Abstract
RNase E is an endoribonuclease that has been studied primarily in Escherichia coli, where it is prominently involved in the processing and degradation of RNA. Homologs of bacterial RNase E are encoded in the nuclear genome of higher plants. RNA degradation in the chloroplast, an organelle that originated from a prokaryote similar to cyanobacteria, occurs via the polyadenylation-assisted degradation pathway. In E. coli, this process is probably initiated with the removal of 5′-end phosphates followed by endonucleolytic cleavage by RNase E. The plant homolog has been proposed to function in a similar way in the chloroplast. Here we show that RNase E of Arabidopsis is located in the soluble fraction of the chloroplast as a high molecular weight complex. In order to characterize its endonucleolytic activity, Arabidopsis RNase E was expressed in bacteria and analyzed. Similar to its E. coli counterpart, the endonucleolytic activity of the Arabidopsis enzyme depends on the number of phosphates at the 5′ end, is inhibited by structured RNA, and preferentially cleaves A/U-rich sequences. The enzyme forms an oligomeric complex of ∼680 kDa. The chloroplast localization and the similarity in the two enzymes' characteristics suggest that plant RNase E participates in the initial endonucleolytic cleavage of the polyadenylation-stimulated RNA degradation process in the chloroplast, perhaps in collaboration with the two other chloroplast endonucleases, RNase J and CSP41.
Keywords: RNA degradation, endoribonuclease, Arabidopsis, chloroplast
INTRODUCTION
RNA degradation is a complex, highly regulated, multistep process for which several pathways and mechanisms have been described in eukaryotes, bacteria, and organelles (Deutscher 2006; Houseley et al. 2006; Garneau et al. 2007). The polyadenylation-assisted RNA degradation pathway is considered the general mechanism of RNA degradation in prokaryotes and organelles, and was also recently described for nuclear-encoded transcripts of yeast and human cells (Dreyfus and Regnier 2002; Kushner 2004; Deutscher 2006; Houseley et al. 2006; Slomovic et al. 2006; Vanacova and Stef 2007). It begins with an endonucleolytic cleavage, followed by exonucleolytic degradation. Alternatively, the cleavage products are polyadenylated and then rapidly degraded by a number of exoribonucleases (Kushner 2002; Houseley et al. 2006; Slomovic et al. 2006). Therefore, the initial endonucleolytic cleavage may constitute a key step in the mRNA decay process in prokaryotes (Cohen and McDowall 1997; Kushner 2002). In addition, the removal of phosphates located at the 5′ end of the transcript was recently proposed to be the step that leads to the endonucleolytic cleavage stage (Celesnik et al. 2007).
RNase E was discovered in Escherichia coli as an rRNA maturation enzyme (Ghora and Apirion 1978) and was later shown to be involved in the processing of numerous other RNAs, such as the antisense regulator of E. coli plasmid replication, RNAI; the precursor of M1 RNA, which is the catalytic subunit of the RNase P; tRNAs; and small noncoding regulatory RNAs and their targets (Kaberdin et al. 1996; Li and Deutscher 2002; Ow and Kushner 2002; Afonyushkin et al. 2005; Morita et al. 2005; Udekwu et al. 2005). In addition, RNase E alters the stability of total RNA molecules and of numerous specific transcripts (Ono and Kuwano 1979; Arraiano et al. 1988; Mackie 1992; Hajnsdorf et al. 1996). Moreover, the enzyme concentration in the cell is regulated by a feedback loop, in which RNase E controls the stability of its own mRNA (Diwa et al. 2000; Sousa et al. 2001; Ow et al. 2002).
The E. coli protein contains 1061 amino acids and has two distinct domains, the amino-terminal catalytic region, and the carboxy-terminal region. The latter serves as a scaffold for assembling the degradosome, a high molecular weight complex that also contains polynucleotide phosphorylase (PNPase), RNA helicase B (Rhl B), and the glycolytic enzyme enolase (Miczak et al. 1996; Py et al. 1996; Vanzo et al. 1998; Carpousis 2007). This E. coli-type degradosome complex, however, is not present in cyanobacteria or spinach chloroplasts (Kaberdin et al. 1998; Baginsky et al. 2001; Rott et al. 2003). E. coli RNase E, which is essential for cell viability, is a single-stranded, nonspecific endonuclease with a preference for cleaving A/U-rich sequences (Mackie 1992; Cohen and McDowall 1997). RNase G is another E. coli endonuclease possessing ∼50% sequence similarity to the RNase E catalytic region and overlapping, but not identical, cleavage specificity (Lee et al. 2002; Ow et al. 2003). Both enzymes were consequently combined into a newly named family of RNase E/G proteins. The cleavage activity of the family members depends on the number of phosphates located at the 5′ end: RNA containing one phosphate is a much better substrate than RNA with three phosphates (Mackie 1998; Jiang et al. 2000; Tock et al. 2000; Celesnik et al. 2007).
Genes encoding RNase E/G-like proteins, as well as ESTs, have been found in many bacteria, cyanobacteria, red and green algae, and the nuclear genomes of higher plants, but not in eukaryotes lacking chloroplasts (Lee and Cohen 2003; Bollenbach et al. 2004). The classification of RNase E-like polypeptides into several groups based on the protein's domain architecture has been proposed (Lee and Cohen 2003). In many bacteria other than E. coli, as well as in the nuclear genomes of several green algae and higher plants, only one member of the RNase E/G is encoded, and it is generally termed RNase E.
In this study, we present the first characterization of a eukaryotic RNase E-like protein. The Arabidopsis RNase E was found to be located in the chloroplast and it exhibits endoribonuclease activity similar to that of E. coli and Synechocystis RNase E proteins, including a dependence on the number of phosphates at the 5′ end. In addition, we found that the protein is active only when it is assembled into a homo-oligomer.
RESULTS
Genes encoding homologs of bacterial RNase E are present in higher plants
RNase E is an endoribonuclease found in many bacteria and some archaea. It plays an important role in the processing and degradation of RNA in E. coli. Analysis of the A. thaliana genome revealed the At2g04270 locus encoding a protein (NP_850987) showing sequence homology with the RNase E/G-like proteins of prokaryotic origin (Fig. 1A). Analysis of the N-terminal 63 amino acids revealed a canonical chloroplast transit peptide that could direct the cytoplasmically translated protein into this organelle (Emanuelsson et al. 2000). Similar proteins are also encoded by the nuclear genomes of other plants, such as rice and tomato (Fig. 1A). Interestingly, three other splice variants are annotated for the At2g04270 locus, of which one has an N terminus predicted to target the protein to the mitochondria. In this study, while performing RACE analysis to obtain the full-length cDNA, we observed only the mRNA presented in Figures 1 and 4 (see below). In addition, no mitochondrial RNase E was identified (see below) and eukaryotic homologs are encoded only in chloroplast-containing organisms. Nevertheless, we cannot exclude the possibility of certain Arabidopsis RNase E mRNA splice variants encoding mitochondrial or other non-chloroplast forms.
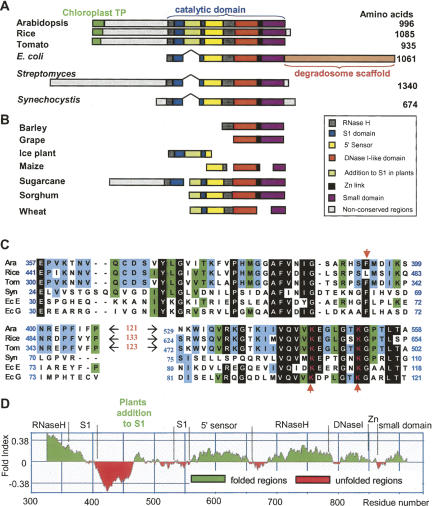
FIGURE 1.
Plant RNase E proteins. (A) The amino acid sequences of Arabidopsis, rice, and tomato RNase E homologs were aligned to those of the E. coli, Streptomyces, and Synechocystis proteins. Regions of significant homology are shown as colored boxes, with catalytic subdomains designated according to Callaghan et al. (2005a). The N terminus of the plant proteins includes sequences predicted to constitute a chloroplast transit peptide (chloroplast TP). In addition, the plant proteins contain N-terminal extensions of several hundreds of amino acids that are not homologous between the plants, as well as a stretch of about 120 amino acids residing inside the S1 domain (colored green), which is not present in any bacterial sequence. (B) The DNA sequence databank was searched for plant ESTs related to RNase E. The position of each domain in the full-length sequences is indicated. When more than one EST was found, the ESTs are shown on one line aligned to the full-length sequence of the Arabidopsis. Sequence accession numbers are as follows: Escherichia coli, P21513; Arabidopsis thaliana, NP_850987; rice (Oryza sativa), NP_001061542; tomato (Lycopersicon esculentum) (this work); Streptomyces, NP_626836; Synechocystis, NP_439978; barley (Hordeum vulgare), TC141965; grape (Vitis vinifera), TC42553; ice plant (Carpobrotus edulis), TC6253; maize (Zea mays), TC308943, TC049636, TC284127; sugarcane (Saccharum officinarum), TC51842, TC68873; sorghum (Sorghum bicolor), TC106341; wheat (Triticum aestivum), TC270983, TC271418. (C) Alignment of the amino acid sequences of the S1 domain of Arabidopsis (Ara), rice, tomato (Tom), Synechocystis (Syn), E. coli RNase E (Ec E), and E. coli RNase G (Ec G). The black background shows the mostly conserved amino acids, which are colored white. The green and blue backgrounds show locations where four or three amino acids are identical. The two conserved lysines that were mutated in this work, as well as the phenylalanine that forms part of the RNA binding surface on the S1 domain (Callaghan et al. 2005a), are marked by red arrowheads. The plant-specific addition to S1 domain is not shown in this alignment since the homology between the three plants is very low. (D) The catalytic domain of the Arabidopsis RNase E catalytic domain (amino acids 327–964 of the full-length protein) was used to draw the fold-index plot (FoldIndex) that estimates the probability of the query sequence to fold into a defined structure. Positive values represent regions likely to be folded, and negative values represent those likely to be intrinsically unfolded. The figure shows that there is a 66-residue region (from Asp428 to Val493), located in the plant-specific S1 domain insertion, which is strongly predicted to be unfolded.
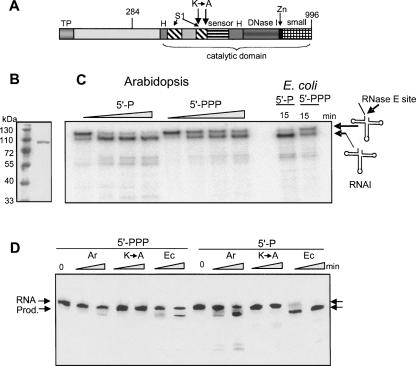
FIGURE 4.
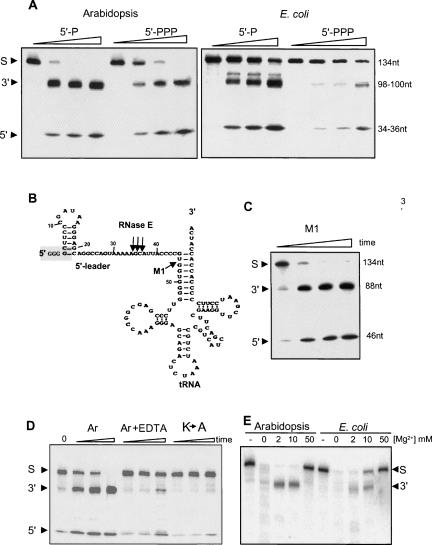
Recombinant Arabidopsis RNase E activity is similar to its bacterial counterpart. (A) Schematic representation of Arabidopsis RNase E. The different domains, as described in Figure 1, are shown: TP, predicted chloroplast transit peptide; H, RNase H domain; S1, S1 domain; sensor, 5′ sensor domain; DNase I, DNase I domain; Zn, Zn link domain; and small, small domain. The location of amino acid 284, which is the start of the truncated sequence which yields a soluble recombinant form of the protein, is indicated. (B) Following expression and purification, the recombinant protein (amino acids 284–996) was fractionated by SDS-PAGE and analyzed by silver staining. Molecular weight markers, shown on the left, were fractionated in the first lane. (C) 5′-end-dependent activity of Arabidopsis RNase E. Recombinant Arabidopsis RNase E or the catalytic part of the E. coli enzyme (amino acids 1–498) were incubated with [32P]-labeled RNA representing the E. coli RNAI harboring either a mono- (5′-P) or tri-phosphate 5′ end (5′-PPP). The enzyme:substrate ratio was 50:1. Samples were withdrawn after 0, 10, 20, and 40 min, and purified RNA was analyzed by denaturing PAGE and autoradiography. Schematic representations of the RNAI and cleaved products are shown on the right. (D) Changing lysines 546 and 552 to alanines significantly inhibited activity. The Arabidopsis RNase E (Ar), in which the two lysines located in the S1 domain were converted into alanines (see panel A), was produced in E. coli and analyzed for cleavage activity of RNAI, as described in C. The enzyme:substrate ratio was 1:1. Incubation times were 5 and 15 min. RNA, full-length RNAI; Prod., cleavage product.
Following the putative chloroplast targeting sequence, the Arabidopsis RNase E as well as the homologs in rice and tomato include a relatively long region of ∼260 amino acids showing no homology with other bacterial or plant RNase E proteins. Such an N-terminal extension is found also in the bacterium Streptomyces coelicolor, where the enzyme has been defined as a type III RNase E (Lee and Cohen 2003). The carboxyl half of the protein contains the multidomain catalytic region and is similar to E. coli RNase E both in sequence and domain architecture (Fig. 1A; Callaghan et al. 2005a). Interestingly, the S1 domain of the plant protein, which is important for RNA cleavage activity, contains an insertion of 121 nonconserved amino acids; the location of this insertion is the same for all plant RNase E proteins (Fig. 1A–C). This places plant RNase E in the type II RNase E proteins, as defined by Lee and Cohen (2003). The EST database was searched for other plant homologs and, as illustrated in Figure 1B, 11 ESTs from seven species were detected, indicating that the RNase E transcript is present in most, if not all, higher plants.
Amino acid alignment of the catalytic domains of the plant, Synechocystis, and E. coli RNase E and RNase G revealed significant homology. For example, both E. coli enzymes displayed 32% identity to the Arabidopsis protein. The alignment of the S1 domains is presented in Figure 1C. As pointed above, aside from the plant-specific insertion the high conservation is evident. Because the plant-specific addition was poorly conserved between the three plants, the fold-index tool, which predicts the likelihood of forming a defined structure (Prilusky et al. 2005), was applied to examine a possible common fold for this domain. As shown in Figure 1D, this region displayed the lowest fold-index values of the catalytic domain, predicting that a globular and defined structure is unlikely to form.
Structure of the Arabidopsis RNase E catalytic domain
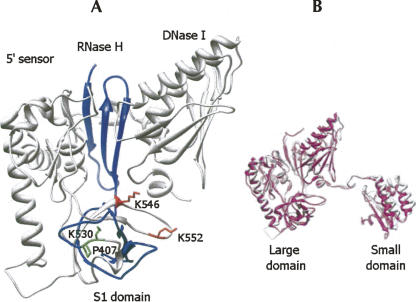
The homology of the Arabidopsis and E. coli catalytic domains enabled us to use the bacterial crystal structure to construct a homology model for the plant enzyme (Schubert et al. 2004; Callaghan et al. 2005a), after removing the 122-amino acid plant-specific addition to S1 domain (see Materials and Methods). Figure 2 presents the predicted Arabidopsis RNase E catalytic domain structure. Its very similar structure to the bacterial enzyme suggests a comparable mode of catalytic activity, which is supported by the biochemical assays described below. The two amino acids bordering the plant-specific insertion are indicated (P407 and K530). Interestingly, the corresponding region in the E. coli structure, composed of amino acids Ala80 to Gly86 (crystal numbering), displayed no defined structural information in the E. coli crystal (Schubert et al. 2004; Callaghan et al. 2005a). Moreover, it is located in the L34 region, as defined by Schubert et al. (2004), where it could readily be accommodated.
FIGURE 2.
Homology-based model of the structure of the catalytic domain. (A) An homology-based model for the catalytic region of Arabidopsis RNase E based on the E. coli X-ray crystal structure (see Materials and Methods). In order to build the model, the plant-specific S1 domain insertion was removed and residues Pro407 and Lys530 were joined together. The region of the RNase H and the S1 domains is shown as blue ribbon. The region following the plant-specific domain toward the carboxy terminus is shown as white ribbon. Residues Pro407 and Lys530, located before and after the plant-specific domain, are shown as green bars. Residues Lys546 and Lys552 (corresponding to the Lys106 and Lys112 of E. coli) are shown as red bars. The small domain of the catalytic part is not presented in this panel. (B) The modeled catalytic region of Arabidopsis RNase E (white) superimposed on the E. coli crystal structure from PDB 2bx2, chain L (pink).
Arabidopsis RNase E is localized in the chloroplast
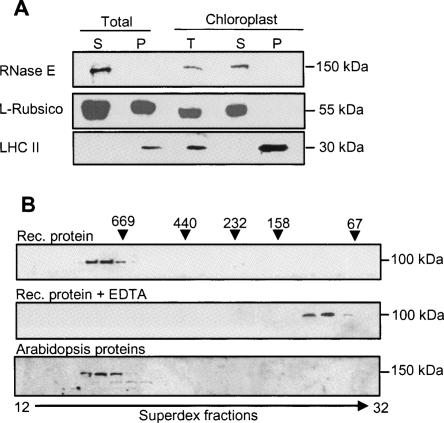
As described above, the Arabidopsis RNase E harbors an N-terminal sequence predicted to be a chloroplast transit peptide. To test whether the protein localizes to the chloroplast, a specific antibody was generated against the recombinant, bacterially produced Arabidopsis RNase E. Total Arabidopsis leaf extract as well as isolated chloroplasts were analyzed for the presence of the native protein. Even though the calculated predicted molecular weight of the native protein is 115 kDa, the protein was detected by SDS-PAGE at a higher molecular weight of 150–160 kDa (Fig. 3). A similar anomaly has been previously reported for E. coli RNase E when SDS-PAGE results were compared with the calculated molecular weight (Casaregola et al. 1992; Cormack et al. 1993). The protein was localized to the soluble fraction of the chloroplast, similar to the Rubisco chloroplast soluble control and unlike the light-harvesting complex II (LHC II) that is embedded in the thylakoids (Fig. 3A). Since the rice and tomato sequences also contain a predicted N-terminal chloroplast targeting transit peptide, chloroplast localization may be a general feature of RNase E proteins in higher plants (Fig. 1A).
FIGURE 3.
Arabidopsis RNase E is located in the chloroplast in a high molecular weight complex. (A) Total Arabidopsis protein extract (soluble [S] and insoluble [P] fractions), together with purified chloroplasts, was separated on a 10% SDS-polyacrylamide gel, transferred to a nitrocellulose membrane, and probed with polyclonal antibodies against Arabidopsis RNase E (upper panel), Rubisco large subunit (middle panel), or LHC II (lower panel). (B) Purified recombinant truncated protein (amino acids 284–996) was fractionated on a Superdex 200 size-exclusion column, and the proteins of each fraction were analyzed by immunoblot with His6-specific antibodies (Rec. protein, upper panel). In the middle panel, 10 mM EDTA was added to the purified recombinant protein prior to fractionation on the same column (Rec. protein + EDTA). (C) Total Arabidopsis proteins were fractionated on the same column, and the proteins of each fraction analyzed by immunoblot using antibodies against RNase E. The elution profile of the following molecular weight markers is indicated along the top: thyroglobulin (669 kDa), ferritin (440 kDa), catalase (232 kDa), aldolase (158 kDa), and bovine serum albumin (67 kDa).
Arabidopsis RNase E forms a high molecular weight complex
In E. coli, the active form of the enzyme is a homotetramer (Callaghan et al. 2005a; Marcaida et al. 2006; Worrall and Luisi 2007). Subunits are held together with the help of zinc ions that bind to a short stretch of amino acids called a zinc-link (Fig. 1). This complex is essential for RNase E activity and is disrupted by the addition of EDTA (Callaghan et al. 2005a,b). In E. coli and several related bacteria, RNase E (but not RNase G) forms the assembly scaffold of the degradosome, a multiprotein complex consisting, in addition to this protein, of PNPase, RNA-helicase, and enolase (Marcaida et al. 2006; Carpousis 2007).
In order to explore whether Arabidopsis RNase E forms a complex with other proteins, the truncated recombinant protein was expressed in bacteria, purified, and studied by size-exclusion chromatography. The truncated protein eluted at a high molecular weight of ∼680 kDa, suggesting an oligomeric complex (Fig. 3B). However, this size is larger than that of the tetrameric complex described in the crystallization studies of E. coli RNase E/G (Callaghan et al. 2005a; Marcaida et al. 2006; Worrall and Luisi 2007). The 680-kDa complex was resistant to a high salt concentration of up to 2 M KCl (not shown) but was disrupted by 10 mM EDTA (Fig. 3B). Next, we analyzed the size of the RNase E complex in the Arabidopsis cell. Arabidopsis protein extract was fractionated on the same size-exclusion column and native RNase E was detected immunologically (Fig. 3B). RNase E eluted at the same molecular weight as the purified recombinant protein of ∼680 kDa. Together, these results indicate that RNase E is present in an oligomeric form in the soluble fraction of the chloroplast. It should be noted, however, that the large size of the complex is close to the size exclusion limit of this column (Superdex 200). Therefore, the inclusion of other proteins in the oligomeric complex that did not significantly change the molecular weight could not be excluded. In addition, this is probably the reason why the truncated recombinant protein (amino acids 284–996) and the full-length one present in the leaf extract eluted in the same fractions very close to the void volume (Fig. 3B).
Arabidopsis RNase E displays endoribonucleolytic activity similar to that of its E. coli counterpart
In order to analyze whether the Arabidopsis RNase E protein is active as an endoribonuclease, the protein was expressed in bacteria and purified. We found that the full-length protein could not be obtained in a soluble form. However, a truncated protein including amino acids 284–996 was soluble in bacteria (Fig. 4A,B). This truncated form contained the entire predicted catalytic region and was therefore expected to be active. In this respect it resembles E. coli RNase E, which investigators find very difficult to work with as a full-length protein; as a result, they frequently use a fragment containing the catalytic part for activity studies. In addition, two highly conserved lysine residues at positions 546 and 552 (E. coli residues 106 and 112), located in the S1 domain on one side of the RNA binding channel and participating in the formation of the catalytic active site by helping the interaction of the RNA backbone, were replaced with alanines. This was done in order to obtain an inactive protein that could serve as a negative control (K→A) (Callaghan et al. 2005b). The preparations of both recombinant enzymes were analyzed by immunoblot using an E. coli RNase E-specific antibody and found to be free of bacterial enzyme contamination. In addition, no residual exonuclease activity was detected when the proteins were incubated with [32P]-RNA and the products analyzed by thin layer chromatography (data not shown). We concluded that the purified, bacterially expressed proteins lacked any residual ribonuclease activity due to bacterial host proteins.
The cleavage of RNAI, a 110-nucleotide (nt)-long noncoding transcript regulating ColE1-type plasmid replication in E. coli, was analyzed first; this is a well characterized substrate of E. coli RNase E (Lin-Chao and Cohen 1991). As shown in Figure 4C, Arabidopsis RNase E produces a cleavage pattern on RNAI similar to that of the E. coli homolog. This cleavage activity was enhanced when the 5′ end of substrate RNA was mono- but not triphosphorylated (Mackie 1998; Jiang et al. 2000; Tock et al. 2000).
To observe RNase E cleavage more clearly, the assay was repeated with an enzyme:substrate ratio of 1:1 (instead of 50:1 in Fig. 4C), making the protein and RNA concentrations 10 nM each (Fig. 4D). In this case, the effect of enhanced activity on mono-phosphorylated 5′-end substrate was much more pronounced. This ratio was used in all subsequent experiments. Only a very small amount (<5%) of 5′-PPP RNA was cleaved during 15 min of incubation, compared to >50% cleavage of 5′-P RNA. No specific cleavage was observed with the K-to-A mutated RNase E, showing that the observed activity was due to recombinant RNase E and not contaminating E. coli ribonucleases (Fig. 4D).
Single-stranded oligoribonucleotide and chloroplast RNA as substrates for Arabidopsis RNase E
Previous characterization of E. coli RNase E showed a cleavage preference for AU-rich sequences present in a single-stranded RNA stretch located near a stem–loop structure (Mackie 1992; McDowall et al. 1995; Kaberdin 2003; Horie et al. 2007). In order to explore Arabidopsis RNase E cleavage activity, a synthetic RNA oligoribonucleotide resembling the RNAI cleavage site was synthesized and used as a substrate. This RNA corresponds to the first 13 nt of RNAI from the 5′ end, with the canonical RNase E cleavage site located between U5 and A6 (Fig. 5, bottom; Horie et al. 2007). The oligo was labeled with either [32P] or [32P]-pCp at the 5′ or 3′ ends, respectively, and incubated with the Arabidopsis RNase E or the E. coli enzyme for comparison. The results showed cleavage by the Arabidopsis and E. coli enzymes at U5, generating the correct 5′ and 3′ fragments (Fig. 5). This cleavage activity was not detected when the K-to-A mutated protein was analyzed (data not shown).
FIGURE 5.
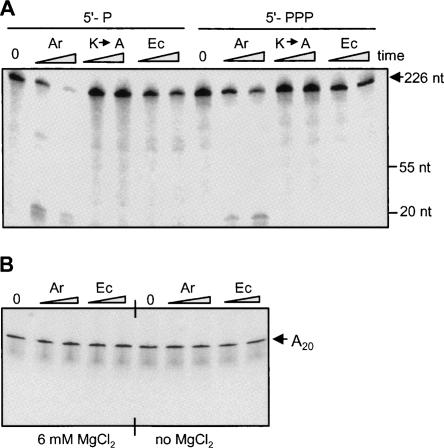
Cleavage of a synthetic RNA oligonucleotide by Arabidopsis RNase E. A 13-nt oligoribonucleotide representing the cleavage site of RNAI, shown at the bottom of the figure, was [32P]-labeled either at the 5′ end (left half of the figure) or the 3′ end (right side of the figure), and incubated with Arabidopsis (Ar) or the E. coli (Ec) enzymes. Following 0, 20, and 40 min of incubation, the RNA was purified and analyzed by denaturing PAGE and autoradiography. An RNA ladder was run on the same gel for an analysis of the precursor and product sizes (M). Additional oligoribonucleotide of 20 nt was fractionated on the same gel to verify the length of the lines of the ladder markers. The signal obtained from this marker is outside the frame shown in the picture. Note that the 3′-end labeling added one nucleotide (pCp). Cleavage products are marked by arrowheads.
Other RNAs were tested for RNase E cleavage to further examine the nature of substrate recognition by the enzyme. When the 226-nt RNA corresponding to the 3′ end of spinach chloroplast petD mRNA was analyzed, the Arabidopsis enzyme digested it to very short fragments; therefore, a specific cleavage site was not observed (Fig. 6A). Similar results were obtained using an RNA representing the 3′ end of the spinach chloroplast psbA transcript (not shown). Interestingly, the E. coli enzyme showed slower cleavage kinetics on this substrate. As with the other substrates, significant inhibition of activity was observed for the K-to-A mutant, and RNAs harboring one phosphate at the 5′ end were digested faster than those with three phosphates (Fig. 6A). When the degradation products of the reaction shown in Figure 6A were analyzed on TLC, no free nucleotides were observed (not shown). Therefore, the spinach petD RNA was fragmented into short oligonucleotides but not into mononucleotides. This experiment revealed that substrates not folded into a tRNA-like structure can be endonucleolytically digested by Arabidopsis RNase E into small oligonucleotides.
FIGURE 6.
Degradation of a transcript corresponding to the chloroplast petD RNA and synthetic A20 oligo by Arabidopsis RNase E. (A) RNA corresponding to the 3′ end of spinach chloroplast petD mRNA harboring either one (5′-P) or three phosphates (5′-PPP) at the 5′ end was incubated with 20 nM of Arabidopsis (Ar), the inactive mutated version (K→A), or E. coli (Ec) enzymes. Following incubation for 5 and 15 min, RNA was purified and analyzed by denaturing PAGE and autoradiography. (B) A synthetic, [5′-32P]-labeled oligonucleotide containing 20 adenosine residues (A20) was incubated with A. thaliana and E. coli RNase E for 30 and 60 min in the presence or absence of 6 mM MgCl2 in the incubation buffer, as indicated. RNA was then isolated and analyzed as described for panel A.
In addition, when oligoribonucleotides composed of A20 or (GU)12 were incubated with RNase E, either with or without Mg2+ in the incubation buffer, no cleavage was detected (Fig. 6B; data not shown). Indeed, of the many RNA substrates tested, these were the only ones not cleaved by RNase E.
Additional substrate for assaying Arabidopsis RNase E
In E. coli, RNase E plays an important role in the processing of tRNA precursors (Li and Deutscher 2002; Ow and Kushner 2002). It has been suggested that RNase E generates the processing intermediates that are subsequently cleaved by RNase P to form the correct tRNA 5′ end (Vioque et al. 1988; Kirsebom and Svard 1992). Indeed, the precursor of the E. coli tRNATyrSu3 (pSu3) is cleaved by the bacterial RNase E upstream of the RNase P cleavage site in vitro and in vivo (Soderbom et al. 2005). The predicted secondary structure of pSu3 is shown in Figure 7B. In order to analyze whether the Arabidopsis enzyme can cleave pSu3 in a similar manner as E. coli RNase E, we generated the corresponding transcript and incubated it with the Arabidopsis enzyme. The results revealed that both enzymes specifically cleave the pSu3 transcript to generate similar 5′ and 3′-end products (Fig. 7A). In fact, this substrate is superior to RNAI for activity analysis since the cleavage is more specific, the 5′-end product is observed on the gel, and the effect of one or three phosphates at the 5′ end is evident (Fig. 7A).
FIGURE 7.
Cleavage of the E. coli tRNATyr precursor substrate by the Arabidopsis and E. coli enzymes. (A) Arabidopsis and E. coli enzymes were incubated with a 131-nt [32P]-labeled RNA corresponding to the precursor of E. coli tRNATyr for 0, 5, 15, and 45 min. The RNAs harbor either one (5′-P) or three (5′-PPP) phosphates at the 5′ end as indicated at the top. Following incubation, RNA was purified and analyzed on denaturing PAGE and autoradiography. The substrate (S) as well as the 3′ and 5′ cleavage products are indicated at the left. (B) A schematic representation of the 131-nt E. coli tRNATyr precursor is adapted from Soderbom et al. (2005). The RNase P canonical cleavage site is indicated (M1). Arabidopsis RNase E cleavage sites, as mapped in the experiments described in Figures 7 and 8, are shown with arrows. Three guanidine residues added to the 5′ end of the RNA substrate are enclosed in a dashed box. (C) Cleavage of the tRNATyr precursor transcript by the M1 RNA of E. coli RNase P. Conditions are as in A. (D) Inhibition of cleavage activity by EDTA treatment and the inactive mutant of Arabidopsis RNase E (K→A). Incubation times were 5, 15, and 45 min. The substrate is the tRNATyr precursor transcript. EDTA treatment was performed by addition to a final concentration of 10 mM. Other conditions are as in A. Ar, Arabidopsis RNase E. (E) Arabidopsis and E. coli enzymes were incubated with the tRNATyr precursor for 30 min in the presence of Mg2+ as indicated. Following incubation, the RNA was purified and analyzed by denaturing PAGE and autoradiography. The 5′ cleavage product is not shown in this figure.
The cleavage of pSu3 by Arabidopsis RNase E was also compared with the canonical processing by M1 RNA, the catalytic subunit of E. coli RNase P (Guerrier-Takada et al. 1983). As shown in Figure 7C, the M1 RNA cleaved at the canonical RNase P site, indicated in Figure 7B by the letters M1, producing two products: a 46-nt 5′ leader and an 88-nt mature tRNA. Similar to the result obtained with the E. coli RNAI substrate, the activity of Arabidopsis RNase E was significantly inhibited in the mutant, where lysines 546 and 552 were converted to alanines (Fig. 7D). The addition of 10 mM EDTA to the reaction mixture, which disrupted the formation of the high molecular weight complex (Fig. 3B), significantly reduced cleavage activity (Fig. 7D).
Interestingly, in cleavage buffer lacking MgCl2, both Arabidopsis and E. coli RNase E nonspecifically cleaved pSu3 RNA to small oligoribonucleotides (Fig. 7E). It should be noted that this effect could be obtained either from a change in enzyme specificity and/or alterations in RNA folding in the absence of Mg2+. In addition, since the enzyme activity is Mg2+ dependent, the trace amounts of metals ions present in the buffer even when no MgCl2 is added were sufficient to activate the enzyme. Accordingly, the addition of EDTA inhibited the activity (Fig. 7D). Cleavage specificity was fully restored by the addition of MgCl2 at 2–10 mM, which approximates the physiological concentration of Mg2+ in bacteria and chloroplasts. Further increasing Mg2+ concentration to 50 mM inhibited the cleavage activity of both E. coli and Arabidopsis enzymes (Fig. 7E).
Mapping the endonucleolytic cleavage sites
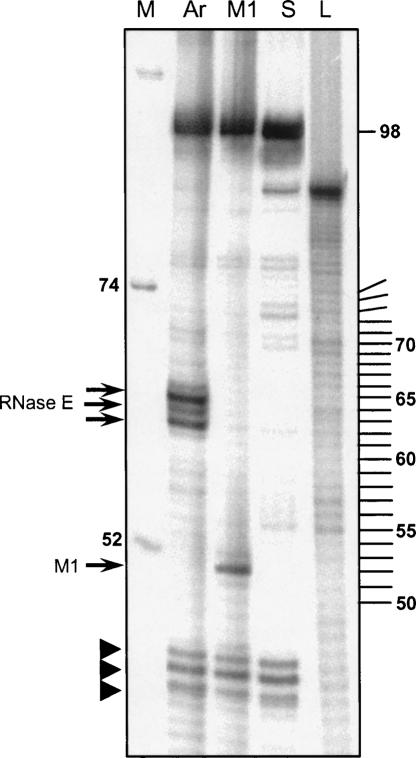
In order to determine the Arabidopsis RNase E cleavage positions in the 5′ leader sequence, primer extension analysis was performed (Fig. 8). The pSu3 RNA was first cleaved by the Arabidopsis RNase E or the M1 RNA. Then, a [32P]-labeled oligonucleotide was annealed to nucleotide numbers 79–98 and elongated by reverse transcription followed by analysis of the extension products by high-resolution gel electrophoresis. As indicated in Figure 8, an extension of the substrate pSu3 RNA before digestion revealed the full-length transcript, as well as several other signals that were most likely generated from effects of the RNA structured region on the extension reaction (Fig. 8, lane S). An analysis of the M1 cleavage products revealed the expected product (Fig. 8, lane M1). An analysis of the Arabidopsis RNase E reaction showed three extension products corresponding to positions 34–36 of pSu3 counting from the 5′ end (Fig. 8, lane Ar). This high-resolution analysis revealed that Arabidopsis RNase E specifically cleaved the pSu3 RNA at three sites located in the adenosine-rich, single-stranded region situated 10 nt 5′ to the RNase P cleavage site. These sites included nucleotide 36, which was previously mapped as the cleavage site of E. coli RNase E (Soderbom et al. 2005).
FIGURE 8.
Mapping RNase E cleavage sites by primer extension. The tRNATyr precursor was first cleaved with either Arabidopsis RNase E (Ar) or M1 RNA and subjected to primer extension analysis. Extension products, together with the RNA ladder (L) and size markers (M), were analyzed by high-resolution PAGE and autoradiography. RNA fragment sizes are shown in nucleotides. Extension products of Arabidopsis RNase E and M1 RNA cleavage fragments are indicated with arrows on the left. In the lane S, noncleaved substrate RNA that was analyzed in the same way is presented. Prematurely terminated RNAs produced by the inhibition of the reverse transcriptase are indicated by arrowheads. The numbers of nucleotides are also indicated on the right.
DISCUSSION
Conservation of RNase E in prokaryotes and photosynthetic organisms
Homologs of RNase E exist in most bacteria, some archaea, as well as in some algae and higher plants (Lee and Cohen 2003; Li et al. 2005; Bollenbach et al. 2007; Horie et al. 2007). Several Gram-negative bacteria contain two proteins of this family, for example, RNase E and RNase G in E. coli, while others such as cyanobacteria contain only one (Kaberdin et al. 1998; Lee and Cohen 2003). There is no RNase E homolog in several Gram-positive bacteria such as Bacillus subtilis and the green alga Chlamydomonas reinhardtii. These organisms contain another endoribonuclease, RNase J, which resembles RNase E and G in its sensitivity to the number of phosphates at the 5′ end and its possible role in cleaving transcripts as part of processing and degradation pathways (Even et al. 2005; Mathy et al. 2007). Several bacteria, including cyanobacteria and archaea, however, contain both RNase E and RNase J (Slomovic et al. 2006; Bollenbach et al. 2007). Interestingly, RNase E homologs are encoded in the chloroplast genomes of red algae, as well as several green algae (Bollenbach et al. 2007; Horie et al. 2007). In higher eukaryotes, genes encoding RNase E homologs are present only in the nuclear genomes of plants and contain putative chloroplast transit peptides. Indeed, results shown here demonstrated chloroplast localization for Arabidopsis, and can be predicted to be the same for other plants. Since genes and ESTs related to RNase E were detected in all 10 plants analyzed here, the presence of this protein seems to be a general characteristic of higher plants. It can therefore be assumed that the nuclear genome acquired the RNase E gene just as it acquired most of the other chloroplast genes, namely, from the genome of the cyanobacteria-like prokaryote that entered the eukaryotic host and ultimately became the chloroplast.
Contrary to the situation in many bacteria and archaea, where either RNase E or RNase J is present, the chloroplast of higher plants seems to contain both enzymes, as do cyanobacteria, relatives of the evolutionary ancestors of the chloroplast (Bollenbach et al. 2007). Moreover, a third endoribonuclease that appears to be unique to the chloroplast, CSP41, functions in this organelle as well. The picture thus emerging for endoribonucleolytic activity in the chloroplast is the concerted action of three types of enzymes, RNase E, RNase J, and CSP41, where each type may contain more than one member (Bollenbach et al. 2007). The division of work, the specificity, and/or the overlapping substrates, as well as the expression during development in different organs, are now being studied in order to obtain the complete picture of the function of each of these enzymes in the initial endonucleolytic cleavage of the polyadenylation-stimulated RNA degradation pathway.
Unique domains of chloroplast RNase E
The chloroplast RNase E of Arabidopsis, rice, and tomato contains an N-terminal extension of ∼260 amino acids (excluding the predicted chloroplast transit peptide). This extension seems to be conserved in its length but not its amino acid sequence. Therefore, even though the related ESTs were not identified for the other plants shown in Figure 1, this extension may be present in every chloroplast RNase E. In this way, chloroplast RNase E belongs to group III of RNase E proteins, as classified by Lee and Cohen (2003). The possible function of this extension remains to be determined. One possibility is that, like the C-terminal extension in the E. coli RNase E, it serves as a platform for binding associated proteins that form a type of “chloroplast degradosome” (Marcaida et al. 2006; Carpousis 2007). However, spinach chloroplast PNPase is not associated with other proteins in an E. coli-type degradosome (Baginsky et al. 2001). Moreover, the results of fractionation on a size-exclusion column showed no significant difference in elution profile between purified recombinant protein and native protein in total leaf extracts (Fig. 3B). Indeed, the fact that the amino acid sequence homology between the different plants is very low in this domain supports the possibility that, if there are proteins associated with the chloroplast RNase E, they are fewer and smaller than those present in the E. coli degradosome. Another possibility is that the N-terminal extension modulates the activity of the catalytic domain. In this work, the properties of truncated versions that include the catalytic parts of the two proteins were compared. It is possible that the activities of the full-length proteins are different, and exploring this requires the ability to produce a soluble, full-length protein, or genetic approaches.
Another difference between plant RNase E and the bacterial enzyme is the insertion of 122 amino acids into the S1 domain; this addition is apparently present in all plant RNase E proteins (Fig. 1). This places plant RNase E in group II, as classified by Lee and Cohen (2003). This extension seems unlikely to give rise to significant structural changes or functional differences, based on the homology model analysis described in Figures 1 and 2. This conclusion is supported by the extensive similarities in activity and behavior between the E. coli and Arabidopsis RNase E enzymes analyzed here. Therefore, the importance of this insertion, whether for the catalytic activity in the chloroplast or as an evolutionary vestige that was deleted in most prokaryotes including cyanobacteria, remains to be determined.
Enzymatic activity of chloroplast RNase E
The endonucleolytic activities of the catalytic parts of the E. coli and Arabidopsis RNase E proteins were found to be very similar. Both activities were sensitive to the number of phosphates at the 5′ end and to substrate secondary structure. In both enzymes, replacing the two conserved lysines located in the S1 domain significantly reduced catalytic activity. Therefore, the catalytic domains of the prokaryotic and chloroplast RNase E apparently retained very similar properties despite their long evolutionary separation.
It is interesting to note that the sensitivity of cleavage activity to the number of phosphates located at the 5′ end of the transcripts is conserved in chloroplast RNase E. While the 5′ end in bacteria is located mainly at the transcription initiation site and therefore contains three phosphates, in the chloroplast of the green algae Chlamydomonas rienhardtii the 5′ end is formed in most cases by a processing event and therefore contains only one phosphate (Bollenbach et al. 2004). While in bacteria the three phosphates are believed to function in protecting the RNA from degradation, this protection in Chlamydomonas chloroplasts is proposed to occur via nuclear-encoded proteins that specifically bind the 5′ untranslated region of chloroplast transcripts (Mayfield 1990; Rochaix 1992; Barkan and Goldschmidt-Clermont 2000; Bollenbach et al. 2004). If this is also the situation in the chloroplast of higher plants, it is not yet defined. If so, it is not obvious why the presence of three 5′ end phosphates significantly inhibits the cleavage activity of the chloroplast enzyme. More research is required to explore whether this phenomenon has a specific function in the chloroplast or, alternatively, is a vestige of the chloroplast's prokaryotic past.
This study reports the first characterization of a eukaryotic RNase E. The catalytic properties were found to be very similar to those of E. coli RNase E and the protein located in the chloroplast. Since the T-DNA null insertion mutant for this protein cannot grow without adding sucrose to the medium (data not shown), Arabidopsis RNase E may be required for chloroplast development, as it is required for viability in E. coli. The major next steps in this research are to understand the specific functions of RNase E and its targeted transcripts in the chloroplast, as well as the interplay of the enzyme with the two other chloroplast endoribonucleases.
MATERIALS AND METHODS
Bioinformatic analysis of RNase E-like sequences and cDNA sequences
Alignment of the full-length and truncated EST RNase E sequences to that of E. coli was performed using ClustalW. The full-length cDNA of tomato RNase E was obtained by screening a cDNA library with a probe corresponding to a truncated EST. Arabidopsis truncated RNase E cDNA was obtained from RIKEN BRC and was extended using a standard RACE protocol in order to obtain the full-length coding region.
The FoldIndex of the 638 residues (from Ile327 to Leu964) was calculated and drawn as described (Prilusky et al. 2005). The plot shows the folding index (Y axis), that estimates the probability of the query sequence to fold into a globular structure.
Structure prediction
The pairwise alignment and the model were produced as follows: an initial pairwise alignment of 638 positions was obtained from HHpred (Soding et al. 2005), with a score of 1165 and a sequence identity of 31%. Within this alignment there is a 122-residue-long region in the Arabidopsis sequence which is missing in the E. coli sequence, and therefore cannot be modeled (the plant addition to S1 domain). This region (from position 408 to 529 in Arabidopsis) was then deleted from the alignment, which was also manually improved using the Jalview editor (Clamp et al. 2004). The resulting pairwise alignment displayed a sequence identity of 32.5% for 519 amino acids in the Arabidopsis sequence and 490 amino acids in the E. coli sequence.
The homology-based model of Arabidopsis RNase E was obtained by feeding this alignment into MODELLER (Sali and Blundell 1993), using the E. coli RNase E crystal structure as the template (PDB ID 2bx2, chain L). Ten models were produced and the one with the best DOPE score was chosen. The images were produced using the UCSF Chimera package (Pettersen et al. 2004). The structural alignment between the Arabidopsis model and the E. coli template was obtained using the Chimera Tool MatchMaker (Meng et al. 2006).
Expression of Arabidopsis RNase E protein in bacteria
Part of the Arabidopsis RNase E cDNA (encoding amino acids 284–996) was PCR-amplified and inserted into the pQE30 expression vector (Qiagen), in frame with an amino-terminal 6×Histidine (6×His) tag. Attempts to express the full-length or longer versions resulted in insoluble proteins. The K-to-A mutant, in which lysines 546 and 552 located in the S1 domain of the catalytic part were changed to alanines, was constructed in the above-mentioned plasmid using the QuickChange site-directed mutagenesis kit (Stratagene). The sequence of the oligonucleotides used is available upon request. The proteins were expressed in the E. coli M15[pREP4,Kan] strain (Qiagen). Expression was induced in log-phase cultures with 1 mM IPTG for 8 h at 20°C.
The cDNA encoding the catalytic part of E. coli RNase E (amino acids 1–498) was inserted into the pET20b expression vector, in frame with a carboxy-terminal 6×His tag. The protein was expressed in E. coli DE-TRX BL21 as described above for Arabidopsis RNase E.
Following expression of the proteins, the cells were harvested, resuspended in binding buffer (50 mM NaH2PO4 at pH 8.0, 300 mM NaCl, 20 mM imidazole, 10% glycerol, 0.1% Tween-20, 2 μM β-mercaptoethanol), and disrupted using a microfluidizer. The recombinant proteins were purified on a Ni-NTA mini-columns followed by MonoQ anion exchange chromatography and dialysis against buffer E (Portnoy and Schuster 2006). The purified protein fractions showed one protein detectable by silver stain and no exonuclease activity. In addition, analyzing immunoblots of purified Arabidopsis RNase E with antibodies against E. coli RNase E (Khemici and Carpousis 2004) indicated no contamination with the E. coli enzyme. Aliquots of the purified recombinant protein were stored at −80°C.
Antibody generation
Antibodies against the Arabidopsis RNase E were produced in rabbits injected with the recombinant protein as previously described (Lisitsky et al. 1997). Antibodies against the E. coli RNase E were a kind gift of A.J. Carpousis (Centre National de la Recherche Scientifique, Toulouse, France).
Size-exclusion chromatography
Purified recombinant RNase E or Arabidopsis soluble proteins were fractionated on a Superdex 200 size-exclusion column (Amersham Biosciences) as previously described (Baginsky et al. 2001; Rott et al. 2003). Fractions were collected and analyzed by immunoblotting using commercial anti-6×His or RNase E antibodies. The column was calibrated with the following protein standards: thyroglobulin, 669 kDa; ferritin, 440 kDa; catalase, 232 kDa; aldolase, 158 kDa; and bovine serum albumin, 67 kDa.
Chloroplast isolation and fractionation
Leaves of 4-wk-old plants were homogenized in buffer containing 20 mM HEPES (pH 7.6), 2 mM EDTA, 1 mM MgCl2, and 0.33 M sorbitol. Chloroplasts were purified by centrifugation through a 10%–50% Percoll gradient at 5000g for 5 min (Meierhoff et al. 2003). The chloroplast layer was collected, disrupted, and separated into insoluble (membranes) and soluble fractions.
Synthetic RNAs
The plasmids used for generating the transcripts resembling E. coli RNAI (Portnoy and Schuster 2006), the 3′ end of spinach chloroplast petD (Lisitsky et al. 1997), the precursor of E. coli tRNATyr (pSu3) (Soderbom et al. 2005), and E. coli catalytic M1 RNA of RNase P (Jarrous et al. 1998) have been described. The 13-nt oligoribonucleotides containing the sequence 5′-ACAGUAUUUGGUA, representing the 5′ sequence of the E. coli RNAI, A20, and (GC)12, were purchased from Sigma.
For uniform RNA labeling, the corresponding plasmid digested with an appropriate restriction enzyme was incubated with T7 RNA polymerase (NEB) and 80 μCi of [α-32P]UTP for 1 h at 37°C (Lisitsky et al. 1997). Alternatively, the corresponding DNA fragments amplified by PCR were used as templates for transcription. Full-length transcripts were gel-purified as previously described (Lisitsky et al. 1997). In order to obtain 5′-monophosphorylated RNA, 15 mM GMP was included in the transcription reaction (Jiang et al. 2000).
5′-end labeling of synthetic RNA was performed first by dephosphorylation for 30 min with 1 unit of shrimp alkaline phosphatase, followed by labeling with 10 units of polynucleotide kinase (Fermentas) and 50 μCi of [γ-32P]ATP for 1 h at 37°C. For 3′-end labeling, 100 ng of RNA was incubated with 50 μCi of 5′-[32P]cytidine-3′, 5′-bis(phosphate) (pCp), and 10 units of T4 RNA ligase. The full-length [32P]-labeled transcripts were purified by denaturing PAGE. An RNA ladder was prepared by alkaline hydrolysis of a 5′-labeled 90-nt oligoribonucleotide.
In vitro activity assays
Cleavage activity assays were performed as previously described (Yehudai-Resheff et al. 2003). Unless otherwise indicated, the reaction consisted of 50 ng recombinant protein (0.5 pmol) and ∼0.5 pmol of RNA (10 nM of each). Following incubation of the purified proteins with synthetic [32P]-RNA, the cleavage products were purified and analyzed by denaturing PAGE and autoradiography.
Primer extension
An oligonucleotide primer harboring the complementary sequence of positions 96–78 of E. coli tRNATyrSu3 precursor (pSu3) (Vioque et al. 1988; Soderbom et al. 2005) was labeled at the 5′ end using [γ-32P]ATP and extended by reverse transcription with the full-length pSu3 transcript, or the same molecule that was previously cleaved either by Arabidopsis RNase E or E. coli M1 RNA. The extension products were analyzed by high-resolution denaturing PAGE and autoradiography.
ACKNOWLEDGMENTS
We thank A.J. Carpousis for the E. coli RNase E antibodies, N. Jarrous for the plasmids producing M1 RNA and pSu3 RNA, E. Lifshitz for the tomato cDNA library, and B. Luisi for his advice on the structure of RNase E and critical reading of the manuscript. This work was supported by grants from the Israel Science Foundation (ISF), the United States–Israel Binational Science Foundation (BSF), and the United States–Israel Binational Agricultural Research and Development (BARD) Fund. A.S. is a recipient of the Nehemia Levtzion Council for Higher Education Scholarship.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.907608.
REFERENCES
- Afonyushkin T., Vecerek B., Moll I., Blasi U., Kaberdin V.R. Both RNase E and RNase III control the stability of sodB mRNA upon translational inhibition by the small regulatory RNA RyhB . Nucleic Acids Res. 2005;33:1678–1689. doi: 10.1093/nar/gki313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arraiano C.M., Yancey S.D., Kushner S.R. Stabilization of discrete mRNA breakdown products in ams pnp rnb multiple mutants of Escherichia coli K-12. J. Bacteriol. 1988;170:4625–4633. doi: 10.1128/jb.170.10.4625-4633.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baginsky S., Shteiman-Kotler A., Liveanu V., Yehudai-Resheff S., Bellaoui M., Settlage R.E., Shabanowitz J., Hunt D.F., Schuster G., Gruissem W. Chloroplast PNPase exists as a homomultimer enzyme complex that is distinct from the Escherichia coli degradosome. RNA. 2001;7:1464–1475. [PMC free article] [PubMed] [Google Scholar]
- Barkan A., Goldschmidt-Clermont M. Participation of nuclear genes in chloroplast gene expression. Biochimie. 2000;82:559–572. doi: 10.1016/s0300-9084(00)00602-7. [DOI] [PubMed] [Google Scholar]
- Bollenbach T.J., Schuster G., Stern D.B. Cooperation of endo- and exoribonucleases in chloroplast mRNA turnover. Prog. Nucleic Acid Res. Mol. Biol. 2004;78:305–337. doi: 10.1016/S0079-6603(04)78008-3. [DOI] [PubMed] [Google Scholar]
- Bollenbach T., Schuster G., Portnoy V., Stern D. Polyadenylation, processing and degradation of chloroplast RNA. Top. Curr. Genet. 2007;19:175–211. [Google Scholar]
- Callaghan A.J., Marcaida M.J., Stead J.A., McDowall K.J., Scott W.G., Luisi B.F. Structure of Escherichia coli RNase E catalytic domain and implications for RNA turnover. Nature. 2005a;437:1187–1191. doi: 10.1038/nature04084. [DOI] [PubMed] [Google Scholar]
- Callaghan A.J., Redko Y., Murphy L.M., Grossmann J.G., Yates D., Garman E., Ilag L.L., Robinson C.V., Symmons M.F., McDowall K.J., et al. “Zn-link”: A metal-sharing interface that organizes the quaternary structure and catalytic site of the endoribonuclease, RNase E. Biochemistry. 2005b;44:4667–4675. doi: 10.1021/bi0478244. [DOI] [PubMed] [Google Scholar]
- Carpousis A.J. The RNA degradosome of Escherichia coli: An mRNA-degrading machine assembled on RNase E. Annu. Rev. Microbiol. 2007;61:71–87. doi: 10.1146/annurev.micro.61.080706.093440. [DOI] [PubMed] [Google Scholar]
- Casaregola S., Jacq A., Laoudj D., McGurk G., Margarson S., Tempete M., Norris V., Holland I.B. Cloning and analysis of the entire Escherichia coli ams gene. ams is identical to hmp1 and encodes a 114 kDa protein that migrates as a 180 kDa protein. J. Mol. Biol. 1992;228:30–40. doi: 10.1016/0022-2836(92)90489-7. [DOI] [PubMed] [Google Scholar]
- Celesnik H., Deana A., Belasco J.G. Initiation of RNA decay in Escherichia coli by 5′ pyrophosphate removal. Mol. Cell. 2007;27:79–90. doi: 10.1016/j.molcel.2007.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clamp M., Cuff J., Searle S.M., Barton G.J. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- Cohen S.N., McDowall K.J. RNase E: Still a wonderfully mysterious enzyme. Mol. Microbiol. 1997;23:1099–1106. doi: 10.1111/j.1365-2958.1997.tb02593.x. [DOI] [PubMed] [Google Scholar]
- Cormack R.S., Genereaux J.L., Mackie G.A. RNase E activity is conferred by a single polypeptide: Overexpression, purification, and properties of the ams/rne/hmp1 gene product. Proc. Natl. Acad. Sci. 1993;90:9006–9010. doi: 10.1073/pnas.90.19.9006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher M.P. Degradation of RNA in bacteria: Comparison of mRNA and stable RNA. Nucleic Acids Res. 2006;34:659–666. doi: 10.1093/nar/gkj472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diwa A., Bricker A.L., Jain C., Belasco J.G. An evolutionarily conserved RNA stem–loop functions as a sensor that directs feedback regulation of RNase E gene expression. Genes & Dev. 2000;14:1249–1260. [PMC free article] [PubMed] [Google Scholar]
- Dreyfus M., Regnier P. The poly(A) tail of mRNAs: Bodyguard in eukaryotes, scavenger in bacteria. Cell. 2002;111:611–613. doi: 10.1016/s0092-8674(02)01137-6. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O., Nielsen H., Brunak S., von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- Even S., Pellegrini O., Zig L., Labas V., Vinh J., Brechemmier-Baey D., Putzer H. Ribonucleases J1 and J2: Two novel endoribonucleases in B.subtilis with functional homology to E.coli RNase E. Nucleic Acids Res. 2005;33:2141–2152. doi: 10.1093/nar/gki505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau N.L., Wilusz J., Wilusz C.J. The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- Ghora B.K., Apirion D. Structural analysis and in vitro processing to p5 rRNA of a 9S RNA molecule isolated from an rne mutant of E. coli . Cell. 1978;15:1055–1066. doi: 10.1016/0092-8674(78)90289-1. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C., Gardiner K., Marsh T., Pace N., Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- Hajnsdorf E., Braun F., Haugel-Nielsen J., Le Derout J., Regnier P. Multiple degradation pathways of the rpsO mRNA of Escherichia coli. RNase E interacts with the 5′ and 3′ extremities of the primary transcript. Biochimie. 1996;78:416–424. doi: 10.1016/0300-9084(96)84748-1. [DOI] [PubMed] [Google Scholar]
- Horie Y., Ito Y., Ono M., Moriwaki N., Kato H., Hamakubo Y., Amano T., Wachi M., Shirai M., Asayama M. Dark-induced mRNA instability involves RNase E/G-type endoribonuclease cleavage at the AU-box and SD sequences in cyanobacteria. Mol. Genet. Genomics. 2007;278:331–346. doi: 10.1007/s00438-007-0254-9. [DOI] [PubMed] [Google Scholar]
- Houseley J., LaCava J., Tollervey D. RNA-quality control by the exosome. Nat. Rev. Mol. Cell Biol. 2006;7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- Jarrous N., Eder P.S., Guerrier-Takada C., Hoog C., Altman S. Autoantigenic properties of some protein subunits of catalytically active complexes of human ribonuclease P. RNA. 1998;4:407–417. [PMC free article] [PubMed] [Google Scholar]
- Jiang X., Diwa A., Belasco J.G. Regions of RNase E important for 5′-end-dependent RNA cleavage and autoregulated synthesis. J. Bacteriol. 2000;182:2468–2475. doi: 10.1128/jb.182.9.2468-2475.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaberdin V.R. Probing the substrate specificity of Escherichia coli RNase E using a novel oligonucleotide-based assay. Nucleic Acids Res. 2003;31:4710–4716. doi: 10.1093/nar/gkg690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaberdin V.R., Chao Y.H., Lin-Chao S. RNase E cleaves at multiple sites in bubble regions of RNA I stem–loops yielding products that dissociate differentially from the enzyme. J. Biol. Chem. 1996;271:13103–13109. doi: 10.1074/jbc.271.22.13103. [DOI] [PubMed] [Google Scholar]
- Kaberdin V.R., Miczak A., Jakobsen J.S., Lin-Chao S., McDowall K.J., von Gabain A. The endoribonucleolytic N-terminal half of Escherichia coli RNase E is evolutionarily conserved in Synechocystis sp. and other bacteria but not the C-terminal half, which is sufficient for degradosome assembly. Proc. Natl. Acad. Sci. 1998;95:11637–11642. doi: 10.1073/pnas.95.20.11637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khemici V., Carpousis A.J. The RNA degradosome and poly(A) polymerase of Escherichia coli are required in vivo for the degradation of small mRNA decay intermediates containing REP-stabilizers. Mol. Microbiol. 2004;51:777–790. doi: 10.1046/j.1365-2958.2003.03862.x. [DOI] [PubMed] [Google Scholar]
- Kirsebom L.A., Svard S.G. The kinetics and specificity of cleavage by RNase P is mainly dependent on the structure of the amino acid acceptor stem. Nucleic Acids Res. 1992;20:425–432. doi: 10.1093/nar/20.3.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushner S.R. mRNA decay in Escherichia coli comes of age. J. Bacteriol. 2002;184:4658–4665. doi: 10.1128/JB.184.17.4658-4665.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushner S.R. mRNA decay in prokaryotes and eukaryotes: Different approaches to a similar problem. IUBMB Life. 2004;56:585–594. doi: 10.1080/15216540400022441. [DOI] [PubMed] [Google Scholar]
- Lee K., Cohen S.N. A Streptomyces coelicolor functional orthologue of Escherichia coli RNase E shows shuffling of catalytic and PNPase-binding domains. Mol. Microbiol. 2003;48:349–360. doi: 10.1046/j.1365-2958.2003.03435.x. [DOI] [PubMed] [Google Scholar]
- Lee K.B., Bernstein J.A., Cohen S.N. RNase G complementation of rne null mutation identifies functional interrelationships with RNase E in Escherichia coli . Mol. Microbiol. 2002;43:1445–1456. doi: 10.1046/j.1365-2958.2002.02848.x. [DOI] [PubMed] [Google Scholar]
- Li Z., Deutscher M.P. RNase E plays an essential role in the maturation of Escherichia coli tRNA precursors. RNA. 2002;8:97–109. doi: 10.1017/s1355838202014929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Gong X., Joshi V.H., Li M. Co-evolution of tRNA 3′ trailer sequences with 3′ processing enzymes in bacteria. RNA. 2005;11:567–577. doi: 10.1261/rna.7287505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin-Chao S., Cohen S.N. The rate of processing and degradation of antisense RNAI regulates the replication of ColE1-type plasmids in vivo. Cell. 1991;65:1233–1242. doi: 10.1016/0092-8674(91)90018-t. [DOI] [PubMed] [Google Scholar]
- Lisitsky I., Kotler A., Schuster G. The mechanism of preferential degradation of polyadenylated RNA in the chloroplast: The exoribonuclease 100RNP/PNPase displays high binding affinity for poly(A) sequence. J. Biol. Chem. 1997;272:17648–17653. doi: 10.1074/jbc.272.28.17648. [DOI] [PubMed] [Google Scholar]
- Mackie G.A. Secondary structure of the mRNA for ribosomal protein S20: Implications for cleavage by ribonuclease E. J. Biol. Chem. 1992;267:1054–1061. [PubMed] [Google Scholar]
- Mackie G.A. Ribonuclease E is a 5′-end-dependent endonuclease. Nature. 1998;395:720–723. doi: 10.1038/27246. [DOI] [PubMed] [Google Scholar]
- Marcaida M.J., DePristo M.A., Chandran V., Carpousis A.J., Luisi B.F. The RNA degradosome: Life in the fast lane of adaptive molecular evolution. Trends Biochem. Sci. 2006;31:359–365. doi: 10.1016/j.tibs.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Mathy N., Benard L., Pellegrini O., Daou R., Wen T., Condon C. 5′-to-3′ exoribonuclease activity in bacteria: Role of RNase J1 in rRNA maturation and 5′ stability of mRNA. Cell. 2007;129:681–692. doi: 10.1016/j.cell.2007.02.051. [DOI] [PubMed] [Google Scholar]
- Mayfield S.P. Chloroplast gene regulation: Interaction of the nuclear and chloroplast genomes in the expression of photosynthetic proteins. Curr. Opin. Cell Biol. 1990;2:509–513. doi: 10.1016/0955-0674(90)90135-2. [DOI] [PubMed] [Google Scholar]
- McDowall K.J., Kaberdin V.R., Wu S.W., Cohen S.N., Lin-Chao S. Site-specific RNase E cleavage of oligonucleotides and inhibition by stem–loops. Nature. 1995;374:287–290. doi: 10.1038/374287a0. [DOI] [PubMed] [Google Scholar]
- Meierhoff K., Felder S., Nakamura T., Bechtold N., Schuster G. HCF152, a PPR protein of Arabidopsis involved in processing of chloroplast psbB-psbT-psbH-petB-petD RNAs. Plant Cell. 2003;15:1480–1495. doi: 10.1105/tpc.010397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng E.C., Pettersen E.F., Couch G.S., Huang C.C., Ferrin T.E. Tools for integrated sequence–structure analysis with UCSF Chimera. BMC Bioinformatics. 2006;7:339. doi: 10.1186/1471-2105-7-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miczak A., Kaberdin V.R., Wei C.L., Lin-Chao S. Proteins associated with RNase E in a multicomponent ribonucleolytic complex. Proc. Natl. Acad. Sci. 1996;93:3865–3869. doi: 10.1073/pnas.93.9.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita T., Maki K., Aiba H. RNase E-based ribonucleoprotein complexes: Mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes & Dev. 2005;19:2176–2186. doi: 10.1101/gad.1330405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono M., Kuwano M. A conditional lethal mutation in an Escherichia coli strain with a longer chemical lifetime of messenger RNA. J. Mol. Biol. 1979;129:343–357. doi: 10.1016/0022-2836(79)90500-x. [DOI] [PubMed] [Google Scholar]
- Ow M.C., Kushner S.R. Initiation of tRNA maturation by RNase E is essential for cell viability in E. coli . Genes & Dev. 2002;16:1102–1115. doi: 10.1101/gad.983502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ow M.C., Liu Q., Mohanty B.K., Andrew M.E., Maples V.F., Kushner S.R. RNase E levels in Escherichia coli are controlled by a complex regulatory system that involves transcription of the rne gene from three promoters. Mol. Microbiol. 2002;43:159–171. doi: 10.1046/j.1365-2958.2002.02726.x. [DOI] [PubMed] [Google Scholar]
- Ow M.C., Perwez T., Kushner S.R. RNase G of Escherichia coli exhibits only limited functional overlap with its essential homologue, RNase E. Mol. Microbiol. 2003;49:607–622. doi: 10.1046/j.1365-2958.2003.03587.x. [DOI] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Portnoy V., Schuster G. RNA polyadenylation and degradation in different Archaea; roles of the exosome and RNase R. Nucleic Acids Res. 2006;34:5923–5931. doi: 10.1093/nar/gkl763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prilusky J., Felder C.E., Zeev-Ben-Mordehai T., Rydberg E.H., Man O., Beckmann J.S., Silman I., Sussman J.L. FoldIndex: A simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics. 2005;21:3435–3438. doi: 10.1093/bioinformatics/bti537. [DOI] [PubMed] [Google Scholar]
- Py P., Higgins C.F., Krisch H.M., Carpousis A.J. A DEAD-box RNA helicase in the Escherichia coli degradosome. Nature. 1996;381:169–172. doi: 10.1038/381169a0. [DOI] [PubMed] [Google Scholar]
- Rochaix J.-D. Post-transcriptional steps in the expression of chloroplast genes. Annu. Rev. Cell Biol. 1992;8:1–28. doi: 10.1146/annurev.cb.08.110192.000245. [DOI] [PubMed] [Google Scholar]
- Rott R., Zipor G., Portnoy V., Liveanu V., Schuster G. RNA polyadenylation and degradation in cyanobacteria are similar to the chloroplast but different from Escherichia coli . J. Biol. Chem. 2003;278:15771–15777. doi: 10.1074/jbc.M211571200. [DOI] [PubMed] [Google Scholar]
- Sali A., Blundell T.L. Comparative protein modeling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Schubert M., Edge R.E., Lario P., Cook M.A., Strynadka N.C., Mackie G.A., McIntosh L.P. Structural characterization of the RNase E S1 domain and identification of its oligonucleotide-binding and dimerization interfaces. J. Mol. Biol. 2004;341:37–54. doi: 10.1016/j.jmb.2004.05.061. [DOI] [PubMed] [Google Scholar]
- Slomovic S., Portnoy V., Liveanu V., Schuster G. RNA polyadenylation in prokaryotes and organelles; different tails tell different tales. Crit. Rev. Plant Sci. 2006;25:65–77. [Google Scholar]
- Soderbom F., Svard S.G., Kirsebom L.A. RNase E cleavage in the 5′ leader of a tRNA precursor. J. Mol. Biol. 2005;352:22–27. doi: 10.1016/j.jmb.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Soding J., Biegert A., Lupas A.N. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa S., Marchand I., Dreyfus M. Autoregulation allows Escherichia coli RNase E to adjust continuously its synthesis to that of its substrates. Mol. Microbiol. 2001;42:867–878. doi: 10.1046/j.1365-2958.2001.02687.x. [DOI] [PubMed] [Google Scholar]
- Tock M.R., Walsh A.P., Carroll G., McDowall K.J. The CafA protein required for the 5′-maturation of 16 S rRNA is a 5′-end-dependent ribonuclease that has context-dependent broad sequence specificity. J. Biol. Chem. 2000;275:8726–8732. doi: 10.1074/jbc.275.12.8726. [DOI] [PubMed] [Google Scholar]
- Udekwu K.I., Darfeuille F., Vogel J., Reimegard J., Holmqvist E., Wagner E.G. Hfq-dependent regulation of OmpA synthesis is mediated by an antisense RNA. Genes & Dev. 2005;19:2355–2366. doi: 10.1101/gad.354405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanacova S., Stef R. The exosome and RNA quality control in the nucleus. EMBO Rep. 2007;8:651–657. doi: 10.1038/sj.embor.7401005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanzo N.F., Li Y.S., Py B., Blum E., Higgins C.F., Raynal L.C., Krisch H.M., Carpousis A.J. Ribonuclease E organizes the protein interactions in the Escherichia coli RNA degradosome. Genes & Dev. 1998;12:2770–2781. doi: 10.1101/gad.12.17.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vioque A., Arnez J., Altman S. Protein-RNA interactions in the RNase P holoenzyme from Escherichia coli . J. Mol. Biol. 1988;202:835–848. doi: 10.1016/0022-2836(88)90562-1. [DOI] [PubMed] [Google Scholar]
- Worrall J.A., Luisi B.F. Information available at cut rates: Structure and mechanism of ribonucleases. Curr. Opin. Struct. Biol. 2007;17:128–137. doi: 10.1016/j.sbi.2006.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yehudai-Resheff S., Portnoy V., Yogev S., Adir N., Schuster G. Domain analysis of the chloroplast polynucleotide phosphorylase reveals discrete functions in RNA degradation, polyadenylation, and sequence homology with Exosome proteins. Plant Cell. 2003;15:2003–2019. doi: 10.1105/tpc.013326. [DOI] [PMC free article] [PubMed] [Google Scholar]