Abstract
The internal loop at the base of domain 3 (D3) is one of the most conserved and catalytically important elements of a group II intron. However, the location and molecular nature of its tertiary interaction partners has remained unknown. By employing a combination of site-directed photo-cross-linking and nucleotide analog interference suppression (NAIS), we show that the domain 3 internal loop (D3IL) interacts with the ε-ε′ duplex, which is an active-site element located near the 5′-splice site in D1. Our data also suggest that the D3IL may interact with the bulge of D5, which is a critical active site component. The results of this and other recent studies indicate that the D3IL participates in a complex network of tertiary interactions involving ε-ε′, the bulge of D5 and J23, and that it helps to optimize active site architecture by supporting interactions among these catalytic motifs. Our results are consistent with the role of D3 as a catalytic effector that enhances intron reactivity through active site stabilization.
Keywords: ribozyme, splicing, catalysis, tertiary structure
INTRODUCTION
Group II introns are large autocatalytic RNAs found in organellar genomes of plants, various microeukaryotic organisms, and some bacterial genomes (Michel and Ferat 1995; Lehmann and Schmidt 2003; Pyle and Lambowitz 2006). The group II intron splicing mechanism is similar to that of the spliceosomal machinery that processes nuclear introns, suggesting that they may have evolved from a common evolutionary ancestor (Pyle and Lambowitz 2006). Some group II introns are mobile genomic elements, which use a reverse-splicing reaction to insert themselves into RNA or double-stranded DNA with high specificity (Lambowitz and Zimmerly 2004; Pyle and Lambowitz 2006). The ability of group II introns to target and migrate into new genes has lead to their development as agents for genetic manipulation in biotechnology and molecular therapy (Lambowitz and Zimmerly 2004).
Based on phylogenetic analysis, group II introns have been divided into several families, designated A1, B1, B2, C, and D, which vary in certain secondary and tertiary structural elements (Toor et al. 2001). All families of group II introns share a conserved secondary structure consisting of six domains (Michel and Ferat 1995; Lehmann and Schmidt 2003; Pyle and Lambowitz 2006). Domains 1 and 5 (D1 and D5) are absolutely indispensable for catalytic function (Qin and Pyle 1998; Lehmann and Schmidt 2003; Pyle and Lambowitz 2006), although removal of other domains leaves the intron with only traces of activity (Koch et al. 1992). Every intronic domain has a specific functional role, ranging from structural stabilization to the contribution of critical active site elements and the binding of cofactor proteins (Qin and Pyle 1998; Lehmann and Schmidt 2003; Pyle and Lambowitz 2006).
Here, we focus on domain 3 (D3), which contains many of the most phylogenetically conserved nucleotides of the intron. Although D3 is not absolutely required for group II catalysis, multiple studies have demonstrated that D3 greatly increases the chemical rate constant for any group II intron ribozyme construct (Xiang et al. 1998; Su et al. 2001; Fedorova et al. 2003). Despite its clear importance in the function and evolution of the intron, information about D3 and its molecular contacts with other intronic domains is very limited. It has been shown that D3 binds the rest of the intron with Kd that is similar to that for the binding of catalytic D5 (Podar et al. 1995). While this indicates that D3 forms a network of strong interactions, phylogenetic analysis is inconsistent with the formation of simple Watson–Crick base-pairings between D3, suggesting that D3 participates in complicated tertiary interaction motifs.
Specific regions of D3 have been implicated in the formation of long-range interactions, including the internal loop and the pentaloop nucleotides (Fig. 1), which have been shown to strongly influence catalytic efficiency (Jestin et al. 1997; Boudvillain and Pyle 1998; Fedorova and Pyle 2005). Clusters of atoms and functional groups in these regions were identified using nucleotide analog interference mapping (NAIM) (Fedorova and Pyle 2005). Modification interference studies using DEPC suggested a tertiary contact between D3 and the bulge of D5 (Jestin et al. 1997), but a specific element in D3 was not implicated by the data. We recently identified the μ-μ′ interaction, which is the first tertiary contact to be identified between D3 and another portion of the intron (Fig. 1). It involves two adenosine residues in the pentaloop of D3 and the 2′-hydroxyl group of G844 in D5 (Fedorova and Pyle 2005), which also participates in the stabilizing κ-κ′ interaction (Fig. 1). Nonetheless, the pentaloop is not a universally conserved feature of D3, and it is therefore likely that the μ-μ′ interaction is present only in a subset of group II introns, which include group IIB1, group IIB2, and bacterial class D (Toor et al. 2001). Interaction partners for remaining substructures in D3, particularly those that are universally conserved, are still unknown.
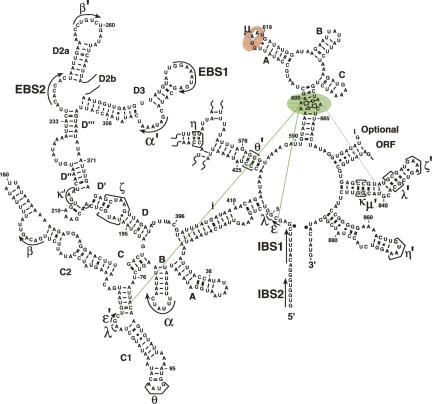
FIGURE 1.
A secondary structure schematic of the ai5γ group II intron. Catalytically important regions of domain 3 (the internal loop (D3IL) and the pentaloop are highlighted in green and red, respectively. The interaction between the D3IL and ε-ε′ is denoted by solid green lines; the possible interaction between the D3IL and the bulge of D5 is denoted by a dotted purple line.
One of the most conserved group II intron motifs is an A-rich internal loop near the base of D3 (the D3IL) (Fig. 1; Michel and Ferat 1995; Pyle and Lambowitz 2006). This substructure is found in all classes of group II introns (Toor et al. 2001). NAIM studies have suggested that the loop structure includes tandem trans-Hoogsteen-sugar edge base pairs (Fedorova and Pyle 2005), which are also phylogenetically conserved throughout group II intron families except for bacterial class B, in which the formation of this structure is possible but not obvious (Toor et al. 2001). It has been previously suggested that the D3IL forms a loop E-like motif in group IIA introns (Leontis and Westhof 1998b); however, it is not known whether this is true for other group II intron classes. Previous hydroxyl radical footprinting studies on IIB introns have shown that the D3IL is as highly internalized as D5 (Swisher et al. 2001), suggesting that it is located within the catalytic core. In this study, we use a combination of NAIS and site-directed photo-cross-linking to identify molecular interaction partners between the D3IL and other regions of the intron. The resulting data reveal the architectural location of D3, and they shed new light on the catalytic role of this important intronic domain.
RESULTS
Mutant design for NAIS experiments
In order to find the interaction partner for the D3IL, we employed a NAIS approach that utilizes the previously described trans-branching reaction for the selection step (Fig. 2A; Chin and Pyle 1995; Boudvillain and Pyle 1998). We prepared exD123 molecules containing mutations within the internal loop (Fig. 2B,C) and then screened for suppression of interference using various nucleotide analogs incorporated into either D1 or D5. Choosing appropriate mutations for the internal loop was challenging because we wished to retain its local architecture while disrupting long-range tertiary interactions.
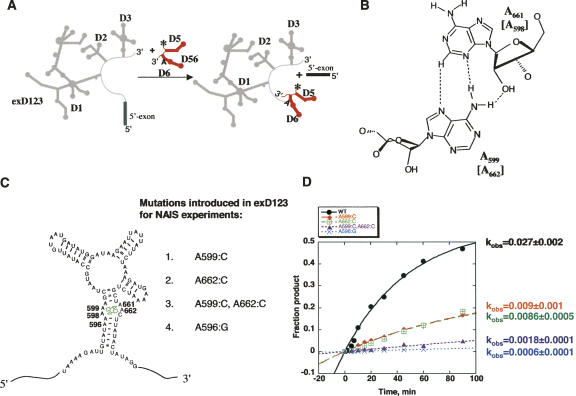
FIGURE 2.
(A) A trans-branching reaction used as a selection step in NAIS experiments. (B) Adenosine residues of the D3 bulge form tandem sheared trans-Hoogsteen-sugar edge base pairs. Mutations in these base-pairs (C) significantly reduce catalytic activity (D).
Previous work has demonstrated that trans-Hoogsteen-sugar edge pairs such as the ones in the D3IL can tolerate certain forms of base substitution (Lescoute et al. 2005). For example, the geometric configuration of tandem trans-Hoogsteen-sugar edge A·A pairs is likely to be retained if these are substituted with C·A pairs (Leontis and Westhof 1998a; Lescoute et al. 2005). We therefore incorporated A:C mutations at positions 599 or 662, thereby converting the sheared A·A pairs to C·A pairs (Fig. 2C). We also created a double mutant A599:C, A662:C, in which both A·A pairs were changed to C·A pairs (Fig. 2C). In addition, since we previously observed a strong 2,6-diaminopurine interference at residue A596, near the internal loop (Fig. 2C; Fedorova and Pyle 2005), we created an A596:G mutant.
In order to determine whether the resulting exD123 mutants were suitable for NAIS experiments, we tested their activity in the trans-branching reaction. This reaction is kinetically well characterized (Chin and Pyle 1995), and it has been successfully used as a selection step in many previous NAIM studies (Boudvillain and Pyle 1998; Boudvillain et al. 2000; Fedorova and Pyle 2005). All mutants exhibited reduced reaction rate compared with the wild-type RNAs (Fig. 2D). Mutation of individual sheared A·A pairs to C·A resulted in threefold losses of activity, whereas the double mutant was 15 times less active than the wild type. These levels of reactivity make the mutants particularly applicable for NAIS experiments, because despite the overall reductions in their activity, it is still possible to generate sufficient amounts of the branched product for iodine sequencing.
NAIS suggests a functional contact between the D3 and D1
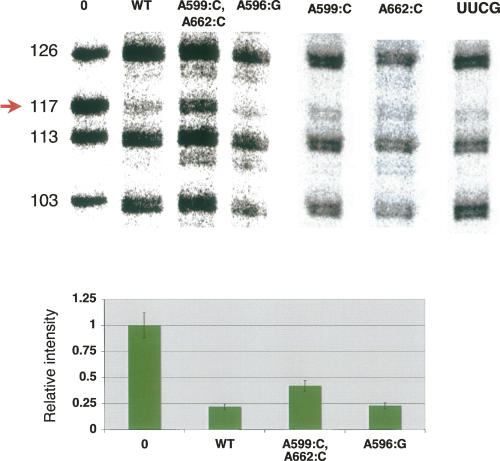
In order to determine whether the internal loop of D3 interacts with D1, we transcribed the wild-type and mutant exD123 RNAs in the presence of various nucleotide analogs and reacted these pools with wild-type D56 RNA. NAIS effects were not observed with mutants A596:G, A599:C, and A662:C. However, when the double mutant (A599:C, A662:C) was transcribed in the presence of a dCαS analog, partial suppression of deoxynucleotide interference was observed at C117 in D1 (Fig. 3). The full suppression of interference could not be observed because of phosphorothioate interference that also occurs at this position (Boudvillain and Pyle 1998). This result suggests that a functional group within the D3IL interacts with the 2′-OH of C117 in D1. The mutation of another catalytically important region of D3, the pentaloop (GUAAU to UUCG), did not suppress deoxynucleotide interference at C117 (Fig. 3), indicating that this suppression is specific to the internal loop of D3. These results are significant because nucleotide C117 is part of the essential ε-ε′ substructure, which is composed of tandem Watson–Crick G-C base pairs.
FIGURE 3.
Double mutation in the tandem sheared base-pair motif in the bulge of D3 causes partial suppression of dCαS interference at C117 in D1. (Top) The autoradiograph of a representative sequencing gel after iodine cleavage of unreacted exD123 RNAs and branched products. Selection step (branching reaction) was carried out in the presence of wild-type 5′-end labeled D56 RNA and unlabeled wild-type and mutant exD123 RNAs transcribed in the presence of 2′-deoxycytidine phosphorothioate. Numbers on the left indicate cytidine positions in exD123. (Bottom) A diagram comparing relative intensity of bands corresponding to C117 in precursor (0) and branched products. The results represent average values obtained from at least three independent experiments, the error did not exceed 15%.
Site-directed cross-linking confirms proximity between the D3IL and ε-ε′
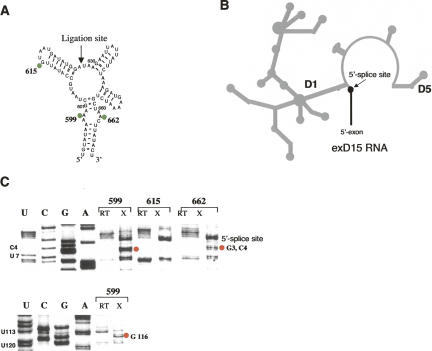
In an effort to obtain independent information on the proximity between residues in D3 and D1, we implemented site-specific photo-cross-linking experiments on a ribozyme construct in which D3 is added as a separate molecule. Previous work has established that catalytic activity of intronic constructs lacking D3 can be restored when D3 is provided in-trans (Podar et al. 1995; Fedorova et al. 2003). For cross-linking studies, we used construct exD15, which contains the 5′-exon, D1, and D5 (Fig. 4B; Fedorova et al. 2003). In this construct, D3 is cleanly deleted, all interdomain joiners are preserved, and short hairpins replace the other intron domains (Fig. 4B; Fedorova et al. 2003). We have previously shown that this construct is catalytically active and undergoes the first step of splicing when separate D3 molecules are added in trans. This reaction exhibits single exponential kinetic behavior, suggesting structural homogeneity of the system (Fedorova et al. 2003). For cross-linking experiments, exD15 RNA was added to D3 RNA, which was chemically synthesized in two halves that were subsequently joined using enzymatic ligation. This strategy enabled us to incorporate single 6-thio-dG moieties at various positions throughout the D3 molecule (Fig. 4A). When 6-thio-dG was incorporated within the D3IL (where it maintains proper conformation of the sheared pairing), specific cross-links were mapped to D1 nucleotides that comprise the ε-ε′ element (nucleotides G3, C4, and G116) (Fig. 4C). These data provide strong evidence that the D3IL is spatially proximal to the ε-ε′ motif, and they are in a good agreement with the NAIS results indicating that the D3IL participates in functional interactions with the backbone of C117 (Fig. 3). Importantly, the D3 construct containing 6-thio-dG at pentaloop nucleotide G615 did not cross-link to ε-ε′ (Fig. 4C), confirming that our results are specific to the D3IL.
FIGURE 4.
UV-cross-linking experiments suggest close proximity of the D3 bulge to ε-ε′ contact in D1. (A) D3 molecules containing a single 6-thio-dG substitution at A 615, A 599, or A 662 (highlighted by green circles) were obtained by enzymatic ligation of two chemically synthesized oligonucleotides at the site indicated by an arrow. (B) A schematic of the exD15 construct used in these studies. (C) Autoradiographs of representative sequencing gels showing specific cross-links of bulge nucleotides 599 and 662 to the residues forming the ε-ε′ contact in D1 (red circles). RT indicates lanes corresponding to the RT analysis of uncross-linked exD15 RNA (control for natural RT stops), X-RT analysis of the exD15 RNA after UV-cross-linking to D3 molecules containing single 6-thio-dG substitutions. Positions of 6-thio-dG substitutions are shown on top. Data represent the results of at least five independent experiments. A figure showing a primary cross-linking gel with indicated cross-linking efficiencies and a larger portion of the sequencing gel is available as supplementary material upon request (please send an e-mail message to anna.pyle@yale.edu).
NAIS experiments suggest a functional connection between D3 and D5
To this point, our studies have indicated that the internal loop of D3 interacts with the ε-ε′ motif in D1. This is particularly significant because ε-ε′ is adjacent to the λ-λ′ interaction that connects D1 and D5. Furthermore, intron nucleotides C4 (a component of ε) and G5 (a component of λ) are known to cross-link specifically to the catalytically essential bulge region of D5 (de Lencastre et al. 2005). These data raise the possibility that the internal loop of D3 also interacts with D5. To screen for D5–D3 tertiary interactions, we carried out NAIS experiments in which phosphorothioate-modified D56 RNAs were reacted with exD123 constructs that contain mutations within the D3IL (Fig. 2C).
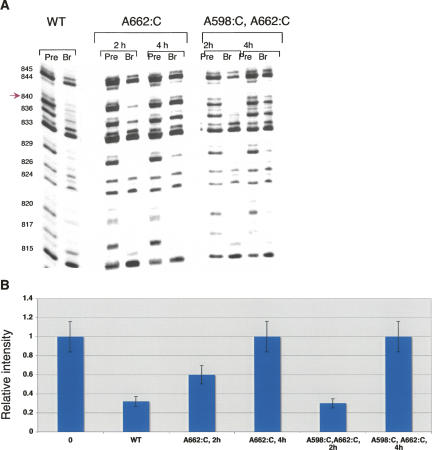
When the trans-branching reaction was conducted for long times (4 h, in which ∼20% of branched product was formed), interference suppressions at G840 were observed for the A662:C mutant and the A598:C, A662:C double mutant (Fig. 5). These data suggest functional interactions between the internal loop of D3 and G840 in the bulge of D5. Notably, when mutations were introduced in the pentaloop of D3, which does not interact with G840, dG interference at this position was not suppressed (Fedorova and Pyle 2005), indicating that this effect is specific to the internal loop of D3. However, when the trans-branching reactions were carried out for shorter times (2 h, in which ∼10% of the branched product is formed), interference suppressions were no longer observed at G840 (Fig. 5). In NAIS studies, the suppression of a nucleotide analog interference is generally attributed to the loss of a hydrogen bond between the nucleotide that has been mutated and the site of interference. However, any NAIS effect attributable to a hydrogen bond should be independent of the reaction time. The fact that deoxynucleotide interference at G840 was observed at short reaction times, while suppression was observed at long reaction times suggests that other interpretations are possible and that interactions between the D3IL and D5 may not be direct. However, the results are consistent with energetic communication between the D3IL and the bulge of D5 through a complex network of interactions that contribute to active-site stabilization.
FIGURE 5.
Mutations in noncanonical A598-A662 base pair cause partial suppression of dG interference at position 840 in D5. (A) Autoradiographs of sequencing gels after iodine cleavage of precursor (pre) and branched (br) RNAs. Selection step (branching reaction) was carried out in the presence of wild-type 5′-end labeled D56 RNA transcribed in the presence of dG phosphorothioate and unlabeled wild-type and mutant exD123 RNAs. Numbers on the left indicate guanosine positions in D5. (B) A diagram comparing relative intensity of bands corresponding to G840 in precursor and branched products. The results represent average values obtained from at least three independent experiments; the error did not exceed 15%.
DISCUSSION
The D3IL is one of the most conserved elements of group II intron secondary structure. Enzymological investigations indicate that the D3IL stimulates chemical reactivity of the intron and chemical probing studies indicate that it is deeply buried within the catalytic core. Despite the central importance of the D3IL in function of the intron, its relative architectural location has remained obscure. Here we provide the first structural insights into the position of the D3IL relative to other intronic motifs, and we demonstrate the existence of specific interaction partners between D3 and elements of the core.
Identification of structural links between conserved structures in D3 and D1
We have established the existence of a long-range tertiary interaction between D3 and D1 using two entirely different methods: site-directed photo-cross-linking, which identifies residues in close proximity to a photoreactive probe nucleotide, and NAIS, which reveals functionally important molecular interactions between nucleotides.
In the cross-linking experiments, purine residues within the D3 internal loop were individually substituted with the photoreactive 6-thio-deoxyguanosine moiety, which provides a probe of the local structure surrounding D3 without compromising reactivity of the molecule. When the highly conserved A599 and A662 residues were substituted with 6-thio-dG, irradiation resulted in highly efficient cross-links between the D3IL and nucleotides that comprise the ε-ε′ motif in D1. The ε-ε′ structure is a highly significant site of interaction because this short duplex is critical for two processes: selection of the 5′-splice site and chemical catalysis (Jacquier and Michel 1990). Given that 6-thio-dG and 4-thioU form cross-links only with molecules at short range (less than 5Å) (Hanna 1989), the results suggest that the D3IL and ε-ε′ are within hydrogen bonding distance of one another. Direct tertiary interaction between these elements was confirmed by NAIS experiments, which revealed that a D3IL mutant can suppress dC interference at C117 in ε′. Notably, this suppression was observed when only 5%–10% of the branched product was formed, which is consistent with hydrogen bonding between the D3IL and C117 and is different from NAIS effects observed in the bulge of D5 (see Results). Taken together, these findings indicate that ε-ε′ and the D3IL are spatially close and functionally connected, forming part of a catalytically important network of tertiary interactions.
Significance and architectural form of the D3IL-D1 interaction
A contact between the D3IL and ε-ε′ is particularly significant in light of recent studies demonstrating that ε-ε′ is arranged in close proximity to the bulge of D5 and the J23 linker (de Lencastre et al. 2005; de Lencastre and Pyle 2008), which are two of the most important active-site components in a group II intron. By extension, an interaction between the internal loop of D3 and ε-ε′ would be expected to play an important role in stabilizing the active-site structure, which is consistent with the fact that the D3IL stimulates the chemical rate constant for catalysis (Bachl and Schmelzer 1990; Boudvillain and Pyle 1998).
Both the ε-ε′ pairing and the tandem sheared pairs in the D3IL are conserved elements of group II intron structure that are observed in almost all classes of group II introns (Toor et al. 2001). Although this implies structural conservation for the D3IL–ε-ε′ interaction among intron families, substantial differences in the local architecture surrounding ε-ε′ suggest that there may be considerable variation in its function. For example, in the group IIB and IID introns ε′ is located within a small asymmetric loop, whereas in group IIA and IIC introns, it is imbedded in a much larger loop substructure (Toor et al. 2001). These local differences suggest that ε-ε′ may not form identical networks of D3 interactions in all group II intron families.
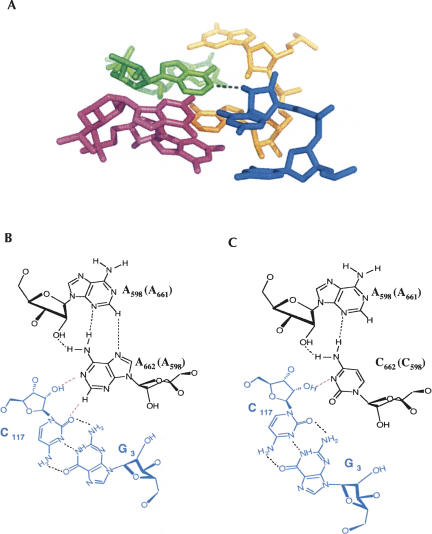
It is interesting to combine all of the available data and thereby project a likely molecular architecture for the D3IL–ε-ε′ interaction within group IIB intron ai5γ. Previous NAIM analysis has suggested that both A599 and A662 form tandem sheared trans-Hoogsteen-sugar edge A·A pairs (Fedorova and Pyle 2005). This motif has been structurally and functionally well characterized in other systems, where the purine bases have been observed to form an unusual cross-strand stacking arrangement (Correll et al. 1997). Indeed, crystallographic studies of the ribosome have revealed a strikingly similar array of a tandem sheared purine–purine pairs interacting with consecutive G-C pairs via trans-sugar edge-sugar edge pairing that involves contacts to the 2′-OH groups of cytosine (Fig. 6A; Ban et al. 2000; Klein et al. 2001). An almost identical set of molecular interactions between the D3IL and ε-ε′ would be consistent with our NAIS results. In addition, since the proposed interaction involves hydrogen bonding between O2 of C and H2 of A (Fig. 6B), the structural motif visualized within the ribosome is also consistent with our previous NAIM data, which shows strong 2,6-diaminopurine interference at both A599 and A662 (Fedorova and Pyle 2005). Importantly, hydrogen bonding of the nucleobase with the 2′-OH of C is preserved upon A:C mutation in the sheared pair, although resulting interaction between the C·A and G-C pairs is expected to be weaker due to a loss of one hydrogen bond (Fig. 6C).
FIGURE 6.
(A) A long-range tertiary interaction between tandem sheared purine–purine pairs and consecutive G-C pairs observed in the structure of the 23S rRNA (Ban et al. 2000). Dashed line denotes a hydrogen bond between H1 of A and 2′-OH of C. (B) A schematic of tertiary interaction involving one of sheared A·A pairs in the internal loop of D3 and the Watson–Crick pair G3-C117 in D1. (C) A schematic of the same interaction upon a mutation of an A·A pair to a C·A.
D3 supports a complex network of essential interactions
D3 does not interact exclusively with residues in D1; it also communicates with catalytically essential regions of D5, such as κ-κ′ (which helps align the active-site, (Fedorova and Pyle 2005) and the D5 bulge. The NAIS experiments presented here clearly indicate some type of energetic communication between the D3IL and the D5 bulge, which is consistent with early DEPC footprinting studies that suggested molecular contacts between the D5 bulge and elements of D3 (Jestin et al. 1997). The data also explain the striking solvent inaccessibility of the D3IL during hydroxyl radical footprinting experiments (Swisher et al. 2001).
While temporal sensitivity of the interference suppression in D5 precludes strict interpretation as a direct tertiary interaction, the data suggest that the D3IL and the D5 bulge may be part of a shared network of contacts that contributes to chemical catalysis.
This notion is supported by site-directed cross-linking studies that place D5 bulge nucleotide C839 in close proximity to a nucleotide G588, which is located within the J2/3 linker (adjacent to the D3IL). The same studies show that D5 bulge nucleotide A838 interacts with C4 of the ε-ε′ motif (de Lencastre et al. 2005; de Lencastre and Pyle 2008). This is remarkable in light of the present study, in which the same motif is implicated through cross-linking and NAIS methods to interact with the D3IL. Taken together, the data indicate that ε-ε′, the D3IL, J2/3, and the D5 bulge are spatially proximal motifs that form complex hub of molecular interactions within the group II intron core.
The role of D3 in the group II intron catalysis
The data on molecular interactions of D3 are consistent with an important, but supporting role for D3 in catalysis by group II introns. Earlier work has shown that the pentaloop of D3 in IIB introns helps buttress the κ−κ′ interaction between D1 and the back, or “binding face,” of D5. In this way, it is expected to stabilize the docking of D5 in a proper register or conformation. Here we show that the D3IL interacts with, and potentially stabilizes, the tiny ε-ε′ motif, which contains atoms that have been demonstrated to play a more direct role in chemical reactivity. Finally, our NAIS results are not strictly consistent with a direct form of molecular interaction between D5 and the D3IL, suggesting instead a form of energetic coupling that may involve other nucleotides or intervening structures. Taken together, these results support the original designation of D3 as a “catalytic effector” that enhances the inherent catalytic power of the ribozyme (Qin and Pyle 1998). This model originally arose from domain deletion studies showing that only D1 and D5 were strictly required for minimal chemical reactivity, although the chemical rate constant was strongly stimulated by the addition of D3, in cis or in trans (Koch et al. 1992; Podar et al. 1995; Fedorova et al. 2003). The effector model is also consistent with recent work on the hierarchical folding pathway for intron ai5γ, which indicates that D1 folds completely before subsequent docking of D5, and completion of the native structure through docking of D3 (Fedorova et al. 2007; Waldsich and Pyle 2007). The structural data provided herein are consistent with this model, indicating that D3 optimizes the active site by helping to correctly position D5 with respect to the splice sites and other active site elements in D1 and D6.
MATERIALS AND METHODS
DNA templates, RNA transcription, and synthesis
The exD123 plasmid (pJDI3′-673) and full splicing construct (pJD20) were kindly provided by Dr. P.S. Perlman (Howard Hughes Medical Institute, Chevy Chase, MD). ExD123 plasmids with mutations in the bulge and pentaloop of D3 (pQL 38 [G615UAAU619:UUCG], pGD 7 [A599:C], pGD8 [A662:C], pGD 9 [A599:C;A662:C], pGD 10 [A598:C;A662:C], pGD 55 [A599:C;A661:C]) and exD15 (pQL) plasmid were created by site-directed mutagenesis (Kunkel et al. 1991) of the wild-type exD123 or the pJD20 construct, respectively. Construct pT7-D56 is described elsewhere (Chin and Pyle 1995).
Wild-type and mutant exD123 plasmids were digested with BamHI, and the pT7-D56 construct was linearized with EcoRV. All RNA transcriptions were carried out as previously described (Pyle and Green 1994; Chin and Pyle 1995). For NAIS experiments, RNAs with randomly incorporated phosphorothioate analogs were prepared by adding NTPαS to the transcription mixture (Gish and Eckstein 1988; Ortoleva-Donnelly et al. 1998). All NTPαS analogs tested in NAIS experiments (CTPαS, GTPαS, dCTPαS, dGTPαS, ITPαS, ATPαS, dATPαS, 7-deaza-ATPαS) were purchased from Glen Research. Of all analogs tested, only those resulting in NAIS effects are discussed in the manuscript.
D56 RNAs (wild-type and containing single-atom substitutions dG840 or I840) used in NAIS experiments and D3 RNAs with a single 6-thio dG substitution at positions 599, 615, or 662 used in photo cross-linking experiments were prepared by enzymatic ligation of two chemically synthesized oligonucleotides as previously described (Fedorova et al. 2005).
All RNAs were gel-purified and resuspended in 10 mM MOPS (pH 6.0), 1 mM EDTA for storage.
Branching kinetics
Kinetic experiments were carried out essentially as described (Chin and Pyle 1995; Fedorova and Pyle 2005). Unlabeled wild-type and mutant exD123 RNAs (1.5 μM final concentration) and 5′-end labeled D56 RNA (10 nM) were denatured separately in MOPS (pH 7.5; 40 mM final concentration) for 1 min at 95°C and then cooled to 42°C. Both RNA solutions were then combined with the simultaneous addition of MgCl2 and NH4Cl (final concentrations, 100 mM and 0.5 M, respectively). Reactions were carried out at 42°C as previously described. Aliquots were taken at specific time points, quenched with denaturing dye solution, chilled, and analyzed as previously described (Chin and Pyle 1995; Boudvillain and Pyle 1998).
NAIS experiments
Branching reactions for NAIS experiments were performed as previously described (Boudvillain and Pyle 1998; Boudvillain et al. 2000; Fedorova and Pyle 2005). In order to screen for NAIS effects in D1, wild-type and mutant exD123 RNAs (1.5 μM final concentration) were transcribed in the presence of NTPαS and reacted with 5′-end labeled wild-type D56 (10 nM final concentration) under splicing conditions [40 mM MOPS at pH 6.8, 100 mM MgCl2, 1M (NH4)2SO4]. The reaction time was adjusted so that only 10%–20% of the branched product was formed. The branched products were purified on a 5% denaturing polyacrylamide gel, eluted from the gel, and ethanol precipitated. For an unreacted control, precursor exD123 RNA was cleaved by a DNAzyme after between nucleotides 97 and 98 in the 5′-exon, so that its size would be comparable to the size of the branched product. The precursor was then 5′-end labeled and, along with the branched products, subjected to iodine sequencing as described.
In NAIS experiments designed to screen for interference suppression in D5, wild-type D56 RNA (10 nM final concentration) was transcribed in the presence of NTPαS, 5′-end labeled and reacted with wild-type and mutant exD123 RNAs (1.5 μM final concentration) under the conditions described above. The unreacted D56 and the branched product were separated on a 5% denaturing polyacrylamide gel, eluted, and ethanol precipitated. Sequencing by iodine cleavage was carried out as described.
Site-directed photo-cross-linking experiments
Photo-cross-linking experiments were carried out as previously described (de Lencastre et al. 2005). Unlabeled exD15 RNA (5 μM final concentration) and 5′-end labeled D3 RNA containing a single 6-thio dG substitution (20 nM final concentration) were separately denatured in 40 mM MOPS (pH 7.5). The two RNAs were cooled to 42°C before combining them and adding MgCl2 and KCl to final concentrations of 100 mM and 1 M, respectively, prior to incubation for 20 min at 42°C to form a complex. This solution was then transferred to a covered polystyrene plate and irradiated at 366 nm for 30 min. The positions of the resulting cross-links were determined by reverse transcription as described.
ACKNOWLEDGMENTS
We thank Gaby Drews for excellent technical assistance. We also thank Dr. Philip S. Perlman for the gift of plasmids pJDI3′-673 and pJD20. O.F. is a Research Specialist and A.M.P. is an Investigator if the Howard Hughes Medical Institute.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.942308.
REFERENCES
- Bachl J., Schmelzer C. Effect of deletions at structural domains of group II intron bI1 on self-splicing in vitro. J. Mol. Biol. 1990;212:113–125. doi: 10.1016/0022-2836(90)90308-9. [DOI] [PubMed] [Google Scholar]
- Ban N., Nissen P., Hansen J., Moore P.B., Steitz T.A. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- Boudvillain M., Pyle A.M. Defining functional groups, core structural features and inter-domain tertiary contacts essential for group II intron self-splicing: A NAIM analysis. EMBO J. 1998;17:7091–7104. doi: 10.1093/emboj/17.23.7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudvillain M., de Lencastre A., Pyle A.M. A new RNA tertiary interaction that links active-site domains of a group II intron and anchors them at the site of catalysis. Nature. 2000;406:315–318. doi: 10.1038/35018589. [DOI] [PubMed] [Google Scholar]
- Chin K., Pyle A.M. Branch-point attack in group II introns is a highly reversible transesterification, providing a possible proofreading mechanism for 5′-splice site selection. RNA. 1995;1:391–406. [PMC free article] [PubMed] [Google Scholar]
- Correll C.C., Freeborn B., Moore P.B., Steitz T.A. Metals, motifs, and recognition in the crystal structure of a 5S rRNA domain. Cell. 1997;91:705–712. doi: 10.1016/s0092-8674(00)80457-2. [DOI] [PubMed] [Google Scholar]
- de Lencastre A., Pyle A.M. Three essential and conserved regions of the group II intron are proximal to the 5-splice site. RNA. 2008;14:11–24. doi: 10.1261/rna.774008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lencastre A., Hamill S., Pyle A.M. A single active-site region for a group II intron. Nat. Struct. Mol. Biol. 2005;12:626–627. doi: 10.1038/nsmb957. [DOI] [PubMed] [Google Scholar]
- Fedorova O., Pyle A.M. Linking the group II intron catalytic domains: tertiary contacts and structural features of Domain 3. EMBO J. 2005;24:6674–6687. doi: 10.1038/sj.emboj.7600852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorova O., Mitros T., Pyle A.M. Domains 2 and 3 interact to form critical elements of the group II intron active site. J. Mol. Biol. 2003;330:197–209. doi: 10.1016/s0022-2836(03)00594-1. [DOI] [PubMed] [Google Scholar]
- Fedorova O., Boudvillain M., Kawaoka J., Pyle A.M. Nucleotide analog interference mapping and suppression: Specific applications in studies of RNA tertiary structure, dynamic helicase mechanism and RNA-protein interactions. In: Hartmann R.K., et al., editors. Handbook of RNA biochemistry. Wiley-VCH; Weinheim, Germany: 2005. pp. 259–293. [Google Scholar]
- Fedorova O., Waldsich C., Pyle A.M. Group II intron folding under near-physiological conditions: Collapsing to the near-native state. 2007. J. Mol. Biol. 2007;366:1099–1114. doi: 10.1016/j.jmb.2006.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gish G., Eckstein F. DNA and RNA sequence determination based on phosphorothioate chemistry. Science. 1988;240:1520–1522. doi: 10.1126/science.2453926. [DOI] [PubMed] [Google Scholar]
- Hanna M.M. Photoaffinity cross-linking methods for studying RNA–protein interactions. Methods Enzymol. 1989;180:383–409. doi: 10.1016/0076-6879(89)80113-2. [DOI] [PubMed] [Google Scholar]
- Jacquier A., Michel F. Base-pairing interactions involving the 5′- and 3′- terminal nucleotides of group II self-splicing introns. J. Mol. Biol. 1990;213:437–447. doi: 10.1016/S0022-2836(05)80206-2. [DOI] [PubMed] [Google Scholar]
- Jestin J.-L., Deme E., Jacquier A. Identification of structural elements critical for inter-domain interactions in a group II self-splicing intron. EMBO J. 1997;16:2945–2954. doi: 10.1093/emboj/16.10.2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein D.J., Schmeing T.M., Moore P.B., Steitz T.A. The kink-turn: A new RNA secondary structure motif. EMBO J. 2001;20:4214–4221. doi: 10.1093/emboj/20.15.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch J.L., Boulanger S.C., Dib-Hajj S.D., Hebbar S.K., Perlman P.S. Group II Introns deleted for multiple substructures retain self-splicing activity. Mol. Cell. Biol. 1992;12:1950–1958. doi: 10.1128/mcb.12.5.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T.A., Bebenek K., McClary J. Efficient site-directed mutagenesis using uracil-containing DNA. Methods Enzymol. 1991;204:125–139. doi: 10.1016/0076-6879(91)04008-c. [DOI] [PubMed] [Google Scholar]
- Lambowitz A.M., Zimmerly S. Mobile group II introns. Annu. Rev. Genet. 2004;38:1–35. doi: 10.1146/annurev.genet.38.072902.091600. [DOI] [PubMed] [Google Scholar]
- Lehmann K., Schmidt U. Group II introns: Structure and catalytic versatility of large natural ribozymes. Crit. Rev. Biochem. Mol. Biol. 2003;38:249–303. doi: 10.1080/713609236. [DOI] [PubMed] [Google Scholar]
- Leontis N.B., Westhof E. The 5S rRNA loop E: Chemical probing and phylogenetic data versus crystal structure. RNA. 1998a;4:1134–1153. doi: 10.1017/s1355838298980566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontis N.B., Westhof E. A common motif organizes the structure of multi-helix loops in 16 S and 23 S ribosomal RNAs. J. Mol. Biol. 1998b;283:571–583. doi: 10.1006/jmbi.1998.2106. [DOI] [PubMed] [Google Scholar]
- Lescoute A., Leontis N.B., Massire C., Westhof E. Recurrent structural RNA motifs, isostericity matrices, and sequence alignments. Nucleic Acids Res. 2005;33:2395–2409. doi: 10.1093/nar/gki535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F., Ferat J.-L. Structure and activities of group II introns. Annu. Rev. Biochem. 1995;64:435–461. doi: 10.1146/annurev.bi.64.070195.002251. [DOI] [PubMed] [Google Scholar]
- Ortoleva-Donnelly L., Sweczak A.A., Gutell R.R., Strobel S.A. The chemical basis of adenosine conservation throughout the Tetrahymena ribozyme. RNA. 1998;4:498–519. doi: 10.1017/s1355838298980086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podar M., Dib-Hajj S., Perlman P.S. A UV-induced Mg2+-dependent cross-link traps an active form of domain 3 of a self-splicing group II intron. RNA. 1995;1:828–840. [PMC free article] [PubMed] [Google Scholar]
- Pyle A.M., Green J.B. Building a kinetic framework for group II intron ribozyme activity: Quantitation of interdomain binding and reaction rate. Biochemistry. 1994;33:2716–2725. doi: 10.1021/bi00175a047. [DOI] [PubMed] [Google Scholar]
- Pyle A.M., Lambowitz A.M. Group II introns: Ribozymes that splice RNA and invade DNA. In: Gesteland R.F., et al., editors. The RNA world. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2006. pp. 469–506. [Google Scholar]
- Qin P.Z., Pyle A.M. The architectural organization and mechanistic function of group II intron structural elements. Curr. Opin. Struct. Biol. 1998;8:301–308. doi: 10.1016/s0959-440x(98)80062-6. [DOI] [PubMed] [Google Scholar]
- Su L.J., Qin P., Michels W., Pyle A. Guiding ribozyme cleavage through motif recognition: The mechanism of cleavage site selection by a group II intron ribozyme. J. Mol. Biol. 2001;306:665–668. doi: 10.1006/jmbi.2000.4323. [DOI] [PubMed] [Google Scholar]
- Swisher J., Duarte C., Su L.J., Pyle A. Visualizing the solvent-inaccessible core of a group II intron ribozyme. EMBO J. 2001;20:2051–2061. doi: 10.1093/emboj/20.8.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toor N., Hausner G., Zimmerly S. Coevolution of group II intron RNA structures with their intron-encoded reverse transcriptases. RNA. 2001;7:1142–1152. doi: 10.1017/s1355838201010251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldsich C., Pyle A.M. A Kinetic intermediate that regulates proper folding of a group II intron RNA. J. Mol. Biol. 2007;375:572–580. doi: 10.1016/j.jmb.2007.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Q., Qin P.Z., Michels W.J., Freeland K., Pyle A.M. The sequence-specificity of a group II intron ribozyme: Multiple mechanisms for promoting unusually high discrimination against mismatched targets. Biochemistry. 1998;37:3839–3849. doi: 10.1021/bi972661n. [DOI] [PubMed] [Google Scholar]