RNase A-treated A3G and GST-NC can be rescued by add-back of RNA (Figs. 3, 4) allowed us to examine in some detail which nucleic acid molecules could support this activity. Our results demonstrate that some highly structured RNAs (i.e., tRNA, as well as rRNA) (Fig. 3B) or fully dsRNAs (Fig. 5A) are unable to support A3G:NC complex formation. In contrast, human Y4 RNA and 7SL RNA were able to support NC:A3G complex formation, even though these RNA are also highly structured (Fig. 3D). The observation that very short ssRNAs can also support NC:A3G complex formation (Fig. 4) may suggest that ssRNA bulges and loops present in Y4 and 7SL RNA are of sufficient length to support this interaction, while tRNA and rRNA may be folded too tightly to be active in this assay. Together, these data are consistent with earlier reports demonstrating that A3G interacts with Y RNAs and 7SL RNA in vivo (Chiu et al. 2006; Wang et al. 2007) yet is incapable of binding fully double-stranded RNAs (Iwatani et al. 2006).
Our results demonstrate that short random ssRNAs of ≥21 nt in length (Fig. 4A) are capable of supporting complex formation (Fig. 4A). Further analysis revealed that 22-nt-long ssRNA oligonucleotides consisting entirely of G and A residues rescued complex formation efficiently (Fig. 4B) while a similar oligonucleotide consisting of G and U residues showed partial activity (note that the rGU-22 oligonucleotide was designed to be only poorly self-complementary). In contrast, 22-nt-long rCU-22 and rCA-22 ssRNAs failed to rescue A3G:NC complex formation (Fig. 4B). This was surprising, as one would predict that a protein with cytidine deaminase activity might prefer to bind to ssRNAs containing C residues. However, we note that Iwatani et al. (2006) have previously reported that A3G preferentially binds ssDNA molecules containing dT or dU—not dC—residues. Alternatively, RNA binding by A3G may be entirely sequence nonspecific and the preference for G residues for ternary complex formation may actually be conferred by NC. Consistent with this latter hypothesis, Navarro et al. (2005), using gel-shift analysis, observed that A3G bound ssRNA nonspecifically, while De Guzman et al. (1998) have reported that RNA binding by HIV-1 NC requires G residues in the target RNA. This suggests a model for NC binding by A3G in which A3G first binds to cellular or viral RNAs and formation of the subsequent RNA:A3:NC ternary complex then requires more specific contacts between NC and both A3G and the A3G-bound RNA. This model could explain previous reports suggesting that HIV-1 genomic RNA is particularly effective at facilitating A3G virion packaging (Khan et al. 2005), even though it is not essential (Schafer et al. 2004; Zennou et al. 2004).
Although A3G is known to bind ssDNAs effectively (Iwatani et al. 2006), several ssDNAs failed to support A3G:NC complex formation (Fig. 5). Perhaps surprisingly, we also failed to see any evidence of inhibition of A3G:NC complex formation when RNase-treated A3G and GST-NC were preincubated with ssDNA prior to the addition of ssRNA to the reaction (data not shown). This result suggests either that the A3G can bind both ssRNA and ssDNA simultaneously or that the affinity of A3G for ssRNA is much higher than its affinity for DNA. However, previous research has suggested that the affinity of A3G for ssRNA and ssDNA is comparable (Iwatani et al. 2006), so this negative result remains difficult to understand.
In total, our data argue that formation of the A3G:NC complex requires short stretches of ssRNA that contain G residues. On this basis, it appears likely that a wide range of cellular RNAs, including mRNAs, but apparently not tRNAs or rRNAs (Fig. 3B), could support A3G virion packaging. As noted above, our data also suggest that human Y RNAs and 7SL RNA, while clearly structured, nevertheless contain short stretches of ssRNA that are sufficient to support NC-A3G complex formation (Fig. 3D). However, these observations do not appear consistent with the hypothesis that 7SL is uniquely capable of supporting Gag binding by A3G (Wang et al. 2007), and previous work (Chiu et al. 2006; Kozak et al. 2006; Gallois-Montbrun et al. 2007) certainly suggests that A3G binds to a very wide range of cellular RNAs in vivo.
The observation that RNA binding is required to render A3G capable of binding to NC (Fig. 2) is also interesting in light of recent data suggesting that HIV-1 preferentially packages newly synthesized A3G, rather than the RNA-bound A3G found in the HMM complex (Soros et al. 2007). While our data do not directly bear on this question, they do argue that this newly synthesized A3G must also bind RNA in order to become competent for virion incorporation.
MATERIALS AND METHODS
Plasmid construction
Plasmids able to express the HA-tagged version of human A3A, wild-type human A3G, as well as the A3G-E1 and A3G-E1+2 mutants, have been previously described (Bogerd et al. 2006, 2007). A bacterial plasmid (pGEX4T-NC) expressing GST fused to the p2:NC:p1 region of HIV-1 Gag was prepared by PCR amplification of the relevant region of HIV-1 Gag, followed by insertion into the BamHI and XhoI sites present in pGEX4T (Pharmacia). A plasmid (pcDNA3.1 human APOBEC3G-Myc-6xHis) that expresses full-length human APOBEC3G, linked to a carboxy-terminal Myc tag and six histidine residues, in mammalian cells has been previously described (Khan et al. 2005). Analogous vectors expressing humanized green fluorescent protein (GFP), firefly luciferase (Luc), or human APOBEC3A (A3A) were prepared by the in-frame substitution of PCR-generated open reading frames (ORFs) encoding these three proteins in place of the A3G ORF.
Protein purification
Escherichia coli strain BL21 was transformed with pGEX4T or pGEX4T-NC and protein expression induced by incubation in the presence of 1 mM IPTG (Invitrogen) for 4 h. Cells were collected and lysed by incubation in PBS supplemented with 1% NP40, 100 mg/mL of DNase I (bovine pancreatic DNase I, Roche), Complete EDTA-free protease inhibitors (Roche), and 5 mg/mL lysozyme (Sigma). The lysates were then sonicated for 1 min and clarified by centrifugation (10 min at 13,000 rpm). The clarified supernatant was then bound to washed glutathione-agarose beads (GE Healthcare) for 30 min at 4°C, and the beads washed extensively with PBS/1% NP40. The washed beads were then left untreated or were incubated with 34 μg/mL of RNase A (Sigma) in buffer A (150 mM NaCl, 10 mM TRIS at pH 7.5, 0.5% NP40) for 1 h at room temperature before being washed extensively with PBS/1% NP40. Bound proteins were then eluted using 10 mM glutathione (Sigma) in 0.5 M NaCl, 50 mM TRIS (pH 8.0). Eluted proteins were concentrated and dialyzed against PBS by centrifugation through Centricon-10 columns (Amicon).
For preparation of recombinant A3G, 2 × 106 293T cells were transfected with 10 μg of the A3G-Myc-His expression vector, using the calcium phosphate procedure. At 44 h after transfection, cells were lysed in His lysis buffer (500 mM NaCl, 20 mM TRIS at pH 7.5, 20 mM imidazole, 100 μg/mL pancreatic DNase I, and 0.5% NP40, supplemented with protease inhibitors). The resultant lysates were sonicated for 1 min and centrifuged for 10 min at 13,000 rpm to remove insoluble material. The supernatants were then bound to Ni-NTA agarose (Qiagen) for 30 min at 4°C and washed extensively with His lysis buffer. Bound samples were left untreated or were incubated with 34 μg/mL of RNase A, as described above, for 1 h at room temperature and were then again extensively washed with His lysis buffer. Bound proteins were then eluted using lysis buffer supplemented to 500 mM imidazole.
Binding assays
Binding assays were performed using either cell lysates containing A3G-HA or A3G-Myc-His, or using partially purified A3G-Myc-His protein. For lysate preparation, 2 × 106 293T cells were transfected with 10 μg of pK/A3G-HA, pK/A3G-E1-HA, pK/A3G-E1+2-HA, pK/A3A-HA, pA3G-Myc-His, pA3A-Myc-His, pLuc-Myc-His, or pGFP-Myc-His using the calcium phosphate procedure. At 44 h after transfection, cells were lysed in 2 mL of buffer A supplemented with protease inhibitors (Roche). After clarification by centrifugation, as described above, the supernatants were used directly in binding assays. In this case, 50 μL of glutathione-agarose beads, 1 mL of the supernatant, and ∼250 ng of purified GST or GST-NC were incubated together at 4°C for 60 min. The agarose beads were washed four times with buffer A and bound proteins eluted using 100 μL of an SDS-acrylamide gel loading buffer containing β-mercaptoethanol. Bound (25% of total) and input (5% of total) proteins were then analyzed by Western blot using a rabbit anti-Myc polyclonal antiserum (Abcam), a mouse monoclonal anti-HA antibody (Covance), or a rabbit anti-GST polyclonal antibody (Santa Cruz), followed by visualization using enhanced chemiluminescence.
When binding assays were performed using the purified proteins described above, ∼250 ng of GST or GST-NC (± RNase A treatment) and ∼125 ng of A3G-Myc-His (± RNase A treatment) were incubated with 50 μL of glutathione-agarose beads for 20 min at room temperature in 1 mL of buffer B (350 mM NaCl, 0.5% NP40, 20 mM TRIS at pH 7.5, 200 μg/mL Bovine Serum Albumin [NEB], 40 units of RNase Out [Invitrogen]). RNA samples were added at the beginning of the incubation period, as indicated in the figures. After incubation, the glutathione-agarose beads were washed four times with buffer B and bound proteins eluted using 100 μl of SDS-acrylamide gel loading buffer containing β-mercaptoethanol. Bound (25% of total) and input (5% of total) proteins were then analyzed by Western blot using a rabbit anti-A3G antiserum (Marin et al. 2003) or a rabbit anti-GST polyclonal antibody.
Nucleic acid samples
Yeast RNA and yeast tRNA were purchased from Ambion. Human total RNA was prepared from Jurkat T-cells using Tri Reagent (Sigma). Human poly(A)+ RNA was purified using oligo dT cellulose using standard techniques. Human Y4 and 7SL RNAs were transcribed in vitro using a T7 polymerase Megascript kit and PCR-generated templates that incorporated the T7 promoter into the sense primer.
RNAs of 66 nt (r66), 36 nt (r36), or 21 nt (r21) were generated using a T7 polymerase MegaScript kit (Ambion) and linearized pGem5Z(f)+ or pGem3Z(f)+ plasmids.
The sequences of the in vitro transcribed RNAs are as follows.
For r66 transcribed from pGEM5Zf(+), linearized with Not I:
5′-GGGCGAAUUGGGCCCGGACGUCGCAUGCUCCCGGCCGCCAUGGCCGCGGAUAUCACUAGUGCGGCC-3′.
For r36 transcribed from pGEM3Zf(+), linearized with Xba I:
5′-GGGCGAAUUCGAGCUCGGUACCCGGGGAUCCUCUAG-3′.
For r21 transcribed from pGEM3Zf(+), linearized with Asp 718:
5′-GGGCGAAUUCGAGCUCGGUAC-3′.
All synthetic RNAs of 22 nt, 15 nt, or 10 nt in length were purchased from IDT:
rGA-22: 5′-GGAAGGGAAAAGGGGAAAGAGG-3′;
rGA-15: 5′-GGAAGGGAAAAGGGG-3′;
rGA-10: 5′-GGAAGGGAAA-3′;
rGU-22: 5′-GGUUGGGUUUUGGGGUUUGUGG-3′;
rCU-22: 5′-CCUCUUUCCCCUUUUCCCUUCC-3′;
rCA-22: 5′-CCACAAACCCCAAAACCCAACC-3′; and
Random-22: 5′-GGGCGAAUUGGGCCCGACGUCG-3′.
RNA duplexes were prepared by resuspending the RNA and/or DNA oligonucleotides in annealing buffer (10 mM Tris at pH 7.6, 50 mM NaCl) and heating the samples to 95°C, followed by slow cooling to room temperature.
Denatured salmon testes DNA (Sigma), m13 single-stranded DNA (NEB), and synthetic DNA oligos (Operon) were purchased:
d66: 5′-GGGCGAATTGGGCCCGGACGTCGCATGCTCCCGGCCGCCATGGCCGCGGATATCACTAGTGCGGCC-3′;
dCT22: 5′-CCTCTTTCCCCTTTTCCCTTCC-3′;
dGA22: 5′-GGAAGGGAAAAGGGGAAAGAGG-3′; and
d22: 5′-GGGCGAATTGGGCCCGACGTCG-3′.
SUPPLEMENTAL DATA
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Dr. Alexandra Schäfer for assistance in the early stages of this research. The following reagents were obtained through the NIH AIDS research and Reference Reagent Program, Division of AIDS, NIAID, NIH: pcDNA3.1 human APOBEC3G-Myc-6xHis from Dr. David Kabat and anti-ApoC17 from Dr. Klaus Strebel. This research was supported by grant AI057099 from the National Institutes of Allergy and Infectious Diseases.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.964708.
REFERENCES
- Aguiar R.S., Lovsin N., Tanuri A., Peterlin B.M. Vpr.A3A chimera inhibits HIV replication. J. Biol. Chem. 2008;283:2518–2525. doi: 10.1074/jbc.M706436200. [DOI] [PubMed] [Google Scholar]
- Alce T.M., Popik W. APOBEC3G is incorporated into virus-like particles by a direct interaction with HIV-1 Gag nucleocapsid protein. J. Biol. Chem. 2004;279:34083–34086. doi: 10.1074/jbc.C400235200. [DOI] [PubMed] [Google Scholar]
- Bogerd H.P., Wiegand H.L., Doehle B.P., Lueders K.K., Cullen B.R. APOBEC3A and APOBEC3B are potent inhibitors of LTR-retrotransposon function in human cells. Nucleic Acids Res. 2006;34:89–95. doi: 10.1093/nar/gkj416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd H.P., Wiegand H.L., Doehle B.P., Cullen B.R. The intrinsic antiretroviral factor APOBEC3B contains two enzymatically active cytidine deaminase domains. Virology. 2007;364:486–493. doi: 10.1016/j.virol.2007.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett A., Spearman P. APOBEC3G multimers are recruited to the plasma membrane for packaging into human immunodeficiency virus type 1 virus-like particles in an RNA-dependent process requiring the NC basic linker. J. Virol. 2007;81:5000–5013. doi: 10.1128/JVI.02237-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cen S., Guo F., Niu M., Saadatmand J., Deflassieux J., Kleiman L. The interaction between HIV-1 Gag and APOBEC3G. J. Biol. Chem. 2004;279:33177–33184. doi: 10.1074/jbc.M402062200. [DOI] [PubMed] [Google Scholar]
- Chen H., Lilley C.E., Yu Q., Lee D.V., Chou J., Narvaiza I., Landau N.R., Weitzman M.D. APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Curr. Biol. 2006;16:480–485. doi: 10.1016/j.cub.2006.01.031. [DOI] [PubMed] [Google Scholar]
- Chiu Y.L., Witkowska H.E., Hall S.C., Santiago M., Soros V.B., Esnault C., Heidmann T., Greene W.C. High-molecular-mass APOBEC3G complexes restrict Alu retrotransposition. Proc. Natl. Acad. Sci. 2006;103:15588–15593. doi: 10.1073/pnas.0604524103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen B.R. Role and mechanism of action of the APOBEC3 family of antiretroviral resistance factors. J. Virol. 2006;80:1067–1076. doi: 10.1128/JVI.80.3.1067-1076.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damgaard C.K., Dyhr-Mikkelsen H., Kjems J. Mapping the RNA binding sites for human immunodeficiency virus type-1 gag and NC proteins within the complete HIV-1 and -2 untranslated leader regions. Nucleic Acids Res. 1998;26:3667–3676. doi: 10.1093/nar/26.16.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Guzman R.N., Wu Z.R., Stalling C.C., Pappalardo L., Borer P.N., Summers M.F. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science. 1998;279:384–388. doi: 10.1126/science.279.5349.384. [DOI] [PubMed] [Google Scholar]
- Derse D., Hill S.A., Princler G., Lloyd P., Heidecker G. Resistance of human T cell leukemia virus type 1 to APOBEC3G restriction is mediated by elements in nucleocapsid. Proc. Natl. Acad. Sci. 2007;104:2915–2920. doi: 10.1073/pnas.0609444104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doehle B.P., Schafer A., Wiegand H.L., Bogerd H.P., Cullen B.R. Differential sensitivity of murine leukemia virus to APOBEC3-mediated inhibition is governed by virion exclusion. J. Virol. 2005;79:8201–8207. doi: 10.1128/JVI.79.13.8201-8207.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doehle B.P., Bogerd H.P., Wiegand H.L., Jouvenet N., Bieniasz P.D., Hunter E., Cullen B.R. The βretrovirus Mason–Pfizer monkey virus selectively excludes simian APOBEC3G from virion particles. J. Virol. 2006;80:12102–12108. doi: 10.1128/JVI.01600-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutko J.A., Schafer A., Kenny A.E., Cullen B.R., Curcio M.J. Inhibition of a yeast LTR retrotransposon by human APOBEC3 cytidine deaminases. Curr. Biol. 2005;15:661–666. doi: 10.1016/j.cub.2005.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnault C., Heidmann O., Delebecque F., Dewannieux M., Ribet D., Hance A.J., Heidmann T., Schwartz O. APOBEC3G cytidine deaminase inhibits retrotransposition of endogenous retroviruses. Nature. 2005;433:430–433. doi: 10.1038/nature03238. [DOI] [PubMed] [Google Scholar]
- Gallois-Montbrun S., Kramer B., Swanson C.M., Byers H., Lynham S., Ward M., Malim M.H. Antiviral protein APOBEC3G localizes to ribonucleoprotein complexes found in P bodies and stress granules. J. Virol. 2007;81:2165–2178. doi: 10.1128/JVI.02287-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goila-Gaur R., Khan M.A., Miyagi E., Kao S., Strebel K. Targeting APOBEC3A to the viral nucleoprotein complex confers antiviral activity. Retrovirology. 2007;4:61. doi: 10.1186/1742-4690-4-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hache G., Liddament M.T., Harris R.S. The retroviral hypermutation specificity of APOBEC3F and APOBEC3G is governed by the C-terminal DNA cytosine deaminase domain. J. Biol. Chem. 2005;280:10920–10924. doi: 10.1074/jbc.M500382200. [DOI] [PubMed] [Google Scholar]
- Harris R.S., Bishop K.N., Sheehy A.M., Craig H.M., Petersen-Mahrt S.K., Watt I.N., Neuberger M.S., Malim M.H. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113:803–809. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- Huthoff H., Malim M.H. Identification of amino acid residues in APOBEC3G required for regulation by human immunodeficiency virus type 1 Vif and Virion encapsidation. J. Virol. 2007;81:3807–3815. doi: 10.1128/JVI.02795-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwatani Y., Takeuchi H., Strebel K., Levin J.G. Biochemical activities of highly purified, catalytically active human APOBEC3G: Correlation with antiviral effect. J. Virol. 2006;80:5992–6002. doi: 10.1128/JVI.02680-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao S., Khan M.A., Miyagi E., Plishka R., Buckler-White A., Strebel K. The human immunodeficiency virus type 1 Vif protein reduces intracellular expression and inhibits packaging of APOBEC3G (CEM15), a cellular inhibitor of virus infectivity. J. Virol. 2003;77:11398–11407. doi: 10.1128/JVI.77.21.11398-11407.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M.A., Kao S., Miyagi E., Takeuchi H., Goila-Gaur R., Opi S., Gipson C.L., Parslow T.G., Ly H., Strebel K. Viral RNA is required for the association of APOBEC3G with human immunodeficiency virus type 1 nucleoprotein complexes. J. Virol. 2005;79:5870–5874. doi: 10.1128/JVI.79.9.5870-5874.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M., Takaori-Kondo A., Shindo K., Abudu A., Fukunaga K., Uchiyama T. APOBEC3G targets specific virus species. J. Virol. 2004;78:8238–8244. doi: 10.1128/JVI.78.15.8238-8244.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak S.L., Marin M., Rose K.M., Bystrom C., Kabat D. The anti-HIV-1 editing enzyme APOBEC3G binds HIV-1 RNA and messenger RNAs that shuttle between polysomes and stress granules. J. Biol. Chem. 2006;281:29105–29119. doi: 10.1074/jbc.M601901200. [DOI] [PubMed] [Google Scholar]
- Lochelt M., Romen F., Bastone P., Muckenfuss H., Kirchner N., Kim Y.B., Truyen U., Rösler U., Battenberg M., Saib A., et al. The antiretroviral activity of APOBEC3 is inhibited by the foamy virus accessory Bet protein. Proc. Natl. Acad. Sci. 2005;102:7982–7987. doi: 10.1073/pnas.0501445102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo K., Liu B., Xiao Z., Yu Y., Yu X., Gorelick R., Yu X.F. Amino-terminal region of the human immunodeficiency virus type 1 nucleocapsid is required for human APOBEC3G packaging. J. Virol. 2004;78:11841–11852. doi: 10.1128/JVI.78.21.11841-11852.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangeat B., Turelli P., Caron G., Friedli M., Perrin L., Trono D. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- Marin M., Rose K.M., Kozak S.L., Kabat D. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 2003;9:1398–1403. doi: 10.1038/nm946. [DOI] [PubMed] [Google Scholar]
- Miyagi E., Opi S., Takeuchi H., Khan M., Goila-Gaur R., Kao S., Strebel K. Enzymatically active APOBEC3G is required for efficient inhibition of human immunodeficiency virus type 1. J. Virol. 2007;81:13346–13353. doi: 10.1128/JVI.01361-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro F., Bollman B., Chen H., Konig R., Yu Q., Chiles K., Landau N.R. Complementary function of the two catalytic domains of APOBEC3G. Virology. 2005;333:374–386. doi: 10.1016/j.virol.2005.01.011. [DOI] [PubMed] [Google Scholar]
- Newman E.N., Holmes R.K., Craig H.M., Klein K.C., Lingappa J.R., Malim M.H., Sheehy A.M. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr. Biol. 2005;15:166–170. doi: 10.1016/j.cub.2004.12.068. [DOI] [PubMed] [Google Scholar]
- Opi S., Kao S., Goila-Gaur R., Khan M.A., Miyagi E., Takeuchi H., Strebel K. Human immunodeficiency virus type 1 Vif inhibits packaging and antiviral activity of a degradation-resistant APOBEC3G variant. J. Virol. 2007;81:8236–8246. doi: 10.1128/JVI.02694-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell R.A., Wiegand H.L., Moore M.D., Schafer A., McClure M.O., Cullen B.R. Foamy virus Bet proteins function as novel inhibitors of the APOBEC3 family of innate antiretroviral defense factors. J. Virol. 2005;79:8724–8731. doi: 10.1128/JVI.79.14.8724-8731.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer A., Bogerd H.P., Cullen B.R. Specific packaging of APOBEC3G into HIV-1 virions is mediated by the nucleocapsid domain of the gag polyprotein precursor. Virology. 2004;328:163–168. doi: 10.1016/j.virol.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Schumacher A.J., Nissley D.V., Harris R.S. APOBEC3G hypermutates genomic DNA and inhibits Ty1 retrotransposition in yeast. Proc. Natl. Acad. Sci. 2005;102:9854–9859. doi: 10.1073/pnas.0501694102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher A.J., Hache G., Macduff D.A., Brown W.L., Harris R.S. The DNA deaminase activity of human APOBEC3G is required for Ty1, MusD and HIV-1 restriction. J. Virol. 2008;82:2652–2660. doi: 10.1128/JVI.02391-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehy A.M., Gaddis N.C., Choi J.D., Malim M.H. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- Shindo K., Takaori-Kondo A., Kobayashi M., Abudu A., Fukunaga K., Uchiyama T. The enzymatic activity of CEM15/Apobec-3G is essential for the regulation of the infectivity of HIV-1 virion but not a sole determinant of its antiviral activity. J. Biol. Chem. 2003;278:44412–44416. doi: 10.1074/jbc.C300376200. [DOI] [PubMed] [Google Scholar]
- Soros V.B., Yonemoto W., Greene W.C. Newly synthesized APOBEC3G is incorporated into HIV virions, inhibited by HIV RNA, and subsequently activated by RNase H. PLoS Pathog. 2007;3:e15. doi: 10.1371/journal.ppat.0030015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svarovskaia E.S., Xu H., Mbisa J.L., Barr R., Gorelick R.J., Ono A., Freed E.O., Hu W.S., Pathak V.K. Human apolipoprotein B mRNA-editing enzyme-catalytic polypeptide-like 3G (APOBEC3G) is incorporated into HIV-1 virions through interactions with viral and nonviral RNAs. J. Biol. Chem. 2004;279:35822–35828. doi: 10.1074/jbc.M405761200. [DOI] [PubMed] [Google Scholar]
- Wang T., Tian C., Zhang W., Luo K., Sarkis P.T., Yu L., Liu B., Yu Y., Yu X.F. 7SL RNA mediates virion packaging of the antiviral cytidine deaminase APOBEC3G. J. Virol. 2007;81:13112–13124. doi: 10.1128/JVI.00892-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichroski M.J., Robb G.B., Rana T.M. Human retroviral host restriction factors APOBEC3G and APOBEC3F localize to mRNA processing bodies. PLoS Pathog. 2006;2:e41. doi: 10.1371/journal.ppat.0020041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegand H.L., Cullen B.R. Inhibition of αretrovirus replication by a range of human APOBEC3 proteins. J. Virol. 2007;81:13694–13699. doi: 10.1128/JVI.01646-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegand H.L., Doehle B.P., Bogerd H.P., Cullen B.R. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J. 2004;23:2451–2458. doi: 10.1038/sj.emboj.7600246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q., Konig R., Pillai S., Chiles K., Kearney M., Palmer S., Richman D., Coffin J.M., Landau N.R. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat. Struct. Mol. Biol. 2004;11:435–442. doi: 10.1038/nsmb758. [DOI] [PubMed] [Google Scholar]
- Zennou V., Perez-Caballero D., Gottlinger H., Bieniasz P.D. APOBEC3G incorporation into human immunodeficiency virus type 1 particles. J. Virol. 2004;78:12058–12061. doi: 10.1128/JVI.78.21.12058-12061.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Yang B., Pomerantz R.J., Zhang C., Arunachalam S.C., Gao L. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature. 2003;424:94–98. doi: 10.1038/nature01707. [DOI] [PMC free article] [PubMed] [Google Scholar]