Abstract
The recently identified glmS ribozyme revealed that RNA enzymes, like protein enzymes, are capable of using small molecules as catalytic cofactors to promote chemical reactions. Flavin mononucleotide (FMN), S-adenosyl methionine (SAM), adenosyl cobalamin (AdoCbl), and thiamine pyrophosphate (TPP) are known ligands for RNA riboswitches in the control of gene expression, but are also catalytically powerful and ubiquitous cofactors in protein enzymes. If RNA, instead of just binding these molecules, could harness the chemical potential of the cofactor, it would significantly expand the enzymatic repertoire of ribozymes. Here we review the chemistry of AdoCbl, SAM, FMN, and TPP in protein enzymology and speculate on how these cofactors might have been used by ribozymes in the prebiotic RNA World or may still find application in modern biology.
Keywords: catalysis, riboswitch, ribozyme
INTRODUCTION
The identification of catalytic RNAs led to the RNA World hypothesis, a point in evolution when RNA acted as both the carrier of genetic information and the catalyst of essential chemical reactions (Gilbert 1986). Most naturally occurring ribozymes promote phosphoryl transfer, including both autolytic cleavage and splicing reactions (Fedor and Williamson 2005). The peptidyl transferase center of the ribosome, which catalyzes ester aminolysis and hydrolysis reactions, is the only naturally occurring exception identified to date (Crick 1968; Noller 1991; Schmeing et al. 2002). For such an RNA World to be viable, RNA must have performed a greater diversity of chemistries than these two rather basic reactions. Efforts to repopulate the RNA World by in vitro selection have resulted in RNA enzymes that catalyze additional reactions (Fiammengo and Jäschke 2005). These include reactions that would have been essential for even the most primitive forms of life, including an RNA-dependent RNA polymerase that is capable of self-replication (Johnston et al. 2001). Other ribozymes have been selected to catalyze carbon–carbon bond formation, isomerization, redox chemistry, and even small molecule synthesis (Prudent et al. 1994; Seelig and Jäschke 1999; Huang et al. 2000; Tsukiji et al. 2003). The experimentally established precedent for this breadth of RNA chemistry is informative regarding the possible mechanisms for the synthesis and breakdown of small molecules by RNA enzymes that would have been a vital component of any RNA-based metabolism (White 1976).
RNA enzymes have persisted into modern genomes despite a selective pressure toward chemistry by protein enzymes, which are better suited for chemical catalysis. The 20 amino acids, some that have neutral pKa's, display a much more diverse set of functional groups than the four nucleotides, all of which have unperturbed pKa's well removed from neutrality. Despite the chemical capacity available to proteins, some chemical reactions still require the use of catalytic cofactors, small molecules with chemical potential that goes beyond the standard amino acids. Although prevalent in protein enzymes, only recently was a catalytic cofactor discovered in a naturally occurring catalytic RNA.
The glmS ribozyme catalyzes nucleolytic RNA cleavage in the presence of glucosamine-6-phosphate (GlcN6P). The glmS ribozyme is upstream of the open reading frame for GlcN6P synthase and is proposed to regulate expression of the protein and GlcN6P production. High concentrations of GlcN6P lead to mRNA cleavage and decreased GlcN6P synthase expression in reporter constructs (Winkler et al. 2004). This role of the glmS ribozyme in gene expression makes it a member of another class of functional RNA molecules, the riboswitches (Barrick et al. 2004). Riboswitches directly sense and regulate levels of cellular metabolites (Winkler and Breaker 2005). In general, ligand binding to the riboswitch stabilizes a conformation that affects transcriptional termination, translational initiation, or alternative splicing. Unlike traditional riboswitches, binding of GlcN6P does not induce a structural rearrangement within the RNA (Hampel and Tinsley 2006; Klein and Ferré-D'Amaré 2006; Cochrane et al. 2007; Tinsley et al. 2007). Instead, GlcN6P directly participates in the chemical reaction (McCarthy et al. 2005; Klein and Ferré-D'Amaré 2006; Cochrane et al. 2007). Within the crystal structures of the glmS ribozyme the primary amine of GlcN6P, the key functional group on the sugar, is within hydrogen bonding distance of the leaving group (Cochrane et al. 2007; Klein et al. 2007). Its near neutral pKa (∼8.2) makes it well suited for participation in acid-base catalysis (McCarthy et al. 2005).
Cofactors are exploited by many protein enzymes to catalyze chemistry that is otherwise difficult. There are several thousand chemical reactions catalyzed by enzymes, many of them requiring the participation of small molecule cofactors (Holliday et al. 2007). These cofactors promote reactions that include carbon–carbon bond making and breaking, oxidation/reduction, and radical chemistry. Common cofactors include adenosine triphosphate (ATP), coenzyme A, riboflavin (from which both flavin adenine dinucleotide and flavin mononucleotide are derived), thiamine pyrophosphate, nicotinamide adenine dinucleotide, coenzyme B12 (parent molecule for both methylcobalamin and adenosyl cobalamin), and S-adenosyl methionine. Many of these cofactors are derived from vitamin molecules, and nearly all of them have a nucleotidyl group.
Intriguingly, naturally occurring riboswitches have been identified that are responsive to several of the most ubiquitous coenzymes found in proteins (Vitreschak et al. 2004; Winkler and Breaker 2005; Barrick and Breaker 2007). To date, 11 different small molecules have been identified as riboswitch targets. These include: adenosyl cobalymin (AdoCbl), flavin mononucleotide (FMN), thiamine pyrophosphate (TPP), S-adenosyl methionine (SAM), guanine (G), adenine (A), preQuosine1 (preQ1), glycine (gly), lysine (lys), GlcN6P, and deoxyguanosine (dG) (Winkler et al. 2002a,b, 2003, 2004; Mandal et al. 2003, 2004; Sudarsan et al. 2003; Mandal and Breaker 2004; Corbino et al. 2005; Fuchs et al. 2006; Kim et al. 2007; Roth et al. 2007). Additional riboswitches have been identified for which the ligand is still unknown (Barrick et al. 2004). This raises the intriguing question: Could RNA, in addition to binding these small molecules, use their chemical diversity to promote reactions beyond what is feasible with only the four nucleotides? If so, the glmS ribozyme, though it catalyzes the same nucleolytic reaction as other ribozymes, may be the first member in a class of naturally occurring RNAs that employ cofactors in catalysis. Such a possibility provokes two questions that constitute the focus of this review. First, what chemistries do riboswitch-bound metabolites make possible when used as cofactors in protein enzymes? And second, how might such chemistries contribute to the viability of a prebiotic RNA world (White 1976)?
ADENOSYLCOBALAMIN
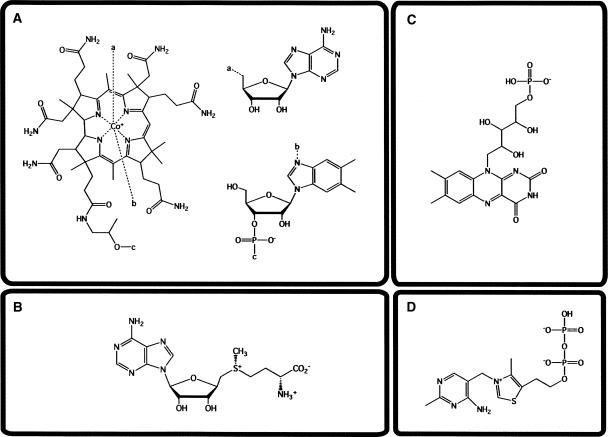
The adenosylcobalamin (AdoCbl) riboswitch is found in the genomes of most prokaryotic organisms, where it controls the levels of AdoCbl by regulating the genes responsible for synthesis or import of the cofactor (Nahvi et al. 2002). AdoCbl (Fig. 1A) is a source of carbon-based free radicals that enzymes use to catalyze a variety of isomerizations (Marsh 1999). AdoCbl is one of two coenzymes derived from vitamin B12, the other being methylcobalamin (MeCbl) (Banerjee 2006). Both coenzymes derive biological function from the ability to form a unique, stable, covalent bond between the central cobalt atom and carbon (Marsh 1999). The energy for homolysis of the bond between the cobalt and the 5′ carbon of adenosine is about 30 kcal/mol, which is weak relative to typical covalent bonds (Ludwig and Matthews 1997). AdoCbl-dependent enzymes use radical chemistry to break carbon–nitrogen, carbon–oxygen, and even carbon–carbon bonds (Marsh 1999).
FIGURE 1.
Structures of the four protein cofactors that are also riboswitch effectors. (A) Adenosyl cobalamin; (B) S-adenosyl methionine; (C) flavin mononucleotide; (D) thiamine pyrophosphate.
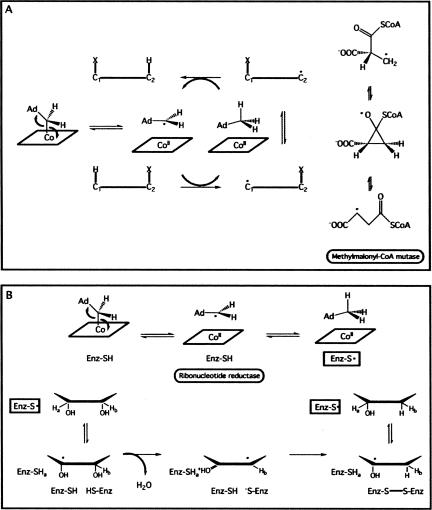
An AdoCbl-dependent ribozyme may have been able to draw nutrients from a variety of sources, which would have been advantageous in a pre-biotic world. Modern AdoCbl-dependent isomerases are found only in microbes and allow anaerobic bacteria to utilize alternative carbon sources (Marsh 1999). All of these rearrangements involve the interchange of a hydrogen with an electron-withdrawing group on the adjacent carbon, facilitating the movement of a hydroxy or amino group to convert substrates containing vicinal diols or amino alcohols into aldehydes following dehydration or deamination (Fig. 2A). In accepting amino acids as the substrates and using pyridoxyl phosphate as a second coenzyme, AdoCbl-dependent aminomutases catalyze the migration of an amino group. Finally, carbon-skeleton mutases use amino or carboxylic acids as substrates and catalyze isomerizations of the carbon skeleton (Halpern 1985). Of these, only the carbon-skeleton mutase, methylmalonyl-CoA mutase, is found in mammals, as well as microbes (Banerjee 2006). Methylmalonyl-CoA mutase transfers a carboxyate group to an adjacent carbon, catalyzing the conversion of succinyl-CoA to methylmalonyl-CoA (Fig. 2A; Marsh 1999).
FIGURE 2.
(A) The mechanistic scheme for the isomerization reaction catalyzed by AdoCbl-dependent enzymes. In the first step, the cobalt–carbon bond undergoes homolysis, leading to the adenosyl radical. The reactive carbon then abstracts a proton and then using a variety, some unknown, of mechanisms catalyzes the migration of a functional group from the adjacent carbon. In the mechanism of methylmalonyl-CoA mutase, a putative oxycyclopropyl intermediate is formed and the carboxylate functional group migrates. Based on material from Marsh (1999). (B) The mechanistic scheme for AdoCbl-dependent ribonucleotide reductase. Again, homolysis of the carbon–cobalt bond leads to radical formation; however, the radical is then transferred to a cysteine side chain in the enzyme. The radical abstracts a proton, causing the 2′-OH to leave via a proton transfer from a second cysteine side chain in the enzyme. Based on material from Marsh (1999).
In the structures of AdoCbl-dependent enzymes, AdoCbl is bound noncovalently in the active site. However, a specific metal–protein interaction is formed, in which a histidine residue replaces one axial ligand to the cobalt. In a ribozyme, this axial ligand could be replaced by a nucleotidyl group, such as the N7 of purines, which originated in the ribozyme and not in AdoCbl. However, in the structure of the selected AdoCbl-binding aptamer this interaction is not seen (Sussman et al. 2000). In fact, in this structure AdoCbl is recognized through hydrophobic interactions with the face of AdoCbl and hydrogen bonding interactions with the amine containing functional groups of AdoCbl (Sussman et al. 2000).
An AdoCbl-dependent ribonucleotide reductase, or its RNA enzyme equivalent, would have been key as the RNA World gave way to a more evolved world where DNA carried genetic information. Ribonucleotide reductase catalyzes the conversion of a ribonucleotide into a deoxyribonucleotide (Fig. 2B). DNA would allow organisms to have longer life spans and facilitate larger, more complex genomes. There are (at least) three different classes of ribonucleotide reductases that differ only in the nature of the coenzyme used to generate free radicals (Reichard 1993). The AdoCbl-dependent ribonucleotide reductases generate free radicals (Fig. 2B), like other AdoCbl-dependent enzymes, by symmetric breaking of the carbon–cobalt bond (Marsh 1999). The radical is then transferred to the enzyme to generate a cysteine radical, a common reactive group in the mechanisms of all ribonucleotide reductases (Reichard 1993; Booker et al. 1994). The cysteine radical removes the 3′ hydrogen of the ribose, generating a radical adjacent to the 2′ carbon. This causes the 2′-OH to become a better leaving group, eliminating and producing an activated substrate radical cation. Two active-site cysteines reduce the substrate leading to a disulphide bridge and substrate radical. The 3′ hydrogen is then replaced to regenerate the cysteine radical and a 2′-deoxyribonucleotide (Stubbe 1989). The action of most, if not all, ribonucleotide reductases is dependent on the formation of this cysteine radical (Reichard 1993). Although there are no side chains comparable to cysteine in RNA, perhaps a modified nucleotide was present in a prebiotic ribozyme, like 4-thio-uridine or 6-thio-guanosine, or perhaps a second cofactor, even a cysteine amino acid, was responsible for facilitating the radical reaction. The free radical chemistry of AdoCbl-dependent ribozymes may have played an instrumental role in the development of an RNA world and its segue into the current biology with DNA-based genomes.
S-ADENOSYL METHIONINE
There are at least three different types of S-adenosyl methionine (SAM) riboswitches; however, all of the known types of the SAM-riboswitch appear to be involved in the same biosynthetic pathways (Winkler et al. 2003; Corbino et al. 2005; Fuchs et al. 2006). SAM-riboswitches are found in many bacteria upstream of genes involved in SAM (Fig. 1B) synthesis or import as well as cysteine and methionine synthesis (Winkler et al. 2003). SAM is synthesized in the reaction of methionine and ATP by SAM synthase (Fontecave et al. 2004).
SAM is the second most ubiquitous protein substrate after adenosine triphosphate (ATP) (Cantoni 1975). Due to the positive charge on the sulfur atom adjacent to the methyl, methylation by a SAM-dependent methyltransferase is a relatively low-energy reaction (Cantoni 1975). SAM is the major methyl donor in biological methylation of carboxyls, alcohols, and amines, making it essential for the viability of all living organisms (Chiang et al. 1996). Methylation by SAM is used in a regulatory capacity in cells to exert control over replication, transcription, and translational fidelity (Loenen 2006). However, SAM is much more than a source of methyl groups and is widely used for its other functional groups in biological synthesis (Fig. 3; Roje 2006). Further, with the aid of an iron–sulfur cluster (4Fe–4S), which functions as a second cofactor, SAM is able to undertake the same types of radical reactions as catalyzed by AdoCbl (Frey and Magnusson 2003). The versatility and universality of this substrate suggests that it arose early in evolution.
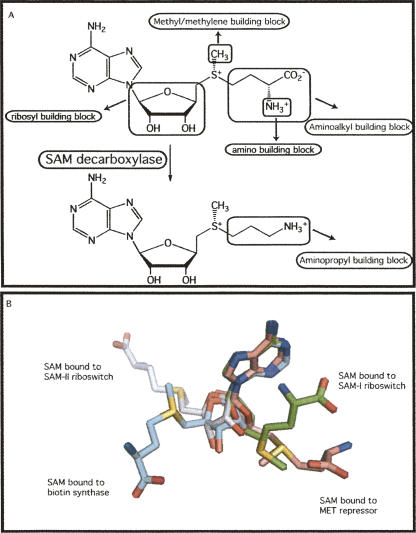
FIGURE 3.
(A) The many chemical precursors of biological synthesis provided by S-adenosyl methionine and decarboxylated S-adenosyl methionine. (B) Superpostion on the adenosine moiety of S-adenosyl methione of the cofactor bound by the SAM-dependent SAM-I riboswitch in green (PDB ID 2GIS) and SAM-II riboswitch in gray (PDB ID 2QWY) and in two SAM-binding proteins, MET repressor in light pink (PDB ID 1CMA) and biotin synthase in light blue (PDB ID 1R30).
A riboyzme capable of using SAM not just as a structural ligand, but also as a substrate in reaction, would have the ability to methylate both itself as well as other RNA molecules. In modern biology, there is extensive methylation of nucleotides in functional RNA molecules. Within eukaryotic ribosomal RNA (rRNA) and transfer RNA (tRNA), there are many conserved positions of methylation (Decatur and Fournier 2003). A class of small nucleolar ribonucleoproteins (snoRNPs), the box C/D snoRNPS, directs and catalyzes this 2′-OH methylation (Decatur and Fournier 2003). Ribose methylation stabilizes RNA molecules and may have led to longer-lived ribozyme structures.
While most of the SAM present in cells is used as a donor of methyl groups, it also acts as a substrate in enzymes that catalyze the transfer of methylene, amino, ribosyl, aminoalkyl, and aminopropyl groups (Fig. 3A; Fontecave et al. 2004). In the pre-biotic RNA world, this might have been a key role for a SAM-dependent ribozyme, as both spermine and spermidine are synthesized from SAM (Yoon et al. 2000). These positively charged, polyvalent amines are known to interact with RNA, DNA, and phospholipids in vivo (Loenen 2006). In vitro, spermine and spermidine are used to stabilize RNA structure through neutralization of the polyanionic backbone (Pyle 2002). In the first step of the reaction, SAM decarboxylase converts SAM into decaborboxylated SAM (dcSAM). Spermidine synthase then uses the aminopropyl group of one molecule of dcSAM to covert putricine into spermidine. Conversion of spermidine to spermine by spermine synthase uses a second molecule of dcSAM.
The structures of two types of SAM-riboswitches bound to SAH or SAM have recently been determined (Montange and Batey 2006; Gilbert et al. 2008). Most SAM-dependent protein enzymes bind SAM in an extended, trans conformation. The SAM-I riboswitch bound the SAH molecule in a compact conformation with the methionine moiety stacked on the adenine ring (Fig. 3B; Somers and Philips 1992; Berkovitch et al. 2004; Montange and Batey 2006; Gilbert et al. 2008). This compact conformation is seen in the structures of some methyltransferases, suggesting that an RNA-based methyltransferase might be able to bind and use SAM in the conformation seen in the riboswitch (Schubert et al. 2003). However, in the SAM-II riboswitch structure, the bound SAM molecule is in an extended conformation, more closely resembling that of the typical binding mode for SAM (Gilbert et al. 2008).
FLAVIN MONONUCLEOTIDE
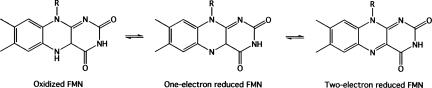
Flavin mononucletoide (FMN) riboswitches are found exclusively upstream of genes involved in riboflavin biosynthesis and transport (Winkler et al. 2002a); riboflavin is the precursor for FMN (Fig. 1C). The flavin moiety, a heterocyclic isoalloxaine chromophore, is a potent redox agent and flavin-dependent enzymes catalyze redox reactions (Massey 1995; De Colibus and Mattevi 2006). Flavin-containing cofactors, FMN and flavin-adenine dinucleotide (FAD), can accept electrons one at a time and exist in three different states, i.e., oxidized, one-electron reduced, and two-electron reduced (Massey 1995). Each of the redox states of FMN has different chemical properties that vary with the protein environment. A ribozyme that was able to harness the chemical capability of FMN would have been able to carry about photosynthetic reactions as well as pyruvate oxidation and nitrogen fixation (Swartz et al. 2001; Sancho 2006).
The oxidized state of FMN is most stable in solution; in fact, FMN cannot access the reduced states unless it is bound to a protein due to the instability of the one-electron reduced state (Fig. 4). A FMN-dependent ribozyme would be able to use mechanisms similar to those used by flavodoxins to stabilize the one-electron reduced state with respect to the oxidized and two-reduced states. Flavodoxins are small, anionic (140–160 amino acid), electronic transfer proteins that contain one molecule of noncovalently bound FMN. First, the one-electron reduced form of FMN can participate in an additional hydrogen bond compared with the oxidized state, thus stabilizing the one-electron reduced state with respect to the oxidized state (Lostao et al. 1997). Second, in flavodoxins (and flavodoxin-like proteins), the isoalloxazine ring is bound between two conserved cyclic amino acids, a tyrosine and a tryptophan (De Colibus and Mattevi 2006). In the structures of flavodoxins, the tyrosine is tightly packed to the face of the isoalloxazine ring and provides much of the affinity of flavodoxins for FMN (Lostao et al. 1997). Although the structure of the FMN-bound riboswitch is not available, the stacking of the isoalloxine ring might be mimicked in a ribozyme by stacking with a cytosine or uracil base. This interaction provides positive interaction energies with all three states of FMN, but those with the two-electron reduced state are much weaker than those with either of the other states. This destabilizes the two-electron reduced state with respect to the other states (Lostao et al. 1997). Further, the overall negative charge of flavodoxins destabilizes the negatively charged two-electron reduced state of FMN (Sancho 2006). The polyanionic RNA backbone would also be able to use this mode of destabilization to access the reduced states of FMN. Using these three modes for stabilization of the one-electron reduced state means that a pathway for electron transfer that is unusable in solution becomes quite efficient in the active site of a flavodoxin or in a hypothetical FMN-dependent ribozyme.
FIGURE 4.
The three redox states of flavin mononucleotide.
Like flavodoxins, an FMN-dependent ribozyme may have served a role in photosynthesis. In the photosystem I (PSI) photosynthetic complex, light capture initiates successive steps of electron transfer, leading to the oxidation of a chlorophyll dimer and the reduction of two terminal acceptors. Both flavodoxin and ferrodoxin, an iron–sulfur (2Fe–2S) complex containing redox enzyme, are subsequently reduced by one of the terminal acceptors (Sétif 2001). These redox enzymes are able to sustain cyclic as well as linear electron flow, a property that makes it possible for ATP synthesis to occur in the absence of photosystem II (Sétif 2001).
While all flavodoxins are responsible for electron transfer reactions, they do not all participate in photosynthesis (Sancho 2006). In Helocibacter pylori, flavodoxin accepts electrons from pyruvate oxydoreductase (POR) in the oxidative cleavage of pyruvate into acetyl-coenzyme A (Kaihovaara et al. 1998). This FMN-dependent enzyme exploits the redox potential of FMN to promote an otherwise unfavorable reaction. Under anaerobic conditions, this pathway also leads to the reduction of a potent antibacterial compound and perhaps contributes to the drug-resistant strains of H. pylori (Kaihovaara et al. 1998). Additionally, the light-oxygen-voltage (LOV) proteins sense blue-light using a bound FMN cofactor (Swartz et al. 2001; Möglich and Moffat 2007). However, unlike other FMN-dependent enzymes, LOV domains form a covalent bond with FMN during the redox cycle. A cysteine in the active site of the LOV domain forms a bond with the C(4a) atom of the isoalloxazine ring following activation by light. While there are no equivalent functional groups in modern ribozymes, the pre-biotic FMN-dependent ribozyme may have included a sulfur-containing nucleotide. This covalent bond is then resolved through a putative acid-base catalyzed reaction in the LOV protein (Möglich and Moffat 2007). Additionally, flavin-containing cofactors play key roles in DNA repair pathways. An FMN-dependent, nucleic acid repair ribozyme would have reduced genomic and enzymatic variability in an RNA world. In some DNA photolyases, 5-deazaflavin or FMN absorb light and then transfer the energy to a second bound flavin cofactor (FAD) that resolves the pyrimidine dimer (Klar et al. 2006). The key chemical component of all of these cofactors is the isoalloxazine ring.
THIAMINE PYROPHOSPHATE
The thiamine pyrophosphate (TPP)-riboswitch exists in all three domains of life, suggesting that this riboswitch arose very early in evolution (Winkler et al. 2002b; Cheah et al. 2007). The TPP-riboswitch is the only riboswitch currently identified in eukaryotes, and is found in conjunction with genes that are involved in thiamine biosynthesis, phosphorylation, and transport (Winkler et al. 2002b; Cheah et al. 2007). It can be located upstream, downstream, or at splice-site junctions in these genes, suggesting that it uses multiple mechanisms for controlling gene expression. TPP (Fig. 1D) is the biologically active form of vitamin B1 and is essential in most, if not all, organisms (Frank et al. 2007). This powerful cofactor is used for making and breaking bonds between carbon and sulfur, nitrogen, oxygen, and carbon. In most modern organisms, TPP-dependent enzymes are required for intermediary anabolic and catabolic metabolism and play roles in the pentose–phosphate pathway and the Krebs cycle (Frank et al. 2007).
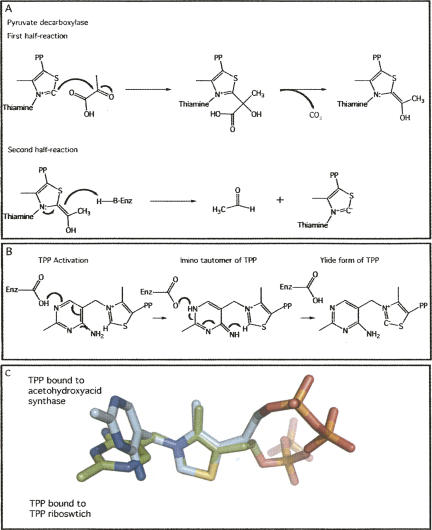
A structured RNA that was able to bind and use TPP for chemistry would have, importantly, been able to decarboxylate pyruvate, a major starting material used in the synthesis of many biomolecules (Malandrinos et al. 2006). Pyruvate decarboxylase (PDC) is one of the most well-studied TPP-dependent enzymes and is representative of its class. PDC catalyzes the penultimate step in the pathway to produce ethanol by anaerobic fermentation in many fungi (Frank et al. 2007). This pathway would certainly have been advantageous in a pre-biotic RNA world. The first substrate of PDC is pyruvate, which is decarboxylated by attack of the TPP C2 carbon on the carbonyl carbon of pyruvate (Fig. 5A; Jordan 2003). Carbon dioxide is released and the covalent coenzyme-substrate intermediate accepts the second substrate, a proton. This leads to the collapse of the covalent complex and release of the aldehyde. The TPP cofactor acts in conjunction with a general acid in the first step of the reaction and a general base in the second step of the reaction. Histidine residues often play these roles in TPP-dependent enzymes (Frank et al. 2007). Despite the fact that RNA has no amino acids with pKa's close to that of histidine, ribozymes are able to carry out reactions that are promoted by histidines in proteins. The reaction catalyzed by PDC is essentially the same as that catalyzed by all TPP-dependent enzymes (Malandrinos et al. 2006). However, the transferase class of TPP-dependent enzymes uses a second substrate other than a proton, one that can sometimes be as large as a protein, to catalyze the transfer of the carboxylate.
FIGURE 5.
(A) The two-step reaction of TPP-dependent pyruvate decarboxylase. In the first step, the C2 ylide carbon attacks the carbonyl carbon, releasing CO2 and forming a coenzyme-substrate intermediate. In the second step, a general base protonates the intermediate leading to reformation of the ylide and aldehyle release. (B) Activation of the ylide form of thiamine pyrophosphate. In the first step, a glutamate stabilizes the imino tautomer of TPP, and then abstracts a proton to lead to the ylide. (C) Superposition of TPP on the thiazolium ring, bound in either the TPP-dependent riboswitch (PDB ID 2HOJ) in green or a TPP-dependent enzyme, acetohydroxyacid synthase (PDB ID 1Z8N), in light blue. The key interaction in the ylide formation, between the 4′ imino and C2 carbon, is shown by a dashed line in the structure of TPP bound to acetohydroxyacid synthase.
The potency of TPP as a coenzyme is greatly enhanced by the architecture of the enzyme active site through interactions with the TPP ligand (Frank et al. 2007). TPP is composed of three basic components, a diphosphate moiety, a thiazolium ring, and a pyrimidine ring (Fig. 1D). All TPP-dependent enzymes require divalent metal ions for coenzyme binding (Malandrinos et al. 2006). This is also true of the TPP-dependent riboswitches, structures of which reveal TPP coordinated by two divalent metals. The reactive carbon in TPP is the C2 carbon of the thiazolium ring, which becomes a potent nucleophile through the formation of a ylide between the N3 and C2 atoms (Fig. 5B; Breslow 1958). This intermediate is created through deprotonation of the C2 by the 4′ imine of the pyrimidine ring, a reaction facilitated by the orientation and coordination of TPP within the enzyme active site (Fig. 5C). In all crystal structures of TPP-dependent enzymes, TPP is in an extended conformation, with the orientation of the thiazolium and pyrimidine rings such that the C2 and N4′ are within hydrogen-bonding distance (Lindqvist et al. 1992; Frank et al. 2007). While TPP is in an extended conformation in the three structures of TPP-dependent riboswitches, the C2 and N4′ are further apart (∼5 Å) in the riboswitches than in the protein structures (∼3.5 Å) (Fig. 5C; Edwards and Ferré-D'Amaré 2006; McCourt et al. 2006; Serganov et al. 2006; Thore et al. 2006). A rotation of the pyrimidine ring with respect to the thiazolium ring would align the two important atoms for ylide formation. Further, in TPP-dependent enzymes, a glutamic acid maintains the N1′ atom in a protonated state, which stabilizes the imino tautomer of the pyrimidine ring. A hydrogen bond between the resulting imino and the backbone carbonyl of a glycine residue serves to localize the lone electron pair on the N4′ imino, priming it to abstract the C2 proton from the thiazolium ring and resulting in ylide formation (Fig. 5B; Malandrinos et al. 2006). The chemistry of TPP-dependent enzymes is dictated by the ability of the TPP to form the ylide, which is controlled by the structure of TPP in the active site of the enzyme. While the current structures of TPP-bound riboswitches would most likely not facilitate the formation of this ylide, it may be that in an RNA-world TPP-dependent ribozyme this orientation was achieved. This architecture would enable a TPP-dependent RNA enzyme to catalyze difficult chemical reactions, such as the making and breaking of carbon–carbon bonds.
AMINO ACIDS
AdoCbl, SAM, FMN, and TPP are the only four riboswitch effectors identified to date that are also used by proteins as cofactors, but it is intriguing to consider what chemistries might be possible through the other metabolites that are recognized by RNA riboswitches (Winkler et al. 2002b; Cheah et al. 2007). The most provocative set of molecules are the amino acids. The majority of current biological chemistries are promoted by amino acid side chains, including lysine, which has been identified as a ligand for a riboswitch (Sudarsan et al. 2003). The chemical advantage provided by lysine, which is conspicuously absent from all nucleotidyl functional groups, is that it carries a positive charge at neutral pH. Within protein-active sites, lysine is used to neutralize the negative charge that it builds up in the transition states of some chemical reactions (Raines 1998). Lysine can also act as a general base, assisting water-mediated nucleophilic attack (Sekimoto et al. 1993). These activities might have aided the phosphoryl transfer reactions that the nucleolytic ribozymes catalyze. One way to develop protein-based catalysis would be to use RNA as a structural scaffold and use amino acids as cofactors to provide catalytic power. Ribozymes that use amino acids as coenzymes may have provided a pathway from an RNA world to a more evolved biological state.
While the glmS ribozyme is the first natural nucleic acid enzyme to use a cofactor to promote chemistry, there are several selected ribozymes and DNAzyme that have this property. A histidine-dependent DNAzyme was selected that is able to cleave an RNA substrate (Roth and Breaker 1998). A ribozyme that uses nicotinamide adenine dinucleotide (NAD) to catalyze redox chemistry in the presence of zinc has also been selected (Tsukiji et al. 2003, 2004). This ribozyme is specific for NAD and catalyzes a reaction equivalent to that of an alcohol dehydrogenase. In the glmS ribozyme, GlcN6P is contributing to a chemical reaction that RNA is known to promote using other mechanisms (Winkler et al. 2004). However, the existence of RNA structures that are designed to specifically bind coenzymes suggests that there may have been RNA catalysts that utilized these small molecules to promote different types of reactions. While there is only one example of a natural cofactor-dependent ribozyme, there is ample precedent to expect that RNAs are able to bind and harness the chemical potential of cofactors, which would provide a significant evolutionary advantage. Certainly, as the RNA world evolved, more and diverse catalytic activities would have been required, some of which may have been facilitated through the use of specifically bound cofactors. These cofactor-dependent ribozymes may still exist in modern genomes, but are sufficiently elusive to current search techniques that they remain to be identified.
ACKNOWLEDGMENTS
We would like to thank R. Breaker and members of the Strobel lab for discussions and comments on the manuscript. This work was supported by NSF grant 0544255 to S.A.S.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.908408.
REFERENCES
- Banerjee R. B12 trafficking in mammals: A for coenzyme escort service. ACS Chem. Biol. 2006;1:149–159. doi: 10.1021/cb6001174. [DOI] [PubMed] [Google Scholar]
- Barrick J.E., Breaker R.R. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biol. 2007;8:R239. doi: 10.1186/gb-2007-8-11-r239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick J., Corbino K., Winkler W., Nahvi A., Mandal M., Collins J., Lee M., Roth A., Sudarsan N., Jona I., et al. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc. Natl. Acad. Sci. 2004;101:6421–6426. doi: 10.1073/pnas.0308014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkovitch F., Nicolet Y., Wan J.T., Jarret J.T., Drennan C.L. Crystal structure of biotin synthase, an S-adenosylmethionine-dependent radical enzyme. Science. 2004;303:76–79. doi: 10.1126/science.1088493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booker S., Licht S., Broderick J., Stubbe J. Coenzyme B12-dependent ribonucleotide reductase: Evidence for the participation of five cysteine residues in ribonucleotide reduction. Biochemistry. 1994;33:12676–12685. doi: 10.1021/bi00208a019. [DOI] [PubMed] [Google Scholar]
- Breslow R. On the mechanism of thiamine action.4. Evidence from studies on model systems. J. Am. Chem. Soc. 1958;80:3719–3726. [Google Scholar]
- Cantoni G. Biological methylation: Selected aspects. Annu. Rev. Biochem. 1975;44:435–451. doi: 10.1146/annurev.bi.44.070175.002251. [DOI] [PubMed] [Google Scholar]
- Cheah M., Wachter A., Sudarsan N., Breaker R. Control of alternative RNA splicing and gene expression by eukaryotic riboswitches. Nature. 2007;447:497–500. doi: 10.1038/nature05769. [DOI] [PubMed] [Google Scholar]
- Chiang P., Gordon R., Tal J., Zeng G., Doctor B., Pardhasaradhi K., McCann P. S-Adenosylmethionine and methylation. FASEB J. 1996;10:471–480. [PubMed] [Google Scholar]
- Cochrane J., Lipchock S., Strobel S. Structural investigation of the glmS ribozyme bound to its catalytic cofactor. Chem. Biol. 2007;14:97–105. doi: 10.1016/j.chembiol.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbino K., Barrick J., Lim J., Welz R., Tucker B., Puskarz I., Mandal M., Rudnick N., Breaker R. Evidence for a second class of S-adenosylmethionine riboswitches and other regulatory RNA motifs in α-proteobacteria. Genome Biol. 2005;6:R70. doi: 10.1186/gb-2005-6-8-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick F.H.C. The origin of the genetic code. J. Mol. Biol. 1968;38:367–379. doi: 10.1016/0022-2836(68)90392-6. [DOI] [PubMed] [Google Scholar]
- Decatur W., Fournier M. RNA-guided nucleotide modification of ribosomal and other RNAs. J. Biol. Chem. 2003;278:695–698. doi: 10.1074/jbc.R200023200. [DOI] [PubMed] [Google Scholar]
- De Colibus L., Mattevi A. New frontiers in structural flavoenzymology. Curr. Opin. Struct. Biol. 2006;16:722–728. doi: 10.1016/j.sbi.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Edwards T., Ferré-D'Amaré A. Crystal structures of the thi-box riboswitch bound to thiamine pyrophosphate analogs reveal adaptive RNA-small molecule recognition. Structure. 2006;14:1459–1468. doi: 10.1016/j.str.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Fedor M., Williamson J. The catalytic diversity of RNAs. Nat. Rev. Mol. Cell Biol. 2005;6:399–412. doi: 10.1038/nrm1647. [DOI] [PubMed] [Google Scholar]
- Fiammengo R., Jäschke A. Nucleic acid enzymes. Curr. Opin. Biotechnol. 2005;16:614–621. doi: 10.1016/j.copbio.2005.10.006. [DOI] [PubMed] [Google Scholar]
- Fontecave M., Atta M., Mulliez E. S-adenosylmethionine: Nothing goes to waste. Trends Biochem. Sci. 2004;29:243–249. doi: 10.1016/j.tibs.2004.03.007. [DOI] [PubMed] [Google Scholar]
- Frank R., Leeper F., Luisi B. Structure, mechanism, and catalytic duality of thiamine-dependent enzymes. Cell. Mol. Life Sci. 2007;64:892–905. doi: 10.1007/s00018-007-6423-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey P., Magnusson O. S-Adenosylmethionine: A wolf in sheep's clothing, or a rich man's adenosylcobalamin? Chem. Rev. 2003;103:2129–2148. doi: 10.1021/cr020422m. [DOI] [PubMed] [Google Scholar]
- Fuchs R.T., Grundy F.J., Henkin T.M. The S MK box is a new SAM-binding RNA for translational regulation of SAM synthetase. Nat. Struct. Mol. Biol. 2006;13:226–233. doi: 10.1038/nsmb1059. [DOI] [PubMed] [Google Scholar]
- Gilbert W. Origin of life - The RNA world. Nature. 1986;319:618. doi: 10.1038/319618a0. [DOI] [Google Scholar]
- Gilbert S.D., Rambo R.P., Van Tyne D., Batey R.T. Structure of the SAM-II riboswitch bound to S-adenosylmethionine. Nat. Struct. Mol. Biol. 2008;15:177–182. doi: 10.1038/nsmb.1371. [DOI] [PubMed] [Google Scholar]
- Halpern J. Mechanisms of coenzyme B12-dependent rearrangements. Science. 1985;227:869–875. doi: 10.1126/science.2857503. [DOI] [PubMed] [Google Scholar]
- Hampel K., Tinsley M. Evidence for preorganization of the glmS ribozyme ligand binding pocket. Biochemistry. 2006;45:7861–7871. doi: 10.1021/bi060337z. [DOI] [PubMed] [Google Scholar]
- Holliday G.L., Almonacid D.E., Mitchell J.B.O., Thornton J.M. The chemistry of protein catalysis. J. Mol. Biol. 2007;372:1261–1277. doi: 10.1016/j.jmb.2007.07.034. [DOI] [PubMed] [Google Scholar]
- Huang F., Bugg C., Yarus M. RNA-catalyzed CoA, NAD, and FAD synthesis from phosphopantetheine, NMN, and FMN. Biochemistry. 2000;39:15548–15555. doi: 10.1021/bi002061f. [DOI] [PubMed] [Google Scholar]
- Johnston W.K., Unrau P.J., Lawrence M.S., Glasner M.E., Bartel D.P. RNA-catalyzed RNA polymerization: Accurate and general RNA-templated primer extension. Science. 2001;292:1319–1325. doi: 10.1126/science.1060786. [DOI] [PubMed] [Google Scholar]
- Jordan F. Current mechanistic understanding of thiamin diphosphate-dependent enzymatic reactions. Nat. Prod. Rep. 2003;20:184–201. doi: 10.1039/b111348h. [DOI] [PubMed] [Google Scholar]
- Kaihovaara P., Höök-Nikanne J., Uusi-Oukari M., Kosunen T., Salaspuro M. Flavodoxin-dependent pyruvate oxidation, acetate production and metronidazole reduction by Helicobacter pylori . J. Antimicrob. Chemother. 1998;41:171–177. doi: 10.1093/jac/41.2.171. [DOI] [PubMed] [Google Scholar]
- Kim J.N., Roth A., Breaker R.R. Guanine riboswitch variants from Mesoplasma florum selectively recognize 2′-deoxyguanosine. Proc. Natl. Acad. Sci. 2007;104:16092–16097. doi: 10.1073/pnas.0705884104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klar T., Kaiser G., Hennecke U., Carell T., Batschauer A., Essen L. Natural and non-natural antenna chromophores in the DNA photolyase from Thermus thermophilus . ChemBioChem. 2006;7:1798–1806. doi: 10.1002/cbic.200600206. [DOI] [PubMed] [Google Scholar]
- Klein D., Ferré-D'Amaré A. Structural basis of glmS ribozyme activation by glucosamine-6-phosphate. Science. 2006;313:1752–1756. doi: 10.1126/science.1129666. [DOI] [PubMed] [Google Scholar]
- Klein D., Wilkinson S., Been M., Ferré-D'Amaré A. Requirement of helix P2.2 and nucleotide G1 for positioning the cleavage Site and cofactor of the glmS ribozyme. J. Mol. Biol. 2007;373:178–189. doi: 10.1016/j.jmb.2007.07.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindqvist Y., Schneider G., Ermler U., Sundström M. Three-dimensional structure of transketolase, a thiamine diphosphate dependent enzyme, at 2.5 Å resolution. EMBO J. 1992;11:2373–2379. doi: 10.1002/j.1460-2075.1992.tb05301.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loenen W. S-adenosylmethionine: Jack of all trades and master of everything? Biochem. Soc. Trans. 2006;34:330–333. doi: 10.1042/BST20060330. [DOI] [PubMed] [Google Scholar]
- Lostao A., Gómez-Moreno C., Mayhew S., Sancho J. Differential stabilization of the three FMN redox forms by tyrosine 94 and tryptophan 57 in flavodoxin from Anabaena and its influence on the redox potentials. Biochemistry. 1997;36:14334–14344. doi: 10.1021/bi971384h. [DOI] [PubMed] [Google Scholar]
- Ludwig M.L., Matthews R.G. Structure-based perspectives on B12-dependent enzymes. Annu. Rev. Biochem. 1997;66:269–313. doi: 10.1146/annurev.biochem.66.1.269. [DOI] [PubMed] [Google Scholar]
- Malandrinos G., Louloudi M., Hadjiliadis N. Thiamine models and perspectives on the mechanism of action of thiamine-dependent enzymes. Chem. Soc. Rev. 2006;35:684–692. doi: 10.1039/b514511m. [DOI] [PubMed] [Google Scholar]
- Mandal M., Breaker R. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nat. Struct. Mol. Biol. 2004;11:29–35. doi: 10.1038/nsmb710. [DOI] [PubMed] [Google Scholar]
- Mandal M., Boese B., Barrick J., Winkler W., Breaker R. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell. 2003;113:577–586. doi: 10.1016/s0092-8674(03)00391-x. [DOI] [PubMed] [Google Scholar]
- Mandal M., Lee M., Barrick J.E., Weinberg Z., Emilsson G.M., Ruzzo W.L., Breaker R.R. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science. 2004;306:275–279. doi: 10.1126/science.1100829. [DOI] [PubMed] [Google Scholar]
- Marsh E. Coenzyme B12 (cobalamin)-dependent enzymes. Essays Biochem. 1999;34:139–154. doi: 10.1042/bse0340139. [DOI] [PubMed] [Google Scholar]
- Massey V. Introduction: Flavoprotein structure and mechanism. FASEB J. 1995;9:473–475. doi: 10.1096/fasebj.9.7.7737454. [DOI] [PubMed] [Google Scholar]
- McCarthy T., Plog M., Floy S., Jansen J., Strauss-Soukup J., Soukup G. Ligand requirements for glmS ribozyme self-cleavage. Chem. Biol. 2005;12:1221–1226. doi: 10.1016/j.chembiol.2005.09.006. [DOI] [PubMed] [Google Scholar]
- McCourt J.A., Pang S.S., King-Scott J., Guddat L.W., Duggleby R.G. Herbicide-binding sites revealed in the structure of plant acetohydroxyacid synthase. Proc. Natl. Acad. Sci. 2006;103:569–573. doi: 10.1073/pnas.0508701103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möglich A., Moffat K. Structural basis for light-dependent signaling in the dimeric LOV domain of the photosensor YtvA. J. Mol. Biol. 2007;373:112–126. doi: 10.1016/j.jmb.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montange R., Batey R. Structure of the S-adenosylmethionine riboswitch regulatory mRNA element. Nature. 2006;441:1172–1175. doi: 10.1038/nature04819. [DOI] [PubMed] [Google Scholar]
- Nahvi A., Sudarsan N., Ebert M., Zou X., Brown K., Breaker R. Genetic control by a metabolite binding mRNA. Chem. Biol. 2002;9:1043. doi: 10.1016/S1074-552(02)00224-7. [DOI] [PubMed] [Google Scholar]
- Noller H.F. Ribosomal RNA and translation. Annu. Rev. Biochem. 1991;60:191–227. doi: 10.1146/annurev.bi.60.070191.001203. [DOI] [PubMed] [Google Scholar]
- Prudent J., Uno T., Schultz P. Expanding the scope of RNA catalysis. Science. 1994;264:1924–1927. doi: 10.1126/science.8009223. [DOI] [PubMed] [Google Scholar]
- Pyle A. Metal ions in the structure and function of RNA. J. Biol. Inorg. Chem. 2002;7:679–690. doi: 10.1007/s00775-002-0387-6. [DOI] [PubMed] [Google Scholar]
- Raines R. Ribonuclease A. Chem. Rev. 1998;98:1045–1066. doi: 10.1021/cr960427h. [DOI] [PubMed] [Google Scholar]
- Reichard P. From RNA to DNA, why so many ribonucleotide reductases? Science. 1993;260:1773–1777. doi: 10.1126/science.8511586. [DOI] [PubMed] [Google Scholar]
- Roje S. S-Adenosyl-L-methionine: Beyond the universal methyl group donor. Phytochemistry. 2006;67:1686–1698. doi: 10.1016/j.phytochem.2006.04.019. [DOI] [PubMed] [Google Scholar]
- Roth A., Breaker R. An amino acid as a cofactor for a catalytic polynucleotide. Proc. Natl. Acad. Sci. 1998;95:6027–6031. doi: 10.1073/pnas.95.11.6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth A., Winkler W.C., Regulski E.E., Lee B.W., Lim J., Jona I., Barrick J.E., Ritwik A., Kim J.N., Weiz R., et al. A riboswitch selective for the queuosine precursor preQ1 contains an unusually small aptamer domain. Nat. Mol. Struct. Biol. 2007;14:308–317. doi: 10.1038/nsmb1224. [DOI] [PubMed] [Google Scholar]
- Sancho J. Flavodoxins: Sequence, folding, binding, function and beyond. Cell. Mol. Life Sci. 2006;63:855–864. doi: 10.1007/s00018-005-5514-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeing T.M., Seila A.C., Hansen J.L., Freeborn B., Strauss-Soukup J.K., Scaringe S.A., Strobel S.A., Moore P.B., Steitz T.A. A pre-translocational intermediate in protein synthesis observed in crystals of enzymatically active 50S subunits. Nat. Struct. Biol. 2002;9:225–230. doi: 10.1038/nsb758. [DOI] [PubMed] [Google Scholar]
- Schubert H., Blumenthal R., Cheng X. Many paths to methyltransfer: A chronicle of convergence. Trends Biochem. Sci. 2003;28:329–335. doi: 10.1016/S0968-0004(03)00090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seelig B., Jäschke A. A small catalytic RNA motif with Diels-Alderase activity. Chem. Biol. 1999;6:167–176. doi: 10.1016/S1074-5521(99)89008-5. [DOI] [PubMed] [Google Scholar]
- Sekimoto T., Matsuyama T., Fukui T., Tanizawa K. Evidence for lysine 80 as general base catalyst of leucine dehydrogenase. J. Biol. Chem. 1993;268:27039–27045. [PubMed] [Google Scholar]
- Serganov A., Polonskaia A., Phan A., Breaker R., Patel D. Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch. Nature. 2006;441:1167–1171. doi: 10.1038/nature04740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sétif P. Ferredoxin and flavodoxin reduction by photosystem I. Biochim. Biophys. Acta. 2001;1507:161–179. doi: 10.1016/s0005-2728(01)00205-5. [DOI] [PubMed] [Google Scholar]
- Somers W.S., Phillips J.E. Crystal structure of the MET repressor-operator complex at 2.8 Å resolution reveals DNA recognition by β-strands. Nature. 1992;359:387–393. doi: 10.1038/359387a0. [DOI] [PubMed] [Google Scholar]
- Stubbe J. Protein radical involvement in biological catalysis? Annu. Rev. Biochem. 1989;58:257–285. doi: 10.1146/annurev.bi.58.070189.001353. [DOI] [PubMed] [Google Scholar]
- Sudarsan N., Wickiser J., Nakamura S., Ebert M., Breaker R. An mRNA structure in bacteria that controls gene expression by binding lysine. Genes & Dev. 2003;17:2688–2697. doi: 10.1101/gad.1140003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman D., Nix J.C., Wilson C. The structural basis for molecular recognition by the vitamin B 12 RNA aptamer. Nat. Struct. Biol. 2000;7:53–57. doi: 10.1038/71253. [DOI] [PubMed] [Google Scholar]
- Swartz T., Corchnoy S., Christie J., Lewis J., Szundi I., Briggs W., Bogomolni R. The photocycle of a flavin-binding domain of the blue light photoreceptor phototropin. J. Biol. Chem. 2001;276:36493–36500. doi: 10.1074/jbc.M103114200. [DOI] [PubMed] [Google Scholar]
- Thore S., Leibundgut M., Ban N. Structure of the eukaryotic thiamine pyrophosphate riboswitch with its regulatory ligand. Science. 2006;312:1208–1211. doi: 10.1126/science.1128451. [DOI] [PubMed] [Google Scholar]
- Tinsley R., Furchak J., Walter N. Trans-acting glmS catalytic riboswitch: Locked and loaded. RNA. 2007;13:468–477. doi: 10.1261/rna.341807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukiji S., Pattnaik S., Suga H. An alcohol dehydrogenase ribozyme. Nat. Struct. Biol. 2003;10:713–717. doi: 10.1038/nsb964. [DOI] [PubMed] [Google Scholar]
- Tsukiji S., Pattnaik S., Suga H. Reduction of an aldehyde by a NADH/Zn2+ -dependent redox active ribozyme. J. Am. Chem. Soc. 2004;126:5044–5045. doi: 10.1021/ja0495213. [DOI] [PubMed] [Google Scholar]
- Vitreschak A.G., Rodionov D.A., Mironov A.A., Gelfand M.S. Riboswitches: The oldest mechanism for the regulation of gene expression? Trends Genet. 2004;20:44–50. doi: 10.1016/j.tig.2003.11.008. [DOI] [PubMed] [Google Scholar]
- White H.B. Coenzymes as fossils of an earlier metabolic state. J. Mol. Evol. 1976;7:101–104. doi: 10.1007/BF01732468. [DOI] [PubMed] [Google Scholar]
- Winkler W., Breaker R. Regulation of bacterial gene expression by riboswitches. Annu. Rev. Microbiol. 2005;59:487–517. doi: 10.1146/annurev.micro.59.030804.121336. [DOI] [PubMed] [Google Scholar]
- Winkler W., Cohen-Chalamish S., Breaker R. An mRNA structure that controls gene expression by binding FMN. Proc. Natl. Acad. Sci. 2002a;99:15908–15913. doi: 10.1073/pnas.212628899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler W., Nahvi A., Breaker R. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002b;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- Winkler W., Nahvi A., Sudarsan N., Barrick J., Breaker R. An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nat. Struct. Biol. 2003;10:701–707. doi: 10.1038/nsb967. [DOI] [PubMed] [Google Scholar]
- Winkler W., Nahvi A., Roth A., Collins J., Breaker R. Control of gene expression by a natural metabolite-responsive ribozyme. Nature. 2004;428:281–286. doi: 10.1038/nature02362. [DOI] [PubMed] [Google Scholar]
- Yoon S., Lee Y., Lee S., Cho Y. Polyamine synthesis in plants: Isolation and characterization of spermidine synthase from soybean (Glycine max) axes. Biochim. Biophys. Acta. 2000;1475:17–26. doi: 10.1016/s0304-4165(00)00039-8. [DOI] [PubMed] [Google Scholar]