Abstract
The human scavenger decapping enzyme, DcpS, functions to hydrolyze the resulting cap structure following cytoplasmic mRNA decay yet is, surprisingly, a nuclear protein by immunofluorescence. Here, we show that DcpS is a nucleocytoplasmic shuttling protein that contains separable nuclear import and Crm-1-dependent export signals. We postulated that the presence of DcpS in both cellular compartments and its ability to hydrolyze cap structure may impact other cellular events dependent on cap-binding proteins. An shRNA-engineered cell line with markedly diminished DcpS levels led to a corresponding reduction in cap-proximal intron splicing of a reporter minigene and endogenous genes. The impaired cap catabolism and resultant imbalanced cap concentrations were postulated to sequester the cap-binding complex (CBC) from its normal splicing function. In support of this explanation, DcpS efficiently displaced the nuclear cap-binding protein Cbp20 from cap structure, and complementation with Cbp20 reversed the reduced splicing, indicating that modulation of splicing by DcpS is mediated through Cbp20. Our studies demonstrate that the significance of DcpS extends beyond its well-characterized role in mRNA decay and involves a broader range of functions in RNA processing including nuclear pre-mRNA splicing.
Keywords: mRNA decapping, mRNA decay, pre-mRNA splicing
INTRODUCTION
Eukaryotic mRNAs possess a cotranscriptional modification at their 5′ end, which consists of an N7 methylated guanine linked to the mRNA by an unusual 5′–5′ linkage (Furuichi et al. 1975). The cap is multifunctional and serves to promote splicing of the first intron (Konarska et al. 1984; Edery and Sonenberg 1985), to transport the mRNA out of the nucleus (Hamm and Mattaj 1990), and to provide translation of the mRNA (Muthukrishnan et al. 1975) and stabilization of the mRNA (Furuichi et al. 1977). The functional determinants of the cap are primarily a consequence of the trans-factors that associate with it. Two of the major cap-binding proteins are the predominantly nuclear cap-binding complex (CBC) (Izaurralde et al. 1994, 1995) and the predominantly cytoplasmic eIF4E (Sonenberg et al. 1978; Marcotrigiano et al. 1997). Association of each protein with the cap confers distinct functional outputs concerning the fate of mRNAs in the nuclear and cytoplasmic compartments. More recently, a third cap-binding protein, scavenger decapping protein DcpS (Liu et al. 2002; Cohen et al. 2004) as well as a novel class of noncanonical cap-binding proteins that preferentially bind the cap only when it is linked to an RNA moiety have been identified and shown to bind the cap and influence its fate. The latter class includes the Dcp2 decapping enzyme (Piccirillo et al. 2003), YB1 (Evdokimova et al. 2001) and PABP (Khanna and Kiledjian 2004) RNA binding proteins, and the decapping inhibitor VCX-A (Jiao et al. 2006).
CBC is one of the first proteins to associate with the 5′ cap of an mRNA (Visa et al. 1996). The CBC heterodimer consists of cap-binding proteins 20 and 80 (Cbp20/80) (Izaurralde et al. 1994; Mazza et al. 2002). Cbp20 is the component that directly contacts the mRNA cap whereas Cbp80 stabilizes this association. This nuclear cap complex functions in multiple processes, including pre-mRNA splicing and RNA export. Following the cotranscriptional association with the 5′ cap, CBC facilitates nascent mRNA splicing through the recruitment of small nuclear RNA-proteins (snRNPs) to facilitate the removal of the first noncoding intron (Izaurralde et al. 1994). Once the mRNA achieves full maturation, CBC performs a subsequent role to ensure the proper transport of RNA from the nucleus to the cytoplasm (Izaurralde et al. 1995). Experiments involving CBC immunodepletion have been the main approach in deciphering its functions. Splicing defects arise in the first intron of a reporter transcript upon CBC depletion, and the compromised splicing has been attributed to a failed formation of a splicing-competent RNP complex (Izaurralde et al. 1994). Similarly, defects in snRNA export arise as a result of CBC depletion (Izaurralde et al. 1995). These studies support the notion that CBC chaperones cap-containing transcripts in its infancy from transcription through export across the nuclear membrane.
The DcpS scavenger-decapping enzyme belongs to a newly identified class of cap-binding proteins that exhibit intrinsic hydrolytic activity. Unlike the Cbp20 and eIF4E cap-binding proteins, DcpS plays a role during an mRNA's terminal stage and exists natively as a dimeric receptor that has the potential to bind two cap ligands simultaneously (Gu et al. 2004). DcpS utilizes an evolutionarily conserved Histidine Triad (HIT) motif (Liu et al. 2002) to hydrolyze the triphosphate linkage of the cap structure and is the only known cap-binding protein that hydrolyzes its substrate (Liu et al. 2002). Despite its purification from cytoplasmic extract (Liu et al. 2002) and its well-characterized cytoplasmic decapping function (Liu et al. 2002; van Dijk et al. 2003; Cohen et al. 2004), DcpS is predominantly if not exclusively a nuclear protein by immunocytochemistry (Salehi et al. 2002; Cougot et al. 2004; Liu et al. 2004).
Eukaryotic mRNAs are primarily degraded by exonucleolytic decay pathways involving decay from either mRNA termini. In the 3′ to 5′ decay pathway, following deadenylation, the mRNA is degraded from the 3′ end by the exosome complex (Mitchell et al. 1997) until a few nucleotides extend off the cap, which is subsequently hydrolyzed by the DcpS enzyme (Wang and Kiledjian 2001). As such, DcpS performs the final step of clearing the residual cap to prevent its potential deleterious accumulation. Previous work illustrates that DcpS can displace eIF4E from the cap structure based on competition assays (Liu et al. 2004), suggesting a regulatory interplay between cap-binding proteins. Recent experiments reveal an auto-regulatory mechanism whereby DcpS positively feeds back onto the 5′–3′ directional mRNA degradation pathway (Liu and Kiledjian 2005). Despite these studies that emphasize cytoplasmic functions, the unexpected DcpS nuclear localization (Cougot et al. 2004; Liu et al. 2004) strongly indicates an unknown nucleus-associated function.
In the current study, we provide findings that functionally define a novel role for nuclear DcpS. We use an interspecies heterokaryon assay to show that DcpS is a nucleocytoplasmic shuttling protein. It employs one canonical basic nuclear localization sequence for nuclear residence, and it requires a conventional leucine-rich sequence for nuclear export. Furthermore, DcpS imposes positive regulation on nuclear splicing mediated by CBC. The collective data depict DcpS as a molecule that modulates cap concentrations for cellular homeostasis of gene expression activities involving at least RNA processing.
RESULTS
DcpS is a shuttling protein
Previous immunofluorescent data have shown the subcellular localization of DcpS to be nuclear (van Dijk et al. 2003; Liu et al. 2004). The observation was surprising given that DcpS has been purified and identified from mammalian cytosolic S130 extract, and its proposed hydrolytic function follows mRNA degradation by the cytoplasmic-resident exosome (Wang and Kiledjian 2001; Liu et al. 2002). The nuclear localization of DcpS and its known cytoplasmic function suggested that DcpS is a nucleocytoplasmic shuttling protein with a dynamic subcellular distribution. To experimentally test this possibility, we utilized a construct expressing GFP-tagged DcpS that localizes to the nucleus at steady state (Fig. 1A) analogous to the endogenous protein (Liu et al. 2004). Interspecies heterokaryon assay that provides a qualitative assessment of a protein's mobility between nuclear and cytoplasmic compartments was used (Piñol-Roma and Dreyfuss 1992). This assay relies on distinct fluorescence properties between human and mouse nuclei, which enable tracking of a protein expressed in one cell type to be detected if localized in another following cell membrane fusion.
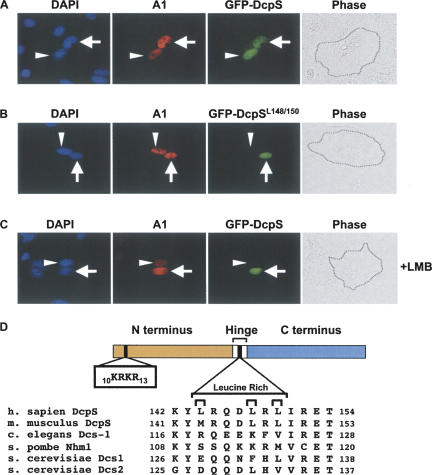
FIGURE 1.
DcpS is a nucleocytoplasmic shuttling protein that requires a leucine-rich Nuclear Export Sequence (NES). Human Hela cells co-transfected with constructs expressing GFP-DcpS and the shuttling myc-hnRNPA1 positive control were fused to mouse NIH3T3 cells. The nuclei are designated by DAPI staining. The Hela cell nuclei is denoted by the arrow and the punctate nuclei indicative of mouse cells is shown by the arrow head. The red staining denotes localization of the myc-hnRNPA1 while the green signal represents GFP-DcpS. The phase contrast of the cells is shown on the right panel with the black dotted lines indicating the fused plasma membrane of the heterokaryons. (A) Similar to the myc-hnRNPA1, GFP-DcpS can exit the human nucleus and be re-imported into the mouse nucleus. (B) A similar interspecies heterokaryon assay is shown except a mutant GFP-DcpS containing substitutions of leucine 148 and 150 to alanines (GFP-DcpSL148/150A) was used. (C) Heterokaryon assay carried out in the presence of leptomycine B (+LMB) is shown. DcpS requires the leucine-rich region for nuclear export in a LMB sensitive manner. (D) A schematic of DcpS is shown with the distinct N- and C-terminal fragments separated by a hinge region. The relative position of the putative N-terminal NLS from amino acids 10 through 13 and putative leucine-rich NES from amino acids 144 through 150 in the hinge region are bracketed with the corresponding sequences expanded. Alignment of the leucine-rich region of DcpS homologs across different species with characterized subcellular localizations are shown at the bottom.
Constructs expressing GFP-tagged DcpS were transfected into human HeLa cells exclusively followed by fusion with mouse 3T3 cells in the presence of cycloheximide to cease ongoing translation. The resulting interspecies heterokaryons (human-mouse fused cells) were further incubated to allow active protein transport to proceed. A construct encoding the well-characterized shuttling hnRNP A1 protein (Piñol-Roma and Dreyfuss 1992) was co-transfected to serve as a positive control for the heterokaryon assay. Similar to the shuttling observed with myc-hnRNP A1, the GFP-DcpS signal appears in both the human and mouse nuclei (Fig. 1A). The presence of the tagged proteins in the mouse nuclei is an outcome originating from protein exported from the human nucleus and subsequently imported into the mouse nucleus. Therefore, DcpS is a shuttling protein that can be exported from the nucleus.
DcpS contains a canonical nuclear export signal
Our demonstration that DcpS is a shuttling protein indicated that this protein contains a nuclear export sequence. Consistent with this property, a putative leucine-rich nuclear export signal was detected at the central hinge domain of the protein (Fig. 1D). To experimentally test the functional relevance of the leucine-rich region in nuclear export, we designed a GFP-tagged DcpS harboring double alanine substitutions of two leucine residues at amino acid positions 148 and 150 (DcpSL148/150A) that are critical for nuclear export function of previously characterized leucine-rich regions (Wen et al. 1995). Figure 1B shows that in contrast to the shuttling exhibited by myc-tagged hnRNP A1, the GFP-DcpSL148/150A signal was exclusively detected in the human cell nucleus of the same heterokaryon, denoting a defect in protein nuclear export. To further substantiate the significance of the leucine-rich region in export, we tested DcpS shuttling in the presence of leptomycin B (LMB) treatment. LMB is an antagonist of Crm1, a protein that recognizes leucine-rich nuclear export sequences and transports them out of the nuclear compartment (Kudo et al. 1999). As shown in Figure 1C, DcpS fails to shuttle within the heterokaryon assay in the presence of LMB and remains in the HeLa nucleus similar to the result observed with the DcpSL148/150A leucine mutant. As expected of the control protein, myc-tagged hnRNP A1 was unaffected by LMB due to its noncanonical nuclear export sequence (Michael et al. 1995). Interestingly, an alignment of the leucine-rich nuclear export signal from several different species where the localization of DcpS has been tested reveals a conservation of this region in the nuclear human and mouse DcpS proteins but diverges in the Caenorhabditis elegans DCS-1 and Saccharomyces cerevisiae Dcs1p, orthologs of DcpS, as well as the catalytically inactive paralog of Dcs1p in S. cerevisiae, Dcs2p (Fig. 1D), that appear to be exclusively cytoplasmic (Malys et al. 2004; Lall et al. 2005). Curiously, the Schizosaccharomyces pombe homolog, Nhm1, is nuclear (Salehi et al. 2002) but also lacks a clear conservation of the leucine-rich sequence (Fig. 1D). Collectively, the data demonstrate that human DcpS is a shuttling protein and supports a role for DcpS in the Crm1-mediated export pathway via its leucine-rich nuclear export signal located in the central hinge domain of the protein and suggest acquisition of the leucine-rich element may be a more recent evolutionary event.
DcpS contains a basic region nuclear localization signal (NLS)
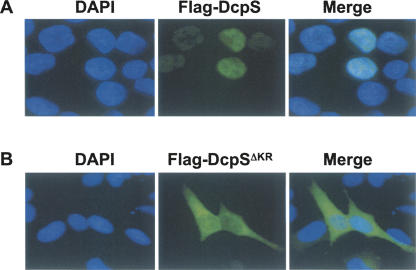
To address the means by which DcpS is imported into the nucleus, we identified its NLS. Inspection of the N-terminal region of DcpS reveals a stretch of basic amino acids (KRKR) that are suggestive of a nuclear localization signal (Fig. 1D). To validate this prediction, we designed a construct expressing a Flag-tagged DcpS containing the four basic amino acid deletion within the N terminus, specifically amino acids 10 through 13 (DcpSΔKR). Consistent with localization of endogenous DcpS (Liu et al. 2004), Flag-tagged DcpS is detected predominantly in the nucleus of HeLa cells transfected with the epitope tagged DcpS expression plasmid (Fig. 2A). In contrast, epitope tagged DcpSΔKR staining was predominantly cytoplasmic, demonstrating that this region is necessary for nuclear localization (Fig. 2B). Therefore, similar to other proteins that utilize a conventional NLS, human DcpS relies on a region of basic amino acids located in the N-terminal domain of the protein for nuclear protein import. Interestingly, the basic NLS and hydrophobic-rich NES of the scavenger decapping protein are conserved exclusively in human and mouse (Gu and Lima 2005). The cytoplasmic C. elegans DcpS homolog Dcs-1 (Lall et al. 2005) and S. cerevisiae DcpS homolog Dcs1 (Malys and McCarthy 2006), lack the first 30 and 31 amino acids, respectively, corresponding to human DcpS, which includes the NLS. In the case of the catalytically inactive S. cerevisiae Dcs2, the NLS region is not conserved, and we are unable to decipher another putative NLS (data not shown). The S. pombe homolog of DcpS, Nhm1, is predominantly nuclear but also present in the cytoplasm (Salehi et al. 2002); however, neither an NLS nor NES could be readily discerned based on examination of the amino acid sequence (Gu and Lima 2005).
FIGURE 2.
Identification of the DcpS nuclear import sequences. Hela cells transfected with either (A) wild-type Flag-DcpS or the (B) mutant construct containing a four amino acid truncation of amino acids 10–13 Flag-DcpSΔKR. The left panel demarcates the position of the nuclei with DAPI staining and the DcpS localization is denoted by the green fluorescence and identified with the use of the anti-Flag monoclonal antibody. A merge of the two panels is shown on the right.
A nuclear role for DcpS in facilitating first intron mRNA splicing
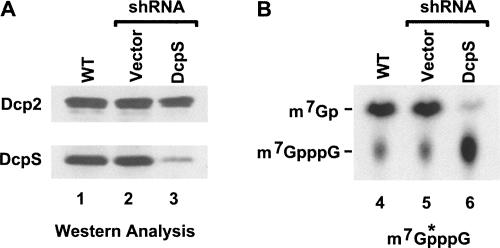
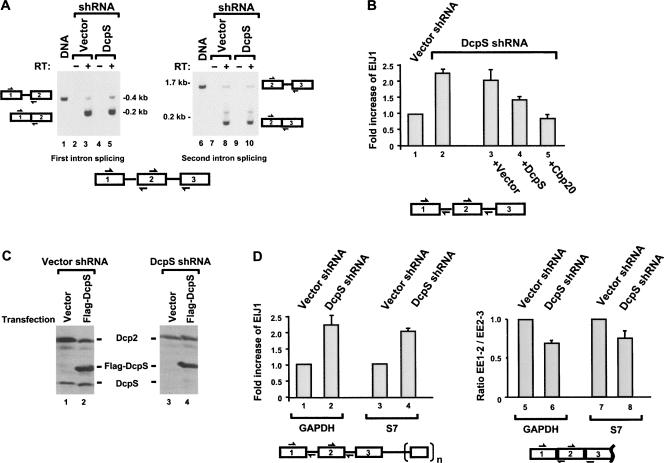
To begin addressing the cellular function of DcpS, we generated a cell line that was constitutively knocked down for DcpS. Human kidney 293T cells were stably transformed with a shRNA expression plasmid specific for DcpS transcribed by a Pol III-directed U6 promoter. Single colonies were expanded and tested for their efficiency of DcpS protein and activity knockdown, and the most significantly silenced clonal cell line was expanded (293TDcpS-KD). As shown in Figure 3A, DcpS levels are reduced in the 293TDcpS-KD cells greater than 90% relative to 293T cells expressing the shRNA empty vector by Western blot (cf. lanes 1–3). Similarly, an in vitro decapping assay confirmed that scavenger decapping activity detected from extract obtained from the knockdown cells was reduced by greater than 80% relative to the control extract (cf. lanes 4–6) in Figure 3B. Aside from its tendency to grow as discrete cells in a nonaggregated form, the 293TDcpS-KD cell line does not pose an additional unique phenotype in comparison to the empty vector cell line. These data demonstrate that the 293TDcpS-KD clonal cell line contains reduced DcpS expression and a corresponding reduction in cap hydrolytic activity.
FIGURE 3.
A knockdown clonal cell line is diminished in DcpS expression and catalysis. A clonal 293T cell line stably transformed with pShag-DcpS expressing shRNA specific to DcpS was used in a western assay (A) and decapping activity assay (B). 293T cells not expressing an shRNA (lane 1), expressing the shRNA vector (lane 2), or shRNA to DcpS (lane 3) are shown with antibodies to either the human Dcp2 protein or DcpS as indicated. DcpS protein levels are reduced in the 293T cells expressing the DcpS-specific shRNA. The right panel shows a decapping assay of labeled cap structure incubated with wild-type 293T cell extract (lane 4), 293T cell extract expressing the empty shRNA vector (lane 5), or 293T cell extract from cells expressing the DcpS-specific shRNA (lane 6). Reaction products were resolved on PEI TLC plates. Standards were developed on the TLC simultaneously, visualized by ultraviolet shadowing and their migration indicated on the left. The labeled cap structure used in the reaction is schematically depicted on the bottom, where the asterisks denote the labeled phosphate.
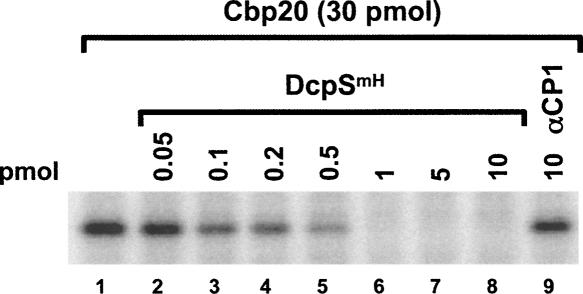
In an effort to begin addressing potential nuclear function(s) of DcpS, we reasoned that the ability of DcpS to hydrolyze cap structure would be essential in modulating the activity of cap-binding proteins since cap structure would otherwise accumulate in the absence of DcpS. Having previously demonstrated that DcpS can efficiently displace eIF4E from cap structure (Liu et al. 2004), we tested whether the same was also true with the nuclear CBC. To begin addressing the relationship between CBC and DcpS, we initially used UV cross-linking to determine whether DcpS could compete the cap-binding potential of the Cbp20 cap-binding component of CBC. Analogous to the ability of DcpS to displace eIF4E from cap structure (Liu et al. 2004), DcpS could readily outcompete Cbp20 from the cap structure (Fig. 4, lanes 2–8) while a control RNA-binding protein, αCP1, could not (lane 9). Although this is an in vitro study with the Cbp20 monomer in the absence of the Cbp80 component, it is suggestive that DcpS activity could be a modulator of CBC access to the cap and, in turn, its function.
FIGURE 4.
DcpS can displace Cbp20 from cap structure. The ability of DcpS to displace Cbp20 from the cap structure was tested. Thirty picomoles of histidine tagged Cbp20 were prebound to 32P-labeled cap structure on ice for 10 min, followed by addition of the indicated amounts of catalytically inactive histidine tagged DcpSmH for an additional 10 min (lanes 2–8). The reactions were subsequently UV cross-linked and resolved by SDS-polyacrylamide gel. The αCP1 RNA-binding protein was used as a control (lane 9). DcpS can efficiently displace Cbp20 from the cap structure at substoichiometric concentrations.
CBC has been shown to serve multiple functions in the nucleus, including facilitation of first intron splicing (Konarska et al. 1984; Edery and Sonenberg 1985; Izaurralde et al. 1994). Exons are defined nucleotide sequences flanked by spliceosomal complexes that assemble at the upstream 3′ and downstream 5′ splice sites. One minor deviation concerns the first exon in which the absence of an upstream spliceosomal component is compensated by the presence of CBC cap recognition (Berget 1995). Depletion of CBC in HeLa extract has been shown to reduce splicing of the cap-proximal intron (Lewis et al. 1996). Given DcpS activity in cap hydrolysis and its ability to displace Cbp20 from cap structure, we reasoned that a reduction of DcpS levels could also result in an accumulation of cap structure in the nucleus, which, in turn, could sequester CBC and reduce splicing of the first intron. To assess whether a reduction of DcpS levels can impact first intron splicing, a rat fibronectin minigene reporter construct that expresses a pre-mRNA consisting of three exons and two introns was transfected into 293TDcpS-KD cells to assess the in vivo splicing efficiency of the first intron relative to the second intron. Following a 36-h post-transfection period, RNA was isolated and splicing analyzed by reverse transcription PCR. A primer pair that anneals to the first and second exons was used to detect the unspliced and spliced RNA forms of the first intron. As shown in Figure 5A, a qualitative difference is detected where a noticeable accumulation of unspliced RNA is detected in the splicing of the first intron in 293TDcpS-KD cells in contrast to that of the control cell line (cf. lanes 3 and 5). However, the splicing reaction in the second intron appears to be largely unaffected between the two cell lines (cf. lanes 8 and 10).
FIGURE 5.
DcpS can influence splicing of the first intron in cells. (A) DcpS affects the splicing efficiency of the rat fibronectin minigene reporter pre-mRNA first intron. Stable 293T cell line of either control empty shRNA or DcpS shRNA background was transfected with the rat fibronectin minigene and was analyzed 36 h post-transfection by reverse transcription PCR assays. Qualitative presence of intron 1 splicing (left panel) or intron 2 splicing (right panel) was determined with primer pairs that monitor the splicing status as shown in the schematic on the side of each panel and summarized at the bottom. The boxes with the numbers denote the exons and the lines represent the introns. DNA size markers are shown to the side of each panel. PCR products corresponding to the unspliced intron 1 and unspliced intron 2 obtained from amplification of the plasmid expressing the rat fibronectin transcript are shown in lanes 1 and 6, respectively. (B) Splicing reactions analogous to A were analyzed by quantitative real-time PCR using the primer sets that span the exon–intron junctions shown schematically below the panel. The EIJ1/EIJ2 ratio obtained from the control shRNA vector was arbitrarily set to one (lane 1) and all other values are presented relative to this control. A twofold increase in the detection of EIJ1 relative to that of EIJ2 is observed in the 293T-DcpSKD cells (lane 2), an indication of a preferential splicing defect in the first intron. Complementation with either the empty vector (lane 3) or vector expressing DcpS (lane 4) or Cbp20 (lane 5) is shown. Quantization of three independent experiments performed in duplicate and the corresponding standard deviation denoted by the error bars are shown. (C) Expression of Flag-DcpS in 293T-DcpSKD cells. Control and 293T-DcpSKD cells were transfected with either a vector construct or Flag-DcpS expression construct as indicated and expression of both endogenous and exogenous DcpS detected by Western analysis using a DcpS antibody. The level of Dcp2 was tested as a control. Expression of exogenous Flag-DcpS is high enough to overcome the DcpS shRNA. (D, left panel) Quantitative real-time PCR was carried out to detect the EIJ1 fold increase relative to that of EIJ2 as in B for the indicated endogenous genes from RNA obtained from the control or DcpS-specific shRNA expressing 293T cell lines. The exons and introns are schematically shown on the bottom with n = 6 for GAPDH and n = 4 for S7. GAPDH and S7 genes exhibit a twofold preferential increase of EIJ1 in 293T-DcpSKD cells (lanes 2,4) relative to control cells (lanes 1,3). (Right panel) Quantitative real-time PCR analysis was carried out analogous to that above except primers that spanned exon–exon 1–2 (EE1–2) and exon–exon 2–3 (EE2–3) were used as indicated in the schematic below and short elongation times were used such that the spliced form was exclusively detected. Quantization of three independent experiments carried out in duplicate and corresponding standard deviation denoted by the error bars are shown.
To obtain a quantitative assessment of the DcpS effect on differential splicing of the reporter pre-mRNA, quantitative real-time PCR was carried out with primer pairs that anneal to an exon and an immediately downstream intron (Fig. 5B). Reverse transcribed RNA from cells transfected with the rat fibronectin reporter construct was monitored to determine first intron splicing efficiency in the 293TDcpS-KD cells or control cells. Relative unspliced RNA levels of the first intron (exon–intron junction 1; EIJ1) were measured in reference to the relative unspliced RNA amount of the second intron, exon–intron junction 2 (EIJ2). Therefore the EIJ1/EIJ2 ratio reflects the fold splicing defect observed of the first intron normalized to that of the second intron upon reduction of DcpS. The data illustrate that splicing of the reporter transcript exhibited a twofold greater splicing defect of the first intron in the 293TDcpS-KD cells relative to the vector control cell line (Fig. 5B). In order to confirm that the effect on splicing is attributed to DcpS, we co-transfected along with the reporter a construct expressing Flag-tagged DcpS to rectify the processing defect. Flag-DcpS is expressed at sufficient levels to overcome the knockdown effect of the DcpS-specific shRNA (Fig. 5C). DcpS overexpression partially corrects the splicing defect in the first intron (Fig. 5B, lane 4) while expression of a control plasmid failed to complement (lane 3). Moreover, Cbp20 overexpression was able to completely reverse the splicing defect (lane 5), indicating that the reduced splicing of intron one was due to the limiting availability of Cbp20 in the DcpS knockdown cells. Taken together, the in vivo splicing experiments consistently indicate that DcpS influences proper first intron splicing of an exogenously expressed gene.
The chimeric rat fibronectin minigene used above contains three exons and two introns. To determine if an influence on first intron splicing could also be seen for an endogenous multi-intron-containing gene, we compared the relative splicing of the first and second introns of two endogenous genes. For this purpose, two relatively abundant mRNAs were used to enable more efficient detection of the pre-mRNA species, GAPDH and the ribosomal S7 genes. The same quantitative real-time PCR strategy was carried out to determine the splicing defect about the first intron in reference to that of the second. Similar to the results obtained with the rat fibronectin minigene, an approximate twofold-splicing defect was observed for the splicing of the first intron relative to the second intron for both genes in the 293TDcpS-KD cells (Fig. 5D, cf. lanes 1–2 and 3–4).
The above analyses were carried out by comparing the relative proportions of EIJ1 to EIJ2. To rule out the formal possibility that higher levels of EIJ1 relative to EIJ2 in the DcpS knockdown cell line reflected increased aberrant pausing or abortive transcription in the 293TDcpS-KD cells, we alternatively conducted real-time PCR on the levels of spliced exonic boundaries. Quantitative real-time PCR was carried out as above except that the primers and PCR parameters were designed to detect the spliced exon–exon junctions exclusively. The fold change of spliced exon–exon 1–2 (EE1–2) mRNA levels relative to spliced exon–exon 2–3 (EE2–3) mRNA levels were determined by real-time PCR. If reduction of DcpS levels results in an increased incidence of transcriptional pauses or termination, a greater level of the upstream spliced exon–exon junction would be detected relative to the next downstream spliced exon–exon junction; that is, the ratio of EE1–2 to EE2–3 would be greater than one. Consistent with a regulation at the level of splicing, rather than transcription pausing, a relative decrease in EE1–2 in relation to EE2–3 was observed (Fig. 5D, cf. lanes 6–5 and 8–7). However, the decrease is not as dramatic as that observed when measuring the pre-mRNA in lanes 1–4 since the abundance of the spliced products in the latter analysis would skew the results relative to the less abundant pre-mRNA. Collectively, these data provide evidence for a novel nuclear function for DcpS in maintaining proper pre-mRNA splicing.
DISCUSSION
The current study demonstrates that DcpS, the scavenger-decapping protein previously thought to exclusively function in mRNA turnover, is a nucleocytoplasmic shuttling protein with broader functionality as a modulator of cap-binding proteins. Heterokaryon assays show that DcpS is a shuttling protein and relies on an N-terminal NLS for import and leucine-rich consensus NES located in the central hinge region for nuclear export. The presence of functional NLS and NES provides a clear explanation of a nuclear protein extending its influence over cytoplasmic events. We show that DcpS can compete Cbp20 from cap structure at substoichiometric concentrations. A splicing reporter assay confirmed the expectation that DcpS modulates CBC-associated functions where splicing activity was notably reduced in the DcpS knockdown cell line 293TDcpS-KD. Rescue experiments show that both DcpS and Cbp20 overexpression can restore the defect, supporting the idea that DcpS positively modulates pre-mRNA processing. More specifically, DcpS regulation preferentially influences the first intron, a result consistent with previous findings of CBC's preferential regulation on the cap-proximal intron. This study extends the primary scope of DcpS regulation beyond mRNA degradation to earlier events involved in RNA maturation. Furthermore, it is interesting that Cbp20 was able to completely restore splicing in the 293TDcpS-KD cell line while DcpS partially restored the splicing defect. The reason for this intriguing discrepancy is not clear and under investigation.
Our demonstration that the primarily nuclear localized DcpS is a shuttling protein with cytoplasmic residency addresses an apparent paradox for the function of DcpS. We, and others, have previously reported a cytoplasmic scavenger-decapping activity (Liu et al. 2002, 2004; van Dijk et al. 2003; Liu and Kiledjian 2005) from a protein residing mainly in the nucleus. The shuttling capacity of DcpS has been confirmed based on the identification of separable nuclear import and export signals (Figs. 1 and 2) in its amino acid sequence and by immunocytochemistry using an interspecies heterokaryon assay (Fig. 1). Nuclear import of DcpS requires a short stretch of basic amino acids at the N terminus of the protein (Fig. 2) consistent with the canonical SV40 large T antigen basic NLS (Kalderon et al. 1984). Interestingly, the DcpS NLS falls within an unstructured N terminus of the protein (Gu et al. 2004) that is dispensable for hydrolytic activity (Liu et al. 2004).
Conforming to the shuttling property of DcpS, a nuclear export signal consisting of a leucine-rich region (Wen et al. 1995) closely matching the consensus leucine-rich NES, LXXXLXL is present in the protein (L denotes leucine and X any amino acid) (Kutay and Guttinger 2005). Confirmation for the functional significance of this region in nuclear export is provided by the substitution of the two highly conserved leucine residues to alanines (L148/150A) that perturbed export (Fig. 1B). As expected for this class of export signals, the exit of DcpS from the nucleus is inhibited by LMB, indicating that DcpS export is Crm1 mediated. Positioning of the leucine-rich export signal lies notably within the hinge region connecting the N- and C-terminal domains of DcpS. One possible consequence for positioning of the NES to the hinge is that the central region provides an interface for protein interaction that would likely lock the N-terminal segment of the DcpS homodimer with respect to the C terminus to inhibit catalytic activity and could be a mechanism to modulate hydrolytic activity during nucleocytoplasmic transport.
The current findings reveal that the predominantly nuclear residency and shuttling capacity of DcpS in mammals are driven by the classical NLS and NES located in the N-terminal and central hinge domains, respectively. However, the absence of a corresponding NLS and NES in the C. elegans and S. cerevisiae orthologs of the human DcpS protein are consistent with the cytoplasmic localization of these proteins (Malys et al. 2004; Lall et al. 2005) and suggest that a nuclear DcpS function may not be uniformly conserved throughout all species. Other than the mammalian DcpS, the exception thus far is the S. pombe Nhm1, which is also nuclear (Salehi et al. 2002). The absence of a leucine-rich NES in Nhm1 suggests this protein either does not shuttle or uses another element to fulfill this function. Whether certain species have evolved to confine the DcpS orthologs in one compartment to restrict its influence while mammals have adapted to utilize DcpS in both compartments remains to be more thoroughly addressed.
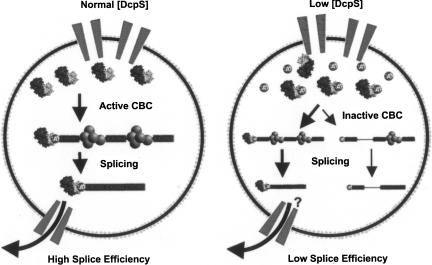
The ability of DcpS to hydrolyze the resulting cap structure end product of 3′ to 5′ exonucleolytic mRNA decay that would otherwise accumulate (Fig. 3B; Liu and Kiledjian 2005) and potentially sequester cap-binding proteins implicates DcpS in cellular functions beyond mRNA decay. We propose DcpS is essential to relieve CBC from cap sequestration to maintain normal levels of protein synthesis and mRNA maturation by maintaining cellular homeostasis by buffering against the potentially adverse effects of cap accumulation. Normally, mRNAs that are targeted into the 3′ to 5′ mRNA decay pathway become degraded to a short oligo-cap structure or cap structure by the exosome (Liu et al. 2002), a product which in turn serves as a substrate for the subsequent hydrolytic activity of DcpS (Wang and Kiledjian 2001). A model can be envisioned whereby DcpS depletion leads to the aberrant accumulation of free cap structure, which otherwise would have been hydrolyzed or captured by DcpS. Consequently, excess cap structure in the nucleus could serve to compete and sequester CBC from their mRNA substrates (Fig. 6, right panel). The net effect of DcpS depletion would be a reduction in the active pool of cap-binding proteins leading to altered phenotypes of cap-binding protein-dependent processes, including pre-mRNA splicing. The established role of CBC in mRNA transport out of the nucleus suggests this process could also be affected by DcpS activity, although this still remains to be tested. Furthermore, the significance of the cap and eIF4E in mRNA translation suggests translation may also be impacted by DcpS through the modulation of eIF4E. Therefore, DcpS appears to be a modulator of cap structure concentrations in a cell, and this activity could in turn impinge on multiple post-transcriptional processes. Further support for this premise is provided with the demonstration that disruption of the DcpS homolog in yeast, Dcs1, leads to an accumulation of cap structure (Liu et al. 2002) and a marked reduction of 5′ to 3′ exonucleolytic activity (Liu and Kiledjian 2005).
FIGURE 6.
Model depicting consequence on nuclear splicing in the presence or reduction of DcpS. A cartoon of a nucleus containing wild-type levels of DcpS is shown on the left panel and a nucleus with reduced DcpS is shown on the right. DcpS reduction leads to an increase in the level of cap structure (sphere). The accumulating cap structure leads to a reduction in the active pool of nuclear CBC (heterodimer subunits) through sequestration, and promotes a splicing defect of the first intron in a subset of mRNAs possibly through incomplete spliceosomal complex (pentamer spheres) assembly on the pre-mRNA.
To our knowledge, the regulatory function of DcpS on splicing presented in this study provides the first cell-based confirmation in mammalian cells for the significance of CBC in splicing of the cap-proximal intron. These results extend previous studies with mammalian in vitro splicing (Konarska et al. 1984; Edery and Sonenberg 1985; Izaurralde et al. 1994) and suggest the regulation of cap structure could be a natural means to influence splicing. The results also provide a framework for future studies to address the potential broader impact on alternative utilization of the first intron in splicing and raise numerous new questions. Approximately 60% of human genes are predicted to contain their translation start site in the first exon (Davuluri et al. 2001). Alterations in the efficiency of intronic inclusion or exclusion could dramatically affect expression from these genes since failure to remove the intron would most likely result in the ribosome encountering a premature stop codon within the intron and subsequent decay of the mRNA by the nonsense-mediated decay pathway (Lejeune and Maquat 2005; Chang et al. 2007). The demonstration that GAPDH and S7 splicing are affected indicates that DcpS can influence endogenous pre-mRNA splicing, and this might be a general function of DcpS. However, it remains to be determined whether all pre-mRNAs are equally impacted by cap structure or whether a certain subset is more susceptible. Further studies are necessary to test the global impact of this previously unanticipated role of DcpS in the potential regulation of first intron splicing.
Although the functional assignment of DcpS extends from general mRNA degradation to pre-mRNA splicing, the importance of the asymmetric distribution whereby DcpS is predominantly nuclear than cytoplasmic remains to be addressed. Identification of a functional role for DcpS in splicing provides a novel avenue to pursue nuclear functions for this protein. An additional function postulated for this family of proteins includes the removal of methylated nucleotides within the nucleus to prevent their misincorporation into RNA during transcription (van Dijk et al. 2003; Malys and McCarthy 2006), although this has yet to be demonstrated. Maintaining the idea of DcpS influence over cap-mediated processes, eIF4E-dependent mRNA translation as well as miRNA-guided translational repression may be additional points of regulatory interest for the cap-hydrolyzing protein. Cytoplasmic sequestration of eIF4E as a consequence of cap structure accumulation would be expected to result in a decrease of mRNA translation. The ability of DcpS to effectively compete eIF4E from cap structure (Liu et al. 2004) supports this hypothesis, and current studies are underway to test whether DcpS has a modulatory role in mRNA translation.
Although the precise mechanism of translational repression by miRNAs is still unclear, inhibition of translation initiation is one likely possibility (Pillai et al. 2005). Interestingly, the recent demonstration that Argonaute 2 (Ago2) is a cap-binding protein raises important questions for the role of DcpS in miRNA-mediated translational suppression. One model posits that the nucleation of Ago2 onto the 3′ UTR of an mRNA through its guide miRNA provides a wider window of opportunity for its cap-binding domain to access the upstream 5′ cap and thus prevent eIF4E from initializing subsequent rounds of ribosome loading (Kiriakidou et al. 2007). Whether the DcpS activity could also influence the cap-binding property of Argonaute 2 and miRNA-mediated translation inhibition remains to be determined.
Collectively, the data support a multifunctional role for the methyl-guanosine cap hydrolyzing human DcpS enzyme at several levels of mRNA metabolism. An important future area to address involves what additional cellular functions are modulated by DcpS and how DcpS activity is regulated. Studies in C. elegans and S. cerevisiae provide initial insight into potential regulation of DcpS. DcpS orthologs in both these organisms are induced in response to stress (Gasch et al. 2000; Kwasnicka et al. 2003; Malys et al. 2004), raising the appealing possibility that DcpS and its downstream functions react to environmental conditions of cellular stress. Future studies will aim to unravel additional targets of DcpS and begin to identify its direct upstream regulators.
MATERIALS AND METHODS
Plasmid constructs
The pcDNA3-Flag-DcpS plasmid that expresses Flag-tagged DcpS in mammalian cells was made by inserting the DcpS open reading frame flanked by a BamHI site at the 5′ end and an XhoI site at the 3′ end into the pcDNA3-Flag (Fenger-Gron et al. 2005) vector. pcDNA3-Flag-DcpSΔKR containing a deletion of amino acids 10–13 of DcpS was derived from pcDNA3-Flag-DcpS construct by using site-directed mutagenesis (Stratagene) according to the manufacturer's instructions. The pEGFP-DcpS plasmid encoding GFP-DcpS was constructed by inserting the DcpS open reading frame into the EcoRI and BamHI sites of the pEGFP-C1 (Clontech) vector. pEGFP-DcpSL148/150A was made by using two rounds of site-directed mutagenesis of the parental pEGFP-DcpS vector. The DcpS-specific shRNA expression construct, pSHAG1-puro-DCPS, was generated by inserting a double-stranded shRNA that encodes the following mature RNA: 5′ TGCAGTTCTCCAATGATATCTACAGCAC 3′ into the BseRI and BamHI sites of the pSHAG1-puro vector (Paddison et al. 2002) modified with the addition of a puromycin selection marker. The pSHAG1 construct was kindly provided by Dr. Gregory J. Hannon (Cold Spring Harbor Laboratory, New York). The pcDNA3-Flag-Cbp20 construct was designed by inserting the Cbp20 open reading frame derived by reverse transcription and PCR-amplification from human K562 cell RNA. The PCR product containing a BamHI and EcoRI site was inserted into the same sites of the pcDNA3-Flag plasmid. pcDNA3-myc-PK-hnRNPA1 was kindly provided by Dr. Serafin Piñol-Roma (CUNY Medical School, New York) and has been previously described (Michael et al. 1995; Siomi and Dreyfuss 1995). The rat fibronectin reporter construct, p773B, was kindly provided by Dr. Samuel Gunderson (Rutgers University, New Jersey) and previously described (Norton and Hynes 1990). The His-Cbp20 construct was kindly provided by Dr. Stephen Cusack and has been previously described (Mazza et al. 2002). Sequences for all primers are listed in Supplemental Table 1.
Cell lines and transfections
HeLa, NIH3T3, and 293T cell lines were grown in DMEM (Invitrogen) supplemented with 10% fetal bovine serum, penicillin–streptomycin, sodium pyruvate under 5% CO2 at 37°C. 293T cells were grown in the presence of 80 μg/mL G418. Unless otherwise stated, all transfections were carried out with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. The 293T cell line stably transformed with the construct expressing an shRNA targeting DcpS (293TDcpS-KD) or expressing the empty pSHAG1-puro vector were generated by transfecting pSHAG1-DcpS or pSHAG1-puro, respectively. After 48 h of transfection, cells were placed under 0.5 mg/mL G418 and 3 μg/mL puromycin selection. Clonal cell lines were isolated by plating the population into a 96-well plate such that one colony occupied a single well under puromycin and G418 selection, and the colony with the most efficient DcpS protein level reduction was expanded and used.
Heterokaryon assay
Heterokaryon assays were carried out as described (Piñol-Roma and Dreyfuss 1992) with minor modifications. Briefly, HeLa cells were co-transfected with 4 μg pcDNA-Flag-DcpS and 4 μg pcDNA3-myc-PK-hnRNPA1 in 60-mm plates. 3T3 cells were seeded onto coverslips in a six-well plate on the same day. At 24 h post-transfection, HeLa cells were trypsinized and seeded onto 3T3 cells at a density 2 times that of the mouse cells. Cell attachment to the coverslip was allowed for 5 h, followed by addition of 100 μg/mL cycloheximide 1 h prior to cell fusion. Human and mouse cells were fused by 50% PEG 3350 (Sigma) for 90 sec. Cells were washed of residual PEG and incubated for an additional 4 h in the presence of cycloheximide prior to visualization by indirect immunofluorescence. Where indicated, 20 ng/mL LMB was added to the cell during the seeding following trypsinization and maintained in LMB throughout the duration of the experiment.
Immunofluorescence and Westerns
Mouse anti-Flag antibody (Sigma) was used at a dilution of 1:10,000 for Western and 1:500 for immunofluorescence. Rabbit anti-myc (Santa Cruz) was used at a dilution of 1:200 for immunofluorescence. Secondary anti-rabbit and anti-mouse (ImmunoJackson) were used at a dilution of 1:200 for immunofluorescence. Cells were fixed and permeabilized as previously described (Liu et al. 2004) and visualized using the Axiovision 4.1 software and an Olympus 100 M Axiovert microscope.
UV cross-linking
UV cross-linking reactions were carried out as described by Liu et al. (2004). Briefly, 30 pmol of His-CBP20 were preincubated with 32P-labeled cap structure (2–5K cpm) in IVDA buffer on ice for 10 min, followed by the addition of the indicated catalytically inactive DcpS mutant containing histidine 277 to asparagine substitution (His-DcpSmH). Following a 10-min incubation, the samples were covalently cross-linked by exposure to a 15 W germicidal UV lamp for 10 min, and the consequent cross-linking samples were resolved on a 12.5% SDS-PAGE and visualized by autoradiography.
Splicing reporter assay
293TDcpS-KD cells were transfected with the p773B rat reporter construct with Lipofectamine 2000 in G418. Cells were harvested 36 h post-transfection and total RNA isolated with Trizol Reagent (Invitrogen). cDNA was generated with MMLV reverse transcriptase (Promega) according to the manufacturer's instructions. Real time PCR was performed with iTaq Supermix (Biorad) according to the manufacturer's instructions. Data was computed by the comparative Ct method as explained previously (Livak and Schmittgen 2001). Sequences for all primer sets are listed in Supplemental Table 1.
SUPPLEMENTAL DATA
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Serafin Piñol-Roma, Stephen Cusack, Samuel Gunderson, Guoliang Qing, and members of the Kiledjian laboratory for providing constructs and/or helpful discussions throughout this work. V.S. was supported in part by a predoctoral fellowship from the New Jersey Commission on Cancer Research, and this work was supported by NIH grant GM67005 to M.K.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1008208.
REFERENCES
- Berget S.M. Exon recognition in vertebrate splicing. J. Biol. Chem. 1995;270:2411–2414. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- Chang Y.F., Imam J.S., Wilkinson M.F. The nonsense-mediated decay RNA surveillance pathway. Annu. Rev. Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- Cohen L.S., Mikhli C., Friedman C., Jankowska-Anyszka M., Stepinski J., Darzynkiewicz E., Davis R.E. Nematode m7GpppG and m3 2,2,7GpppG decapping: Activities in Ascaris embryos and characterization of C. elegans scavenger DcpS. RNA. 2004;10:1609–1624. doi: 10.1261/rna.7690504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cougot N., Babajko S., Seraphin B. Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol. 2004;165:31–40. doi: 10.1083/jcb.200309008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davuluri R.V., Grosse I., Zhang M.Q. Computational identification of promoters and first exons in the human genome. Nat. Genet. 2001;29:412–417. doi: 10.1038/ng780. [DOI] [PubMed] [Google Scholar]
- Edery I., Sonenberg N. Cap-dependent RNA splicing in a HeLa nuclear extract. Proc. Natl. Acad. Sci. 1985;82:7590–7594. doi: 10.1073/pnas.82.22.7590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evdokimova V., Ruzanov P., Imataka H., Raught B., Svitkin Y., Ovchinnikov L.P., Sonenberg N. The major mRNA-associated protein YB-1 is a potent 5′ cap-dependent mRNA stabilizer. EMBO J. 2001;20:5491–5502. doi: 10.1093/emboj/20.19.5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenger-Gron M., Fillman C., Norrild B., Lykke-Andersen J. Multiple processing body factors and the ARE-binding protein TTP activate mRNA decapping. Mol. Cell. 2005;20:905–915. doi: 10.1016/j.molcel.2005.10.031. [DOI] [PubMed] [Google Scholar]
- Furuichi Y., Morgan M., Shatkin A.J., Jelinek W., Salditt-Georgieff M., Darnell J.E. Methylated, blocked 5 termini in HeLa cell mRNA. Proc. Natl. Acad. Sci. 1975;72:1904–1908. doi: 10.1073/pnas.72.5.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuichi Y., LaFiandra A., Shatkin A.J. 5′-Terminal structure and mRNA stability. Nature. 1977;266:235–239. doi: 10.1038/266235a0. [DOI] [PubMed] [Google Scholar]
- Gasch A.P., Spellman P.T., Kao C.M., Carmel-Harel O., Eisen M.B., Storz G., Botstein D., Brown P.O. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu M., Lima C.D. Processing the message: Structural insights into capping and decapping mRNA. Curr. Opin. Struct. Biol. 2005;15:99–106. doi: 10.1016/j.sbi.2005.01.009. [DOI] [PubMed] [Google Scholar]
- Gu M., Fabrega C., Liu S.W., Liu H., Kiledjian M., Lima C.D. Insights into the structure, mechanism, and regulation of scavenger mRNA decapping activity. Mol. Cell. 2004;14:67–80. doi: 10.1016/s1097-2765(04)00180-7. [DOI] [PubMed] [Google Scholar]
- Hamm J., Mattaj I.W. Monomethylated cap structures facilitate RNA export from the nucleus. Cell. 1990;63:109–118. doi: 10.1016/0092-8674(90)90292-m. [DOI] [PubMed] [Google Scholar]
- Izaurralde E., Lewis J., McGuigan C., Jankowska M., Darzynkiewicz E., Mattaj I.W. A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell. 1994;78:657–668. doi: 10.1016/0092-8674(94)90530-4. [DOI] [PubMed] [Google Scholar]
- Izaurralde E., Lewis J., Gamberi C., Jarmolowski A., McGuigan C., Mattaj I.W. A cap-binding protein complex mediating U snRNA export. Nature. 1995;376:709–712. doi: 10.1038/376709a0. [DOI] [PubMed] [Google Scholar]
- Jiao X., Wang Z., Kiledjian M. Identification of an mRNA-decapping regulator implicated in X-linked mental retardation. Mol. Cell. 2006;24:713–722. doi: 10.1016/j.molcel.2006.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalderon D., Richardson W.D., Markham A.F., Smith A.E. Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature. 1984;311:33–38. doi: 10.1038/311033a0. [DOI] [PubMed] [Google Scholar]
- Khanna R., Kiledjian M. Poly(A)-binding-protein-mediated regulation of hDcp2 decapping in vitro. EMBO J. 2004;23:1968–1976. doi: 10.1038/sj.emboj.7600213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiriakidou M., Tan G.S., Lamprinaki S., De Planell-Saguer M., Nelson P.T., Mourelatos Z. An mRNA m7G cap binding-like motif within human Ago2 represses translation. Cell. 2007;129:1141–1151. doi: 10.1016/j.cell.2007.05.016. [DOI] [PubMed] [Google Scholar]
- Konarska M.M., Padgett R.A., Sharp P.A. Recognition of cap structure in splicing in vitro of mRNA precursors. Cell. 1984;38:731–736. doi: 10.1016/0092-8674(84)90268-x. [DOI] [PubMed] [Google Scholar]
- Kudo N., Matsumori N., Taoka H., Fujiwara D., Schreiner E.P., Wolff B., Yoshida M., Horinouchi S. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc. Natl. Acad. Sci. 1999;96:9112–9117. doi: 10.1073/pnas.96.16.9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay U., Guttinger S. Leucine-rich nuclear-export signals: Born to be weak. Trends Cell Biol. 2005;15:121–124. doi: 10.1016/j.tcb.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Kwasnicka D.A., Krakowiak A., Thacker C., Brenner C., Vincent S.R. Coordinate expression of NADPH-dependent flavin reductase, Fre-1, and Hint-related 7meGMP-directed hydrolase, DCS-1. J. Biol. Chem. 2003;278:39051–39058. doi: 10.1074/jbc.M306355200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lall S., Piano F., Davis R.E. Caenorhabditis elegans decapping proteins: Localization and functional analysis of Dcp1, Dcp2, and DcpS during embryogenesis. Mol. Biol. Cell. 2005;16:5880–5890. doi: 10.1091/mbc.E05-07-0622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune F., Maquat L.E. Mechanistic links between nonsense-mediated mRNA decay and pre-mRNA splicing in mammalian cells. Curr. Opin. Cell Biol. 2005;17:309–315. doi: 10.1016/j.ceb.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Lewis J.D., Izaurralde E., Jarmolowski A., McGuigan C., Mattaj I.W. A nuclear cap-binding complex facilitates association of U1 snRNP with the cap-proximal 5′ splice site. Genes & Dev. 1996;10:1683–1698. doi: 10.1101/gad.10.13.1683. [DOI] [PubMed] [Google Scholar]
- Liu H., Kiledjian M. Scavenger decapping activity facilitates 5′ to 3′ mRNA decay. Mol. Cell. Biol. 2005;25:9764–9772. doi: 10.1128/MCB.25.22.9764-9772.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Rodgers N.D., Jiao X., Kiledjian M. The scavenger mRNA decapping enzyme DcpS is a member of the HIT family of pyrophosphatases. EMBO J. 2002;21:4699–4708. doi: 10.1093/emboj/cdf448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S.W., Jiao X., Liu H., Gu M., Lima C.D., Kiledjian M. Functional analysis of mRNA scavenger decapping enzymes. RNA. 2004;10:1412–1422. doi: 10.1261/rna.7660804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-Δ Δ C T method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Malys N., McCarthy J.E. Dcs2, a novel stress-induced modulator of m7GpppX pyrophosphatase activity that locates to P bodies. J. Mol. Biol. 2006;363:370–382. doi: 10.1016/j.jmb.2006.08.015. [DOI] [PubMed] [Google Scholar]
- Malys N., Carroll K., Miyan J., Tollervey D., McCarthy J.E. The ‘scavenger’ m7GpppX pyrophosphatase activity of Dcs1 modulates nutrient-induced responses in yeast. Nucleic Acids Res. 2004;32:3590–3600. doi: 10.1093/nar/gkh687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotrigiano J., Gingras A.C., Sonenberg N., Burley S.K. Cocrystal structure of the messenger RNA 5′ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell. 1997;13:951–961. doi: 10.1016/s0092-8674(00)80280-9. [DOI] [PubMed] [Google Scholar]
- Mazza C., Segref A., Mattaj I.W., Cusack S. Large-scale induced fit recognition of an m7GpppG cap analogue by the human nuclear cap-binding complex. EMBO J. 2002;21:5548–5557. doi: 10.1093/emboj/cdf538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael W.M., Choi M., Dreyfuss G. A nuclear export signal in hnRNP A1: A signal-mediated, temperature-dependent nuclear protein export pathway. Cell. 1995;83:415–422. doi: 10.1016/0092-8674(95)90119-1. [DOI] [PubMed] [Google Scholar]
- Mitchell P., Petfalski E., Shevchenko A., Mann M., Tollervey D. The exosome: A conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- Muthukrishnan S., Both G.W., Furuichi Y., Shatkin A.J. 5′-Terminal 7-methylguanosine in eukaryotic mRNA is required for translation. Nature. 1975;255:33–37. doi: 10.1038/255033a0. [DOI] [PubMed] [Google Scholar]
- Norton P.A., Hynes R.O. In vitro splicing of fibronectin pre-mRNAs. Nucleic Acids Res. 1990;18:4089–4097. doi: 10.1093/nar/18.14.4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddison P.J., Caudy A.A., Bernstein E., Hannon G.J., Conklin D.S. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes & Dev. 2002;16:948–958. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccirillo C., Khanna R., Kiledjian M. Functional characterization of the mammalian mRNA decapping enzyme hDcp2. RNA. 2003;9:1138–1147. doi: 10.1261/rna.5690503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai R.S., Bhattacharyya S.N., Artus C.G., Zoller T., Cougot N., Basyuk E., Bertrand E., Filipowicz W. Inhibition of translational initiation by Let-7 microRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- Piñol-Roma S., Dreyfuss G. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature. 1992;355:730–732. doi: 10.1038/355730a0. [DOI] [PubMed] [Google Scholar]
- Salehi Z., Geffers L., Vilela C., Birkenhager R., Ptushkina M., Berthelot K., Ferro M., Gaskell S., Hagan I., Stapley B., et al. A nuclear protein in Schizosaccharomyces pombe with homology to the human tumour suppressor Fhit has decapping activity. Mol. Microbiol. 2002;46:49–62. doi: 10.1046/j.1365-2958.2002.03151.x. [DOI] [PubMed] [Google Scholar]
- Siomi H., Dreyfuss G. A nuclear localization domain in the hnRNP A1 protein. J. Cell Biol. 1995;129:551–560. doi: 10.1083/jcb.129.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N., Morgan M.A., Merrick W.C., Shatkin A.J. A polypeptide in eukaryotic initiation factors that crosslinks specifically to the 5′-terminal cap in mRNA. Proc. Natl. Acad. Sci. 1978;75:4843–4847. doi: 10.1073/pnas.75.10.4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk E., Le Hir H., Seraphin B. DcpS can act in the 5′-3′ mRNA decay pathway in addition to the 3′-5′ pathway. Proc. Natl. Acad. Sci. 2003;100:12081–12086. doi: 10.1073/pnas.1635192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visa N., Izaurralde E., Ferreira J., Daneholt B., Mattaj I.W. A nuclear cap-binding complex binds Balbiani ring pre-mRNA cotranscriptionally and accompanies the ribonucleoprotein particle during nuclear export. J. Cell Biol. 1996;133:5–14. doi: 10.1083/jcb.133.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Kiledjian M. Functional link between the mammalian exosome and mRNA decapping. Cell. 2001;107:751–762. doi: 10.1016/s0092-8674(01)00592-x. [DOI] [PubMed] [Google Scholar]
- Wen W., Meinkoth J.L., Tsien R.Y., Taylor S.S. Identification of a signal for rapid export of proteins from the nucleus. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]