Abstract
Trypanosome mitochondrial mRNAs achieve their coding sequences through RNA editing. This process, catalyzed by ∼20S protein complexes, involves large numbers of uridylate (U) insertions and deletions within mRNA precursors. Here we analyze the role of the essential TbMP42 protein (band VI/KREPA2) by individually examining each step of the U-deletional and U-insertional editing cycles, using reactions in the approximately linear range. We examined control extracts and RNA interference (RNAi) extracts prepared soon after TbMP42 was depleted (when primary effects should be most evident) and three days later (when precedent shows secondary effects can become prominent). This analysis shows TbMP42 is critical for cleavage of editing substrates by both the U-deletional and U-insertional endonucleases. However, on simple substrates that assess cleavage independent of editing features, TbMP42 is similarly required only for the U-deletional endonuclease, indicating TbMP42 affects the two editing endonucleases differently. Supplementing RNAi extract with recombinant TbMP42 partly restores these cleavage activities. Notably, we find that all the other editing steps (the 3′-U-exonuclease [3′-U-exo] and ligation steps of U-deletion and the terminal-U-transferase [TUTase] and ligation steps of U-insertion) remain at control levels upon RNAi induction, and hence are not dependent on TbMP42. This contrasts with an earlier report that TbMP42 is a 3′-U-exo that may act in U-deletion and additionally is critical for the TUTase and/or ligation steps of U-insertion, observations our data suggest reflect indirect effects of TbMP42 depletion. Thus, trypanosomes require TbMP42 for both endonucleolytic cleavage steps of RNA editing, but not for any of the subsequent steps of the editing cycles.
Keywords: RNA editing, trypanosome, RNA interference, U-deletion, U-insertion, TbMP42
INTRODUCTION
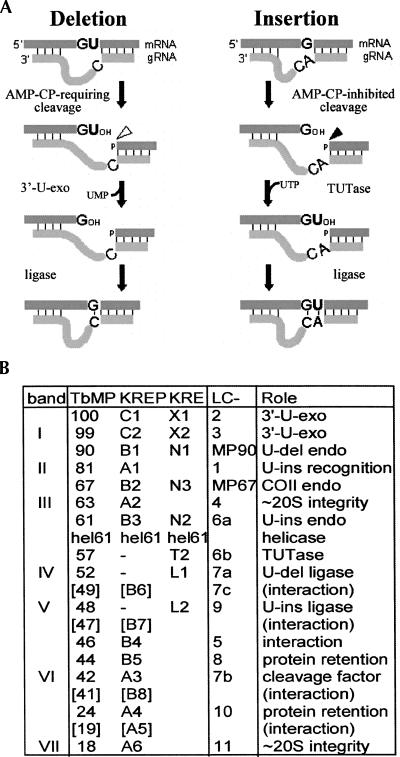
In trypanosomes, many mitochondrial transcripts are edited to generate mature mRNAs by the deletion and insertion of U's at numerous specific locations (for reviews, see Stuart and Panigrahi 2002; Simpson et al. 2003; Stuart et al. 2005). Such editing can create over three-quarters of the codons and nearly double the length of an mRNA (Feagin et al. 1988). The location and extent of the U-deletions and U-insertions are directed by guide RNAs (gRNAs; ∼40–70 nucleotides [nt]) that are complementary to regions of the mature mRNA and thus mismatch the pre-mRNA at each site to be edited (Blum et al. 1990). Initially, the 5′ portion of the gRNA base pairs with the pre-mRNA, forming an “anchor duplex” that extends to the first gRNA/pre-mRNA mismatch, which defines the first editing site. A mismatched region beginning with U's in the mRNA strand signifies a U-deletion, while a mismatched region beginning with non-U's (generally purines) signifies a U-insertion. For both kinds of editing cycles, the pre-mRNA is first endonucleolytically cleaved at the start of the mismatched region (see Fig. 1A; Blum et al. 1990; see also Cruz-Reyes et al. 1998b; Carnes et al. 2005; Trotter et al. 2005). Then U's are either removed or added at the 3′ end of the upstream cleavage fragment by a 3′-U-exonuclease (3′-U-exo) (Blum et al. 1990; see also Rusche et al. 1997) or a terminal-U-transferase (TUTase) (Bakalara et al. 1989; Aphasizhev et al. 2003c; Ernst et al. 2003). Finally, the transcript is resealed by an RNA ligase (Rusche et al. 1997, 2001; McManus et al. 2001; Schnaufer et al. 2001). After each successful editing cycle, the anchor duplex extends up to the next mismatch, which defines the next site to be processed, and editing thereby progresses 3′ to 5′ along the pre-mRNA.
FIGURE 1.
Mechanism of RNA editing and the editing proteins. (A) Editing cycles, as described in the text, with the responsible enzymes indicated. The G and A represent either purine and the C a pyrimidine. G-U pairing is allowed between the gRNA and the pre-mRNA. (B) Various nomenclatures for editing proteins, including the original “band” designations of T. brucei proteins from Rusche et al. (1997) (see Law et al. 2007 for additional verification of the reproducibility of this purification), the “TbMP” designation (representing T. brucei mitochondrial protein followed by the molecular weight of its cytoplasmic precursor) from Panigrahi et al. (2001a), and a more recent kinetoplastid RNA editing protein nomenclature (“KREPX#,” representing kinetoplastid RNA editing protein followed by a letter for the class of protein and a number indicating approximate ascending size within the group), that is being replaced with a KREØ# functional nomenclature (KRE as above, followed by a letter for the biological role of protein and a number indicating approximate ascending size within the group) from the Stuart laboratory (see references in the text; for reviews, see Worthey et al. 2003; Simpson et al. 2004; Stuart et al. 2005; Kang et al. 2006; Panigrahi et al. 2006). The proteins shown in brackets appear quite unabundant and/or are only obtained from some purification procedures (e.g., Panigrahi et al. 2006). In some references, the initial “K” is omitted, and, additionally, TbMP52 has also been referred to as DREL (Cruz-Reyes et al. 2002) and TbMP48 as IREL (Cruz-Reyes et al. 2002). Purification of editing complexes from L. tarentolae yields many homologous proteins, initially named LmLC# (Simpson et al. 2004) then shortened to LC# (Kang et al. 2006). For uniformity with the bulk of the editing literature, after their initial mention, we use the TbMP# designation. The final column summarizes the demonstrated (and proposed) roles in editing, as cited in the Introduction and the above references. mHel61p helicase of the ∼20S complex may aid removal of gRNAs after editing (Missel et al. 1997); also proteins separate from the ∼20S complex can affect editing, including TbMP108/KRET1 (that adds U-tails onto gRNAs) (Aphasizhev et al. 2002, 2003c), TbgBP21 and TbgBP25 (that stimulate RNA annealing) (Müller et al. 2001; Müller and Göringer 2002; Aphasizhev et al. 2003b; Schumacher et al. 2006), REAP1 (a putative mRNA-binding protein) (Madison-Antenucci et al. 1998), and RBP16 (a CYb RNA factor) (Pelletier and Read 2003).
The type of editing cycle is determined already as U-deletional or U-insertional by the endonucleolytic cleavage step. These cleavages are, respectively, stimulated or inhibited by adenosine polyphosphates (Cruz-Reyes et al. 1998b) and use different nucleases (Cruz-Reyes et al. 1998b; Carnes et al. 2005, 2008; Trotter et al. 2005). The main determinate specifying the kind of cleavage appears to be the unpaired U or non-U abutting the anchor duplex (see Fig. 1A), since cleavages of similar RNA substrates lacking the unpaired gRNA region, termed “basic cleavages,” retain the same adenosine polyphosphate specificity as occurs at normal editing sites (diagrammed in Fig. 5, below; cf. Figs. 1, 4 and Fig. 5, below; Law et al. 2005, 2007). Furthermore, the use of such basic enzymatic assays can discern differences between the fundamental activity of an editing enzyme and its ability to act normally at an editing site.
FIGURE 5.
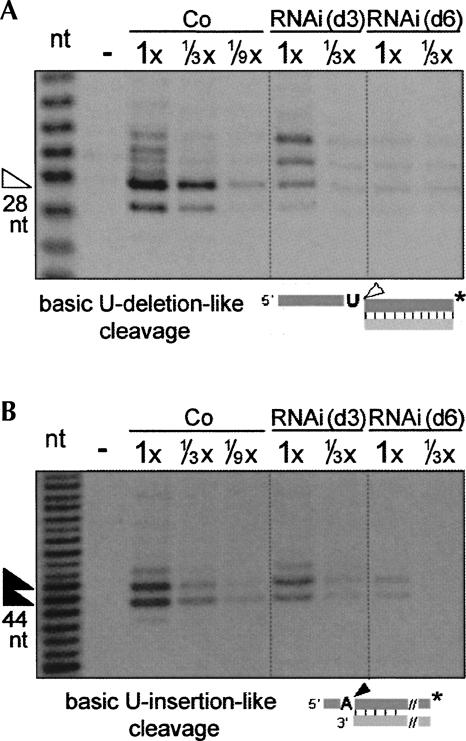
TbMP42 is required for retention of the basic U-deletional-like cleavage but not similarly for the basic U-insertional-like cleavage. (A) The basic U-deletion-like cleavage and (B) the basic U-insertion-like cleavage activities of the respective editing endonucleases, with products marked by arrowheads in the figures and assessed using the indicated substrates. In A, a nonediting single-strand nuclease also cleaves 1- and 2-nt upstream from the indicated site, especially in the control extract, and other nuclease activities generate two additional upper bands that are enhanced upon TbMP42 depletion; this could arise because the editing complex is less stable (see Fig. 8) and protects that upstream region less well from attack by single-strand-specific nucleases present in these crude cell extracts. In B, there are two main products due to heterogeneity in the gRNA length (Law et al. 2005). Control experiments confirm that the cleavages shown by the arrowheads retain their normal AMP-CP sensitivities (data not shown).
FIGURE 4.
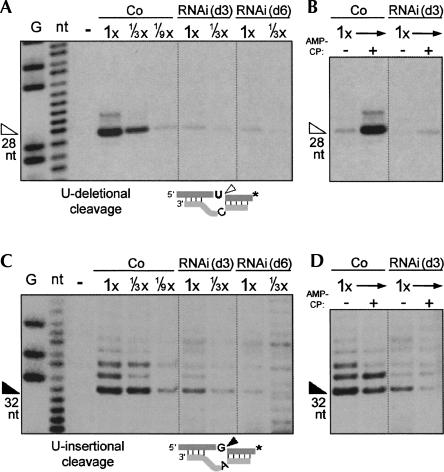
TbMP42 is essential for both U-deletional and U-insertional cleavages. Assessment of (A,B) U-deletional cleavage activity and (C,D) U-insertional cleavage activity using the editing substrates diagrammed below. (B,D) Verification of adenosine nucleotide specificity (see Cruz-Reyes et al. 1998b), using the nonhydrolyzable ADP analog, AMP-CP. These assays, as those of Figures 5 and 6, use the rapid extracts prepared from control or RNAi cells induced for 3 or 6 d. The weaker upstream cleavages are routinely observed (see Huang et al. 2002; Law et al. 2005, 2007). The analyzed protein amount is indicated, with 1× = 2.4 μg. G and nt indicate ladders showing guanosine residues and all nucleotides, from treatment of this mRNA with RNase TI and NaOH; − indicates no extract; arrowheads indicate the expected product (size shown below). In substrate diagrams of this and subsequent figures, * represents the 32P label of the pre-mRNA strand.
Biochemical purifications of editing activities have demonstrated that they reside in protein complexes of ∼20S (Pollard et al. 1992; Kable et al. 1996; Seiwert et al. 1996; Rusche et al. 1997; Panigrahi et al. 2006; Carnes et al. 2008). The initial procedure used to purify the editing complex from Trypanosoma brucei (Rusche et al. 1997) yields seven major staining proteins (designated bands I through VII, as well as minor protein bands that were not individually noted) (see Fig. 1B; Rusche et al. 1997). In later purification procedures (from another T. brucei line and from a Leishmania tarentolae line) (Panigrahi et al. 2001a, 2003a,b; Aphasizhev et al. 2003a; Gao et al. 2005; Stuart et al. 2005), ∼10–15 additional copurifying proteins were reported; they include the balance of the editing enzymes and appear to be in nonstochiometric amounts that vary in relative abundance in different preparations (see, e.g., Panigrahi et al. 2006). These ∼20 proteins were designated TbMP# from T. brucei (Panigrahi et al. 2001a) or LC-# from L. tarentolae (Aphasizhev et al. 2003a), and later renamed KREPX#, which is transitioning to a KREØ# functional designation (summarized in Fig. 1B; Stuart et al. 2005). Notably, editing complexes purified by the initial procedure (Rusche et al. 1997; see also Zhelonkina et al. 2006) catalyze full cycles of U-deletion and U-insertion more actively than other reported preparations (Cruz-Reyes et al. 1998c, 2002) despite those additional proteins appearing as only minor species. The differences in the protein profiles between the various editing complex preparations do not appear to be due to a selective loss of essential proteins during purification, since TbMP57 and TbMP63 (an apparently minor and major protein, respectively) were found at the same relative abundance in cell lysate as in the seven-major-protein complex (Zhelonkina et al. 2006). Instead, the differences could reflect the cell lines used for the purifications (discussed by Law et al. 2007). Specifically, the T. brucei complexes with seven major proteins or ∼20 proteins of apparently variable abundance were isolated from different cell lines and in different laboratories: 667 cells (Rusche et al. 1997) or IsTar 1.7a cells (Panigrahi et al. 2003a, and references therein), respectively, but isolation in the latter laboratory from yet another T. brucei line (29.13 cells) yielded a simple protein profile (Schnaufer et al. 2003) very similar to that originally purified from 667 cells. There also may be different populations of editing complexes that utilize many proteins in common but have a few protein differences (Panigrahi et al. 2006; Carnes et al. 2008, and references therein).
U-deletion and U-insertion involve parallel reaction steps that utilize distinct activities (Fig. 1A; Cruz-Reyes and Sollner-Webb 1996; Rusche et al. 1997; Cruz-Reyes et al. 1998b), and the relevant enzymes have now been largely identified (Fig. 1B). The U-deletional and U-insertional cleavage steps are catalyzed by TbMP90/KREN1 and TbMP61/KREN2, respectively (Carnes et al. 2005; Trotter et al. 2005; Kang et al. 2006) (or by TbMP67/KREPB2/KREN3 for the U-insertion that uses a cis-located gRNA; Carnes et al. 2008), although it has additionally been inferred that TbMP42/KREPA3/band VI may catalyze editing endonucleolytic cleavage (Brecht et al. 2005). The U-removal step is catalyzed by TbMP100/KREX1 (Worthey et al. 2003; Kang et al. 2005) and TbMP99/KREPC2/KREX2/band I (Schnaufer et al. 2003; Worthey et al. 2003; Rogers et al. 2007) and/or possibly TbMP42/KREPA3/band VI (Brecht et al. 2005); the U-addition step is catalyzed by the TbMP57/KRET2 TUTase (Aphasizhev et al. 2003c; Ernst et al. 2003). Finally, the ligation steps are catalyzed by the TbMP52/KREL1/band IV and TbMP48/KREL2/band V RNA ligases (Rusche et al. 1997, 2001; Schnaufer et al. 2001; see also Sabatini and Hajduk 1995; McManus et al. 2001), which preferentially seal in U-deletion and U-insertion, respectively (Huang et al. 2001; Cruz-Reyes et al. 2002; Schnaufer et al. 2003; see also Gao and Simpson 2003).
Editing complexes also contain several proteins with potential OB fold and/or zinc finger interaction domains (Schnaufer et al. 2003; Worthey et al. 2003) that are critical for their structure and/or activity. These include TbMP63/KREPA2/band III, which is required for editing complex integrity, for retention of the U-deletional ligase (TbMP52), and for the U-deletional and U-insertional cleavage activities (Huang et al. 2002), and TbMP81/KREPA1/band II, which is required for protein and/or substrate recognition at each step of U-insertion (Law et al. 2005), including for retention of the U-insertional ligase (TbMP48) (Drozdz et al. 2002; O'Hearn et al. 2003). Further refining these roles, recombinant TbMP63 associates with and stimulates the U-deletional ligase (TbMP52), while recombinant TbMP81 associates with and stimulates the U-insertional ligase (TbMP48) and the TbMP57 TUTase (Ernst et al. 2003; Schnaufer et al. 2003). In addition, overexpressing TAP-tagged ligases yielded two different ∼5–10S subcomplexes that contain TbMP63, TbMP52, and TbMP99 or TbMP81, TbMP48, and TbMP57 and, importantly, catalyze the final two steps of U-deletion or U-insertion, respectively (Schnaufer et al. 2003). Additional OB fold proteins include TbMP18/KREPA6/band VII (Law et al. 2007) and TbMP46/KREPB4 (Babbarwal et al. 2007), which appear critical for holding these subcomplexes together in the 20S complex and for both the U-deletional and U-insertional cleavages, as well as TbMP44/KREPB5 (Wang et al. 2003) and TbMP24/KREPA4 (Salavati et al. 2006), which appear essential for retention of all examined proteins of the ∼20S complex. Finally, the OB fold protein TbMP42/KREPA3/band VI was reported to be critical for trypanosome growth and required for the TUTase and/or ligase steps of precleaved U-insertion; it was also reported to have endonuclease and 3′-U-exonuclease activities that were inferred to catalyze those respective steps in U-deletion (Brecht et al. 2005). A few additional proteins have been reported in some preparations (see Fig. 1B), apparently at low levels, but their analysis has not been reported.
The roles of these major editing proteins were studied largely by specifically depleting them, using RNAi, and examining the effect on editing. Activities that diminish in consort with the depleted protein depend on that protein, likely directly. However, some activities remain present when the protein is initially depleted but diminish several days later, after the cells stop growing (Law et al. 2005, 2007); these activities are likely to only indirectly require the depleted protein.
We now report new studies on TbMP42, the last of the major proteins whose role appears unresolved, showing it is essential for allowing the editing endonucleases to function in U-deletion and U-insertion while the final two steps of U-deletion and U-insertion are not similarly dependent on TbMP42, quite different than the previous suggestions (Brecht et al. 2005). We also provide potential explanations for the previous, differing conclusions.
RESULTS
Depletion of TbMP42 by RNAi
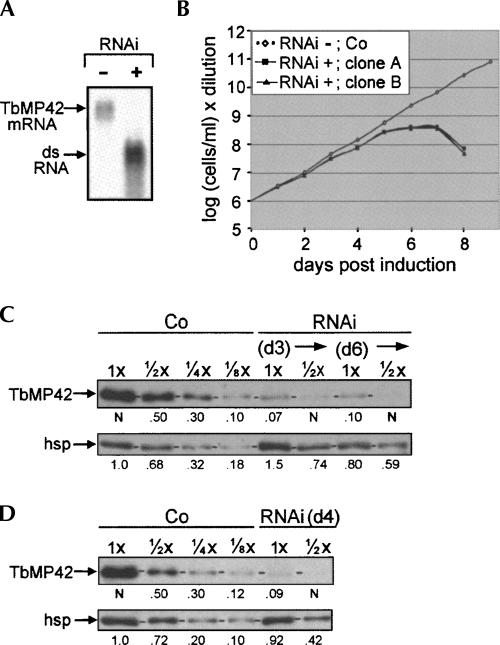
TbMP42, originally identified as band VI of the editing complex (Rusche et al. 1997), is a component of all reported preparations of editing complex (Rusche et al. 1997; Panigrahi et al. 2001b, 2003b, 2006; Aphasizhev et al. 2003a; Carnes et al. 2008). Previously, TbMP42 had been reported to have important roles in the U-addition and/or ligation steps of U-insertion and to be the enzyme that may catalyze the editing cleavage and U-removal steps of U-deletion (Brecht et al. 2005). We now present additional studies using trypanosome extracts depleted of TbMP42 by RNAi that completely revise this view. The same two clonal TbMP42 RNAi cell lines were used for all analyses reported below, including the editing assays, and gave confirmatory results. In these and additional examined TbMP42 RNAi lines, the TbMP42 mRNA was efficiently depleted by 24 h post-induction (Fig. 2A). The cells propagate normally for ∼5 d post-induction, then cease expanding (as also observed by Brecht et al. 2005), and after ∼2 more days our induced cells die (Fig. 2B), demonstrating TbMP42 is essential for viability.
FIGURE 2.
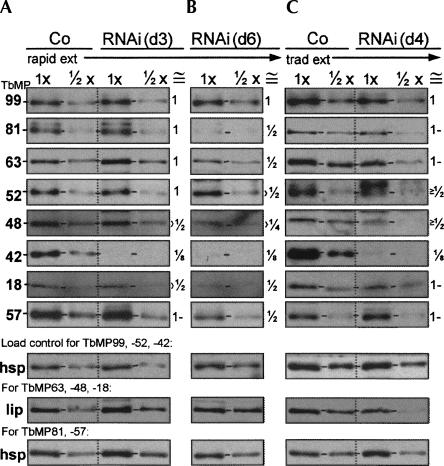
TbMP42 RNAi cell lines. (A) Northern blot from noninduced (−) or 24 h induced (+) RNAi cells showing the ∼1.5-kb TbMP42 mRNA and the (denatured) ∼0.7-kb RNA from the RNAi construct. (B) Growth curves of independent, clonal RNAi cell lines (RNAi) and control 29.13 cells (Co), following Tet addition. (C,D) Western blots using antibodies against TbMP42 and an hsp70 load control protein (see Materials and Methods; Law et al. 2005, 2007) to analyze (C) rapid cell extracts prepared from control cells or RNAi cells at day 3 (d3) and day 6 (d6) of induction and (D) traditional extracts prepared from control cells or RNAi cells at day 4 (d4) of induction. The protein amount analyzed is indicated, with 1× = 3 μg; the numbers below the lanes indicate the relative protein abundances, quantified as by Law et al. (2007), with TbMP42 values reported relative to lane loading (determined by the mitochondrial control protein hsp70) (see Materials and Methods). Horizontal dash marks on these and subsequent gels indicate the position of the examined protein.
The extent of TbMP42 depletion was determined by Western blotting using rapid cell extracts prepared at day 3 or day 6 of RNAi induction (Fig. 2C) and traditional extract prepared at day 4 of induction (Fig. 2D). Because Western blots can exhibit a highly nonlinear response, signals were quantified by comparison with similarly intense signals from dilutions of control extracts (the values were also corrected for variations in loading, using the mitochondrial hsp or lip control proteins, as in Law et al. 2007) (Fig. 2C,D). The TbMP42 signal at days 3, 4, and 6 of RNAi induction matches that of approximately one-eighth the amount of control extract (cells similarly treated for 3 d), indicating the RNAi reduces TbMP42 protein to about one-eighth the control level by day 3 of induction and does not appreciably further reduce it after longer times (Fig. 2C,D).
TbMP42 is not required for stability of the other major editing proteins
To determine the effect of TbMP42 depletion on other editing proteins, Western blots of the RNAi extracts were performed and detected using antibodies to each of the seven major proteins of our editing complex (Rusche et al. 1997) as well as to the TbMP57 TUTase (Fig. 3A–C). Quantification of these Western blots, as in Figure 2C,D, showed that at day 3 of RNAi induction, when TbMP42 is approximately one-eighth the control level, the levels of these other proteins are largely unaffected (Fig. 3A; see the ≅ column). Even at day 6, considerably after cell propagation has ceased, these proteins remain at approximately one-half the control level (Fig. 3B). Similar levels of protein retention were observed in the traditional extract (Fig. 3, cf. A and C). Thus, the presence of these other editing proteins is not dependent on TbMP42.
FIGURE 3.
Retention of other editing proteins upon depletion of TbMP42. Western blot analyses, as in Figure 2C,D, using antibodies against the indicated proteins (left), including load control proteins (hsp and lip) used for the indicated gels, to probe extracts of control 29.13 cells and RNAi cells, prepared by the rapid extract protocol at (A) day 3 and (B) day 6 of tet administration and (C) by the traditional extract protocol at day 4. For each antibody and type of extract, the RNAi and control samples were run in the same gel and blotted together. The protein amount analyzed is indicated, with 1× = 3 μg. The approximate abundance of each editing protein relative to the control level, after correction for the signal of a load control antibody, is presented in the approximately equals (≅) column to the right of each panel.
Effect of TbMP42 RNAi on editing activities
To assess the effects of TbMP42 depletion on editing, each of the U-deletional and U-insertional editing steps (Fig. 1A) was examined using the rapid extracts at day 3 (before growth inhibition, when primary effects of TbMP42 loss should be most evident) and at day 6 (after growth inhibition, when precedent has shown secondary effects can arise; Law et al. 2005, 2007) (see Materials and Methods). These experiments assessed multiple extract concentrations to assure evaluation in the approximately linear range for each assay and thus to allow meaningful quantification by comparison with control extracts. The analyses were preformed using duplicate extracts from two clonal RNAi cell lines, which gave similar results.
The cleavage steps of editing
U-deletional cleavage (Fig. 4A,B) and U-insertional cleavage (Fig. 4C,D) were assessed separately from the latter two steps of editing by adding PPi to inhibit the ligation step and using 3′-end-labeled mRNA to score the downstream fragment that is not affected by the second editing step. At day 3 of RNAi induction, both these kinds of cleavage are greatly reduced (Fig. 4A,C). Nonetheless, the residual activities retain their normal response to AMP-CP (Fig. 4B,D), which affirms that those editing cleavages are being scored. (Specifically, the U-deletional and U-insertional activities being stimulated and inhibited by this nucleotide, respectively; see Introduction and Cruz-Reyes et al. [1998b].) U-deletional cleavage appears depleted in parallel with the TbMP42 protein (reduced to approximately one-ninth of the control level; compare 1× of RNAi extract to 1/9× of the control extract), while U-insertional cleavage is slightly less affected at day 3 (reduced to approximately one-sixth of the control level; Fig. 4C). However, both kinds of cleavage become diminished to one-ninth or less of the control level by day 6 (Fig. 4A,C). Taken together, the data suggest that TbMP42 loss may directly affect U-deletional cleavage since this cleavage activity decreases rapidly upon RNAi induction, but may indirectly affect U-insertional cleavage since this cleavage activity decreases at a slower rate. Nonetheless, both U-deletional cleavage and U-insertional cleavage are highly dependent on TbMP42.
We next assessed the basic activities of the U-deletional and U-insertional editing endonucleases (see Introduction and Law et al. 2005, 2007) to discern whether loss of TbMP42 affects the enzymes themselves or their specific action at an editing site (as was shown for TbMP81 by Law et al. 2005). These basic assays score cleavage at simple 5′ single strand/3′ double strand junctions that resemble the pre-mRNA strand at the end of an anchor duplex, with the pre-mRNA residue just upstream of the duplex a U to assess basic U-deletion-like cleavage or a purine to assess basic U-insertion-like cleavage (Law et al. 2005, 2007, see diagrams in Fig. 5). In TbMP42 RNAi extract, this basic U-deletion-like cleavage (the band indicated by the hollow arrow in Fig. 5A; see legend) is reduced almost as much as cleavage at the U-deletional editing site (Fig. 4A). In contrast, the basic U-insertion-like cleavage (the sum of the two bands indicated by the solid arrows in Fig. 5B; see legend) remains within approximately twofold of the control level at day 3 of induction, when cleavage at the U-insertional editing site is reduced to a much greater extent (Fig. 4C). Verifying that the catalysis is by the respective editing endonucleases, those residual activities are stimulated and inhibited by AMP-CP, respectively (data not shown). These results demonstrate that the U-deletional endonuclease (which Trotter et al. [2005] showed is TbMP90) requires TbMP42 for its retention or basic activity in trypanosomes, and hence also for its specific action at an editing site. Our results also demonstrate that the U-insertional endonuclease (which Carnes et al. [2005] showed is TbMP61) initially remains largely present and exhibits basic activity without TbMP42 for several days. Thus, TbMP42 affects the two editing endonuclease cleavages differently.
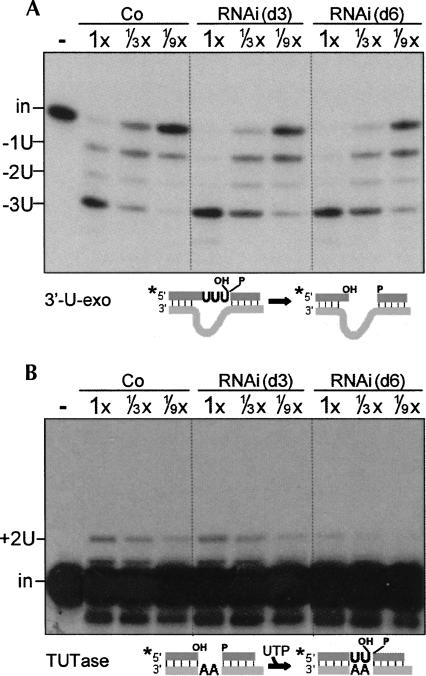
The 3′-U-exo step of U-deletion and the TUTase step of U-insertion
To specifically assess the second steps of editing, U-removal and U-addition, we used precleaved editing substrates to bypass the cleavage step and PPi to inhibit the ligation step. TbMP42 RNAi causes no decrease in the 3′-U-exo step (Fig. 6A). Indeed, this activity consistently appears slightly more robust in TbMP42 RNAi extracts than in control extracts (Fig. 6A), as was also observed upon TbMP18 depletion by RNAi (Law et al. 2007). The TUTase step also remains as robust as in control extracts at day 3 of induction (Fig. 6B), when TbMP42 is already greatly reduced (Fig. 2). Also, both the TUTase and 3′-U-exo activities are also fully retained in the traditional extract prepared at day 4 of induction (data not shown). However, by day 6 of induction, when secondary effects have been shown to arise upon RNAi of other editing proteins (Law et al. 2005, 2007) and when the basic activity of the U-insertional endonuclease becomes lost upon RNAi of TbMP42 (Fig. 5), the TUTase activity is also reduced, approximately ninefold (Fig. 6B). This implies that the TUTase activity initially remains present upon depletion of TbMP42, but that it has a reduced stability in vivo without TbMP42. We conclude that neither the 3′-U-exo nor the TUTase requires TbMP42 for activity.
FIGURE 6.
TbMP42 is not required for the 3′-U-exo or TUTase activities. (A) Specific 3′-U-exo activity and (B) specific TUTase activity assessed using the indicated precleaved editing substrates. The precleaved substrate in A (Law et al. 2005) guides removal of 3 U's; this assay is U specific, as the next residue in this substrate pre-mRNA, a single-stranded A, is not significantly removed. The precleaved substrate in B (Igo et al. 2000) guides insertion of 2 U's. The band below the input RNA was seen previously (Law et al. 2007) and may results from an unrelated single-strand nuclease acting on some small, unannealed fraction of the labeled upstream oligo. In this and subsequent experiments using precleaved substrates, the labeled input oligo (in) and U removal products (−1U, −2U, −3U) or U-addition product (+2U) are indicated.
Ligation in U-deletion and U-insertion
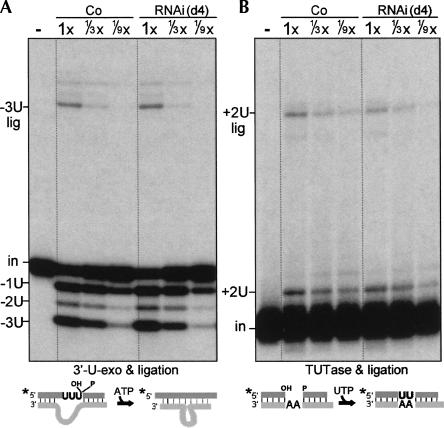
The ligation steps of U-deletion and U-insertion were assessed in combination with their prior 3′-U-exo and TUTase steps, which remain active upon TbMP42 depletion (Fig. 6), using the same precleaved substrates as in Figure 6 but reaction conditions to favor ligation (see Materials and Methods and Law et al. 2007). In precleaved U-deletion reactions, the rapid extracts prepared at day 3 (as well as day 6) of induction and the traditional extract prepared at day 4 all produced ligated product with similar efficiency to the control extract (Fig. 7A, −3U lig band; data not shown). Similarly, in precleaved U-insertion reactions, the extracts prepared at day 3 and day 4 of induction produce nearly the control level of ligated product (Fig. 7B, +2U lig band; data not shown). (The slight reduction in the level of +2 ligated product appears consistent with a slight reduction in the abundance of the TbMP48, which Cruz-Reyes et al. [2002] and Schnaufer et al. [2003] showed is the ligase specific for U-insertion [Fig. 3].) We conclude that TbMP42 is not required for the ligation steps of U-deletion or U-insertion or for their coordination with the previous 3′-U-exo and TUTase steps of the respective editing pathways. Underscoring the stability of these activities from the TbMP42 RNAi cells, they also are retained in the traditional extract (Fig. 7), despite these editing activities being diminished during such lengthy extract preparations from cells lacking other editing proteins (Law et al. 2005, 2007).
FIGURE 7.
TbMP42 is not required for U-deletional or U-insertional ligation. (A) Precleaved U-deletion (3′-U-exo plus ligation) and (B) precleaved U-insertion (TUTase plus ligation) assessed using the substrates of Figure 6 and the traditional extracts from control and RNAi cells induced for 4 d. 1× = 1.2 and 0.8 μg protein in A and B, respectively. The fully edited (−3U lig and +2U lig) products are indicated. Three-day rapid extracts yield comparable results (data not shown).
Stability of the TbMP42-depleted editing complex
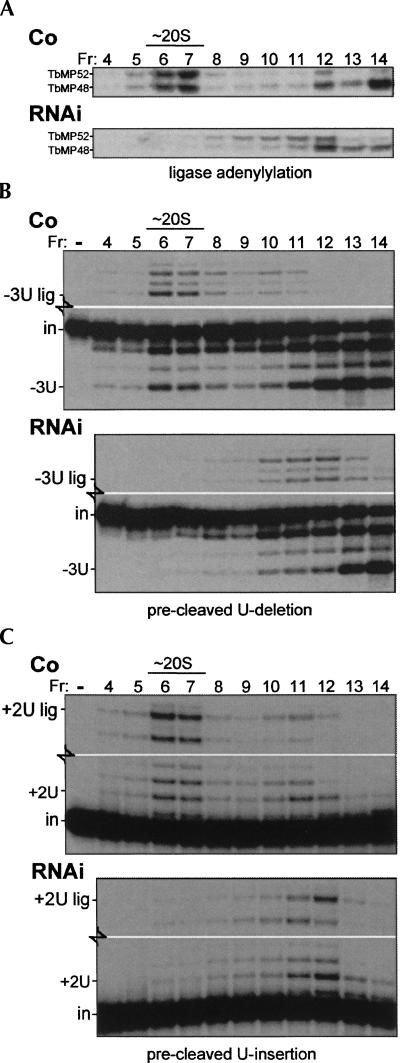
Upon depletion of TbMP42, the residual editing complex is reported to sediment like the editing complex from control cell extract (Brecht et al. 2005). This conclusion is underscored by our finding that the other editing proteins are largely unaffected by TbMP42 depletion at day 3 of induction (Fig. 3), since proteins from dissociated editing complex appear to be less stable in vivo (Huang et al. 2002; O'Hearn et al. 2003; Law et al. 2007). Nonetheless, the ∼50% decrease in abundance of these other editing proteins by day 6 of TbMP42 RNAi induction (Fig. 3B) suggests that TbMP42-depleted editing complexes have a somewhat reduced longevity in vivo. To examine the in vitro stability of the TbMP42-depleted editing complex, traditional extracts from control and TbMP42 RNAi cells were subjected to glycerol gradient centrifugation at hypotonic conditions. Under these conditions, editing complexes from control extracts remain appreciably at ∼20S, as indicated by ligase adenylylation (Fig. 8A) and precleaved editing assays (Fig. 8B,C). In contrast, when TbMP42 RNAi cell extract was similarly sedimented, ligase adenylylation (Fig. 8A) and both precleaved editing activities (Fig. 8B,C) were barely detectable in the ∼20S region and instead appear spread throughout the ∼15S to ∼5S region of the gradient (Fig. 8A–C). The selective instability of editing complex lacking TbMP42 under these somewhat destabilizing hypotonic conditions (Fig. 8) and the gradual decrease of the other editing proteins following TbMP42 depletion (Fig. 3B) suggest that although TbMP42 is not absolutely required for integrity of the ∼20S editing complex (Brecht et al. 2005), it does augment the stability of this complex, both in vitro and in vivo.
FIGURE 8.
TbMP18 is critical for stability of the editing complex. Glycerol gradient sedimentation analysis using the traditional extracts shown in Figures 2D, 3C, and 7. The indicated fractions (Fr 16 = top) were assayed for (A) the ligase proteins, by adenylylation, (B) precleaved U-deletion, as in Figure 7A, and (C) precleaved U-insertion, as in Figure 7B. In B and C, blank middle regions of these gels are not shown. Assessment of the individual 3′-U-exo and TUTase activities in these gradient fractions (reactions with PPi; not shown) gave results similar to those shown in the lower regions of B and C.
Although editing complexes lacking TbMP42 exhibit fragmentation under hypotonic in vitro conditions (Fig. 8), the ligases that seal the precleaved U-deletion and U-insertion reactions continue to function in a coordinated manner, acting preferentially on the products of the previous (3′-U-exo or TUTase) step of that editing cascade (Fig. 8B,C). For example, in the precleaved U-deletion reaction of Figure 8B, the ligase generates −3U lig RNA from −3U RNA with similar efficiency in the ∼15–5S fractions of the RNAi gradient (lower panel) as it had in the ∼20S fractions of the control gradient (Fig. 8B, upper panel). This concerted action suggests that the activities of each precleaved editing cascade remain associated, even following in vitro fragmentation of the editing complexes, and adds support that authentic editing activities are being assessed.
Supplementation with recombinant TbMP42 increases endonucleolytic cleavage
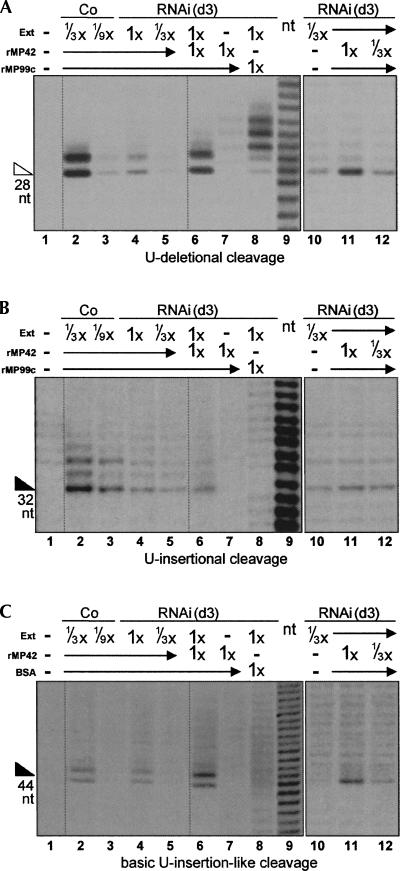
Since TbMP42 is important for U-deletional and U-insertional cleavages and stabilizes the editing complex, we next investigated whether recombinant TbMP42 could restore editing activities in an extract where the endogenous TbMP42 had been depleted by 3 d of RNAi induction. Indeed, recombinant TbMP42 (rTbMP42), renatured as in Brecht et al. (2005), significantly increases the observed level of U-deletional cleavage (Fig. 9A, cf. lanes 6 and 4, cf. lanes 11 and 10). Importantly, this specific cleavage is not catalyzed by rTbMP42 alone (Fig. 9A, lane 7) and is not increased when the RNAi extract is supplemented with BSA or with a different, similarly prepared and similar size recombinant protein (Fig. 9A, lane 8; data not shown). (While various preparations of recombinant proteins do catalyze some cleavages within the upstream single-stranded region of substrates [e.g., Fig. 9, lanes 8], those cleavages do not appear to be related to editing; see Fig. 9 legend and Discussion.) The ability of rTbMP42 to increase U-deletional cleavage activity in TbMP42 RNAi extract was observed in multiple independent experiments, with several different renatured preparations of rTbMP42 (Fig. 9A, cf. lanes 6 and 4; data not shown), and as expected, the extent of increase is dependent on the amount of added protein (Fig. 9A, lanes 10–12).
FIGURE 9.
Supplementation of TbMP42 RNAi cleavage reactions with rTbMP42. (A) U-deletional cleavage, (B) U-insertional cleavage, and (C) basic U-insertional cleavage were assessed as in Figures 4 and 5, with the reactions catalyzed by the indicated amounts of control extract or RNAi extract (1× = 2.4 μg of extract protein) alone (lanes 1–5,10), or supplemented with rTbMP42 (“rMP42”; lanes 6,11,12), or a with control protein (lanes 8; a recombinant 50-kDa segment of TbMP99 in A and B, “rMP99”; BSA in C), or by rTbMP42 alone (lane 7). Lane 9 is a nucleotide ladder. Lanes 10–12 are from a different experiment than lanes 1–9; the mRNA used for the experiment of lanes 1–9 in panels A and C had a 1-nt 3′ length heterogeneity and thus generated two product bands from the 3′ end-labeled substrate. The recombinant TbMP42 protein preparations not only stimulate editing cleavages by the RNAi extract (lanes 6), but alone they also direct some cleavage in the single-stranded regions upstream of the editing sites (lanes 7; see also lanes 8 for such cleavage by nonspecific recombinant protein preparations; substrates diagrammed in Figs. 4 and 5). However, those upstream cleavages are unlikely to be editing related since they generate 5′OH termini (the fragments co-migrate with an alkali-generated ladder of the same sequence [lane 9] and are out of phase with fragments from the editing cleavage; see, especially, Fig. 9B, lane 7 vs. lanes 2–6); in contrast, editing intermediates have 5′-P termini (see Cruz-Reyes et al. 1998a, and references therein).
rTbMP42 also stimulates the U-insertional cleavage reaction, but to a considerably lesser extent (Fig. 9B, cf. lanes 6 and 4, cf. lanes 11 and 10), and again this specific cleavage is not catalyzed by rTbMP42 alone (Fig. 9B, lane 7) and is not increased when the RNAi extract is supplemented with BSA or a different recombinant protein (Fig. 9B, lane 8; data not shown). Since the TbMP42 RNAi extract has diminished U-insertional cleavage due partly to its reduced basic cleavage activity (Fig. 5B) and partly to reduced recognition of the editing substrate by this endonuclease (combined data of Fig. 4B and Fig. 5B), we measured how the basic U-insertional cleavage activity of TbMP42 RNAi extracts is affected by supplementing with rTbMP42 (Fig. 9C). Indeed, there is a notable stimulation of the basic U-insertion-like cleavage activity (Fig. 9C, cf. lanes 6 and 4) that is dose dependent (Fig. 9C, lanes 10–12) and does not arise from the recombinant protein alone (Fig. 9C, lane 7) or from adding BSA or a different recombinant protein to the RNA extract (Fig. 9C, lane 8; data not shown). Thus, the added rTbMP42 significantly augments the basic (and specific) U-deletional cleavage activity (Fig. 9A; data not shown) and the basic U-insertional cleavage activity (Fig. 9C) of the 3 d induced TbMP42 RNAi extract but apparently does not equally enhance its capacity to recognize a U-insertional editing site (Fig. 9B).
DISCUSSION
This study assesses the role of TbMP42 by scoring the effect of its RNAi-mediated depletion on each step of editing (summarized in Fig. 10) and on the other major editing proteins. A previous RNAi study (Brecht et al. 2005) reported that TbMP42 is needed for cell growth and for U-insertion on a precleaved substrate, and that the TbMP42 protein also possessed a U-exonuclease and a single-strand endonuclease activity that could be part of the editing process, although the individual editing steps were not assessed. In the current study, we present data that greatly revise this view of TbMP42. We show induction of RNAi reduced TbMP42 protein to approximately one-eighth the control level (by day 3) (Fig. 2C,D), which caused not only growth inhibition (by day 5) (Fig. 2B) but then cell death (by day 8) (Fig. 2B), demonstrating that this protein is essential for viability. We find TbMP42 depletion greatly diminishes the cleavage steps in both U-deletion and U-insertion by day 3 of RNAi (Fig. 4), at times when our earlier studies have shown that direct effects of RNAi on editing are the most apparent (Law et al. 2007). In striking contrast with the previous report on TbMP42 (Brecht et al. 2005), we find extracts from TbMP42-depleted cells catalyze the coordinated steps of precleaved U-deletion and precleaved U-insertion at virtually control levels (Figs. 6, 7, when assessed within a few days of TbMP42 depletion), indicating that the 3′-U-exo, TUTase, and ligase activities are not directly dependent on TbMP42. A potential explanation for the discrepancy with the earlier publication is that we find that TUTase activity becomes diminished by nearly 90% at longer times following TbMP42 depletion, after cell growth has ceased (Figs. 6B, 10), suggesting that this activity is unstable in vivo in cells lacking TbMP42. Indeed, already at early times following TbMP42 depletion, the residual editing complex appears to have a reduced stability since its components, which sediment at ∼20S under isotonic conditions (Brecht et al. 2005), instead sediment as smaller associations under low salt conditions (Fig. 8). (Nonetheless, those editing activities continue to function in a coordinated manner [Fig. 8].) Furthermore, the secondary loss of catalytic activity at editing sites appears quite selective for the TUTase activity, since the bulk of the 3′-U-exo activity (Fig. 6) and both ligase activities (Fig. 7), as well as the TbMP57 TUTase protein and the other major proteins of the editing complex (Fig. 3), largely remain at day 6 of RNAi induction. Thus, TbMP42 is required for the endonuclease cleavages of the trypanosome's editing complex but not for the other activities of editing, and it enhances the stability of the editing complex.
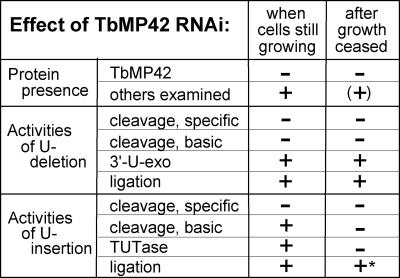
FIGURE 10.
Summary of the data from Figures 2–9. Retention of indicated protein or activity in RNAi extract at day 3 of induction (when cells are still growing) and at day 6 of induction (after growth ceased). “+” Indicates at least one-half as much as the control extract; “−” indicates less than one-fifth as much as the control extract. “*” represents the U-insertional ligase efficiency assessed in combination with the remaining TUTase product (as in Fig. 7).
Interestingly, TbMP42 depletion affects the U-deletional and U-insertional cleavage steps in different manners, as shown by the different effects on their basic activities (Fig. 5), using previously documented assays (Law et al. 2005, 2007). Specifically, in extracts depleted of TbMP42, the basic cleavage activity of the U-deletional endonuclease (Fig. 5A) is reduced approximately as much as this nuclease's activity at an editing site (Fig. 4A), indicating that TbMP42 is needed for any cleavage by the TbMP90 endonuclease in trypanosome extracts. (However, studies using recombinant proteins have shown that rTbMP90 cleaves an editing site in the presence of rTbMP63, without rTbMP42 [Kang et al. 2006], suggesting that in vivo TbMP42 likely serves to retain the presence of the active TbMP90.) In contrast to its effects on the U-deletional endonuclease, depletion of TbMP42 (at 3 d of RNAi induction) does not affect the basic cleavage activity of the U-insertional endonuclease (Fig. 5B) nearly as much as it affects its activity at an editing site (Fig. 4C). This may reflect a difference in stability of the U-deletional and the U-insertional endonucleases of the editing complexes upon depletion of TbMP42. Furthermore, TbMP42 appears to play a key role in substrate recognition by the TbMP61 endonuclease but less so in that nucleases' retention or actual cleavage mechanism. A role for TbMP42 in substrate recognition by the U-insertional endonuclease is reminiscent of the function of TbMP81 (Law et al. 2005). It has been proposed that TbMP81 facilitates substrate recognition by associating with the U-insertional enzymes and editing substrate using its OB fold domain (Ernst et al. 2003; Schnaufer et al. 2003; Worthey et al. 2003; Law et al. 2005), and TbMP42 might similarly utilize its OB fold domain to augment recognition of editing substrates by the U-insertional endonuclease. Additionally, the role of TbMP42 in augmenting stability of the editing complex (Figs. 6B, 8) could involve its zinc fingers, as has been shown for TbMP63 (Kang et al. 2004).
When added to reactions containing extracts depleted of TbMP42, rTbMP42 increases activity of the U-deletional endonuclease (Fig. 9A) and of the U-insertional endonuclease to a lesser extent (Fig. 9B). These effects are consistent with roles in augmenting stability of the editing complex and/or augmenting substrate recognition by the editing enzymes. In a potentially related effect, rTbMP42 was previously reported to restore precleaved U-insertion activity when added to extracts depleted of TbMP42 (Brecht et al. 2005). In light of our demonstration that precleaved U-insertion activity remains at virtually the control level when direct effects of the TbMP42 RNAi are already observed (Figs. 6B, 7B) and only diminishes subsequently, after the cells cease growing and secondary effects become more apparent (Fig. 6B; data not shown), the restoration of precleaved U-insertion observed by Brecht et al. (2005) appears likely to also have resulted from an indirect role of TbMP42. Nonetheless, the stimulatory effects of supplementing the RNAi extract with rTbMP42 attest to the importance of this protein in the editing complex.
In contrast to our conclusion that TbMP42 plays a noncatalytic essential role, it was previously suggested that TbMP42 catalyzes the endonucleolytic cleavages and the U-exonucleolytic cleavage of editing (Brecht et al. 2005). The inference that TbMP42 might catalyze the endonucleolytic cleavages was based on observing cleavage of a model U-deletional editing substrate a few nucleotides upstream of its expected editing cleavage site, within a single-stranded region, using either rTbMP42 preparations or control cell extract, and reduced amounts of this cleavage using cell extract lacking TbMP42 (Brecht et al. 2005). We also observe cleavage at such single-stranded positions a few nucleotides upstream of the expected U-deletional cleavage site catalyzed by some recombinant protein preparations (Fig. 9A, lanes 7,8), but the following observations suggest this cleavage is not specific to the TbMP42, nor is it involved in RNA editing. First, no appreciable amounts of these single-strand cleavages are observed using our cell extracts (Figs. 4A; Fig. 9A, lanes 2–5). Second, we observe only very low levels of these single-stranded cleavages with our preparations of rTbMP42 (Fig. 9A, lane 7), yet much more of them from other recombinant protein preparations, both from our preparation of a different protein (Fig. 9A, lane 8) and from a preparation of rTbMP42 kindly provided to us by Dr. Uli Goringer (Darmstadt University of Technology, Darmstadt, Germany) (data not shown). Third, alternate editing substrates also show such spurious cleavage from various recombinant protein preparations (Fig. 9B,C; data not shown). Fourth, these single-strand cleavages form 5′-OH termini (see legend to Fig. 9), not the 5′-P termini of editing intermediates, suggesting that they do not represent an editing endonuclease. Fifth, other proteins have been reported to be the endonucleases of editing, based on active site mutation as well as RNAi-mediated depletion (TbMP90 and TbMP61 catalyzing cleavage of normal U-deletion and U-insertion, and TbMP67 catalyzing cleavage of a U-insertion with cis-located gRNA; Carnes et al. 2005, 2008; Trotter et al. 2005). In addition, the U-insertional endonuclease remains largely present in extracts depleted of TbMP42 (Fig. 5B), indicating that that enzyme does not require TbMP42. Thus the single-strand endoribonuclease activity reported for preparations of rTbMP42 (Brecht et al. 2005) may not be due to the TbMP42 protein and appears to not catalyze the endonucleolytic cleavage steps of editing.
rTbMP42 was additionally reported to possess exoribonuclease activity, with preference for oligo-U over oligo-A, suggesting it may catalyze the 3′-U-exo step of editing (Brecht et al. 2005). However, we find that extracts depleted of TbMP42 exhibit no defect in the 3′-U-exo step (Fig. 6) or in precleaved U-deletion (Fig. 7), and Schnaufer et al. (2003) observed active precleaved U-deletion from protein associations that contain minimal TbMP42. Furthermore, TbMP99 (Schnaufer et al. 2003; Worthey et al. 2003) and/or TbMP100 (Kang et al. 2005; Rogers et al. 2007) have been reported to be enzymes catalyzing the 3′-U-exo activity of U-deletion. Nonetheless, editing could utilize another 3′-U-exo activity. Indeed, there is ample precedent for such protein multiplicity, as the editing machinery uses two endonucleases (Carnes et al. 2005; Trotter et al. 2005), two TUTases (Aphasizhev et al. 2002; Ernst et al. 2003), and two RNA ligases (Cruz-Reyes et al. 2002; Schnaufer et al. 2003), but for distinct functions (although the ligases also provide redundancy for sealing U-insertion) (Drozdz et al. 2002; Gao and Simpson 2003; O'Hearn et al. 2003). Thus, the 3′-U-exo function reported for TbMP42 (Brecht et al. 2005) could be utilized in editing, possibly providing a redundant 3′-U removal activity for U-deletion and/or a nonredundant role, such as the removal of excess U's that the TUTase might add in U-insertion (Byrne et al. 1996; Alfonzo et al. 1997; McManus et al. 2000; Igo et al. 2002) and the TUTase has been shown to add U's in U-deletion (Zhelonkina et al. 2006).
The roles of all but the least abundant members of the ∼20 proteins that copurify in preparations of the trypanosome editing complex have now been assessed (summarized in Fig. 1B), largely through biochemical analysis of extracts from cells in which the protein has been depleted using RNAi (and occasionally complemented by activity assays using the recombinant protein). Almost all the examined proteins are critical for various catalytic steps in the U-deletion and/or U-insertion cycles, either as the enzymes themselves or as accessory factors that help retain an enzyme's basic activity or facilitate recognition of an editing site. TbMP42 serves both such functions. It is only the second accessory factor reported to facilitate an enzyme's recognition of an editing site, since, like TbMP81, it appears to help the TbMP61 endonuclease to act at U-insertional editing sites (Figs. 4C, 5B). The noted RNA-binding activity of TbMP42 (Brecht et al. 2005) might provide this editing site-specific recognition, much as proposed for TbMP81 (Schnaufer et al. 2003). However, unlike TbMP81, we find TbMP42 is additionally needed to retain the basic activity of the U-deletional endonuclease (Fig. 5A). Thus TbMP42 appears to serve unique and important roles in the editing complex that are largely different than previously thought for this protein.
MATERIALS AND METHODS
Cloning the TbMP42 gene and preparation of RNAi cell lines
Band VI was isolated from an SDS-PAGE gel of the purified T. brucei editing complexes, and after trypsin digestion, peptides were sequenced by Edman degradation (as in Huang et al. 2001; Rusche et al. 2001; O'Hearn et al. 2003; Law et al. 2007). Four resultant peptides of 28, 21, and 10 amino acids identified a presumptive contig in the then partly complete T. brucei genomic sequence, and it was PCR amplified, yielding a 1182-nt sequence identical to the subsequently reported TbMP42 sequence (Panigrahi et al. 2001b). For RNAi, a 686-bp PCR product starting with GGAATACCGT was cloned into the tetracycline (Tet) inducible pZJM RNAi vector (Wang et al. 2000; as in Huang et al. 2002; O'Hearn et al. 2003) and was transfected into procyclic T. brucei 29.13 cells, which constitutively express T7 RNA polymerase and Tet repressor proteins (Wirtz et al. 1999). After extreme dilution (to ∼0.3 live cells per well), clonal cell lines that express the TbMP42 double-stranded RNA upon Tet induction were selected (as in Huang et al. 2002; O'Hearn et al. 2003; Law et al. 2007).
Extract preparation and glycerol gradient sedimentation
Most assays used small-scale extracts (from ∼2 × 108 cells), rapidly prepared (∼1.5 h post-lysis) (Rusche et al. 2001; Huang et al. 2002; Law et al. 2005) to focus on the primary effects of the protein and minimize secondary loss of editing activities (Law et al. 2005, 2007). Sedimentation analysis used traditional trypanosome mitochondrial extracts (Harris et al. 1990; Sabatini et al. 1998) (from ∼4 × 1010 cells) that require ∼6–8 h post-lysis, during which time some editing activities can diminish from certain RNAi cells (e.g., Law et al. [2005, 2007] also found here for the basic U-insertional endonuclease and TUTase activities in Figures 5B and 6B). We refer to these extracts as “rapid” and “traditional” (Law et al. 2007).
Glycerol gradient analysis was performed as described (Law et al. 2007) except we collected 16 fractions.
Northern and Western blotting
The pZJM insert was used to probe Northern blots of RNA from 2.5 × 107 cells (as in O'Hearn et al. 2003). For semi-quantitative Western analysis, extract dilutions in twofold increments (≤3 μg protein) were run on SDS-PAGE mini gels (Bio-Rad), transferred to nitrocellulose membranes (Bio-Rad), probed with antibodies and detected using the ECL-Plus chemiluminescent reagents (Amersham) (described in O'Hearn et al. 2003; Law et al. 2007), and quantified (Zhelonkina et al. 2006). The antibodies, including ones recognizing load control mitochondrial proteins, lipoamide dehydrogenase (“lip”) and heat shock protein 70 (“hsp”), were described (O'Hearn et al. 2003; Law et al. 2007).
Activity assays
The “specific” and “basic” assays were as previously described (Law et al. 2007, and references therein) and analyzed on 40 cm, 9% (w/v) (Figs. 4, 5, 9) or 15% (w/v) (Figs. 6–8) polyacrylamide/7.5 M urea gels. In brief, “specific” editing assays score the individual steps of U-deletion and U-insertion. They use standard T. brucei editing substrates: ATPase 6 (A6) pre-mRNA m[0,4] annealed to gRNA D33′, which guides the removal of 3 U's at editing site 1 (ES1), or annealed to gRNA I47G, which guides the addition of 2 U's at ES2, or three oligoribonucleotides (oligos) annealed to mimic a precleaved U-insertional editing site or a precleaved U-deletional editing site. The U-deletional or U-insertional cleavage step were assayed using 3′-end-labeled pre-mRNA and potassium pyrophosphate (PPi) plus either AMP-CP or no nucleotide, respectively. The 3′-U-exo and TUTase steps were assayed using PPi and precleaved substrates, with the upstream oligo 5′-end labeled; the 3′-U-exo assays also contained excess single-stranded RNA, and the TUTase assays also contained 0.15 mM UTP. U-deletional and U-insertional ligations were assessed in combination with their previous U-removal or U-addition steps, using their precleaved substrates and no PPi but supplemented with ATP (0.3 or 0 mM), UTP (0 or 2.4 mM) and yeast torula RNA (4.5 or 0 ng), respectively. The glycerol gradient fractions were assessed similarly. The “basic” cleavage assays, in which the endonucleases are assessed independent of their ability to act at an editing site, used substrates that retain minimal features required for cleavage activity: the pre-mRNA substrate annealed to an RNA that provides an anchor region but no guiding region or tether, as described (Law et al. 2005, 2007).
Recombinant TbMP42 preparation
Recombinant TbMP42 protein was expressed with a C-terminal His6 tag using the pET29a vector (Novagen) in Escherichia coli BL21 cells and was purified under denaturing conditions, after guanidine solubilization of the inclusion bodies, on Probond Nickel columns according to the manufacturer's instructions (Invitrogen). The recombinant protein was then renatured as in Brecht et al. (2005) immediately prior to use.
ACKNOWLEDGMENTS
We greatly thank H. Ulrich Göringer and Moritz Niemann in Uli's laboratory for very helpful discussions and critical reading of the manuscript, as well as for providing rTbMP42, which affirmed the data shown in Figure 9. We also thank Paul Englund and present and past members of his and our laboratories for helpful discussions, David Reim at the Wistar Institute for sequencing the tryptic peptides, Ken Piller for the procyclic strain TREU 667 cDNA library, Paul Englund for the pZJM vector and heat shock protein 70 antibody, and R. Luise Krauth-Siegel for lipoamide dehydrogenase antibody. We also thank the reviewers for helpful suggestions. This work was supported by NIH grant GM34231 to B.S.-W.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.899508.
REFERENCES
- Alfonzo J.D., Thiemann O., Simpson L. The mechanism of U-insertion/deletion RNA editing in kinetoplastid mitochondria. Nucleic Acids Res. 1997;25:3751–3759. doi: 10.1093/nar/25.19.3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizhev R., Sbicego S., Peris M., Jang S.H., Aphasizheva I., Simpson A.M., Rivlin A., Simpson L. Trypanosome mitochondrial 3′ terminal uridylyl transferase (TUTase): The key enzyme in U-insertion/deletion RNA editing. Cell. 2002;108:637–648. doi: 10.1016/s0092-8674(02)00647-5. [DOI] [PubMed] [Google Scholar]
- Aphasizhev R., Aphasizheva I., Nelson R.E., Gao G., Simpson A.M., Kang X., Falick A.M., Sbicego S., Simpson L. Isolation of a U-insertion/deletion editing complex from Leishmania tarentolae mitochondria. EMBO J. 2003a;22:913–924. doi: 10.1093/emboj/cdg083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizhev R., Aphasizheva I., Nelson R.E., Simpson L. A 100-kDa complex of two RNA-binding proteins from mitochondria of Leishmania tarentolae catalyzes RNA annealing and interacts with several RNA editing components. RNA. 2003b;9:62–76. doi: 10.1261/rna.2134303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizhev R., Aphasizheva I., Simpson L. A tale of two TUTases. Proc. Natl. Acad. Sci. 2003c;100:10617–10622. doi: 10.1073/pnas.1833120100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babbarwal V.K., Fleck M., Ernst N.L., Schnaufer A., Stuart K. An essential role of KREPB4 in RNA editing and structural integrity of the editosome in Trypanosoma brucei . RNA. 2007;13:737–744. doi: 10.1261/rna.327707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakalara N., Simpson A.M., Simpson L. The Leishmania kinetoplast-mitochondrion contains terminal uridylyltransferase and RNA ligase activities. J. Biol. Chem. 1989;264:18679–18686. [PubMed] [Google Scholar]
- Blum B., Bakalara N., Simpson L. A model for RNA editing in kinetoplastid mitochondria: “Guide” RNA molecules transcribed from maxicircle DNA provide the edited information. Cell. 1990;60:189–198. doi: 10.1016/0092-8674(90)90735-w. [DOI] [PubMed] [Google Scholar]
- Brecht M., Niemann M., Schlüter E., Müller U.F., Stuart K., Göringer H.U. TbMP42, a protein component of the RNA editing complex in African trypanosomes, has endo-exoribonuclease activity. Mol. Cell. 2005;17:621–630. doi: 10.1016/j.molcel.2005.01.018. [DOI] [PubMed] [Google Scholar]
- Byrne E.M., Connell G.J., Simpson L. Guide RNA-directed uridine insertion RNA editing in vitro. EMBO J. 1996;15:6758–6765. [PMC free article] [PubMed] [Google Scholar]
- Carnes J., Trotter J.R., Ernst N.L., Steinberg A., Stuart K. An essential RNase III insertion editing endonuclease in Trypanosoma brucei . Proc. Natl. Acad. Sci. 2005;102:16614–16619. doi: 10.1073/pnas.0506133102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnes J., Trotter J.R., Peltan A., Fleck M., Stuart K. RNA editing in Trypanosoma brucei requires three different editosomes. Mol. Cell. Biol. 2008;28:122–130. doi: 10.1128/MCB.01374-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Reyes J., Sollner-Webb B. Trypanosome U-deletional RNA editing involves guide RNA-directed endonuclease cleavage, terminal U exonuclease, and RNA ligase activities. Proc. Natl. Acad. Sci. 1996;93:8901–8906. doi: 10.1073/pnas.93.17.8901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Reyes J., Piller K.J., Rusche L.N., Mukherjee M., Sollner-Webb B. Unexpected electrophoretic migration of RNA with different 3′ termini causes a RNA sizing ambiguity that can be resolved using nuclease P1-generated sequencing ladders. Biochemistry. 1998a;37:6059–6064. doi: 10.1021/bi972868g. [DOI] [PubMed] [Google Scholar]
- Cruz-Reyes J., Rusche L.N., Piller K.J., Sollner-Webb B. T. brucei RNA editing: Adenosine nucleotides inversely affect U-deletion and U-insertion reactions at mRNA cleavage. Mol. Cell. 1998b;1:401–409. doi: 10.1016/s1097-2765(00)80040-4. [DOI] [PubMed] [Google Scholar]
- Cruz-Reyes J., Rusche L.N., Sollner-Webb B. Trypanosoma brucei U insertion and U-deletion activities co-purify with an enzymatic editing complex but are differentially optimized. Nucleic Acids Res. 1998c;26:3634–3639. doi: 10.1093/nar/26.16.3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Reyes J., Zhelonkina A.G., Huang C.E., Sollner-Webb B. Distinct functions of two RNA ligases in active Trypanosoma brucei RNA editing complexes. Mol. Cell. Biol. 2002;22:4652–4660. doi: 10.1128/MCB.22.13.4652-4660.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drozdz M., Palazzo S.S., Salavati R., O'Rear J., Clayton C., Stuart K. TbMP81 is required for RNA editing in Trypanosoma brucei . EMBO J. 2002;21:1791–1799. doi: 10.1093/emboj/21.7.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst N.L., Panicucci B., Igo R.P., Jr, Panigrahi A.K., Salavati R., Stuart K. TbMP57 is a 3′ terminal uridylyl transferase (TUTase) of the Trypanosoma brucei editosome. Mol. Cell. 2003;11:1525–1536. doi: 10.1016/s1097-2765(03)00185-0. [DOI] [PubMed] [Google Scholar]
- Feagin J.E., Abraham J.M., Stuart K. Extensive editing of the cytochrome c oxidase III transcript in Trypanosoma brucei . Cell. 1988;53:413–422. doi: 10.1016/0092-8674(88)90161-4. [DOI] [PubMed] [Google Scholar]
- Gao G., Simpson L. Is the Trypanosoma brucei REL1 RNA ligase specific for U-deletion RNA editing, and is the REL2 RNA ligase specific for U-insertion editing? J. Biol. Chem. 2003;278:27570–27574. doi: 10.1074/jbc.M303317200. [DOI] [PubMed] [Google Scholar]
- Gao G., Simpson A.M., Kang X., Rogers K., Nebohacova M., Li F., Simpson L. Functional complementation of Trypanosoma brucei RNA in vitro editing with recombinant RNA ligase. Proc. Natl. Acad. Sci. 2005;102:4712–4717. doi: 10.1073/pnas.0500553102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris M.E., Moore D.R., Hajduk S.L. Addition of uridines to edited RNAs in trypanosome mitochondria occurs independently of transcription. J. Biol. Chem. 1990;265:11368–11376. [PubMed] [Google Scholar]
- Huang C.E., Cruz-Reyes J., Zhelonkina A.G., O'Hearn S., Wirtz E., Sollner-Webb B. Roles for ligases in the RNA editing complex of Trypanosoma brucei: Band IV is needed for U-deletion and RNA repair. EMBO J. 2001;20:4694–4703. doi: 10.1093/emboj/20.17.4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C.E., O'Hearn S.F., Sollner-Webb B. Assembly and function of the RNA editing complex in Trypanosoma brucei requires band III protein. Mol. Cell. Biol. 2002;22:3194–3203. doi: 10.1128/MCB.22.9.3194-3203.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igo R.P., Jr, Palazzo S.S., Burgess M.L., Panigrahi A.K., Stuart K. Uridylate addition and RNA ligation contribute to the specificity of kinetoplastid insertion RNA editing. Mol. Cell. Biol. 2000;20:8447–8457. doi: 10.1128/mcb.20.22.8447-8457.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igo R.P., Jr, Weston D.S., Ernst N.L., Panigrahi A.K., Salavati R., Stuart K. Role of uridylate-specific exoribonuclease activity in Trypanosoma brucei RNA editing. Eukaryot. Cell. 2002;1:112–118. doi: 10.1128/EC.1.1.112-118.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kable M.L., Seiwert S.D., Heidmann S., Stuart K. RNA editing: A mechanism for gRNA-specified uridylate insertion into precursor mRNA. Science. 1996;273:1189–1195. doi: 10.1126/science.273.5279.1189. [DOI] [PubMed] [Google Scholar]
- Kang X., Falick A.M., Nelson R.E., Gao G., Rogers K., Aphasizhev R., Simpson L. Disruption of the zinc finger motifs in the Leishmania tarentolae LC-4 (=TbMP63) L-complex editing protein affects the stability of the L-complex. J. Biol. Chem. 2004;279:3893–3899. doi: 10.1074/jbc.M310185200. [DOI] [PubMed] [Google Scholar]
- Kang X., Rogers K., Gao G., Falick A.M., Zhou S., Simpson L. Reconstitution of uridine-deletion precleaved RNA editing with two recombinant enzymes. Proc. Natl. Acad. Sci. 2005;102:1017–1022. doi: 10.1073/pnas.0409275102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang X., Gao G., Rogers K., Falick A.M., Zhou S., Simpson L. Reconstitution of full-round uridine-deletion RNA editing with three recombinant proteins. Proc. Natl. Acad. Sci. 2006;19:13944–13949. doi: 10.1073/pnas.0604476103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law J.A., Huang C.E., O'Hearn S.F., Sollner-Webb B. In Trypanosoma brucei RNA editing, band II enables recognition specifically at each step of the U insertion cycle. Mol. Cell. Biol. 2005;25:2785–2794. doi: 10.1128/MCB.25.7.2785-2794.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law J.A., O'Hearn S., Sollner-Webb B. In Trypanosoma brucei RNA editing, TbMP18 (band VII) is critical for editosome integrity and for both insertional and deletional cleavages. Mol. Cell. Biol. 2007;27:777–787. doi: 10.1128/MCB.01460-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison-Antenucci S., Sabatini R.S., Pollard V.W., Hajduk S.L. Kinetoplastid RNA-editing-associated protein 1 (REAP-1): A novel editing complex protein with repetitive domains. EMBO J. 1998;17:6368–6376. doi: 10.1093/emboj/17.21.6368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus M.T., Adler B.K., Pollard V.W., Hajduk S.L. Trypanosoma brucei guide RNA poly(U) tail formation is stabilized by cognate mRNA. Mol. Cell. Biol. 2000;20:883–891. doi: 10.1128/mcb.20.3.883-891.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus M.T., Shimamura M., Grams J., Hajduk S.L. Identification of candidate mitochondrial RNA editing ligases from Trypanosoma brucei . RNA. 2001;7:167–175. doi: 10.1017/s1355838201002072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missel A., Souza A.E., Nörskau G., Göringer H.U. Disruption of a gene encoding a novel mitochondrial DEAD-box protein in Trypanosoma brucei affects edited mRNAs. Mol. Cell. Biol. 1997;17:4895–4903. doi: 10.1128/mcb.17.9.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller U.F., Göringer H.U. Mechanism of the gBP21-mediated RNA/RNA annealing reaction: Matchmaking and charge reduction. Nucleic Acids Res. 2002;30:447–455. doi: 10.1093/nar/30.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller U.F., Lambert L., Göringer H.U. Annealing of RNA editing substrates facilitated by guide RNA-binding protein gBP21. EMBO J. 2001;20:1394–1404. doi: 10.1093/emboj/20.6.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hearn S.F., Huang C.E., Hemann M., Zhelonkina A., Sollner-Webb B. Trypanosoma brucei RNA editing complex: Band II is structurally critical and maintains band V ligase, which is nonessential. Mol. Cell. Biol. 2003;23:7909–7919. doi: 10.1128/MCB.23.21.7909-7919.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi A.K., Gygi S.P., Ernst N.L., Igo R.P., Jr, Palazzo S.S., Schnaufer A., Weston D.S., Carmean N., Salavati R., Aebersold R., et al. Association of two novel proteins, TbMP52 and TbMP48, with the Trypanosoma brucei RNA editing complex. Mol. Cell. Biol. 2001a;21:380–389. doi: 10.1128/MCB.21.2.380-389.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi A.K., Schnaufer A., Carmean N., Igo R.P., Jr, Gygi S.P., Ernst N.L., Palazzo S.S., Weston D.S., Aebersold R., Salavati R., et al. Four related proteins of the Trypanosoma brucei RNA editing complex. Mol. Cell. Biol. 2001b;21:6833–6840. doi: 10.1128/MCB.21.20.6833-6840.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi A.K., Allen T.E., Stuart K., Haynes P.A., Gygi S.P. Mass spectrometric analysis of the editosome and other multiprotein complexes in Trypanosoma brucei . J. Am. Soc. Mass Spectrom. 2003a;14:728–735. doi: 10.1016/S1044-0305(03)00126-0. [DOI] [PubMed] [Google Scholar]
- Panigrahi A.K., Schnaufer A., Ernst N.L., Wang B., Carmean N., Salavati R., Stuart K. Identification of novel components of Trypanosoma brucei editosomes. RNA. 2003b;9:484–492. doi: 10.1261/rna.2194603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi A.K., Ernst N.L., Domingo G.J., Fleck M., Salavati R., Stuart K.D. Compositionally and functionally distinct editosomes in Trypanosoma brucei . RNA. 2006;12:1038–1049. doi: 10.1261/rna.45506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier M., Read L.K. RBP16 is a multifunctional gene regulatory protein involved in editing and stabilization of specific mitochondrial mRNAs in Trypanosoma brucei . RNA. 2003;9:457–468. doi: 10.1261/rna.2160803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard V.W., Harris M.E., Hajduk S.L. Native mRNA editing complexes from Trypanosoma brucei mitochondria. EMBO J. 1992;11:4429–4438. doi: 10.1002/j.1460-2075.1992.tb05543.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers K., Gao G., Simpson L. U-specific 3′–5′ exoribonucleases involved in U-deletion RNA editing in trypanosomatid mitochondria. J. Biol. Chem. 2007;282:29073–29080. doi: 10.1074/jbc.M704551200. [DOI] [PubMed] [Google Scholar]
- Rusche L.N., Cruz-Reyes J., Piller K.J., Sollner-Webb B. Purification of a functional enzymatic editing complex from Trypanosoma brucei mitochondria. EMBO J. 1997;16:4069–4081. doi: 10.1093/emboj/16.13.4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusche L.N., Huang C.E., Piller K.J., Hemann M., Wirtz E., Sollner-Webb B. The two RNA ligases of the Trypanosoma brucei RNA editing complex: Cloning the essential band IV gene and identifying the band V gene. Mol. Cell. Biol. 2001;21:979–989. doi: 10.1128/MCB.21.4.979-989.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini R., Hajduk S.L. RNA ligase and its involvement in guide RNA/mRNA chimera formation. Evidence for a cleavage-ligation mechanism of Trypanosoma brucei mRNA editing. J. Biol. Chem. 1995;270:7233–7240. doi: 10.1074/jbc.270.13.7233. [DOI] [PubMed] [Google Scholar]
- Sabatini R.S., Adler B.K., Madison-Antenucci S., McManus M.T., Hajduk S.L. Biochemical methods for analysis of kinetoplastid RNA editing. Methods. 1998;15:15–26. doi: 10.1006/meth.1998.0602. [DOI] [PubMed] [Google Scholar]
- Salavati R., Ernst N.L., O'Rear J., Gilliam T., Tarun S., Jr, Stuart K. KREPA4, an RNA binding protein essential for editosome integrity and survival of Trypanosoma brucei . RNA. 2006;12:819–831. doi: 10.1261/rna.2244106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnaufer A., Panigrahi A.K., Panicucci B., Igo R.P., Jr, Wirtz E., Salavati R., Stuart K. An RNA ligase essential for RNA editing and survival of the bloodstream form of Trypanosoma brucei . Science. 2001;291:2159–2162. doi: 10.1126/science.1058955. [DOI] [PubMed] [Google Scholar]
- Schnaufer A., Ernst N.L., Palazzo S.S., O'Rear J., Salavati R., Stuart K. Separate insertion and deletion subcomplexes of the Trypanosoma brucei RNA editing complex. Mol. Cell. 2003;12:307–319. doi: 10.1016/s1097-2765(03)00286-7. [DOI] [PubMed] [Google Scholar]
- Schumacher M.A., Karamooz E., Zikova A., Trantirek L., Lukes J. Crystal structures of T. brucei MRP1/MRP2 guide-RNA binding complex reveal RNA matchmaking mechanism. Cell. 2006;126:701–711. doi: 10.1016/j.cell.2006.06.047. [DOI] [PubMed] [Google Scholar]
- Seiwert S.D., Heidmann S., Stuart K. Direct visualization of uridylate deletion in vitro suggests a mechanism for kinetoplastid RNA editing. Cell. 1996;84:831–841. doi: 10.1016/s0092-8674(00)81062-4. [DOI] [PubMed] [Google Scholar]
- Simpson L., Sbicego S., Aphasizhev R. Uridine insertion/deletion RNA editing in trypanosome mitochondria: A complex business. RNA. 2003;9:265–276. doi: 10.1261/rna.2178403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson L., Aphasizhev R., Gao G., Kang X. Mitochondrial proteins and complexes in Leishmania and Trypanosoma involved in U-insertion/deletion RNA editing. RNA. 2004;10:159–170. doi: 10.1261/rna.5170704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart K., Panigrahi A.K. RNA editing: Complexity and complications. Mol. Microbiol. 2002;45:591–596. doi: 10.1046/j.1365-2958.2002.03028.x. [DOI] [PubMed] [Google Scholar]
- Stuart K.D., Schnaufer A., Ernst N.L., Panigrahi A.K. Complex management: RNA editing in trypanosomes. Trends Biochem. Sci. 2005;30:97–105. doi: 10.1016/j.tibs.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Trotter J.R., Ernst N.L., Carnes J., Panicucci B., Stuart K. A deletion site editing endonuclease in Trypanosoma brucei . Mol. Cell. 2005;20:403–412. doi: 10.1016/j.molcel.2005.09.016. [DOI] [PubMed] [Google Scholar]
- Wang Z., Morris J.C., Drew M.E., Englund P.T. Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J. Biol. Chem. 2000;275:40174–40179. doi: 10.1074/jbc.M008405200. [DOI] [PubMed] [Google Scholar]
- Wang B., Ernst N.L., Palazzo S.S., Panigrahi A.K., Salavati R., Stuart K. TbMP44 is essential for RNA editing and structural integrity of the editosome in Trypanosoma brucei . Eukaryot. Cell. 2003;2:578–587. doi: 10.1128/EC.2.3.578-587.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz E., Leal S., Ochatt C., Cross G.A. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei . Mol. Biochem. Parasitol. 1999;99:89–101. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]
- Worthey E.A., Schnaufer A., Mian I.S., Stuart K., Salavati R. Comparative analysis of editosome proteins in trypanosomatids. Nucleic Acids Res. 2003;31:6392–6408. doi: 10.1093/nar/gkg870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhelonkina A.G., O'Hearn S.F., Law J.A., Cruz-Reyes J., Huang C.E., Alatortsev V.S., Sollner-Webb B. T. brucei RNA editing: Action of the U-insertional TUTase within a U-deletion cycle. RNA. 2006;12:476–487. doi: 10.1261/rna.2243206. [DOI] [PMC free article] [PubMed] [Google Scholar]