Abstract
PIWI proteins are expressed in germ cells in a wide variety of metazoans, where they participate in the synthesis and function of a novel class of small RNAs called PIWI associated RNAs (piRNAs). One function of piRNAs is to preserve the integrity of the germline genome by silencing transposons, though they also participate in epigenetic and post-transcriptional gene regulation. In the planarian Schmidtea mediterranea, the PIWI proteins SMEDWI-1 and SMEDWI-2 are expressed in neoblasts and SMEDWI-2 is required for regeneration and homeostasis. Here, we identify a diverse population of ∼32-nucleotide small RNAs that strongly resemble vertebrate and insect piRNAs and map to hundreds of thousands of loci in the S. mediterranea genome. The expression of these RNAs occurs predominantly in neoblasts and is not restricted to the germline. RNAi knockdown of either SMEDWI-2 or a newly identified PIWI protein, SMEDWI-3, impairs regeneration and homeostasis and decreases the levels of both piRNAs and neoblasts. Therefore, SMEDWI-2 and SMEDWI-3 are required for piRNA expression, regeneration, and neoblast function in S. mediterranea.
Keywords: PIWI proteins, small RNAs, piRNAs, planarians, stem cells
INTRODUCTION
Planarians are an excellent model system for studying the biology of totipotent stem cells (Newmark and Sanchez Alvarado 2002). Planarians possess a population of stem cells called neoblasts that comprise ∼30% of the cells in an adult planarian and have the ability to give rise to all somatic cells and germ cells (Baguñá et al. 1989; Wang et al. 2007). In addition to their role in regeneration, neoblasts are required for homeostasis in intact animals, where they serve to replace damaged or nonfunctional cells (Newmark and Sanchez Alvarado 2002).
Many genes have been identified that are required for regeneration and/or neoblast maintenance in the planarian Schmidtea mediterranea. A large-scale RNAi screen found that 240 of the 1065 targeted genes display some type of regeneration phenotype (Reddien et al. 2005a). One of the best characterized genes that is required for regeneration is smedwi-2 (Reddien et al. 2005b). Smedwi-1 and smedwi-2 encode proteins that belong to the PIWI/Argonaute family of proteins, and both are expressed in neoblasts (Reddien et al. 2005b). RNAi-mediated knockdown of SMEDWI-2, but not SMEDWI-1, blocks regeneration by reducing the ability of neoblasts to replace lost tissue during regeneration and maintain homeostasis (Reddien et al. 2005b). However, the molecular mechanism by which SMEDWI-2 functions in planarians is unknown.
Metazoan germ cells have recently been shown to express a novel class of small RNA molecules called piRNAs that are bound by members of the PIWI subclass of the Argonaute superfamily of proteins (Lin 2007; O'Donnell and Boeke 2007; Zamore 2007). In Drosophila, the PIWI subclass of proteins are encoded by three genes—Piwi, Aubergine (Aub), and Argonaute 3 (Ago3)—that collaborate in the synthesis of piRNAs that are frequently derived from transposon-containing genomic loci (Vagin et al. 2006; Brennecke et al. 2007). Specifically, PIWI and/or AUB form complexes with antisense piRNAs that direct the slicing of sense strand transposon transcripts (Brennecke et al. 2007; Gunawardane et al. 2007). The sliced sense strands are then bound by AGO3, and this complex directs the slicing of antisense transposon transcripts (Brennecke et al. 2007). This amplification loop is thought to generate large quantity of piRNA:PIWI complexes that serve to silence transposons and, thus, prevent these mobile DNA elements from destroying the germline genome (Girard et al. 2006; Lau et al. 2006; Vagin et al. 2006; Aravin et al. 2007; Brennecke et al. 2007; Carmell et al. 2007; Houwing et al. 2007).
PIWI proteins and piRNAs appear to have functions in addition to repressing transposition. Whereas only ∼20% of mammalian piRNAs are derived from repetitive sequences, transposons, or retrotransposons (Kim 2006), most mammalian piRNAs actually map to unique genomic locations including intronic and exonic sequences. The existence of piRNAs that map to exonic sequences suggests that piRNAs, like miRNAs, potentially regulate gene expression post-transcriptionally. In fact, Drosophila PIWI and AUB and mouse MIWI have been shown to positively regulate mRNA stability and translation (Wilson et al. 1996; Deng and Lin 2002; Grivna et al. 2006; Megosh et al. 2006). Moreover, PIWI plays an important role in the epigenetic control of gene expression in Drosophila (Pal-Bhadra et al. 2004; Grimaud et al. 2006; Brower-Toland et al. 2007; Yin and Lin 2007). Thus, in addition to transposon silencing, piRNAs and PIWIs appear to be involved in the epigenetic control of gene expression, the control of mRNA stability and translation regulation.
Here, we have identified a diverse population of ∼32-nucleotide (nt) small RNAs that strongly resemble piRNAs. The S. mediterranea piRNAs map to hundreds of thousands of locations within the genome that correspond to a wide variety of sequences, including repetitive elements, introns and exons of both known and predicted genes, and sequences of unknown function. In contrast to other metazoans, the expression of planarian piRNAs is not restricted to germ cells but rather predominantly occurs in the pluripotent neoblasts. We also identified SMEDWI-3—a new member of the PIWI/Argonaute family—and demonstrate that both smedwi-2 and smedwi-3 are required for normal neoblast function and that piRNA levels decrease upon smedwi-2 or smedwi-3 RNAi. These results establish that smedwi-2 and -3 play a key role in maintaining the ability of pluripotent stem cells to regenerate lost or aged tissue in planarians and in piRNA synthesis and/or expression.
RESULTS
Small RNA sequencing and mapping
To explore the role of small RNAs in regeneration and neoblast function, we sequenced small RNA libraries from intact and regenerating sexually mature planarians. Specifically, total RNA was isolated from 20 intact animals and 20 regenerating animals 3 d after amputation. Next, the RNA from each group of animals was resolved on a denaturing gel and size selected to enrich for RNAs ∼15 to ∼30 nt in length. Linkers were ligated to the 5′ and 3′ ends of the RNAs, and each population was amplified by RT-PCR and subjected to high-throughput pyrosequencing (Margulies et al. 2005). After filtering to remove low quality sequences and size markers, we obtained 14,449 and 39,491 sequences (a total of 53,940 reads) from the intact and regenerating samples, respectively. This comprised a set of 23,106 unique reads, of which 8669 were from intact animals and 15,684 were obtained from regenerating animals; 1247 sequences were present in both samples.
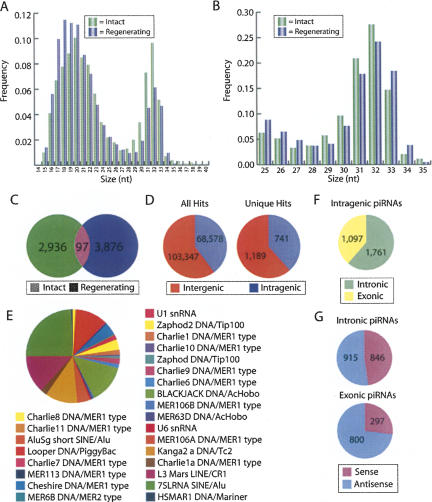
We observed two distinct size populations of RNAs with peaks at ∼20 nt and ∼32 nt (Fig. 1A). The ∼20-nt RNAs contained all of the previously characterized miRNAs (Palakodeti et al. 2006) as well as a number of previously uncharacterized miRNAs and will be described in detail elsewhere. The population of ∼32-nt small RNAs (between 25 and 35 nt) represented a remarkably diverse collection of 8448 total reads comprising 6909 unique sequences that were typically obtained only once or twice (Fig. 1A,B; Supplemental Table 1). Only 97 of these sequences (1.4%) were present in both the intact and regenerating samples (Fig. 1C), most likely due to low sampling of this extraordinarily diverse RNA population, rather than regeneration-specific expression (see below). The size and diversity of this population, as well as their characteristics described below, are reminiscent of the PIWI-associated RNAs identified in other organisms (Girard et al. 2006; Grivna et al. 2006; Lau et al. 2006; Vagin et al. 2006; Aravin et al. 2007; Brennecke et al. 2007; Houwing et al. 2007; Klattenhoff et al. 2007), and we will therefore refer to them as piRNAs, though we have not yet demonstrated a direct physical interaction between these RNAs and any of the S. mediterranea PIWI proteins.
FIGURE 1.
The small RNA repertoire of Schmidtea mediterranea. The frequency of small RNAs sequenced from intact and regenerating animals was calculated for all small RNAs together (A) and for the 25–35-nt size class (piRNAs) alone (B). (C) Venn diagram indicating the number of unique piRNAs sequenced from intact and regenerating animals and the overlap between the samples. (D) Distribution of piRNAs that perfectly match the S. mediterranea draft genome assembly among predicted intergenic and intragenic regions. (E) The distribution of piRNAs among known classes of repetitive elements. (F) Distribution of the intragenic piRNAs that map to predicted exons and introns. (G) Distribution of the exonic and intronic piRNAs that map in the sense and antisense orientations.
Of the 6909 piRNA sequences, 4254 map perfectly to 171,925 locations in the draft assembly of the S. mediterranea genome. Of the 4254 perfectly matching piRNAs, 1930 map to only a single location in the unannotated draft assembly (Fig. 1D). In contrast, 1321, 727, 250, and 26 piRNAs map to 2–10, 11–100, 101–1000, and more than 1000 locations, respectively. Thus, 2324 of the 4254 perfectly matching piRNAs map to more than one location (an average of 73 locations) in the draft genome assembly. Despite the fact that most piRNAs map to multiple genomic locations, only 35 and 277 of the small RNAs mapped directly to or within 1000 nt, respectively, of regions annotated by Repeatmasker as a putative repeat (excluding ribosomal RNAs, tRNAs, simple repeats, and regions of low complexity), suggesting that the S. mediterranea genome contains many previously uncharacterized repetitive elements. Figure 1E illustrates the classes of known repeats with which the piRNAs are associated.
We also examined whether any of the piRNAs mapped to genes. To do this we generated a preliminary annotation of known and predicted genes in the draft assembly of the genome (see Materials and Methods). Based on our preliminary gene annotation, ∼60% and 40% of the piRNAs map to intergenic and intragenic sequences, respectively (Fig. 1D). Moreover, we find that 1761 of the intragenic piRNAs map to predicted introns while 1097 of the intragenic piRNAs map to predicted exons (Fig. 1F). Strikingly, whereas the intronic piRNAs are evenly distributed to both the sense and antisense orientation, 73% of the exonic piRNAs map in the antisense orientation (Fig. 1G). These results suggest that piRNAs have the potential to post-transcriptionally regulate gene expression by binding directly to mRNAs.
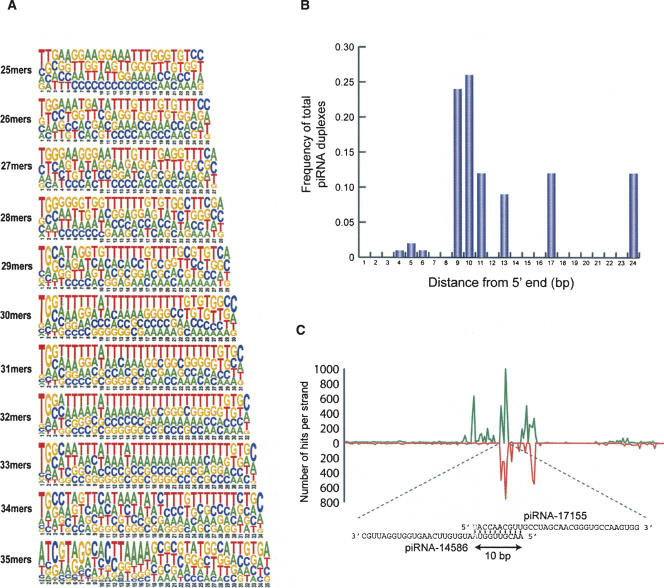
In other organisms, piRNAs have been shown to overlap one another for 10 nt at their 5′ ends and have a preference for U at +1 and A at +10 (Brennecke et al. 2007; Gunawardane et al. 2007; Houwing et al. 2007). We examined whether any of the S. mediterranea piRNAs had sequence preferences reminiscent of previously characterized piRNAs. As shown in Figure 2A, the S. mediterranea small RNAs display a strong preference for U at +1, particularly for the 31–33-nt species. In addition, these RNAs display a weak, but observable preference for A at +10, suggesting that some of the S. mediterranea piRNAs may overlap by 10 nt at their 5′ ends. Consistent with this idea, we found 31 piRNA duplexes that overlap at their 5′ ends and collectively map to 3898 genomic loci. Importantly, the majority of these piRNA duplexes overlap at their 5′ ends by 9 or 10 nt (Fig. 2B). These characteristics are consistent with the proposed ping-pong model for piRNA synthesis in which a PIWI protein complexed with a piRNA containing a U at +1 binds to and cleaves a complementary RNA 10 nt from the 5′ end of the piRNA (Brennecke et al. 2007; Gunawardane et al. 2007).
FIGURE 2.
Characteristics of the S. mediterranea piRNAs. (A) Pictograms of the nucleotide frequency among all sequenced RNAs of each size class. The height of the letters indicates the nucleotide frequency at that position. (B) Plot of the frequency of the size of the overlap among all identified piRNA duplexes. (C) The distribution of piRNA sequences for the novel repeat characterized by the piRNA-17155/piRNA-14586 duplex. The number of piRNAs mapping to all genomic loci defined by the piRNA-17155/piRNA-14586 duplex are plotted as a function of distance (1000 bp are shown). The piRNA duplex is located in the middle of the X axis and 500 bp on either side are shown. The green line indicates the number of piRNAs mapping to the sense strand while the red line indicates the number of piRNAs mapping to the antisense strand.
The overlapping piRNA duplexes map to between 4 and nearly 1500 different locations in the genome. For each genomic region that perfectly matched a piRNA duplex, we identified other piRNAs that map within 500 base pairs (bp) on either side of the piRNA duplex. This allowed us to derive a composite view of the piRNA distribution for the loci defined by these piRNA duplexes. For example, Figure 2C depicts the number, location, and orientations of all piRNAs that map to the genomic segments defined by the piRNA-17155/piRNA-14586 duplex. This analysis shows that the sequences flanking the regions defined by the piRNA duplexes we identified are rich in piRNAs derived from both strands. The large number of loci to which these piRNA duplexes map, and the large number of piRNAs these loci contain, suggest that they may correspond to unknown repetitive and possibly transposable elements.
piRNAs are expressed in neoblasts
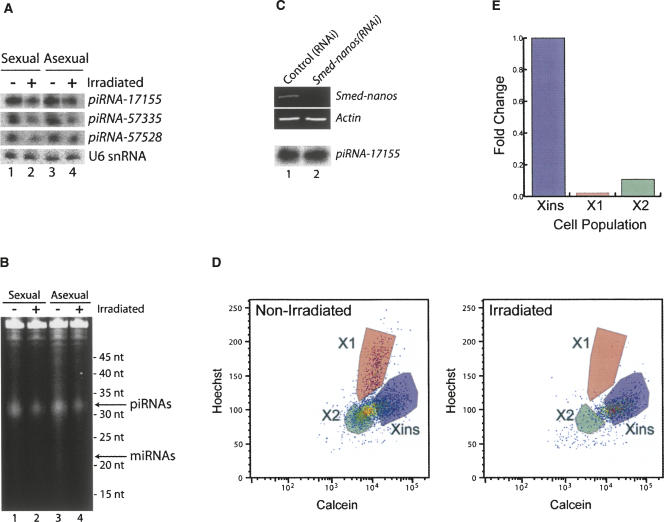
In all other organisms examined to date, piRNAs have been shown to be expressed only in germ cells (Girard et al. 2006; Vagin et al. 2006; Brennecke et al. 2007; Carmell et al. 2007; Houwing et al. 2007; Lin 2007; O'Donnell and Boeke 2007; Zamore 2007). To test whether the S. mediterranea piRNAs are expressed in germ cells, we compared piRNA levels in total RNA isolated from sexually mature planarians, which contain testes and ovaries, with that of asexual planarians, which contain a limited number of presumptive germ cells (Wang et al. 2007), but completely lack testes and ovaries. Surprisingly, we observed no difference in the relative abundance of the piRNAs between sexual and asexual animals by Northern blotting for three distinct piRNAs (Fig. 3A), or by SYBR gold staining (Fig. 3B) or 5′ end labeling (data not shown) to examine the entire population of piRNAs. As asexual planarians lack testes and ovaries, these results strongly suggest that the majority of piRNAs are expressed in cells other than germ cells.
FIGURE 3.
S. mediterranea piRNAs are predominantly expressed in neoblasts. Total RNA was isolated from sexual and asexual animals that were either untreated or were γ-irradiated and analyzed by Northern blotting to detect piRNA-17155, piRNA-57335, piRNA-57528, or U6 snRNA as a control (A) or SYBR gold staining to examine bulk piRNAs (B). (C) Animals were fed RNAi food containing either no insert (control(RNAi)) or the Smed-nanos insert (Smed-nanos(RNAi)). RNA was isolated from the animals 14 d after the initial feeding, and the mRNA levels of Smed-nanos and Actin were monitored by RT-PCR (top). In addition, piRNA levels were monitored by Northern blotting to detect piRNA-17155 (bottom). (D) FACS analysis to monitor the neoblast population in untreated or γ-irradiated animals. (E) Quantitation of the data in D to depict the fold change in the Xins, X1, and X2 cell populations.
Asexual planarians do, however, contain a limited number of presumptive germ cells that, while unable to generate testes or ovaries, could potentially express the majority of the planarian piRNAs. Smed-nanos is expressed in the presumptive germ cells and is required for their viability (Wang et al. 2007). We therefore tested whether the piRNAs are specifically expressed in these presumptive germ cells by analyzing piRNA expression in asexual animals depleted of Smed-nanos by RNAi, which destroys the presumptive germ cells (Wang et al. 2007). While the level of Smed-nanos mRNA is significantly lower in the Smed-nanos(RNAi) animals than in the control(RNAi) animals, we found that piRNA levels are unaffected (Fig. 3C). These results further support the conclusion that the bulk of the piRNAs are expressed in cells other than the germ cells.
We next tested whether the piRNAs were expressed in the neoblasts. Sexual and asexual worms were subjected to γ-irradiation, which selectively eliminates the neoblasts (Reddien et al. 2005b). As previously reported (Reddien et al. 2005b), irradiated animals displayed a curling phenotype and lysed 15–18 d after irradiation (data not shown). We also performed FACS analysis (Reddien et al. 2005b; Hayashi et al. 2006) to verify that this phenotype was due to the depletion of neoblasts. We observed a population of γ-irradiation insensitive cells (Xins) corresponding to differentiated cells and two other cell populations (X1 and X2) that are both highly sensitive to γ-irradiation and correspond to proliferating and nonproliferating neoblasts, respectively (Fig. 3D,E; Reddien et al. 2005b; Hayashi et al. 2006). Strikingly, the irradiated animals have significantly lower levels of neoblasts than the unirradiated animals, while the differentiated cells were relatively unaffected (Fig. 3D,E). Concomitantly, we observed a significant decrease in the levels of piRNAs in both sexual and asexual worms upon irradiation. This was observed by Northern blotting (Fig. 3A), SYBR gold staining (Fig. 3B), and 5′ end labeling (data not shown). Together, our observations that the piRNA levels are identical between sexual and asexual animals, are not impacted upon depletion of Smed-nanos, but decrease significantly upon elimination of the neoblasts by γ-irradiation suggest that planarian piRNAs are predominantly expressed in neoblasts. Nonetheless, as residual piRNA expression is observed in irradiated animals the expression of piRNAs in other cell types cannot be ruled out.
SMEDWI-3: A new member of the PIWI protein family expressed in neoblasts
The smedwi-1 and smedwi-2 genes encode PIWI protein homologs that are specifically expressed in neoblasts (Reddien et al. 2005b). Depletion of smedwi-2 by RNAi results in regeneration and homeostasis defects because neoblasts fail to generate progeny that can replace lost tissue (Reddien et al. 2005b). The observations that PIWI proteins are key players in piRNA synthesis and function in organisms such as Drosophila, mice, and zebrafish (Girard et al. 2006; Grivna et al. 2006; Lau et al. 2006; Vagin et al. 2006; Aravin et al. 2007; Brennecke et al. 2007; Houwing et al. 2007; Klattenhoff et al. 2007) and that the piRNAs and smedwi-1 and smedwi-2 genes are expressed in neoblasts in S. mediterranea suggested that smedwi-1 and smedwi-2 may play a role in piRNA synthesis and/or function in planarians.
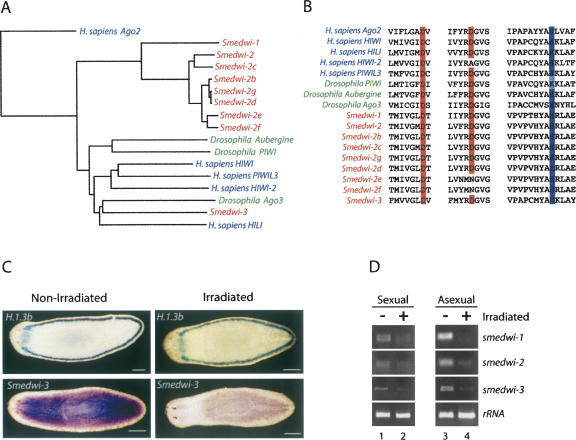
A search of the draft assembly of the S. mediterranea genome revealed seven additional genes encoding members of the PIWI/Argonaute family of proteins that clearly belong to the PIWI subfamily (Fig. 4A,B). Six of these genes (smedwi-2b, c, d, e, f, and g) are highly similar to smedwi-2 at the nucleotide level (Fig. 4A), and it is therefore difficult to determine whether or not these are pseudogenes or are recently duplicated genes that are in fact expressed. However, we have detected smedwi-2c and smedwi-2d transcripts by RT-PCR and sequencing (data not shown).
FIGURE 4.
Smedwi-3 encodes a PIWI protein that is expressed in neoblasts. (A) A phylogenetic tree depicting the relationship between the PIWI proteins encoded in the S. mediterranea, D. melanogaster, and H. sapiens genomes. The tree was derived from a multiple sequence alignment of the PIWI domains of each protein and was rooted with the PIWI domain of H. sapiens Ago2. (B) Alignment of the regions of the PIWI domains from the S. mediterranea, D. melanogaster, and H. sapiens PIWI proteins that contain the catalytic residues required for slicer activity. (C) Whole-mount in situ hybridizations depicting the expression pattern of smedwi-3 and H.1.3b (which is expressed in marginal adhesive gland cells, but not neoblasts) in sexual animals (n = 24). Animals on the left were not irradiated while animals on the right were γ-irradiated (scale bars, 0.2 mm). (D) The expression levels of smedwi-1, -2, and -3 and 16S rRNA were examined by RT-PCR using RNA isolated from untreated or γ-irradiated sexual and asexual animals.
The seventh additional PIWI encoding gene is clearly distinct from smedwi-1 and smedwi-2 and will be referred to as smedwi-3. The smedwi-3 gene contains six exons, spans 3205 nt of genomic DNA, and expresses an mRNA of ∼3,000 nt (data not shown). SMEDWI-3 is most similar to human HIWI (34% identity/54% similarity) and Drosophila Ago3 (32% identity/51% similarity) (Fig. 4A). All of the SMEDWI proteins except for SMEDWI-2e and SMEDWI-2f possess the catalytic residues within the PIWI domain that are required for slicer activity (Liu et al. 2004; Song et al. 2004; Tolia and Joshua-Tor 2007; Fig. 4B) and are therefore good candidates for participating in piRNA synthesis. Whole-mount in situ hybridization in sexual animals revealed that smedwi-3 is broadly expressed, but is expressed at lower levels in the pharynx and anterior of the photoreceptors than in the remainder of the animal (Fig. 4C). This pattern of expression differs somewhat from the expression pattern of smedwi-2 previously reported in asexual animals (Reddien et al. 2005b) but is nearly identical to the expression pattern observed in sexual animals (Wang et al. 2007). Importantly, the expression of smedwi-3 is dramatically reduced in irradiated sexual animals while the expression of H.1.3b, which is not expressed in neoblasts (Sanchez Alvarado et al. 2002), is unaffected (Fig. 4C). Additionally, RT-PCR experiments revealed that the levels of smedwi-1, -2, and -3 mRNAs are significantly decreased in both sexual and asexual animals that were γ-irradiated (Fig. 4D). These results demonstrate that smedwi-3 encodes a newly identified PIWI protein that is expressed in neoblasts.
smedwi-2 and smedwi-3 are required for neoblast function, regeneration, homeostasis, and piRNA synthesis
To test whether smedwi-3 is required for neoblast function, regeneration, or homeostasis, we performed RNAi on smedwi-1, -2, and -3 (Fig. 5A; Newmark et al. 2003; Reddien et al. 2005a). Briefly, the experiment was initiated by feeding groups of animals each type of RNAi food. The animals were fed again on day 5, amputated into three pieces, and allowed to regenerate for 10 d. The animals that successfully regenerated were fed again and amputated into two pieces and allowed to regenerate for another 10 d. Additionally, a few animals were removed at various times to analyze the neoblast populations by FACS. Finally, RT-PCR was performed to demonstrate that the RNAi treatments were both effective and specific (Fig. 5B).
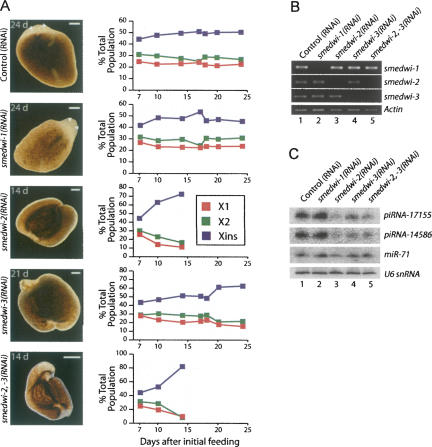
FIGURE 5.
smedwi-2 and smedwi-3 are required for regeneration, neoblast function, and piRNA synthesis. (A) Phenotypes of animals depleted of smedwi-1 (n = 72), -2 (n = 46), and -3 (n = 63) alone or smedwi-2 and -3 together (n = 24) by RNAi (right) (scale bars, 0.2 mm). Quantitation of FACS analysis of the X1, X2, and Xins populations in the animals depleted of smedwi-1, -2, and -3 alone or smedwi-2 and -3 together at the days indicated following the initial RNAi feeding. Each data point represents the average of individual measurements taken from two animals. (B) The levels of smedwi-1, -2, and -3 mRNAs were monitored by RT-PCR in each of the RNAi treated animals. (C) Northern blot analysis to examine the levels of piRNA-17115, piRNA-14586, and sme-miR-71 in the control, smedwi-1, -2, -3, and -2,-3 (RNAi) animals.
All of the control(RNAi) (n = 66) and smedwi-1(RNAi) (n = 72) animals (Reddien et al. 2005b) fully regenerated and displayed no apparent phenotype during the first or second round of regeneration (Fig. 5A). As previously reported (Reddien et al. 2005b), nearly all of the smedwi-2(RNAi) animals (n = 42/46) displayed profound regeneration defects at the end of the first round of regeneration (day 14) reminiscent of the phenotype induced by γ-irradiation, including curling, blastema regression, and the inability to regenerate (Reddien et al. 2005b), and by day 17 all (n = 46/46) of the smedwi-2(RNAi) worms had lysed (Fig. 5A). In contrast, only 30% (n = 14/46) of the smedwi-3(RNAi) animals displayed regeneration defects after the first round of regeneration (day 14), and these were less severe than those observed for the smedwi-2(RNAi) animals. On day 16—1 d following the third RNAi feeding and amputation—all of the smedwi-3(RNAi) animals (n = 71), including those that had displayed regeneration defects at day 14, had formed initial blastemas. However, regeneration defects such as curling and regression of the head and tail blastemas began to become apparent on day 17 (n = 5/71), were prevalent by day 20 (n = 55/63), and all of the smedwi-3(RNAi) animals had lysed by day 24 (Fig. 5A). Though the onset of the phenotypes exhibited by the smedwi-3(RNAi) animals was delayed with respect to the smedwi-2(RNAi) animals, the nature of the phenotypes were quite similar. Accordingly, the phenotypes of animals that were codepleted of both smedwi-2 and smedwi-3 (smedwi-2,-3(RNAi)) were nearly indistinguishable, though slightly more severe, than those displayed by animals depleted of only smedwi-2(RNAi) (Fig. 5A).
We also used FACS to examine the impact of depleting smedwi-1, -2, and -3 on the neoblast populations at different time points during regeneration. As reported previously (Reddien et al. 2005b), the smedwi-2(RNAi) animals showed a significant decrease in the X1 and X2 neoblast populations and an increase in the Xins population on day 14 (Fig. 5A). In contrast, smedwi-3(RNAi) animals displayed little change in the cell population until day 20, at which point there was a significant decrease in the X1 and X2 populations and an increase in the Xins population (Fig. 5A). However, the effect of smedwi-3 depletion on the neoblast population was less dramatic than that observed in the smedwi-2(RNAi) animals. The impact of depleting both smedwi-2 and smedwi-3 on the neoblast populations was even more dramatic than when smedwi-2 was depleted alone (Fig. 5A). Importantly, the neoblast populations did not change in either the control(RNAi) nor the smedwi-1(RNAi) animals (Fig. 5A). Together, these results confirm previous reports that smedwi-2, but not smedwi-1, is required for regeneration and neoblast function (Reddien et al. 2005b) and demonstrate that smedwi-3 also plays an important role in neoblasts to facilitate regeneration.
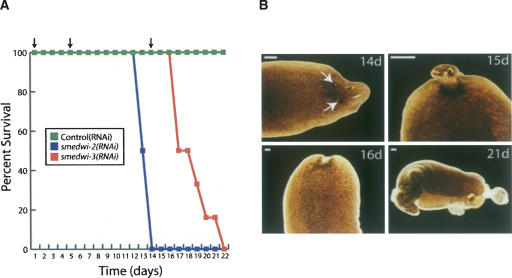
We also studied the roles of smedwi-3 in homeostasis by RNAi feeding in the absence of amputation (Fig. 6). As reported previously (Reddien et al. 2005b), although control(RNAi) and smedwi-1(RNAi) animals showed no effects on homeostasis throughout the entire experiment, by day 13 the animals depleted of smedwi-2 (n = 5/6) displayed tissue regression anterior of the photoreceptors (data not shown). Moreover, by day 15 all of the smedwi-2(RNAi) animals had curled and lysed. In contrast, all animals depleted of smedwi-3 (n = 12/12) developed lesions in the anterior region by day 15 (Fig. 6B), followed by head regression by day 16 and lysis by day 21. The formation of lesions has been attributed to a possible dysfunction of differentiated cells (Reddien et al. 2005a). Thus, the appearance of lesions in the smedwi-3(RNAi) animals indicates that smedwi-3 may play a critical role in differentiated cells in addition to that in neoblasts.
FIGURE 6.
The roles of the smedwi genes during homeostasis. (A) Survival curve for control(RNAi), smedwi-2(RNAi), and smedwi-3(RNAi) animals (n = 12) during homeostasis. (B) Phenotypes of the smedwi-3(RNAi) animals during homeostasis (n = 12) at the days indicated following the initial RNAi feeding. The arrows in the upper left panel indicate the lesions that appear in these animals (scale bars, 0.5 mm).
Finally, we examined piRNA expression in animals depleted of smedwi-1, -2, and -3. The piRNA levels were significantly reduced in the smedwi-2(RNAi), smedwi-3(RNAi), and the smedwi-2,-3(RNAi) animals 14 d after beginning the RNAi experiment, but not in smedwi-1(RNAi) or control(RNAi) animals (Fig. 5C). In contrast, miRNA levels were relatively unaffected by depletion of any of the smedwi genes (Fig. 5C). Thus, smedwi-2 and smedwi-3 are required for piRNA expression.
DISCUSSION
smedwi-3 is required for regeneration and homeostasis
The smedwi-1 and smedwi-2 mRNAs are specifically expressed in neoblasts, and smedwi-2 appears to function by promoting the generation of cells required for regeneration and homeostasis (Reddien et al. 2005b). Here we identify a third PIWI protein encoding gene, smedwi-3, that is expressed in neoblasts and is required for regeneration and homeostasis. Our observations suggest that in addition to its requirement for neoblast function, smedwi-3 plays a role in the differentiation of neoblast progeny. This is based on the fact that, while smedwi-3 is expressed in neoblasts and required for regeneration, lesions appear in the smedwi-3(RNAi) animals during homeostasis. It has been suggested that the appearance of lesions is due to defects in differentiated cells (Reddien et al. 2005a). Thus it is possible that smedwi-3 is transcribed in neoblasts but that the SMEDWI-3 protein persists and functions in differentiating neoblast progeny. Interestingly, a similar role has been suggested for smedwi-1 (Guo et al. 2006). Though smedwi-1 mRNA is expressed in neoblasts (Reddien et al. 2005b), SMEDWI-1 protein is present in neoblasts that have exited the cell cycle and no longer express smedwi-1 mRNA (Guo et al. 2006). Although more work will be required to determine the role of smedwi-3 in detail, our results clearly demonstrate that at least two PIWI proteins, SMEDWI-2 and -3, are required for neoblast function, regeneration, and homeostasis.
We also identified six additional PIWI encoding loci in the S. mediterranea genome and have named these smedwi-2b, -2c, -2d, -2e, -2f, and -2g, as they are all highly related to smedwi-2 but located on different contigs in the draft genome assembly (Fig. 3A). Thus, the smedwi-2 gene family appears to have undergone several recent duplications, underscoring the importance of the SMEDWI proteins in neoblast function. Though we have not studied the smedwi-2 gene family members in much detail, and it is possible that some of them are pseudogenes, RT-PCR and sequencing has revealed that at least smedwi-2c and smedwi-2d are transcribed (data not shown). Given the extraordinarily high degree of nucleotide sequence similarity between the smedwi-2 gene family members, it is possible that the smedwi-2(RNAi) phenotype is due to the codepletion of multiple smedwi-2 gene family members (Supplemental Fig. 1). While determining this will require additional work, if true, it would suggest that some or all members of the smedwi-2 gene family are functionally redundant, which may be a safeguard to ensure the integrity of the neoblast genome. It will be intriguing to explore the specific functions of these additional smedwi-2 related genes.
Evidence that the ∼32 nt RNAs are piRNAs
Though we have not yet shown that the ∼32 nt RNA population we have identified physically interact with any of the SMEDWI proteins, our data strongly suggest that these RNAs are in fact piRNAs, as they share many of the characteristics of piRNAs identified in other organisms (Girard et al. 2006; Grivna et al. 2006; Lau et al. 2006; Vagin et al. 2006; Aravin et al. 2007; Brennecke et al. 2007; Houwing et al. 2007; Klattenhoff et al. 2007). First, this RNA population is extraordinarily diverse (see below). Second, these RNAs are larger in size than the miRNAs (Palakodeti et al. 2006). Third, these RNAs map to hundreds of thousands of repetitive sequences located throughout the genome. Fourth, we have identified several cases of piRNA duplexes that overlap for 10 bp at their 5′ end and the RNAs have a strong preference for U at +1. Fifth, piRNA levels significantly decrease upon RNAi depletion of either smedwi-2 or smedwi-3. Finally, piRNAs have been shown to contain a 2′-O-methyl group on the 3′ terminal nucleotide in several species (Horwich et al. 2007; Houwing et al. 2007; Kirino and Mourelatos 2007; Ohara et al. 2007; Saito et al. 2007) and we have obtained evidence that the 32-nt RNAs are modified at their 3′ ends, though we have not yet determined the nature of this modification. Thus, while we have not formally proven that these RNAs physically interact with the SMEDWI proteins, these RNAs have all other characteristics that have been attributed to piRNAs in other species.
The diversity of planarian piRNAs
Though we identified nearly 7000 distinct piRNAs species, they were actually contaminants of the small RNA libraries we made for deep sequencing of the planarian miRNAs. We size selected the RNAs to enrich for molecules between ∼15 and ∼30 nt. Accordingly, of the 53,940 individual sequence reads we obtained, only ∼10% were larger than 30 nt. While the piRNAs are the minority of the RNAs we sequenced, the fact that the piRNAs, but not the miRNAs, are clearly visible on SYBR gold stained agarose gels (Fig. 3B) suggests that most of the small RNAs in planarians are actually piRNAs, not miRNAs. This is further underscored by the negligible overlap in the piRNAs sequenced from intact and regenerating animals. Thus, the overall population of piRNAs is significantly more diverse than our sequencing results indicate. Deeper sequencing of the piRNA population will provide additional insight into their diversity, distribution, biogenesis, and function.
Mechanism of piRNA synthesis
We identified a number of piRNA pairs that overlap by 10 bp, as has been observed in other species (Brennecke et al. 2007; Gunawardane et al. 2007; Houwing et al. 2007). Moreover, as in other organisms, the planarian piRNAs have a strong preference for U at +1 and a weak but detectable preference for A at +10. Though we lack evidence that SMEDWI-2 and SMEDWI-3 directly interact with the planarian piRNAs, the observations that SMEDWI-2 and SMEDWI-3 contain the catalytic residues required for slicer activity and that depletion of SMEDWI-2 or SMEDWI-3 results in a significant reduction in the levels of piRNAs suggest that planarian piRNAs synthesis is mediated by SMEDWI-2 and -3. It will be interesting to determine whether SMEDWI-2 and SMEDWI-3, as well as the other SMEDWI proteins, are involved in the synthesis of or interact with specific classes of piRNAs.
Potential roles for planarian piRNAs
A large fraction of the piRNAs we identified map to repetitive elements distributed throughout the genome, as was described for other organisms, in particular Drosophila melanogaster (Brennecke et al. 2007; Gunawardane et al. 2007). This strongly suggests that piRNAs and PIWI proteins play an important role in transposon silencing in planarians. Unfortunately, we have not yet identified transposon-derived transcripts that increase in the smedwi-2(RNAi) or smedwi-3(RNAi) animals. However, because the characterization of the S. mediterranea genome is just beginning, it is not yet known which of the hundreds to thousands of transposons the piRNAs map to are actually transcribed. Moreover, this experiment is technically challenging because the transposon transcripts should increase in the neoblasts, the very cells that are affected upon depleting SMEDWI-2 and SMEDWI-3. Thus, while our data suggest that piRNAs and PIWI proteins likely play a role in transposon silencing in planarians, confirmation of this will require a more thorough knowledge of the landscape and activity of the genome.
piRNAs have also been shown to play a role in the epigenetic and post-transcriptional control of gene expression in Drosophila. Although a large fraction of the piRNAs we identified map to repetitive elements, the sequences of many of these elements do not correspond to any known transoposable elements. Some of these sequences may represent heterochromatic regions of the genome that could be involved in epigenetic regulation. Unfortunately, this is difficult to assess yet given our current understanding of the landscape of the planarian genome.
Interestingly, many of the piRNAs that map to unique sequences map within exons of predicted genes. Moreover, whereas the intronic piRNAs are evenly distributed between the sense and antisense orientations of the genes they reside within, the majority of the exonic piRNAs map in the antisense orientation. This strongly suggests that piRNAs may also play a role in the post-transcriptional control of gene expression in planarians, in particular the control of translation or mRNA stability.
Distinctions between piRNAs and PIWI proteins in planarians and other metazoans
In other metazoans, piRNAs and PIWI proteins are expressed in germ cells and some adult stem cells (Girard et al. 2006; Grivna et al. 2006; Lau et al. 2006; Vagin et al. 2006; Brennecke et al. 2007; Carmell et al. 2007; Houwing et al. 2007). In planarians, however, piRNAs and PIWI proteins are primarily expressed in neoblasts, and smedwi-2 (Reddien et al. 2005b) and smedwi-3 are required for proper neoblast function. This suggests that the piRNAs themselves play a key role in neoblast function, which is distinct from the findings in other eukaryotes, as planarian neoblasts are truly totipotent stem cells that have the ability to give rise to all cell types in the entire organism, including germ cells (Baguñá et al. 1989). Future studies aimed at determining the precise functions of these molecules in planarians should provide significant new insight into the biology of neoblasts and regeneration.
MATERIALS AND METHODS
Planarian cultures
Hermaphroditic sexual (Zayas et al. 2005) and asexual (Sanchez Alvarado et al. 2002) strains of S. mediterranea used for these experiments were kind gifts from P. Newmark (University of Illinois) and A. Sánchez Alvarado (University of Utah/HHMI). The animals were maintained essentially as described (Cebrià and Newmark 2005) at 22°C in ddH2O supplemented with 1.6 mM NaCl, 1.0 mM CaCl2, 1.0 mM MgSO4, 0.1 mM MgCl2, 0.1 mM KCl, 1.2 mM NaHCO3 and fed homogenized organic beef liver. All the animals were starved 1 wk prior to any experiments.
Small RNA cloning and sequencing
Two groups of 20 sexual S. mediterranea animals were either kept intact or were amputated into small fragments and allowed to regenerate for 3 d. Total RNA was isolated from each group using Trizol (Invitrogen) as described by the manufacturer. Trace amounts of [32P]-labeled RNA markers of 18 (AGCGUGUAGGGAUCCAAA) and 30 (TAGGTTGCTAAGCTATTCCTGTGCCTGTCC) nt were added to each RNA sample and subsequently resolved on a 15% denaturing polyacrylamide gel. RNAs in the size range of ∼16–30 were excised from the gel, eluted, and ligated to cloning linkers and amplified by RT-PCR as described previously (Palakodeti et al. 2006). The resulting RT-PCR products were reamplified with the 454 cloning primers A (5′-GCCTCCCTCGCGCCATCAGATCGTAGGCACCTGAAA) and B (5′-GCCTTGCCAGCCCGCTCAGATTGATGGTGCCTACA). The PCR products were gel extracted using Qiagen gel extraction columns and subjected to pyrosequencing (454 Life Sciences).
Analysis of the piRNA distribution in the Schmidtea mediterranea genome
A preliminary annotation of the draft genome assembly (version 3.1) of the S. mediterranea genome (http://genome.wustl.edu/pub/organism/Invertebrates/Schmidtea_mediterannea/assembly/Schmidtea_mediterranea-3.1) was performed to facilitate mapping of the piRNAs. We first generated a set of S. mediterranea peptide sequences by assembling 27,164 S. mediterranea ESTs (Sanchez Alvarado et al. 2002; Zayas et al. 2005) into 9849 EST clusters with CAP3 (Huang and Madan 1999) and selecting the largest possible open reading frame for each cluster. Peptide sequences for Saccharomyces cerevisiae, Caenorhabditis elegans, D. melanogaster, Mus musculus, and Homo sapiens were obtained from Ensembl (http://www.ensembl.org), and all sequences were combined to generate a single peptide database. Repeatmasker (http://repeatmasker.org) was used to mask repetitive sequences and low complexity regions in the genome assembly. BLAST (Altschul et al. 1997) was then used to map the repeat masked genome assembly to the combined peptide database and the output reformatted for analysis with Genomescan. Genomescan (Yeh et al. 2001) used orthologous peptide information from the combined peptide library to predict exonic and intronic regions in the genome assembly. A total of 107,389 genic loci were predicted. This overestimate of the number of genes is due to a variety of factors such as imperfect prediction and gaps and duplications in the draft assembly. Megablast was used to map the piRNAs to the draft S. mediterranea genome assembly.
γ-Irridiation
Animals were γ-irradiated with a cesium source essentially as described (Guo et al. 2006) except that we found that two 30 Gy treatments were required to observe the phenotypes previously reported (Reddien et al. 2005b; Guo et al. 2006). FACS analysis was performed 2 d after the second irradiation dose.
Fluorescence-activated cell sorting analysis
FACS analysis of the neoblast populations was carried out based on protocols that had previously been described (Reddien et al. 2005b; Hayashi et al. 2006). Planarians were cut into small pieces with a sterile scalpel on ice in cold calcium, magnesium-free medium (CMF) (15 mM HEPES, 400 mg/L NaH2PO4, 800 mg/L NaCl, 1200 mg/L KCl, 800 mg/L NaHCO3, 240 mg/L glucose at pH 7.3) and then washed twice with CMF. The fragments were then soaked in CMF containing 0.25% (w/v) Trypsin (GIBCO) and 1% BSA (Fisher Scientific) at 4°C for ∼18 h to maximize penetration of the enzyme with little trypsin activity. The fragments were then rocked for 20 min at 20°C and completely dissociated into single cells by gentle pipetting. The cell mixture was then sequentially filtered through 40-μm and 20-μm nylon filters (Millipore). Cells were collected by centrifugation, resuspended in fresh CMF supplemented with 1% BSA and 0.5 μg/mL Calcein AM (Sigma), and incubated at room temperature for 20 min. Cells were collected and resuspended in CMF supplemented with 1% BSA, 18 μg/mL Hoechst 3342 (Sigma), and 2 μg/mL propidium iodide (Sigma). The samples were then analyzed with a LSR II flow cytometer (Becton-Dickinson).
Northern blotting
Thirty micrograms of total RNA were resolved on a denaturing polyacrylamide gel, transfered to nitrocellulose, and hybridized as described previously (Palakodeti et al. 2006). Three 10-min post-hybridization washes were performed at room temperature in 2× SSC and 0.1% SDS. The membranes were then visualized with a Cyclone PhosphorImager (Perkin Elmer).
Identification and cloning of smedwi-3
Putative PIWI protein encoding genes were identified by performing tblastn searches of the version 3.1 assembly of the S. mediterranea genome with PIWI proteins from other organisms and with SMEDWI-1 and SMEDWI-2. The smedwi-3 cDNA was obtained by RT-PCR cloning of an internal fragment, followed by 5′ and 3′ RACE using the FirstChoice RLM-RACE kit (Ambion, Inc.). A cDNA containing the entire coding region was isolated by RT-PCR with primers at the 5′ and 3′ ends of the RACE products. The accession number for the smedwi-3 cDNA is EU586258.
Whole-mount in situ hybridization
In situ hybridization was performed essentially as described (Umesono et al. 1999; Oviedo and Levin 2007) by treating animals with 2% HCl made in 5/8 Holtfreters solution (2.188 g NaCl, 0.031g KCl, 0.063 g CaCl2·2H2O, 0.125 g NaHCO3 in 1 L of ddH2O). Animals were then fixed in Carnoy's fixative (6:3:1 ethanol:chloroform:acetic acid) for 2 h at 4°C. Rehydration was done with an ethanol:PBST (PBS, 0.1% Triton X-100) dilution series followed by two PBST washes of 15 min each at 4°C. Animals were treated with proteinase K (20 μg/mL) for 5–6 min at 37°C, followed by fixation and rehydration as described above. Prehybridization was done at 50°C for 3 h in prehybridization solution (50% formamide, 5× SSC at pH 7.0, 100 μg/mL heparin, 0.05% Tween-20, 0.05% Triton X-100, 5 mM DTT, 1 mg/mL yeast RNA, 1× Denhardt's). Hybridization was performed at 50°C for 16–18 h with digoxigenin (DIG)-conjugated riboprobes in prehybridization solution supplemented with 5% dextran sulfate. Animals were washed 5 times in 50% formamide, 5× SSC, and 0.1% Tween-20 for 1 h each at 50°C, followed by two washes with MABT (100 mM maleic acid, 150 mM NaCl, 0.1% Tween-20 at pH 7.5). Animals were blocked in MABT supplemented with 1% blocking reagent (Roche) for 1 h at room temperature and then incubated with a sheep anti-DIG antibody conjugated to alkaline phosphatase (Roche) at 1:2000 in the blocking solution for 4 h at room temperature. Animals were washed 8 times with MABT and the signal was developed using BM purple (Roche).
Semiquantitative RT-PCR
First strand cDNA was synthesized from 500 ng of total RNA using SuperScriptII (Invitrogen). PCR amplification was performed for 20, 25, and 30 cycles and the samples resolved on agarose gels stained with ethidium bromide. The primers used were as follows:
smedwi-1 forward, GGAACCGTATTGAAACGTGAGC;
smedwi-1 reverse, CCCCAAACTTCCAATCATCAGG;
smedwi-2 forward, GTCATCGTAAAGAAAAGAGTCGGC;
smedwi-2 reverse, CCACAACAGTTCCAGGATTTGG;
smedwi-3 forward, TTGGAGCCGTAGTTGGTTC;
smedwi-3 reverse, AACTGAACCTGATGCGGAG;
rRNA forward, TCTTAGTTGGTGGAGCGATTTGTC;
rRNA reverse, TGGATTCTTTAGCAGGTTACCCG;
Smed-nanos forward, CCTGAATCATTGAAGATGGC; and
Smed-nanos reverse, CCAAGAGTGGATTGTGACATGC.
RNA interference
RNAi experiments were performed essentially as described previously (Reddien et al. 2005a). cDNA fragments for the targeted genes were cloned into pDONRdT7 (Reddien et al. 2005a) using a BP reaction (Invitrogen) and the resulting clones transformed into Escherichia coli strain HT115 (Timmons et al. 2001). Overnight cultures grown in 2XYT media containing kanamycin (25 μg/mL) and tetracycline (12.5 μg/mL) were diluted 1:100 in fresh media, grown to OD0.4 at 37°C, and induced with 1 mM IPTG for 2 h. We pelleted 2.5 mL of bacterial culture and resuspended it in 25 μL of 1:1 homogenized beef liver and water, mixed with 1 μL of red food coloring and 10.4 μL of 2% ultra-low gelling agarose (Sigma) to form pellets that were fed to 10 animals. After 1 h of feeding, the animals were transferred to a new container containing fresh media. After 4 d, the animals were fed RNAi food again and were amputated anterior and posterior of the pharynx to generate three pieces. After 9 d of regeneration, animals were fed RNAi food again and were amputated as described above. Phenotypes were only scored if the majority of animals in each population displayed similar effects.
SUPPLEMENTAL DATA
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank E. Sontheimer, A. Pasquinelli, H. Furneaux, A. Das, and members of the Graveley laboratory for discussions and critically reading the manuscript. We also thank Alejandro Sánchez-Alvarado for providing asexual planarians and the pDONRT7 plasmid and Phil Newmark for providing sexual planarians and the H.1.3b cDNA clone. Genomic sequence data were produced by the Washington University Genome Sequencing Center in St. Louis. This work was generously supported by the Raymond and Beverly Sackler Fund for the Arts and Sciences (B.R.G.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1085008.
REFERENCES
- Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin A.A., Sachidanandam R., Girard A., Fejes-Toth K., Hannon G.J. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- Baguñá J., Saló E., Auladell C. Regeneration and pattern formation in planarians III. Evidence that neoblasts are totipotent stem cells and the source of blastema cells. Development. 1989;107:77–86. [Google Scholar]
- Brennecke J., Aravin A.A., Stark A., Dus M., Kellis M., Sachidanandam R., Hannon G.J. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila . Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Brower-Toland B., Findley S.D., Jiang L., Liu L., Yin H., Dus M., Zhou P., Elgin S.C., Lin H. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes & Dev. 2007;21:2300–2311. doi: 10.1101/gad.1564307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell M.A., Girard A., van de Kant H.J., Bourc'his D., Bestor T.H., de Rooij D.G., Hannon G.J. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev. Cell. 2007;12:503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Cebrià F., Newmark P.A. Planarian homologs of netrin and netrin receptor are required for proper regeneration of the central nervous system and the maintenance of nervous system architecture. Development. 2005;132:3691–3703. doi: 10.1242/dev.01941. [DOI] [PubMed] [Google Scholar]
- Deng W., Lin H. miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev. Cell. 2002;2:819–830. doi: 10.1016/s1534-5807(02)00165-x. [DOI] [PubMed] [Google Scholar]
- Girard A., Sachidanandam R., Hannon G.J., Carmell M.A. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- Grimaud C., Bantignies F., Pal-Bhadra M., Ghana P., Bhadra U., Cavalli G. RNAi components are required for nuclear clustering of Polycomb group response elements. Cell. 2006;124:957–971. doi: 10.1016/j.cell.2006.01.036. [DOI] [PubMed] [Google Scholar]
- Grivna S.T., Beyret E., Wang Z., Lin H. A novel class of small RNAs in mouse spermatogenic cells. Genes & Dev. 2006;20:1709–1714. doi: 10.1101/gad.1434406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane L.S., Saito K., Nishida K.M., Miyoshi K., Kawamura Y., Nagami T., Siomi H., Siomi M.C. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila . Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- Guo T., Peters A.H., Newmark P.A. A Bruno-like gene is required for stem cell maintenance in planarians. Dev. Cell. 2006;11:159–169. doi: 10.1016/j.devcel.2006.06.004. [DOI] [PubMed] [Google Scholar]
- Hayashi T., Asami M., Higuchi S., Shibata N., Agata K. Isolation of planarian X-ray-sensitive stem cells by fluorescence-activated cell sorting. Dev. Growth Differ. 2006;48:371–380. doi: 10.1111/j.1440-169X.2006.00876.x. [DOI] [PubMed] [Google Scholar]
- Horwich M.D., Li C., Matranga C., Vagin V., Farley G., Wang P., Zamore P.D. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr. Biol. 2007;17:1265–1272. doi: 10.1016/j.cub.2007.06.030. [DOI] [PubMed] [Google Scholar]
- Houwing S., Kamminga L.M., Berezikov E., Cronembold D., Girard A., van den Elst H., Filippov D.V., Blaser H., Raz E., Moens C.B., et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell. 2007;129:69–82. doi: 10.1016/j.cell.2007.03.026. [DOI] [PubMed] [Google Scholar]
- Huang X., Madan A. CAP3: A DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim V.N. Small RNAs just got bigger: Piwi-interacting RNAs (piRNAs) in mammalian testes. Genes & Dev. 2006;20:1993–1997. doi: 10.1101/gad.1456106. [DOI] [PubMed] [Google Scholar]
- Kirino Y., Mourelatos Z. Mouse Piwi-interacting RNAs are 2′-O-methylated at their 3′ termini. Nat. Struct. Mol. Biol. 2007;14:347–348. doi: 10.1038/nsmb1218. [DOI] [PubMed] [Google Scholar]
- Klattenhoff C., Bratu D.P., McGinnis-Schultz N., Koppetsch B.S., Cook H.A., Theurkauf W.E. Drosophila rasiRNA pathway mutations disrupt embryonic axis specification through activation of an ATR/Chk2 DNA damage response. Dev. Cell. 2007;12:45–55. doi: 10.1016/j.devcel.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Lau N.C., Seto A.G., Kim J., Kuramochi-Miyagawa S., Nakano T., Bartel D.P., Kingston R.E. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- Lin H. piRNAs in the germ line. Science. 2007;316:397. doi: 10.1126/science.1137543. [DOI] [PubMed] [Google Scholar]
- Liu J., Carmell M.A., Rivas F.V., Marsden C.G., Thomson J.M., Song J.J., Hammond S.M., Joshua-Tor L., Hannon G.J. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- Margulies M., Egholm M., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megosh H.B., Cox D.N., Campbell C., Lin H. The role of PIWI and the miRNA machinery in Drosophila germline determination. Curr. Biol. 2006;16:1884–1894. doi: 10.1016/j.cub.2006.08.051. [DOI] [PubMed] [Google Scholar]
- Newmark P.A., Sanchez Alvarado A. Not your father's planarian: A classic model enters the era of functional genomics. Nat. Rev. Genet. 2002;3:210–219. doi: 10.1038/nrg759. [DOI] [PubMed] [Google Scholar]
- Newmark P.A., Reddien P.W., Cebria F., Sanchez Alvarado A. Ingestion of bacterially expressed double-stranded RNA inhibits gene expression in planarians. Proc. Natl. Acad. Sci. 2003;100(Suppl. 1):11861–11865. doi: 10.1073/pnas.1834205100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell K.A., Boeke J.D. Mighty Piwis defend the germline against genome intruders. Cell. 2007;129:37–44. doi: 10.1016/j.cell.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohara T., Sakaguchi Y., Suzuki T., Ueda H., Miyauchi K., Suzuki T. The 3′ termini of mouse Piwi-interacting RNAs are 2′-O-methylated. Nat. Struct. Mol. Biol. 2007;14:349–350. doi: 10.1038/nsmb1220. [DOI] [PubMed] [Google Scholar]
- Oviedo N.J., Levin M. smedinx-11 is a planarian stem cell gap junction gene required for regeneration and homeostasis. Development. 2007;134:3121–3131. doi: 10.1242/dev.006635. [DOI] [PubMed] [Google Scholar]
- Pal-Bhadra M., Leibovitch B.A., Gandhi S.G., Rao M., Bhadra U., Birchler J.A., Elgin S.C. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science. 2004;303:669–672. doi: 10.1126/science.1092653. [DOI] [PubMed] [Google Scholar]
- Palakodeti D., Smielewska M., Graveley B.R. MicroRNAs from the planarian Schmidtea mediterranea: A model system for stem cell biology. RNA. 2006;12:1640–1649. doi: 10.1261/rna.117206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien P.W., Bermange A.L., Murfitt K.J., Jennings J.R., Sanchez Alvarado A. Identification of genes needed for regeneration, stem cell function, and tissue homeostasis by systematic gene perturbation in planaria. Dev. Cell. 2005a;8:635–649. doi: 10.1016/j.devcel.2005.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien P.W., Oviedo N.J., Jennings J.R., Jenkin J.C., Sanchez Alvarado A. SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science. 2005b;310:1327–1330. doi: 10.1126/science.1116110. [DOI] [PubMed] [Google Scholar]
- Saito K., Sakaguchi Y., Suzuki T., Suzuki T., Siomi H., Siomi M.C. Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi-interacting RNAs at their 3′ ends. Genes & Dev. 2007;21:1603–1608. doi: 10.1101/gad.1563607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez Alvarado A., Newmark P.A., Robb S.M., Juste R. The Schmidtea mediterranea database as a molecular resource for studying platyhelminthes, stem cells and regeneration. Development. 2002;129:5659–5665. doi: 10.1242/dev.00167. [DOI] [PubMed] [Google Scholar]
- Song J.J., Smith S.K., Hannon G.J., Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- Timmons L., Court D.L., Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans . Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- Tolia N.H., Joshua-Tor L. Slicer and the argonautes. Nat Chem Biol. 2007;3:36–43. doi: 10.1038/nchembio848. [DOI] [PubMed] [Google Scholar]
- Umesono Y., Watanabe K., Agata K. Distinct structural domains in the planarian brain defined by the expression of evolutionarily conserved homeobox genes. Dev. Genes Evol. 1999;209:31–39. doi: 10.1007/s004270050224. [DOI] [PubMed] [Google Scholar]
- Vagin V.V., Sigova A., Li C., Seitz H., Gvozdev V., Zamore P.D. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- Wang Y., Zayas R.M., Guo T., Newmark P.A. nanos function is essential for development and regeneration of planarian germ cells. Proc. Natl. Acad. Sci. 2007;104:5901–5906. doi: 10.1073/pnas.0609708104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson J.E., Connell J.E., Macdonald P.M. Aubergine enhances oskar translation in the Drosophila ovary. Development. 1996;122:1631–1639. doi: 10.1242/dev.122.5.1631. [DOI] [PubMed] [Google Scholar]
- Yeh R.F., Lim L.P., Burge C.B. Computational inference of homologous gene structures in the human genome. Genome Res. 2001;11:803–816. doi: 10.1101/gr.175701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H., Lin H. An epigenetic activation role of Piwi and a Piwi-associated piRNA in Drosophila melanogaster . Nature. 2007;450:304–308. doi: 10.1038/nature06263. [DOI] [PubMed] [Google Scholar]
- Zamore P.D. RNA silencing: Genomic defence with a slice of pi. Nature. 2007;446:864–865. doi: 10.1038/446864a. [DOI] [PubMed] [Google Scholar]
- Zayas R.M., Hernandez A., Habermann B., Wang Y., Stary J.M., Newmark P.A. The planarian Schmidtea mediterranea as a model for epigenetic germ cell specification: Analysis of ESTs from the hermaphroditic strain. Proc. Natl. Acad. Sci. 2005;102:18491–18496. doi: 10.1073/pnas.0509507102. [DOI] [PMC free article] [PubMed] [Google Scholar]