Abstract
This letter describes the architecture and transcriptional output of a novel noncoding RNA gene in mouse and rat. The mRNA-like piRNA (mpiR) gene, lies between the Perp and KIAA1244 genes on mouse chromosome 10 and rat chromosome 1. In mouse, the mpiR gene is associated with the production of at least 13 different alternatively spliced and polyadenylated transcripts ranging from 500 nt to over 6 kb. Although these transcripts are structurally similar to conventional mRNAs, only short polypeptides are predicted on each of the three possible reading frames. Intron 2 is unique in that it harbors a novel low copy repeat with homology with the 3′-UTR of the lin-28 gene, while Exon 4 contains an unusual cluster of nine sequence modules that are dispersed throughout the mouse genome. The mpiR gene is expressed at low levels in somatic tissues, but is transcriptionally up-regulated in the testis at day 14 post-partum, a time that coincides with the pachytene stage of meiosis I. Bisulfite methylation analysis shows that expression in brain, liver, and testis is correlated with the methylation status of the promoter region. In addition to mRNA-like transcripts, the mpiR gene is also a precursor to testis-specific piRNAs, and these can be detected by both Northern and PCR-based approaches. Remarkably, piRNAs originate from two specific regions of the gene, one corresponding to Intron 2 and the other to Exon 4. Overall, this work provides a picture of a novel, lineage-specific, noncoding RNA gene and describes its processing into both mRNA-like and piRNA products.
Keywords: noncoding RNA, piRNA, testis, repetitive element, alternative splicing, gene expression
INTRODUCTION
Whole genome tiling arrays and high-throughput cDNA sequencing projects have shown that genomes are widely transcribed (Okazaki et al. 2002; Bertone et al. 2004; Carninci et al. 2005; Cheng et al. 2005; Birney et al. 2007; Kapranov et al. 2007). In addition to genes that encode recognized classes of RNA, such as mRNAs, tRNAs, rRNAs, snRNAs, snoRNAs, and so forth, there are a number of RNA-encoding genes for which categorization and identification of individual genes is still ongoing. These genes produce long noncoding RNAs (ncRNA) (Numata et al. 2003; Frith et al. 2006; Furuno et al. 2006) as well as diverse microRNAs, piRNAs, and other small transcripts. There is considerable interest in determining the structural and functional characteristics of genes that encode unconventional RNA species and in understanding their contributions to cellular regulation (for review, see Zamore and Haley 2005; Mattick and Makunin 2006; Carninci and Hayashizaki 2007; Nilsen 2007).
Although the function for many noncoding-type RNAs is still unknown, a number of roles are emerging. First, they are in some cases processed into small (micro) RNAs that act as sequence-specific addressing marks for the control of gene expression. An example of this class is the germ-cell-specific piRNAs (Aravin et al. 2006, 2007; Grivna et al. 2006; Girard et al. 2006; Lau et al. 2006; Vagin et al. 2006; Watanabe et al. 2006), which have roles in the defense against viruses and transposons (Saito et al. 2006; Brennecke et al. 2007; Carmell et al. 2007; Houwing et al. 2007). Second, some ncRNAs may encode polypeptides that are smaller than those produced by conventional mRNAs (Kondo et al. 2007). Third, some ncRNA genes produce transcripts that regulate imprinting and gene silencing, while others such as Xist and Tsix regulate X chromosome inactivation. Finally, a growing number of ncRNAs regulate specific phenomena, including TUG1 RNA-dependent regulation of retinal differentiation (Young et al. 2005), ncRNA-dependent regulation of HOX gene expression (Rinn et al. 2007), NRON-dependent regulation of NFAT (Willingham et al. 2005), bxd ncRNA-dependent regulation of Ubx gene expression (Petruk et al. 2006), locus control region RNA-dependent regulation of globin gene activation (Ho et al. 2006), and ncRNA-dependent regulation of the DHFR gene (Martianov et al. 2007).
In this letter we have examined the structure, expression, and processing of transcripts generated by an mRNA-like piRNA (mpiR) gene. The gene was identified by the discovery of a set of testis-specific ESTs complementary to the 3′-UTR of the gene that encodes the germ-cell transcription factor ALF (DeJong 2006). The mpiR gene, located on mouse chromosome 10, consists of four exons, a variety of conventional repetitive elements, and two novel low copy number repeats. RACE and RT-PCR analyses show that the gene is alternatively spliced to produce at least 13 different mRNA-like species, all of which have limited or no coding potential. The gene is expressed at low levels in somatic tissues but is up-regulated to high levels in the testis. Related to its testis expression is the fact that mpiR is associated with the production of germ-cell-specific piRNAs. Remarkably, such piRNAs are not distributed throughout the mpiR gene but are largely confined to Intron 2 and Exon 4. Overall, the results describe the detailed structure and tissue-specific transcriptional output of a novel noncoding RNA gene.
RESULTS AND DISCUSSION
BLAST searches aimed at identifying RNAs that might target the germ-cell-specific ALF transcription factor gene resulted in the identification of several unique testis ESTs complementary to the 3′-end (Figs. 1; Supplemental Fig. S1). The genomic region from which these ESTs originate is between the p53 effector related to pmp22 (Perp) and KIAA1244 genes at the A2–A3 boundary on mouse chromosome 10 (Fig. 1B). Database searches showed the region was transcribed into a variety of unorganized transcripts, suggesting that it might harbor one or more testis-specific genes (Supplemental Fig. S1). In order to define the structure of the gene or genes within this locus, we synthesized backward and forward primers to the dispersed ESTs and used these for RT-PCR analysis of testis and liver cDNA (Supplemental Fig. S1). Sequencing of over 100 RT-PCR products allowed us to organize the transcripts into 13 distinct variants (mpiR v.1 to mpiR v.13, where “v” indicates “variant”) (Fig. 1B). For reasons described later, we refer to this locus as the mpiR gene.
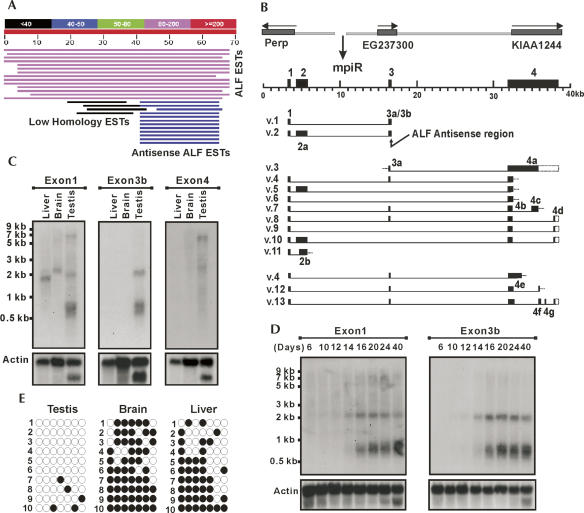
FIGURE 1.
Identification, organization, and expression of the mpiR gene. (A) BLAST search of mouse ESTs using the last 70 nt of the 3′-UTR of mouse ALF mRNA reveal a class of antisense-oriented testis-specific ESTs. (B) The mpiR locus from which the antisense ESTs are derived is located between the Perp, EG237300, and KIAA1244 genes on mouse chromosome 10A2/A3. Sequencing of RT-PCR products derived with multiple primer sets revealed over 13 distinct splicing products (v.1 to v.11 from testis; v.4, v.12, and v.13 from liver) originate from the mpiR locus. The 12 unique exons are noted along with the location of the ALF antisense region. (C) Northern analysis of mpiR expression revealed a variety of transcript isoforms in different tissues whose detection depended on the exon used as the probe. The results reveal complex tissue-specific splicing of the mpiR gene. (D) Developmental Northern blot of mpiR expression shows up-regulation of the 0.7- and 2.0-kb mpiR transcripts in pre-pubertal mice at about day 14, a time that corresponds to the pachytene stage of meiosis I. (E) Bisulfite methylation analysis of the mpiR promoter region demonstrates hypomethylation in testis compared to brain and liver.
The mpiR gene is composed of four main exons that span ∼40 kb of genomic DNA (Fig. 1B). Exon 1 is common to all transcripts. Exon 2 is divided into two alternatively spliced forms, 2a and 2b. Exon 3 is also divided into two forms, 3a and 3a+3b; whenever Exon 3a is used, the transcript is always spliced to include Exon 4. However, when this splicing event does not occur, the transcript terminates with a 3a+3b form of Exon 3 which retains complementarity to ALF mRNA. Exon 4 has a full-length size of just over 6.5 kb but can be spliced using at least seven different sub-exons (4a through 4g). RACE experiments defined two ends to mpiR transcripts: one that terminates at Exon 3b and the other that terminates at Exon 4 (Supplemental Fig. S1). All but one of the splicing patterns follows the GT-AG rule, and the polarity of splicing confirms the direction of transcription across the locus. BLAST searches and the identification of genomic sequences between the Perp and KIAA1244 genes in the rat, dog, cow, and human genomes showed that only the rat possessed a gene similar to mpiR, indicating the gene is likely restricted to the rodent lineage.
To determine how mpiR was expressed, we performed Northern blot analysis with probes from several different exons (Fig. 1C,D). Hybridization with an Exon 1 probe showed bands in liver (∼1.8 kb), brain (∼2.1 kb), and testis (∼0.7, ∼2.0, and ∼7.0 kb). The results also showed that expression of mpiR in testis involves distinct splicing isoforms and occurs at higher levels than in somatic tissues. These points are illustrated in part by the strongly hybridizing 0.7-kb transcript and by the selective detection of the 2.0-kb transcript in testis using the Exon 3b-specific probe. To further evaluate the regulated expression of mpiR, we analyzed developmentally timed RNA from the testis of pre-pubertal and post-pubertal mice. The results showed a dramatic increase in the 0.7- and 2.0-kb transcripts at about day 14 post-partum (Fig. 1D), a time that corresponds to the appearance of male germ cells that are in the pachytene stage of prophase I. Overall, the results reveal a deliberate pattern of transcript expression and splicing and suggest the potential for tissue-specific physiological roles.
We also asked if expression was associated with differential methylation of the promoter, as described for other testis-specific genes. PCR amplification and sequencing of bisulfite-treated genomic DNA fragments that spanned a 340-base-pair (bp) promoter region showed an average ∼69% CpG methylation in brain and liver tissues where mpiR expression was low, and little to no methylation (7% average) in testis (Fig. 1E). These results are consistent with an association between testis-specific hypomethylation and an active transcriptional state.
Because mpiR appeared to be a novel RNA-type gene, and because the architecture of these genes is still poorly understood, it was analyzed in terms of its repetitive element distribution, its coding potential, and its homology with the corresponding rat gene. The distribution of LINEs, SINEs, LTRs, and other elements was compared to the exon–intron organization using the RepeatMasker program (Fig. 2A). The results show that while Exon 2 was composed almost entirely of LINEs and other repetitive elements, splicing events that involved Exons 1, 3, and 4 generated transcripts that were free of such elements. Further comparison of sequences surrounding Exon 3 in the mouse and rat mpiR genes showed conserved positioning of repetitive and unique, single copy DNA in this region (Fig. 2A; Supplemental Fig. S2).
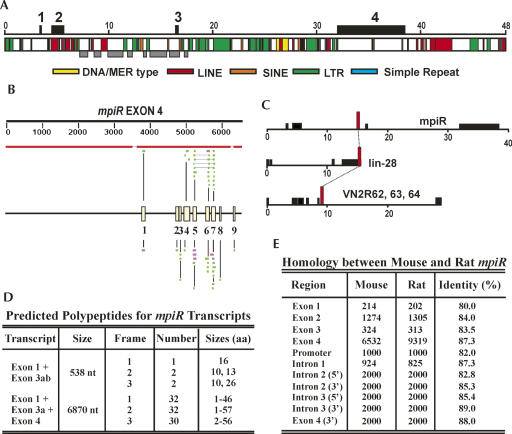
FIGURE 2.
Repetitive element distribution, homology, and coding potential of mpiR. (A) Distribution of repetitive elements across a 48-kb mpiR-containing region from mouse shows Exons 1, 3, and 4 are largely composed of single copy sequences. Exon positions are shown above, and gray boxes identify a region within Intron 2 and Exon 3 where the relative spacing of unique (single copy) and repetitive elements is similar between the mouse and rat mpiR genes. (B) BLASTN (top) and cross-species megaBLAST (bottom) searches using an Exon 4 query reveal fragmented matches toward the 3′-end. The organization and spacing of the nine elements are illustrated by the yellow boxes in the center of the diagram. More than 70 partial or complete copies of these modules are present dispersed throughout the mouse genome on at least 16 chromosomes. (C) Intron 2 of mpiR contained a region homologous to sequences from the 3′-UTR of the lin-28 gene and from Intron 6 of three vomeronasal receptor genes. (D) Representative mpiR transcripts show predicted polypeptides no larger than 57 amino acids. (E) Comparison of sequences from the mouse and rat mpiR genes revealed similar sequence homology from flanking regions, from introns, or from exons.
BLAST searches with mpiR sequences showed that the downstream 2.5-kb region of Exon 4 was divided into discrete modules that were present at other locations throughout the mouse genome (Fig. 2B). These elements could be organized into nine subdomains with sizes ranging from 48 to 132 nt, and over 70 such units could be identified on at least 16 different chromosomes (Supplemental Fig. S2). Some of these genomic copies were similar in organization to those described in mpiR, some contained changes in the spacing between the modules, and some consisted of solitary units. Alignments among each type of element allowed us to determine nine distinct “core” sequences (Supplemental Fig. S3). Surprisingly, however, none of these were recognized by RepeatMasker, nor were they present in available databases of tRNAs, snoRNAs, snRNAs, and microRNAs, indicating that they are a previously unrecognized class of low copy repeats.
BLAT searches using the July 2007 build of the whole mouse genome on the UCSC Genome Bioinformatics Server also revealed a novel ∼350-bp element in Intron 2 that was present in an estimated 5–10 copies throughout the mouse genome. Interestingly, this element constitutes the last ∼350 nucleotides (nt) of the 3′-UTR of the lin-28 gene and is also present within Intron 6 of three clustered vomeronasal receptor genes 2r62, 2r63, and 2r64 (Fig. 2C). These, and several other dispersed copies of this sequence, were highly homologous (Supplemental Fig. S4).
Although mpiR transcripts are spliced and polyadenylated like conventional mRNAs, several lines of evidence suggest that they might be noncoding. First, reading frames that begin with methionine in the v.1 and v.4 transcripts ranged in size from a minimum of 1 to a maximum of 57 (Fig. 2D). Second, ATG codons located in Exon 1 and Exon 3a, near the putative 5′-cap site, showed a poor fit to the A/GNNATGG Kozak consensus, and the polypeptides predicted from these ATGs terminated soon after they began. Third, none of the peptides predicted on any of the three reading frames, including those that did not begin with methionine, were similar to protein or nucleotide sequence databases. Fourth, Exon 2 was composed almost entirely of repetitive DNA, and the inclusion of this sequence would be inconsistent with the production of unique open reading frame. Finally, pairwise comparison of exons, promoter, and intron sequences between the mouse and rat mpiR genes showed very little difference in sequence conservation regardless of the location being analyzed (Fig. 1E). This result suggests that the exons in mpiR are not under strong selection pressure to maintain a conserved coding sequence. Although it is possible that mpiR transcripts might be spliced, initiated, or edited to encode a longer polypeptide, or that very short peptides are translated as observed in a Drosophila RNA gene (Kondo et al. 2007), the current data suggest it is probably noncoding.
Mouse testis contains a large collection of small Piwi-interacting RNAs (piRNAs) which display strand-specific clustering at ∼100 discrete genomic loci. Here we asked if there might be a relationship between these piRNAs and the noncoding mpiR transcripts abundant in the testis. In fact, mouse mpiR gene queries revealed numerous piRNA matches (Fig. 3A), and it was determined that mpiR corresponds to piRNA cluster 44 in mouse and to clusters 41/51 in rat (Lau et al. 2006). A more focused analysis revealed a number of interesting points. First, the matches were not distributed across the entire locus but were concentrated in two regions, one corresponding to Intron 2 and the other to Exon 4 (Fig. 3A). Second, the distribution of piRNA hits in the mouse and rat genes was similar except that the rat gene displayed many more hits within Intron 2, including some which mapped at the 5′-most side of Intron 3 (Fig. 3C). Third, piRNA matches landed in both repetitive and nonrepetitive genomic DNA. For instance, many piRNAs are from Exon 4, which is largely free of repetitive elements (Fig. 3A,C), and a repeat-masked Intron 2 from rat mpiR retained many of the piRNA matches observed with an unmasked query (Supplemental Fig. S2). The results show that at least one function of the noncoding mpiR gene is the generation of testis-specific piRNAs.
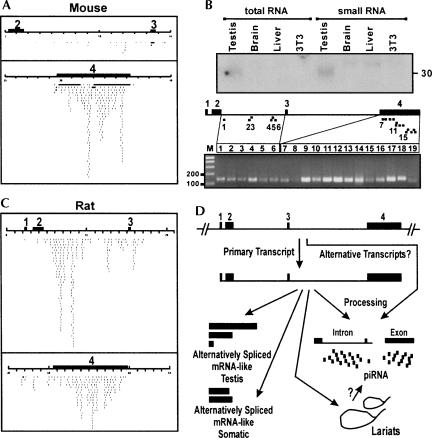
FIGURE 3.
Analysis of piRNA production from the mpiR gene. (A) The mouse mpiR gene displays matches to testis-specific mouse piRNAs that are largely restricted to Intron 2 and Exon 4. (B) An Exon 4 probe detects piRNA-sized (∼ 30 nt) small RNAs in total RNA from testis but not from other tissues. Small RNA preparations also revealed slightly larger but much weaker bands in brain and liver. The lower diagram shows the PCR-based detection of piRNAs from across Intron 2 and Exon 4. (C) The rat mpiR gene displays abundant matches to rat piRNAs in Intron 2 and Exon 4 regions similar to those seen with the mouse gene. (D) Model depicts an mpiR gene composed of four exons which undergoes alternative splicing to form a variety of mRNA-like species. One or more RNAs from this gene, possibly including the primary transcript, intron lariats, or mRNA-like transcripts themselves, undergo processing to generate Intron 2- and Exon 4-specific piRNAs.
To experimentally confirm the relationship between the mpiR gene and piRNA production, an Exon 4-specific probe was used in Northern analyses to detect small RNAs from liver, brain, testis, and NIH3T3 cells. The results revealed cross-hybridizing small RNAs in testis of the predicted size (∼30 nt) (Fig. 3B), as well as slightly larger but much weaker signals in brain and liver. A second approach to detect mpiR-specific piRNAs involved PCR-based amplification (Fig. 3B). The results revealed the existence of piRNA products along the length of Intron 2 and Exon 4. Primers that matched abundant piRNAs tended to give the strongest signals, and we observed consistently stronger signals with primers against Exon 4 compared to those against Intron 2, in agreement with the relative abundance of database-predicted piRNA hits (Fig. 3A). Intron 2-specific primers that did not correspond to a specific piRNA match gave signals in this assay that were similar in intensity to those obtained with primers that matched solitary piRNAs (Fig. 3B, e.g., cf. lanes 1 and 3 [which do not match known piRNAs] and lanes 2 and 6 [which match single hit piRNAs]). This result is in agreement with the fact that piRNA library sequencing has not reached saturation.
We have examined the distribution of piRNA hits with respect to (1) the locations of alternatively spliced Exons 4a–4g; (2) the location of the novel Exon 4 repeats; (3) the distribution of standard repetitive elements; (4) predicted RNA structural features such as hairpins; (5) the distribution of Exon 4-derived EST fragments; and (6) the region of conserved repetitive element spacing in Intron 2. In every case, piRNA hits extended beyond each of these sequence features. Reports of an association between small RNAs and transposon silencing based on studies on the Drosophila flamenco locus provide clues as to the function of germline piRNAs (Brennecke et al. 2007), but it appears that more information is required before these functions can be fully appreciated using the mouse mpiR gene as a model. The preferential use of an intron and an alternatively spliced exon for piRNA production, as reported here, demonstrates a unique sequence selectivity to processing across the transcript. Finally, we speculate that the fragmented nature of ESTs which map to Exon 4 (Supplemental Fig. S1) may represent a consequence of piRNA processing and might be a feature shared with other piRNA genes.
A summary of the organizational and transcriptional features of the mpiR gene is illustrated in Figure 3D. First, mpiR is expressed in both somatic and germline tissues in a variety of alternatively spliced and polyadenylated forms. Second, the gene is up-regulated in testis at the time of the pachytene stage of meiotic prophase I. Third, mpiR is composed of at least three distinct sequence types: (1) single copy (unique) sequences; (2) repetitive elements such as LINES, SINES, and LTRs; and (3) at least two novel low copy elements. Fourth, the gene is specific to the rodent lineage and is either absent or unrecognizably divergent in human. Finally, numerous matches to piRNAs mapped to both intronic and exonic regions of the gene. Overall, the results define a novel noncoding RNA-type gene with a surprisingly complex transcriptional output. In future studies we hope to ask how the organization of the mpiR gene compares with other long ncRNAs and whether it can be used to dissect early steps in piRNA biogenesis.
MATERIALS AND METHODS
RT-PCR and cDNA cloning
To detect full-length mpiR transcripts and to identify new splice variants, oligo(dT) primed first-strand cDNA from mouse testis and liver poly(A) RNA was used as a template for PCR reactions with mpiR-specific primers (Supplemental Fig. S1). Over 100 PCR products were cloned and sequenced.
Northern blotting and PCR-based piRNA detection
Poly(A) RNAs isolated from adult mouse brain, liver, and testis and from testis at post-natal days 6, 10, 12, 14, 16, 20, 24, and 40 were used for the Northern analysis of mpiR expression. Small RNAs (∼20 μg) from mouse testis, brain, liver, and NIH3T3 cells (Watanabe et al. 2006) were separated on 15% denaturing polyacrylamide gels (Grivna et al. 2006). Hybridizations were carried out at 52°C with in vitro transcribed and [α-32P]UTP-labeled RNA probes spanning a 1-kb region of Exon 4.
PCR-based detection of piRNAs (Ro et al. 2006) was performed with small RNA samples isolated using the mirVana miRNA isolation kit (Ambion). RNAs were then polyadenylated and used to synthesize small RNA cDNA (srcDNA) with SuperScript II Reverse Transcriptase (Invitrogen) and 1 μg of RTQ-primer. Individual piRNAs were detected by PCR using the RTQ-UNir primer along with mpiR-specific primers.
DNA methylation
Genomic DNA (1.5 μg) from mouse testis, liver, and brain was processed using the EpiTect Bisulfite Kit (Qiagen). Modified DNA was used as a template for the PCR reactions with mpiR-specific primers 1 (5′-TTTGAAAGTATTTAGTTGAGGTGAGGG) and 2 (5′-TACATTACCTCACAATTACCCTAACAA) that spanned a 340-bp region of the mpiR promoter that contained seven CpGs.
Bioinformatic/sequence analysis
NCBI Blast searches (http://www.ncbi.nlm.nih.gov/), including megaBLAST, cross-species megaBLAST, and BLASTN programs, were used to identify the mpiR-derived ESTs complementary to the ALF 3′-UTR as well as for other homology searches (Altschul et al. 1990). Typically, genomes were searched using the genome (reference only) database, cDNAs were identified with the RefSeq RNA and ESTs databases, and piRNAs were detected with the Non-RefSeq RNA database. Some homology searches were performed by BLAST-like alignment tool (BLAT) available at the UCSC Genome Browser (http://genome.ucsc.edu/) (Kent 2002). Specific sequence alignments were also performed using the DNAstar program. RepeatMasker (http://www.repeatmasker.org/) was used to identify known repetitive elements and to prepare repeat-masked sequences.
DATA DEPOSITION
Sequences reported in this paper have been deposited in GenBank. These include the annotated mouse mpiR gene (TPA: BK006463) and mouse mpiR cDNAs (EU523858 to EU523869 and TPA: BK006464).
SUPPLEMENTAL DATA
Supplemental material can be found at http://www.utdallas.edu/biology/faculty/research/dejong.html.
ACKNOWLEDGMENTS
This work was supported by a grant from the NICHD to J.D.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.974608.
REFERENCES
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Aravin A., Gaidatzis D., Pfeffer S., Lagos-Quintana M., Landgraf P., Iovino N., Morris P., Brownstein M.J., Kuramochi-Miyagawa S., Nakano T., et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- Aravin A.A., Sachidanandam R., Girard A., Fejes-Toth K., Hannon G.J. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- Bertone P., Stolc V., Royce T.E., Rozowsky J.S., Urban A.E., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- Birney E., Stamatoyannopoulos J.A., Dutta A., Guigo R., Gingeras T.R., Margulies E.H., Weng Z., Snyder M., Dermitzakis E.T., Thurman R.E., et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J., Aravin A.A., Stark A., Dus M., Kellis M., Sachidanandam R., Hannon G.J. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila . Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Carmell M.A., Girard A., van de Kant H.J.G., Bourc'his D., Bestor T.H., de Rooij D.G., Hannon G.J. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev. Cell. 2007;12:503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Carninci P., Hayashizaki Y. Noncoding RNA transcription beyond annotated genes. Curr. Opin. Genet. Dev. 2007;17:139–144. doi: 10.1016/j.gde.2007.02.008. [DOI] [PubMed] [Google Scholar]
- Carninci P., Kasukawa T., Katayama S., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Cheng J., Kapranov P., Drenkow J., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- DeJong J. Basic mechanisms for the control of germ cell gene expression. Gene. 2006;366:39–50. doi: 10.1016/j.gene.2005.10.012. [DOI] [PubMed] [Google Scholar]
- Frith M.C., Wilming L.G., Forrest A., Kawaji H., Tan S.L., Wahlestedt C., Bajic V.B., Kai C., Kawai J., Carninci P., et al. Pseudo-messenger RNA: Phantoms of the transcriptome. PLoS Genet. 2006;2:e23. doi: 10.1371/journal.pgen.0020023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuno M., Pang K.C., Ninomiya N., Fukuda S., Frith M.C., Bult C., Kai C., Kawai J., Carninci P., Hayashizaki Y., et al. Clusters of internally primed transcripts reveal novel long noncoding RNAs. PLoS Genet. 2006;2:e37. doi: 10.1371/journal.pgen.0020037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard A., Sachidanandam R., Hannon G.J., Carmell M.A. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- Grivna S.T., Beyret E., Wang Z., Lin H. A novel class of small RNAs in mouse spermatogenic cells. Genes & Dev. 2006;20:1709–1714. doi: 10.1101/gad.1434406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houwing S., Kamminga L.M., Berezikov E., Cronembold D., Girard A., van den Elst H., Filippov D.V., Blaser H., Raz E., Moens C.B., et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in zebrafish. Cell. 2007;129:69–82. doi: 10.1016/j.cell.2007.03.026. [DOI] [PubMed] [Google Scholar]
- Ho Y., Elefant F., Liebhaber S.A., Cooke N.E. Locus control region transcription plays an active role in long-range gene activation. Mol. Cell. 2006;23:365–375. doi: 10.1016/j.molcel.2006.05.041. [DOI] [PubMed] [Google Scholar]
- Kapranov P., Cheng J., Dike S., Nix D.A., Duttagupta R., Willingham A.T., Stadler P.F., Hertel J., Hackermuller J., Hofacker I.L., et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- Kent W.J. BLAT—The BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T., Hashimoto Y., Kato K., Inagaki S., Hayashi S., Kageyama Y. Small peptide regulators of actin-based cell morphogenesis encoded by a polycistronic mRNA. Nat. Cell Biol. 2007;9:660–665. doi: 10.1038/ncb1595. [DOI] [PubMed] [Google Scholar]
- Lau N.C., Seto A.G., Kim J., Kuramochi-Miyagawa S., Nakano T., Bartel D.P., Kingston R.E. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- Martianov I., Ramadass A., Serra Barros A., Chow N., Akoulitchev A. Repression of the human dihydrofolate reductase gene by a noncoding interfering transcript. Nature. 2007;445:666–670. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- Mattick J.S., Makunin I.V. Noncoding RNA. Hum. Mol. Genet. 2006;15:R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- Nilsen T.W. RNA 1997–2007: A remarkable decade of discovery. Mol. Cell. 2007;14:715–720. doi: 10.1016/j.molcel.2007.11.017. [DOI] [PubMed] [Google Scholar]
- Numata K., Kanai A., Saito R., Kondo S., Adachi J., Wilming L.G., Hume D.A., Hayashizaki Y., RIKEN GER Group, GSL Members et al. Identification of putative noncoding RNAs among the RIKEN mouse full-length cDNA collection. Genome Res. 2003;13:1301–1306. doi: 10.1101/gr.1011603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki Y., Furuno M., Kasukawa T., Adachi J., Bono H., Kondo S., Nikaido N., Osato N., Saito R., Suzuki H., et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- Petruk S., Sedkov Y., Riley K.M., Hodgson J., Schweisguth F., Hirose S., Jaynes J.B., Brock H.W., Mazo A. Transcription of bxd noncoding RNAs promoted by Trithorax represses Ubx in cis by transcriptional interference. Cell. 2006;127:1209–1221. doi: 10.1016/j.cell.2006.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn J.L., Kertesz M., Wang J.K., Squazzo S.L., Xu X., Brugmann S.A., Goodnough L.H., Helms J.A., Farnham P.J., Segal E., et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ro S., Park C., Jin J., Sanders K.M., Yan W. A PCR-based method for detection and quantification of small RNAs. Biochem. Biophys. Res. Commun. 2006;351:756–763. doi: 10.1016/j.bbrc.2006.10.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K., Nishida K.M., Mori T., Kawamura Y., Miyoshi K., Nagami T., Siomi H., Siomi M.C. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes & Dev. 2006;20:2214–2222. doi: 10.1101/gad.1454806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin V.V., Sigova A., Li C., Seitz H., Gvozdev V., Zamore P.D. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- Watanabe T., Takeda A., Tsukiyama T., Mise K., Okuno T., Sasaki H., Minami N., Imai H. Identification and characterization of two novel classes of small RNAs in the mouse germline: Retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes & Dev. 2006;20:1732–1743. doi: 10.1101/gad.1425706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willingham A.T., Orth A.P., Batalov S., Peters E.C., Wen B.G., Aza-Blanc P., Hogenesch J.B., Schultz P.G. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science. 2005;309:1570–1573. doi: 10.1126/science.1115901. [DOI] [PubMed] [Google Scholar]
- Young T.L., Matsuda T., Cepko C.L. The noncoding RNA taurine upregulated gene 1 is required for differentiation of the murine retina. Curr. Biol. 2005;15:501–512. doi: 10.1016/j.cub.2005.02.027. [DOI] [PubMed] [Google Scholar]
- Zamore P.D., Haley B. Ribo-gnome: The big world of small RNAs. Science. 2005;309:1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]