Abstract
Potential applications for functional RNAs are rapidly expanding, not only to address functions based on primary nucleotide sequences, but also by RNA aptamer, which can suppress the activity of any target molecule. Aptamers are short DNA or RNA folded molecules that can be selected in vitro on the basis of their high affinity for a target molecule. Here, we demonstrate the ability of RNA aptamers to recognize—and bind to—human IgG with high specificity and affinity. An optimized 23-nucleotide aptamer, Apt8-2, was prepared, and was shown to bind to the Fc domain of human IgG, but not to other IgG's, with high affinity. Apt8-2 was observed to compete with protein A, but not with the Fcγ receptor, for IgG binding. NMR chemical-shift analyses localized the aptamer-binding sites on the Fc subdomain, which partially overlaps the protein A binding site but not the Fcγ receptor binding site. The tertiary structures of the predicted recognition sites on the Fc domain differ significantly between human IgG and other species of IgGs; this, in part, accounts for the high specificity of the selected aptamer. Apt8-2 can therefore be used as a protein A alternative for affinity purification of human IgG and therapeutic antibodies. Using Apt8-2 would have several potential advantages, raising the possibility of developing new applications based on aptamer design.
Keywords: RNA aptamer, immunoglobulin G Fc domain, NMR chemical shift, antibody purification, protein A
INTRODUCTION
Laboratory-engineered therapeutic antibodies are designed to recognize and bind to an extracellular protein. Each therapeutic antibody presumably binds to a different antigen, and, in general, it can be used alone, in combination with chemotherapy, or as a carrier for toxins or radiation. Recent advancements in relevant technologies have facilitated the development of monoclonal antibody therapies. In 2006, 21 antibody-based therapeutics were approved by the US Food and Drug Administration (FDA), more than 400 are in clinical trials, and the antibody therapeutic market was expected to reach in excess of 22 billion dollars in 2007.
During purification of therapeutic antibodies, impurities, including host cell proteins, DNA, antibody variants, and small molecules, as well as potential contaminants such as endotoxins and viral particles, must be removed (Fahrner et al. 2001). Since many of the monoclonal antibody therapies require high doses and/or continued administration, economical and quality-controlled large-scale production of these antibodies is of great importance. At present, protein A affinity chromatography is the common procedure used in purification of antibodies, because it selectively and efficiently binds antibodies in complex solutions, such as harvested cell culture media (Fahrner et al. 2001; Ghose et al. 2005). Protein A is a natural product of Staphylococcus aureus; it binds the Fc portion of a variety of mammalian IgG molecules (Huse et al. 2002).
The key disadvantages of protein A chromatography include cost, quality control difficulties, resin stability, and acidic elution procedures. Protein A is obtained from genetically modified bacteria through complex and expensive procedures. Therefore, protein A resin is over 30 times more expensive than other ion exchange resins, and may account for >35% of the total raw material costs for large-scale recovery of IgG. Also, since protein A molecules may cause immunogenic or other physiological responses in humans, any contaminating ligand leaked from the base matrix must be removed during processing (Gagnon 1996). Another drawback of using protein A for affinity purification is that the acidic conditions required for eluting bound antibodies can impair the antibody's conformation and activity (Tsumoto et al. 2004; Ghose et al. 2005; Cromwell et al. 2006).
To overcome the disadvantages, several synthetic ligands have been proposed as replacements for protein A in the affinity purification of antibodies; these include the use of a thiophilic ligand (Porath et al. 1985), histidyl ligand (Elkak and Vjiayalakshmi 1991), Avid A1 (Ngo and Khatter 1990), or peptides or nonpeptides designed to mimic protein A (Fassina et al. 2001; for review, see Huse et al. 2002). However, to our knowledge none of these have as yet become protein A alternatives at the manufacturing level.
The concept of using single-stranded nucleic acids (aptamers) as affinity molecules for protein binding was initially described in 1990 (Ellington and Szostak 1990, 1992; Tuerk and Gold 1990). The concept is based on the ability of short (20–80 mer) sequences to fold, in the presence of a target, into unique, three-dimensional structures that bind the target with high affinity and specificity. Aptamers are generated by a process that combines combinatorial chemistry with in vitro evolution, known as SELEX (systematic evolution of ligands by exponential enrichment), from a complex library of randomized sequences of typically 1014 different molecules (Oguro et al. 2003; Klussmann 2006; Miyakawa et al. 2006; Ohuchi et al. 2006; Keefe and Schaub 2008). It has been demonstrated that aptamers could be used as reagents for affinity purification (Romig et al. 1999; Blank et al. 2001). Moreover, the first aptamer-based therapeutic, Pegaptanib (Macugen), targeting vascular endothelial growth factor, was approved by the FDA in December 2004 for the treatment of age-related macular degeneration (AMD) (Ng et al. 2006; Zhou and Wang 2006). Considering the basic principle of aptamer selection, the high potential of RNA to create a vast set of tertiary structures is conceivable from both the “RNA world” hypothesis (Gesteland et al. 1999) and the concept of “molecular mimicry” between RNA and protein (Nakamura 2001).
In this study, we isolated RNA aptamers that bind the Fc portion of human IgG for the purpose of creating a protein A alternative for the mass purification of therapeutic antibodies. The thoroughly optimized 23-mer aptamer is highly specific to human IgG, but not to any other species IgGs, opening a new avenue for aptamer-based industrial development. We demonstrate here the structural and molecular basis for the high specificity of a RNA aptamer.
RESULTS
Selection of IgG aptamers
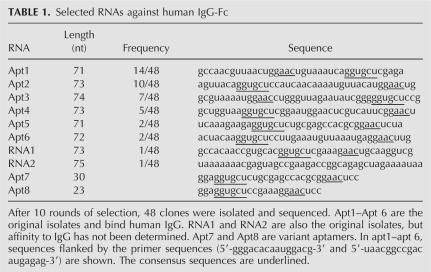
RNA aptamers against the Fc portion of human IgG1 subclass (hIgG1-Fc) were selected from a library of RNA sequences by modified SELEX with 2′-fluoro pyrimidines. After 10 rounds of selection, we obtained six sets of RNA sequences of 71–74 nucleotides (nt) long, Apt1 through Apt6 (Table 1). All of these sequences, 40 out of 48 sequences, contain consensus motifs, 5′-GGUGCU-3′ and 5′-AAC-3′. Two sequences were single and the other six sequences were not determined. The MFOLD prediction (Zuker 2003) suggests that all examined sequences can adopt relevant secondary structures containing a GGUGCU bulging loop paired with ACC as one low-energy possibility (Fig. 1). The affinity of the selected aptamers to hIgG1-Fc was examined by surface plasmon resonance (SPR) using BIAcore 2000. Divalent kinetic parameters for each aptamer were estimated by curve fitting of the sensorgrams (Table 2; Nakajima et al. 2001), reflecting the nuclear magnetic resonance (NMR) observation that two aptamers bind to one Fc (homodimer) unit (described below).
TABLE 1.
Selected RNAs against human IgG-Fc
FIGURE 1.
MFOLD prediction of Fc aptamer secondary structures. The consensus bulging structure is outlined with dotted squares. 2′-fluoro nucleotides are indicated with circles. Uppercase letters denote ribonucleotides, while lowercase letters denote deoxyribonucleotides. AptNMR is used for NMR analysis.
TABLE 2.
SPR kinetic constants of aptamers for binding IgG1
Optimized aptamer, Apt8-2
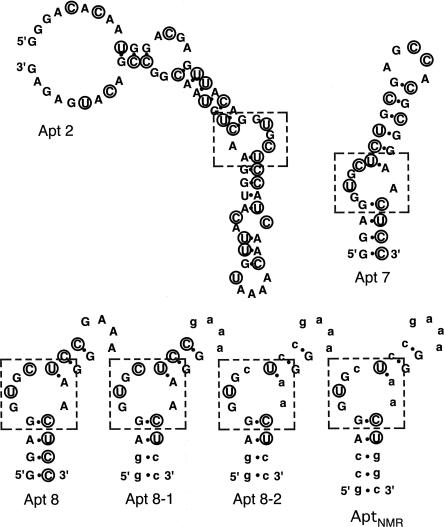
Several truncated and modified aptamers were designed and their affinity to hIgG1 was measured by SPR. Apt7 is a truncated Apt2 of 30 nucleotides, and binds hIgG1 with equal efficiency. Apt8 is a 23-nt variant of Apt7 with a GAAA tetra-loop substitution that also binds hIgG1 with high affinity (see Fig 1; Table 2). Apt8 contains nine 2′-fluoro pyrimidines, six Cs, and three Us, which confer protection against nuclease degradation. To further enhance the stability and/or to reduce the cost of chemical synthesis at the manufacturing level, ribose 2′-deoxy substitutions were systematically introduced into each of the 23 nucleotides and the resulting affinity to IgG1 was measured by SPR. Apt8-1 and Apt8-2, which have eight and 13 substituted 2′-deoxy nucleotides, respectively (Fig. 1), are two such derivatives that retain high affinity to IgG1 (see Fig. 2A). An in vitro stability experiment revealed that the half-life of Apt8-2 conjugated with an amino group at the 5′ end was 4 h in human serum at 25°C, which is much longer than that of natural RNA (data not shown).
FIGURE 2.
SPR sensorgrams. 3′-poly(A)-tailed RNA aptamers were immobilized to the streptavidin sensor chip via hybridization to a 5′-biotinylated oligo(dT), and the chip was injected with the indicated samples. Experimental conditions and procedures are described in Materials and Methods. (A) Affinity of Apt8, 8-1, 8-2, and mutated apt8 (U6C) to hIgG1 (400 nM). (B) Affinity of apt7 to hIgG1 complexes with FcγR or protein A. hIgG1 was preincubated with an excess of FcγR or protein A prior to injection. FcγR and protein A were also injected as control samples. (C) Affinity of apt7 to Remicade (500 nM). Running buffers used during the operation were the same buffer as used for aptamer selection (i.e., “binding buffer”) except for the concentration of NaCl used, 0.145 M or 1 M. (D) Affinity of apt8-2 to anti-CD26 humanized antibody and IgA (control).
Indispensability of the GGUGCU bulging motif
To investigate the requirement of the GGUGCU bulge motif, we reselected for hIgG1 affinity molecules from a library derived from the Apt8-2 backbone, but with the GGUGCU fully randomized. After three rounds of selection, 48 clones were isolated and sequenced. A total of 36 preserved the GGUGCU, while the remaining 12 that lost the GGUGCU exhibited poor affinity to hIgG1, as determined by SPR. Of 48 clones from pools prior to the third selection, only one clone contained GGUGCU, 46 were distinct sequences, and two were the same. MFOLD predicted that only one sequence, GGUGAU, passed the criteria of forming a GGUGCU-like bulge structure. SPR analysis indicated that the GGUGAU variant bound hIgG1, although the affinity was lower than Apt8-2 (data not shown). In view of the appearance of the alternative sequence, GGUGAU, other potential bulging sequences 6-nt long, i.e., GAUGCU, GCUGCU, GGCGCU, GGUACU, and GGUUCU (in which substitutions are underlined), were substituted for GGUGCU in Apt8. These variants, however, did not bind to hIgG1 (data not shown).
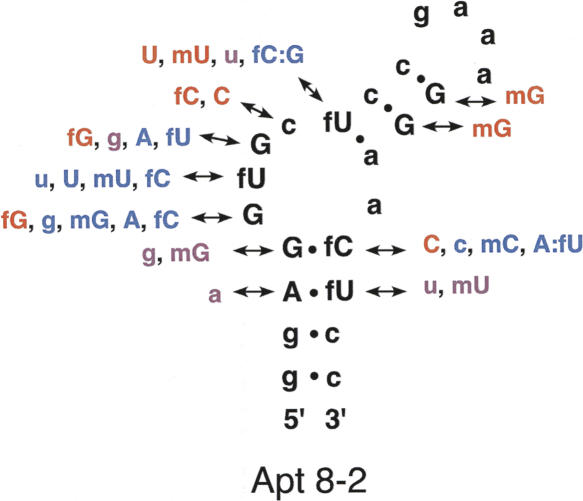
We further performed extensive substitution analysis, coupled with SPR, using synthetic Apt8-2 derivatives, in which each nucleotide of GGUGCU was systematically changed to a 2′ modification or another base, as indicated in Figure 3. Importantly, the 2′-fluoro U (fU) at position 6 (counted from the 5′ end of Apt8) is indispensable since its replacement to 2′-fluoro C (fC), 2′-hydroxy U (rU), 2′-deoxy U (dU), or 2′-O-methyl U (mU) completely abolished the affinity to hIgG1 (see Figs. 2A, 3). In contrast, the 2′ group in other nucleotides is somewhat flexible: For example, the 2′-fluoro U9 could be replaced by rU or 2′-O-methyl U without significant loss of affinity (see Fig. 3). These findings highlight the importance of the GGUGCU bulging motif and the indispensability of 2′-fluoro U6.
FIGURE 3.
Functional annotation of base substitutions and ribose 2′ modifications in Apt8-2. The indicated variations were introduced into Apt8-2 systematically by chemical synthesis, and the affinity of these modified oligonucleotides to hIgG1 was monitored by SPR. Symbols: uppercase letters, ribonucleotide; lowercase letters, deoxyribonucleotide; f, 2′-fluoro; m, 2′-O-methyl. A:fu and fC:G indicate substitutions of the corresponding base pairs, G:fC and fU:a, respectively. The affinity of each substitution, relative to the original Apt8-2 is indicated: red, equivalent; purple, drastically reduced; blue, no affinity. These were evaluated with BIAcore sensorgrams.
High specificity of Apt2 and Apt8 to human IgG
The affinity of Apt2 and Apt8 to different hIgG subclasses and other mammalian IgGs was examined by SPR. Both aptamers bound to all hIgG subclasses, hIgG1 through hIgG4, but not to other IgGs examined so far, such as those from mouse, rat, rabbit, bovine, chicken, canine, feline, and guinea pig (data not shown). This high selectivity is in sharp contrast to protein A, which shows much less stringent affinity and binds to these IgGs, except for chicken IgG, rat IgG2a, rat IgG2b, and bovine IgG1 (Huse et al. 2002).
Multiple sequence comparisons of Fc fragments
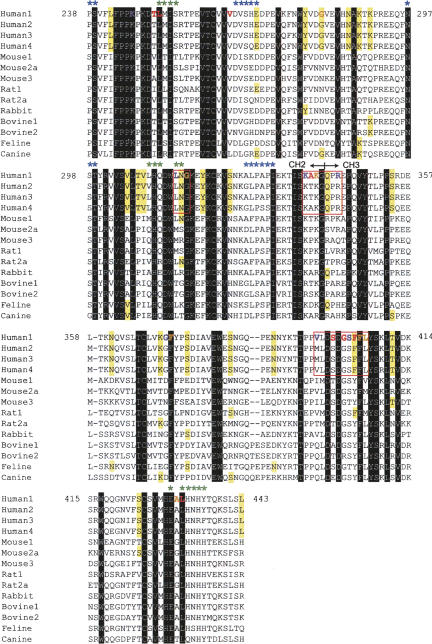
The sequences from 14 different IgG-Fc fragments from different animals or subclasses (examined above) were compared (Fig. 4). The hIgG1-Fc (between P238 and L443) shares 95% amino acid identity to hIgG2 ∼ 4 Fc fragments, and >64% identity to other animal species. It is thus, rather surprising that none of the selected aptamers bind mouse IgG1-Fc despite its 96.6% similarity to hIgG1-Fc. The multiple sequence alignment highlights several amino acids that are unique to, or favored by hIgG1 through hIgG4 (residues that are found, at most, twice in 10 nonhuman IgG Fc sequences are marked in Fig. 4). Of 24 of these unique residues, G281 and F404 show an NMR chemical shift perturbation upon binding to the aptamer (described below). Therefore, we reason that multiple amino acids, including those unique to human Fc, might synergistically contribute to the specific interaction with the aptamer (discussed shortly).
FIGURE 4.
Sequence alignments of selected IgG Fc fragments and amino acids involved in binding to the aptamer, protein A, and FcγR. Multiple sequences of Fc domains from different organisms and subclasses were aligned with GENETYX Ver. 8 software. Fc species: Human1, human IgG1 (P238-L443, Accession no. BC006402); Human2, human IgG2 (AF449616); Human3, human IgG3 (BC033178); Human4, human IgG4 (BC025985); Mouse1, mouse IgG1 (AB097849); Mouse2a, mouse IgG2a (AB097847); Mouse3, mouse IgG3 (D14625); Rat1, rat IgG1 (BC107452); Rat2a, rat IgG2a (BC088240); Rabbit, rabbit IgG (N00008); Bovine1, bovine IgG1 (S82409); Bovine2, bovine IgG2 (S82407); Feline, feline IgG (AB016711); and Canine, canine IgG (AF354264). Amino acids conserved in all of the sequences are highlighted in black, and those conserved in human IgG subclasses, but rarely in other species (at most two of 10 species), are highlighted in yellow. Amino acids that showed NMR chemical shifts upon binding to the aptamer are indicated in red for large shifts, orange for medium shifts, and purple for perturbed residues that were not quantitatively assigned. Three amino acid clusters, sites 1–3, which are suggested as the primary binding sites for the aptamer, are outlined in red squares (see text for explanation). The binding sites of protein A and FcγR are marked by green and blue asterisks, respectively. The CH2–CH3 domain junction is also indicated.
SPR competition experiments of the aptamer with protein A and Fcγ receptor
The Fc portion of IgG-coated antigens (immune complexes) binds the receptor protein (FcγR) on the surface of macrophages or monocytes, and initiates a series of signaling events that result in the phagocytosis/destruction of the immune complex. The binding site of FcγR on Fc is distinct from that of Apt7 since the SPR signal of an Apt7-immobilized sensor chip increased drastically upon the addition of hIgG1-FcγR (50 kDa) complexes (Fig. 2B). This is in contrast to what was observed with known anti-IgE aptamers, which block IgE binding to FcεR (Wiegand et al. 1996). On the other hand, the SPR signal only slightly increased, if at all, upon the addition of hIgG1-protein A (46 kDa) complexes, far below the level theoretically expected (Fig. 2B). Therefore, the aptamer-binding site on Fc does not overlap with FcγR, but rather overlaps, at least in part, with protein A. These findings are consistent with the known separate binding sites of FcγR and protein A on the tertiary structure of Fc (Deisenhofer 1981; Kato et al. 2000; Sondermann et al. 2000) (shown below in Fig. 6B).
FIGURE 6.
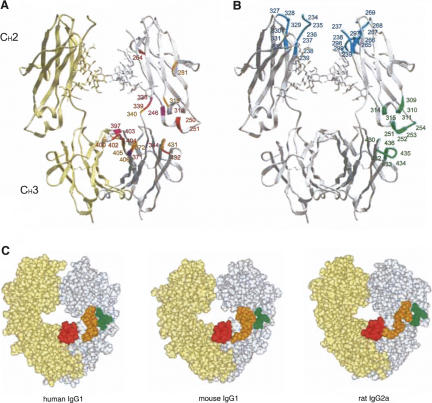
The structural organization of the Fc domain of human IgG1 (PDB code: 2DTQ) (Yamaguchi et al. 2006) and the interface of the interaction with the aptamer, protein A, and FcγR. Shown are the CH2 and CH3 domains. (A) Spatial localization of amino acids perturbed upon addition of AptNMR. Amino acids that showed NMR chemical shifts upon binding to the aptamer are indicated in red for large shifts, orange for medium shifts, and purple for residues that were perturbed but not quantitatively assigned due to overlap or line broadening of the signals. The assigned residues are marked only on one strand of the CH2 and CH3 domains for clarity. (B) Amino acids responsible for binding FcγR and protein A are shown in cyan and green, respectively (Deisenhofer 1981; Kato et al. 2000; Sondermann et al. 2000). (C) Space-filling model of Fc structures of human IgG1 (left), mouse IgG1 (middle; PDB code 1IGY) (Harris et al. 1998), and rat IgG2 (right; PDB code 1I1C) (Martin et al. 2001). Shown by different colors in one strand of Fc are the three sites in human IgG1-Fc responsible for the specific interaction with the aptamer (indicated also in Fig. 4) and their equivalent sites in the mouse and rat Fc: site 1, green; site 2, orange; site 3, red. The diagrams were created with the program ICM (MolSoft).
NMR probing of the aptamer-binding site on Fc
To identify the aptamer-binding site on the Fc fragment, we performed titration experiments using 1H-15N heteronuclear single quantum coherence (HSQC) spectra of NMR using an Apt8-2 derivative, AptNMR (see Fig. 1). Several chemical-shift changes were observed upon titration of Fc with AptNMR (Fig. 5). For example, the T250 resonance completely disappears from its original position and appears at another position in the spectrum when a two molar equivalent of aptamer (to Fc) is added, suggesting a 1:2 (Fc:aptamer) stoichiometric interaction.
FIGURE 5.
Superposition of the 1H-15N HSQC spectra of the 15N-G, I, L, K, T, V, A, M, C, H, W, Y, F, S-labeled Fc fragment (black) and that of the aptamer-bound Fc fragment (red). Relevant amino acid positions are assigned.
These chemical shifts were quantified for each residue according to the equation [((ΔHN)2 + (ΔN/5)2)/2]1/2 (Pellecchia et al. 1999), where ΔHN and ΔN represent the difference in proton and nitrogen chemical-shift changes between the free and the aptamer-bound forms. Large chemical shifts ([((ΔHN)2 + (ΔN/5)2)/2]1/2 > 0.06 ppm) were observed for T250, L251, V264, A339, S400, G402, F404, and L432 (marked in red in Fig. 4), while medium chemical shifts (0.06 > [((ΔHN)2 + (ΔN/5)2)/2]1/2 > 0.04 ppm) were observed for G281, G316, K340, F372, F405, L406, and A431(marked in orange in Fig. 4). Although the chemical-shift changes for K246, L314, K338, R344, G371, V397, and S403 (marked in purple in Fig. 4) were not quantitatively assigned due to overlapping or line broadening of the signals, these signals were obviously perturbed upon binding to the aptamer.
The amino acids showing chemical-shift changes are mapped on the crystal structure of human IgG1-Fc (marked by red in Fig. 6A) and on the amino acid sequence (Fig. 4). These residues cluster on the CH3 domain and the boundary between CH2 and CH3, which is the known binding site of protein A (marked by green in Fig. 6B). Of these protein A-interacting residues, signals from L251, L314, and L432 were perturbed by the aptamer. In contrast, the FcγR-binding site is clearly separated from the aptamer-binding site (marked by blue in Fig. 6B). These findings are consistent with the SPR study on protein A and FcγR binding to immobilized Apt8. Interestingly, the extensive surface of the CH3 domain seems to be involved in aptamer binding. This mode of interaction is not known with natural ligands of IgG.
Aptamer-based affinity purification of IgG and therapeutic antibodies under neutral elution conditions
Apt7, Apt8-2, and its two variants bearing minor modifications (see legend of Fig. 7) were immobilized to resin by amino coupling at 5′ or 3′ termini to test the affinity purification of IgG from human sera. Aptamer–IgG complexes were pulled down, washed, and bound IgGs were eluted with neutral buffer EB1 (200 mM KCl, 10 mM EDTA and 10 mM Tris•HCl, pH 7.6). The purity of IgG was investigated by SDS polyacrylamide gel electrophoresis (SDS-PAGE), which was equivalent to that achieved by currently available protein A resin (Fig. 7A). With the 3′ amino-conjugated aptamer purified by HPLC, which was immobilized at about 1 μmol/mL-gel, IgG was purified at about 30 mg/mL-gel. The capacity of protein A resin was also about 30 mg/mL-gel. Consistent with the SPR analysis, the aptamer resin failed to pulldown IgG from mouse sera (data not shown). Efficient elution was reproducibly confirmed with several neutral buffers containing 10 mM EDTA or 0.2 M KCl (see Fig. 7B), but not with 1 M NaCl, 1 M urea, or pH 3 citrate (see Fig. 7C). Aptamer resin can be regenerated multiple times without significant loss of activity by washing with 6 M urea (at room temperature or 65°C), or by heating at 85°C for 5 min in pure water (data not shown).
FIGURE 7.
Purification of IgG with Fc-aptamer resin. (A) SDS-PAGE of affinity purified IgG from human sera. The same amount of serum was applied to each affinity resin, and bound IgG was either eluted with EB1 in lanes 1–4, or solubilized by SDS-sample buffer in lane 5. (Note that elution with pH 3 citrate from rProtein A resin gives rise to essentially the same pattern as lane 5.) Lane M, molecular marker; lane 1, Apt8-2; lane 2, Apt8 variant containing 2′-O-methyl nucleotides at positions G1, G2, G12, A13, A14, A15, C22, and C23; lane 3, Apt8-2 variant containing 2′-O-methyl nucleotides at position U9; lane 4, Apt7; lane 5, rProtein A; lane 6, hIgG1 (standard); lane 7, human serum. The purity was calculated as a percentage of total stain using ImagJ. (B) Buffer conditions that efficiently elute IgG from aptamer resin. Human sera was applied to Apt8-2 affinity resin and bound hIgG was recovered using neutral elution buffers: lane 1, EB1; lane 2, EB2 (200 mM KCl and 10 mM Tris•HCl, pH 7.6); lane 3, EB3 (10 mM EDTA, 300 mM NaCl, and 10 mM Tris•HCl, pH 7.6); lane 4, EB4 (10 mM EDTA and 10 mM Tris•HCl, pH 7.6). Under each of these conditions, it was reproducibly confirmed that IgG could be efficiently recovered from sera with >80% purity. ( C ) Buffer conditions that did not efficiently elute IgG from aptamer resin. IgG released from (lanes 1–4) or remained bound (lanes 5–8) to the Apt7 resin was analyzed by SDS-PAGE after heating in SDS sample buffer. Elution buffers: lanes 1 and 5, 1 M NaCl; lanes 2 and 6, 1 M KCl (under this condition IgG was lost as aggregates); lanes 3 and 7, 1 M Urea; lanes 4 and 8, pH 3 citrate. (D) CamPath (anti-CD52 humanized antibody) was purified from a CHO cell supernatant with Apt8-2 affinity resin using EB2 buffer, and subjected to HPLC chromatography with a SE-HPLC column (G3000swxl, TOSOH).
Antibodies for therapeutic use are either chimeric or humanized. We examined three therapeutic antibodies, Remicade (Infliximab, FDA-approved chimeric antibody against TNFα), humanized anti-CD26 (in clinical trials), and CamPath (Alemtuzumab, FDA-approved humanized antibody against CD52), for their affinity to the aptamer. Pure antibody preparations of Remicade and anti-CD26 bound strongly to the Apt8-2 immobilized sensor chip of SPR, and little-bound Remicade could be removed, even with 1 M NaCl solution (Fig. 2C,D). CamPath was supplied as a harvested cell (CHO) culture fluid. The IgG fraction was affinity purified with the Apt8-2 resin, eluted with neutral buffer containing 300 mM KCl and 10 mM Tris•HCl, pH 7.6, and was analyzed by high-performance liquid chromatography (HPLC). The data clearly indicate that the CamPath antibody can be purified to near homogeneity from the CHO culture fluid (Fig. 7D). Additionally, the aptamer can bind to Fc fusion proteins such as CD28 and RANK, which are potential therapeutics (data not shown).
DISCUSSION
This is the first report of an aptamer to human IgG Fc, as well as of an application of aptamer to process-scale production of therapeutic antibodies. The optimized aptamer, Apt8-2, is 23-nt long and contains four 2′-fluoro U/C and 13 2′-deoxy nucleotides. NMR chemical-shift analysis indicated that two aptamer molecules bind to one IgG-Fc. Reselection and site-directed substitution studies clearly highlighted the requirement of the GGUGCU bulging motif for high affinity to hIgG-Fc. Importantly, the central U of this motif (i.e., U6 from the 5′ end of Apt8-2) must be 2′-fluoro, since any other base or 2′ modifications could not substitute functionally for the 2′-fluoro U6. Thus, the 2′-fluorine of U6 likely plays a crucial role in forming or stabilizing a structure that fits Fc via hydrogen bonding to amino acid(s) or other chemical interactions. Other positions were less stringent, with substitutions or modifications more generally accepted. Nucleotides outside of the GGUGCU motif are also somewhat replaceable, such as in the stem or loop region, as long as the predicted secondary structure is similar to Apt7. Accordingly, a GAAA tetra loop was substituted into Apt7, giving rise to the shortest aptamer, Apt8, which is appropriate for chemical synthesis. It is anticipated that the cost of RNA production will be greatly reduced in a few years, which will be important to make process-scale production of aptamer-based ligands more cost effective.
Acid elution of bound IgG is one of the key disadvantages of protein A resin for manufacturing therapeutic antibodies, since some antibodies are known to adopt an inactive conformation (Tsumoto et al. 2004; Ghose et al. 2005; Cromwell et al. 2006). In this study, we showed that the Apt8-2 affinity resin is able to purify IgG from human sera at equivalent efficacy and purity to protein A resin. Importantly, the Apt8-2 resin is able to release bound IgG under neutral conditions using an EDTA- or 0.2 M KCl-based elution buffer. EDTA is a chelating agent and probably alters the three-dimensional structure of the aptamer by removing divalent metal ions Mg2+ and Ca2+, thereby eluting Fc. Interestingly, monovalent cations, K+ and Na+, have opposite effects on affinity. Bound IgG is released with 0.2 M and 1 M KCl, but not with 0.145 M and 1 M NaCl (see Fig. 7). We assume that the proper combination of monovalent and divalent cations is a prerequisite for the active conformation of aptamer, and an excess of K+ leads to inactive conformation.
The SPR analysis suggests that Apt8-2 does not bind Fc at the same site as FcγR binding, but rather binds at the site of protein A binding. Previous crystallographic and NMR studies have shown that protein A binds at the boundary between CH2 and CH3 of hIgG-Fc, while FcγR binds at the hinge region of IgG-Fc (see Fig. 6B) (Deisenhofer 1981; Kato et al. 2000; Sondermann et al. 2000). Consistently, the current NMR study revealed that the aptamer binding caused a perturbation of the signals from L251, L314, and L432, which are known to interact with protein A and are apart from the FcγR-binding site (Fig. 6A). Importantly, the predicted aptamer-binding area on hIgG-Fc seems to be broader than the protein A-binding area. This might account, at least in part, for the aptamer's high and specific affinity to hIgG-Fc relative to protein A. The chemical-shift analysis highlights three primary sites involved in the interaction, i.e., L314-G316 (site 1), K338-R344 (site 2), and V397-L406 (site 3) (marked by red squares in Fig. 4). Most of the amino acids that did not show a chemical shift within these sites are probably due to non-15N-labeling of the relevant residues (Q, E, D, N, P). These three sites are located on the surface of the Fc domain, and their structural organizations are not identical between human Fc and other species (Fig. 6C). We speculate that the aptamer discriminates primarily site 2, since it is topologically most different among different species IgGs. Moreover, of these, N315, Q342, and F404 are relatively unique to human Fc (Fig. 4). These findings indicate that the aptamer recognizes both the structure and residues, thereby achieving high specificity to human Fc.
In sum, RNA aptamer has the potent ability to achieve not only high affinity but also hyperspecificity to target protein. The IgG-Fc aptamer-conjugated resin showed high performance in the antibody purification. The purity and efficiency are comparable to those with protein A resin. Importantly, antibody can be purified under neutral condition, unlike protein A resin, which requires acidic elution. Therefore, the anti-Fc RNA aptamer developed in this study could overcome several disadvantages of protein A resin and certainly provides us with a novel opportunity to develop a protein A alternative for the mass production of therapeutic antibodies.
MATERIALS AND METHODS
Materials
Human IgG1 through IgG4 were purchased from Calbiochem; mouse IgG1 and IgG2a, human serum, and mouse serum from Chemicon; mouse IgG2b, rat IgG2a, rat IgG2b, and rabbit IgG from Zymed Laboratories; mouse IgG3, bovine IgG1, bovine IgG2, and cat IgG from Bethyl; rat IgG1, RANK-Fc fusion protein (hIgG1-Fc Pro100-Lys330) and CD28-Fc fusion protein from R&D Systems; IgGs from chicken and dog from Rockland; rat IgG2c from UK-Seratec; guinea pig IgG from Biogenesis; Remicade (Infliximab, 10 mg/mL) from Centocor B.V. Anti-CD26 humanized antibody was a kind gift from Chikao Morimoto (Institute of Medical Science, University of Tokyo), and CHO cell culture fluid containing CamPath (Alemtuzumab, anti-CD52 humanized antibody) was a kind gift from G. Hale and H. Waldmann (the Therapeutic Antibody Center, Oxford). rProtein A-conjugated resin and NHS-activated Sepharose 4 resin were purchased from Amersham Bioscience, Tresyl-TOYOPEARL resin from TOSOH, Ni-NTA affinity resin from Qiagen, and BD TalonRT affinity resin from BD science. RNA was prepared either by in vitro transcription using a DuraScribe T7 Transcription kit (Epicentre) according to the manufacturer's instructions or by chemical synthesis (Gene Design, Inc.).
Selection-amplification of hIgG aptamers
The preparation of a pool of 5 × 1014 different RNA molecules randomized over 40 nucleotides (40N RNA pool) and affinity RNA selection amplification was carried out essentially as described previously (Oguro et al. 2003; Miyakawa et al. 2006) with the exception of the target protein being histidine-tagged RANK-Fc fusion protein. The sequence of the first template was 5′-ctctcatgtcggccgtta-(40N)-cgtccattgtgtccctatagtgagtcgtatta-3′, and the primers used were 5′-taatacgactcactatagggacacaatggacg-3′ and 5′-ctctcatgtcggccgtta-3′. The DuraScribe T7 Transcription kit produced RNA containing 2′-fluoro pyrimidine nucleotides. The binding buffer used for selection was 145 mM sodium chloride, 5.4 mM potassium chloride, 0.8 mM magnesium chloride, 1.8 mM calcium chloride, and 20 mM Tris•HCl (pH 7.6). The target protein was immobilized on Ni-NTA affinity resin (Qiagen) or BD TalonRT affinity resin (BD Science), and protein–RNA complexes were released with 100 mM imidazole. After 10 rounds of selection, aptamer cDNAs were cloned into pGEM-T Easy vector (Promega) and sequenced. Optimizing SELEX at the GGUGCU bulge sequence was conducted with 2′-fluoro C/U using apt8 as the base template, 5′-tgtcggccgttacagttccggtttcccgg-(6N)-tgtaactcgtccattgtccc-3′, where 6N was six random sequences instead of GGUGCU, and primers 5′-taatacgactcactatagggacaatggacgagttac-3′ and 5′-tgtcggccgttacagttc-3′. A total of 40 μg of hIgG1 were immobilized in 40 μL of NHS-activated Sepharose resin.
Surface plasmon resonance assay
The SPR assays were performed essentially as described previously using the BIAcore 2000 instrument (BIAcore) (Oguro et al. 2003; Miyakawa et al. 2006). The aptamer templates were amplified and tagged at the 3′-end with dT16 by PCR using 5′-template primer (5′-taatacgactcactatagggacacaatggacg-3′) and dT16 tagging 3′-template primer (5′-ttttttttttttttttctctcatgtcggccgtta-3′). These DNA templates were then transcribed to poly(A)-tailed RNAs. A 5′-biotinylated dT16 oligomer was bound to the surface of the streptavidin sensor chip (BIAcore) of flow cells 1 and 2. The poly(A)-tailed RNA was immobilized to about 1000 RUs in flow cell 2 by complementary hybridization to the dT16 oligomer. IgG solution (of different concentration between 500 and 12.5 nM) was injected to the flow cells 1 and 2 of the sensor chip. Data was obtained by subtracting the flow cell 1 data from the flow cell 2 data, thereby showing the net interaction between RNA and protein. To regenerate the sensor chip, bound material was completely removed by injecting 6 M urea. Kinetic constants were estimated by using BIAevaluation 3.0 software (BIAcore) (Nakajima et al. 2001).
NMR measurements
AptNMR (see Fig. 1) was purified by denaturing PAGE and extensive desalting. The aptamer was then annealed by heating for 5 min at 95°C followed by snap-cooling on ice, and was dissolved in 5 mM sodium phosphate (pH 6.0) containing 50 mM NaCl and 5 mM MgCl2 in 10% D2O. The final concentration of the aptamer was 3.2 mM. The Fc fragments of amino acids selectively labeled human IgG1s (15N-G, I, L, K, T, V, A, M, C, H, W, Y, F, S-label, 15N-V, W, R-label, and 15N-K-label) were prepared as described previously (Yamaguchi et al. 2006; Matsumiya et al. 2007). The Fc fragment was dissolved in 5 mM sodium phosphate (pH 6.0) containing 50 mM NaCl and 5 mM MgCl2 in 10% D2O. The final concentration of the Fc fragment was 0.32 mM. The two-dimensional 1H-15N HSQC spectra (Bodenhausen and Ruben 1980) were measured on a Bruker DMX-500 spectrometer equipped with a 5-mm triple-resonance cryo-probe. The probe temperature was set to 37°C or 52°C. 1H chemical shifts were given from external sodium 2,2-dimetyl-2-silapentane-5- sulfonate, while 15N chemical shifts were from liquid ammonia at 21.6 ppm. Spectral assignments have been described previously (Yamaguchi et al. 2006). The concentrated aptamer (3.2 mM) was added stepwise to the 0.32 mM Fc fragment and the titration was monitored by collecting two-dimensional 1H-15N HSQC spectra at each titration point. The final protein-to-RNA ratio was 1:2.
Pulldown experiment
Amino-modified aptamers were immobilized to Tresyl TOYOPEAL resin or NHS-activated Sepharose 4 Fast Flow according to the manufacturer's instructions. An amino group was attached to the aptamer's 5′ or 3′ end through a C18 linker. The ligand density was about 1 μmol/mL-gel. Most purification experiments were conducted with 10 μL of resin in a PCR-tube. Several 10 μL of human serum were added to it and incubated for 15 min at room temperature. After washing the resin with the binding buffer several times, IgG was eluted with a neutral elution buffer and analyzed by SDS-PAGE. The aptamer resin was regenerated by washing with 6 M urea at room temperature or 65°C for 15 min, or heating at 85°C for 5 min with pure water. For comparison, IgG precipitation with rProtein A resin was performed with a similar method as shown above except that bound IgG was eluted with pH 3 citrate or solubilized with SDS-sample buffer by heating at 65°C for 15 min. Proteins were analyzed by SDS-PAGE and the purity was calculated using ImageJ (Abramoff et al. 2004).
ACKNOWLEDGMENTS
We thank C. Morimoto for the anti-CD26 humanized antibody; G. Hale and H. Waldmann for a CHO cell culture fluid containing CamPath; and Kyowa Hakko Kogyo Co., Ltd. for KM3060 cells producing a monoclonal human IgG1 antibody for the NMR study. We thank S. Ohuchi and all members of Ribomic, Inc. for discussion; and C.G. Crist, S.T. Cload, and C. Wilson for critical reading of the manuscript and valuable comments. This work was supported in part by grants from the following: The Ministry of Education, Sports, Culture, Science and Technology of Japan (MEXT); the Program for Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation (NIBIO); and Core Research for Evolution Science and Technology (CREST) grants (K.K. and Y.N.) from the Japan Science and Technology Agency.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1005808.
REFERENCES
- Abramoff M.D., Magelhaes P.J., Ram S.J. Image processing with imageJ. Biophotonics Int. 2004;11:36–42. [Google Scholar]
- Blank M., Weinschenk T., Priemer M., Schluesener H. Systematic evolution of a DNA aptamer binding to rat brain tumor microvessels. Selective targeting of endothelial regulatory protein pigpen. J. Biol. Chem. 2001;276:16464–16468. doi: 10.1074/jbc.M100347200. [DOI] [PubMed] [Google Scholar]
- Bodenhausen G., Ruben D.J. Natural abundance nitrogen-15NMR by enhanced heteronuclear spectroscopy. Chem. Phys. Lett. 1980;69:185–189. [Google Scholar]
- Cromwell M.E.M., Hilario E., Jacobson F. Protein aggregation and bioprocessing. AAPS J. 2006;8:E572–E579. doi: 10.1208/aapsj080366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisenhofer J. Crystallographic refinement and atomic models of a human Fc fragment and its complex with fragment B of protein A from Staphylococcus aureus at 2.9 and 2.8 Å resolution. Biochemistry. 1981;20:2361–2370. [PubMed] [Google Scholar]
- Elkak A., Vjiayalakshmi M.A. Study of the separation of mouse monoclonal-antibodies by pseudobioaffnity chromatography using matrix-linked histidine and histamine. J. Chromatogr. 1991;570:29–41. doi: 10.1016/0378-4347(91)80198-l. [DOI] [PubMed] [Google Scholar]
- Ellington A.D., Szostak J.W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- Ellington A.D., Szostak J.W. Selection in vitro of single stranded DNA molecules that fold into specific ligand-binding structures. Nature. 1992;355:850–852. doi: 10.1038/355850a0. [DOI] [PubMed] [Google Scholar]
- Fahrner R.L., Knudsen H.L., Basey C.D., Galan W., Feuerhelm D., Vanderlaan M., Blank G.S. Industrial purification of pharmaceutical antibodies: Development, operation, and validation of chromatography processes. Biotechnol. Genet. Eng. Rev. 2001;18:301–327. doi: 10.1080/02648725.2001.10648017. [DOI] [PubMed] [Google Scholar]
- Fassina G., Ruvo M., Palombo G., Verdoliva A., Marino M. Novel ligands for the affinity-chromatographic purification of antibodies. J. Biochem. Biophys. Methods. 2001;49:481–490. doi: 10.1016/s0165-022x(01)00215-9. [DOI] [PubMed] [Google Scholar]
- Gagnon P. Purification tools for monoclonal antibodies. Validated Biosystems; Tucson, AZ: 1996. [Google Scholar]
- Gesteland R.F., Cech T.R., Atkins J.F. Cold Spring Harbor Laboratory Press; New York: 1999. The RNA world. [Google Scholar]
- Ghose S., Allen M., Hubbard B., Brooks C., Cramer S.M. Antibody variable region interactions with Protein A: Implications for the development of generic purification processes. Biotechnol. Bioeng. 2005;92:665–673. doi: 10.1002/bit.20729. [DOI] [PubMed] [Google Scholar]
- Harris L.J., Skaletsky E., McPherson A. Crystallographic structure of an intact IgG1 monoclonal antibody. J. Mol. Biol. 1998;275:861–872. doi: 10.1006/jmbi.1997.1508. [DOI] [PubMed] [Google Scholar]
- Huse K., Bohme H.-J., Scholz G.H. Purification of antibodies by affinity chromatography. J. Biochem. Biophys. Methods. 2002;51:217–231. doi: 10.1016/s0165-022x(02)00017-9. [DOI] [PubMed] [Google Scholar]
- Kato K., Sautes-Fridman C., Yamada W., Kobayashi K., Uchiyama S., Kim H., Enokizono J., Galenha A., Kobayashi Y., Fridman W.H., et al. Structural basis of the interaction between IgG and Fcγ receptors. J. Mol. Biol. 2000;295:213–224. doi: 10.1006/jmbi.1999.3351. [DOI] [PubMed] [Google Scholar]
- Keefe A.D., Schaub R.G. Aptamers as candidate therapeutics for cardiovascular indications. Curr. Opin. Pharmacol. 2008;8:1–6. doi: 10.1016/j.coph.2007.12.005. [DOI] [PubMed] [Google Scholar]
- Klussmann S. The aptamer handbook. WILEY-VCH; Weinheim, Germany: 2006. [Google Scholar]
- Martin W.L., West A.P., Jr, Gan L., Bjorkman P.J. Crystal structure at 2.8 Å of an FcRn/heterodimeric Fc complex: Mechanism of pH-dependent binding. Mol. Cell. 2001;7:867–877. doi: 10.1016/s1097-2765(01)00230-1. [DOI] [PubMed] [Google Scholar]
- Matsumiya S., Yamaguchi Y., Saito J., Nagano M., Sasakawa H., Otaki S., Satoh M., Shitara K., Kato K. Structural comparison of fucosylated and nonfucosylated fc fragments of human immunoglobulin g1. J. Mol. Biol. 2007;368:767–779. doi: 10.1016/j.jmb.2007.02.034. [DOI] [PubMed] [Google Scholar]
- Miyakawa S., Oguro A., Ohtsu T., Imataka H., Sonenberg N., Nakamura Y. RNA aptamers to mammalian initiation factor 4G inhibit cap-dependent translation by blocking the formation of initiation factor complexes. RNA. 2006;12:1825–1834. doi: 10.1261/rna.2169406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima H., Kiyokawa N., Katagiri Y.U., Taguchi T., Suzuki T., Sekino T., Mimori K., Ebata T., Saito M., Nakao H., et al. Kinetic analysis of binding between Shiga toxin and receptor glycolipid Gb3Cer by surface plasmon resonance. J. Biol. Chem. 2001;276:42915–42922. doi: 10.1074/jbc.M106015200. [DOI] [PubMed] [Google Scholar]
- Nakamura Y. Molecular mimicry between protein and tRNA. J. Mol. Evol. 2001;53:282–289. doi: 10.1007/s002390010218. [DOI] [PubMed] [Google Scholar]
- Ng E.W., Shima D.T., Calias P., Cunningham E.T., Jr, Guyer D.R., Adamis A.P. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat. Rev. Drug Discov. 2006;5:123–132. doi: 10.1038/nrd1955. [DOI] [PubMed] [Google Scholar]
- Ngo T.T., Khatter N. Chemistry and preparation of affinity ligands useful in immunoglobulin isolation and serum-protein separation. J. Chromatogr. 1990;510:281–291. doi: 10.1016/s0021-9673(01)93762-8. [DOI] [PubMed] [Google Scholar]
- Oguro A., Ohtsu T., Svitkin Y.V., Sonenberg N., Nakamura Y. RNA aptamers to initiation factor 4A helicase hinder cap-dependent translation by blocking ATP hydrolysis. RNA. 2003;9:394–407. doi: 10.1261/rna.2161303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohuchi S.P., Ohtsu T., Nakamura Y. Selection of RNA aptamers against recombinant transforming growth factor-β type III receptor displayed on cell surface. Biochimie. 2006;88:897–904. doi: 10.1016/j.biochi.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Pellecchia M., Sebbel P., Hermanns U., Wuthrich K., Glockshuber R. Pilus chaperone FimC-adhesin FimH interactions mapped by TROSY-NMR. Nat. Struct. Biol. 1999;6:336–339. doi: 10.1038/7573. [DOI] [PubMed] [Google Scholar]
- Porath J., Maisano F., Belew M. Thiophilic adsorption—a new method for protein fractionation. FEBS Lett. 1985;185:306–310. doi: 10.1016/0014-5793(85)80928-5. [DOI] [PubMed] [Google Scholar]
- Romig T.S., Bell C., Drolet D.W. Aptamer affinity chromatography: Combinatorial chemistry applied to protein purification. J. Chromatogr. B Biomed. Sci. Appl. 1999;731:275–284. [PubMed] [Google Scholar]
- Sondermann P., Huber R., Oosthuizen V., Jacob U. The 3.2 Å crystal structure of the human IgG1 Fc fragment–FcγRIII complex. Nature. 2000;406:267–273. doi: 10.1038/35018508. [DOI] [PubMed] [Google Scholar]
- Tsumoto K., Umetsu M., Kumagai I., Ejima D., Philo J.S., Arakawa T. Role of arginine in protein refolding, solubilization, and purification. Biotechnol. Prog. 2004;20:1301–1308. doi: 10.1021/bp0498793. [DOI] [PubMed] [Google Scholar]
- Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- Wiegand T.W., Williams P.B., Dreskin S.C., Jouvin M.H., Kinet J.P., Tasset D. High affinity oligonucleotide ligands to human IgE inhibit binding to Fc ε receptor 1. J. Immunol. 1996;157:221–230. [PubMed] [Google Scholar]
- Yamaguchi Y., Nishimura M., Nagano M., Yagi H., Sasakawa H., Uchida K., Shitara K., Kato K. Glycoform-dependent conformational alteration of the Fc region of human immunoglobulin G1 as revealed by NMR spectroscopy. Biochim. Biophys. Acta. 2006;1760:693–700. doi: 10.1016/j.bbagen.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Zhou B., Wang B. Pegaptanib for the treatment of age-related macular degeneration. Exp. Eye Res. 2006;83:615–619. doi: 10.1016/j.exer.2006.02.010. [DOI] [PubMed] [Google Scholar]
- Zuker M. MFOLD web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]