Abstract
RNase P from Bacillus subtilis cleaves in vitro the adenine riboswitch upstream of pbuE, which codes for an adenine efflux pump. The guanine riboswitch, encoded upstream of xpt-pbuX operon, is not cleaved. The cleavage sites do not occur at any predicted structures that should be recognized by RNase P in the theoretical model of the adenine riboswitch. However, it is possible to draw alternative secondary structure models that match the apparent requirements for RNase P substrates at these cleavage sites. Support for these models is provided by appropriate mutagenesis experiments. Adenine showed no effect on the cleavage in vitro of the pbuE adenine riboswitch by RNase P holoenzyme from B. subtilis. The results of genetic experiments performed in B. subtilis support the cleavage of adenine riboswitch by RNase P in vivo and suggest that it induces the stabilization of pbuE mRNA under normal conditions.
Keywords: transient structures, RNA enzyme, RNA processing, purines
INTRODUCTION
Ribonuclease P (RNase P) is an endonuclease known primarily for its role in tRNA maturation. It cleaves the 5′ leader sequence of tRNA precursors in the three domains of life (Altman 2007). Escherichia coli RNase P also cleaves the precursors to 4.5S RNA (Bothwell et al. 1976; Guerrier-Takada and Altman 1984) and tmRNA (Komine et al. 1994) as well as CI RNA (Forti et al. 1995), and within the intergenic region of several operon mRNAs (Li and Altman 2003, 2004). RNase P has also been shown to cleave coenzyme B12 riboswitches from E. coli and Bacillus subtilis. This latter phenomenon may be widespread. However, not all riboswitches (Altman et al. 2005) are substrates for RNase P.
The RNase P cleavage of operon and riboswitch RNAs is not understood in terms of the physiological function of these endoribonuclease events. They may be “accidental” cleavages of misfolded RNAs before they are in their native states or they may play precise roles in the regulation of protein amounts inside cells. The lac operon cleavage, in which RNase P cuts between the lacY and lacA genes that ultimately results in a loss in the stoichiometric yield of the A protein, seems to be of the latter kind (Li and Altman 2004).
We report the results of RNase P cleavage of adenine and guanine riboswitches upstream of the pbuE gene (previously known as ydhl) and the xpt-pbuX operon in B. subtilis, respectively. The pbuE gene codes for a purine efflux pump, as suggested from sequence comparison and genetic studies (Johansen et al. 2003). The transcription of pbuE is under the control of the adenine riboswitch (Mandal and Breaker 2004). In the absence of adenine, the riboswitch adopts an “off state” structure that includes a transcription terminator. This transcription control mechanism couples the excretion of adenine to its intracellular concentration and allows the cell to maintain a stable pool of adenine.
The xpt-pbuX operon includes two overlapping genes that are implicated in the metabolism of guanine (Christiansen et al. 1997). The transcription of these genes is under the control of a guanine riboswitch present in the 5′ upstream sequence (Mandal et al. 2003).
We show that the pbuE adenine riboswitch is a substrate for B. subtilis RNase P holoenzyme in vitro, but the xpt-pbuX guanine riboswitch is not. Secondary structure models near the cleavage sites recognized by RNase P are proposed. These models are validated by appropriate site-specific mutagenesis. We further provide support for the occurrence of this cleavage in vivo by genetic studies performed in B. subtilis.
RESULTS
RNase P cleaves in vitro the pbuE adenine riboswitch but not the xpt-pbuX guanine riboswitch
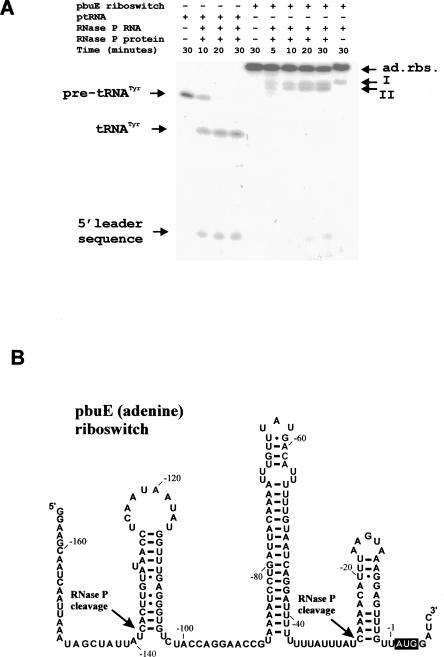
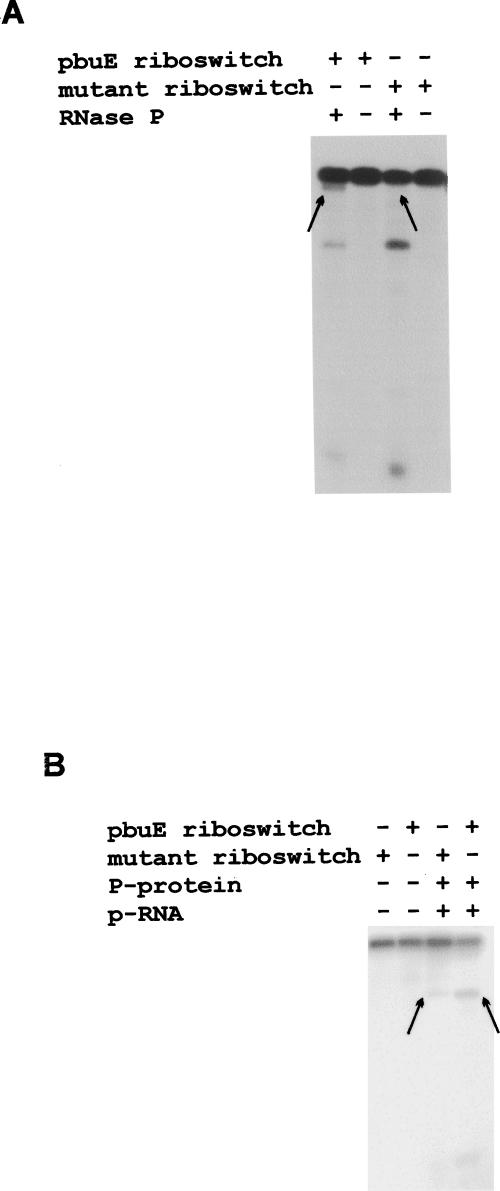
5′ UTR regions upstream of pbuE (−165 to +6: +1 is the A of AUG) and xpt-pbuX (−241 to +35), which encode the adenine and guanine riboswitches, respectively, were PCR amplified from B. subtilis genomic DNA and cloned under a T7 promoter in pUC19 (Materials and Methods). The purified plasmid DNA was linearized with BamHI. Following transcription in vitro, the products were purified and assayed for cleavage by the B. subtilis RNase P holoenzyme. No products were detected for the xpt-pbuX guanine riboswitch substrate after 30 min of incubation at 37°C in BP buffer in the presence of 0.5 nM of RNase P holoenzyme (not shown). Using 0.25 nM of RNase P holoenzyme in the same buffer and temperature conditions, and after 10 min, two cleavages were detected in the pbuE adenine riboswitch. These cleavages generated two products that differ slightly in size (Fig. 1) and two other small products, very difficult to visualize because of their size and low content of G (the radiolabeled nucleotide) in their sequence. The riboswitch cleavage is ∼20-fold less efficient than the cleavage of the precursor to tRNATyr (Fig. 1).
FIGURE 1.
RNase P holoenzyme cleaves the pbuE adenine riboswitch. (A) RNase P assays of the pbuE adenine riboswitch. Internally labeled adenine riboswitch, prepared as described in Materials and Methods, was incubated at 37°C in BP buffer (0.2 M NH4Cl, 50 mM MgCl2, 50 mM Tris-HCl pH 8) for 5, 10, 20, and 30 min in the presence of 0.25 nM of B. subtilis RNase P holoenzyme. Substrate with no RNase P served as a negative control. Internally labeled tRNATyr precursor was used as a control for this experiment. For the P RNA only assay, the substrate was incubated in 300 mM MgCl2, 2.5 M NH4Acetate, 0.01% SDS, 50 mM Tris-HCl pH 8. The upper band in each lane represents the uncleaved substrate. The products on the gel were quantified with a FujiX phosphorimager and ImageGauge software. Cleavage rates were calculated during the linear portion of the kinetic curves. Comparisons showed that the rate of cleavage on the tRNA precursor is about 20-fold more efficient than that of the adenine riboswitch. Cleavage at site I is 20% more efficient than cleavage at site II. (B) Secondary structure model of pbuE adenine riboswitch. The transcription terminator helix, position −89 to position −38, has been previously described by Mandal and Breaker (2004). The boxed AUG is the translation initiation codon of pbuE gene. RNase P cleavage sites are indicated by arrows, and the corresponding transient helices were determined as described in the text. Numbering of the nucleotides is based on the translation initiation site of pbuE gene; the first A of the AUG codon corresponds to +1.
Mapping of the cleavage sites
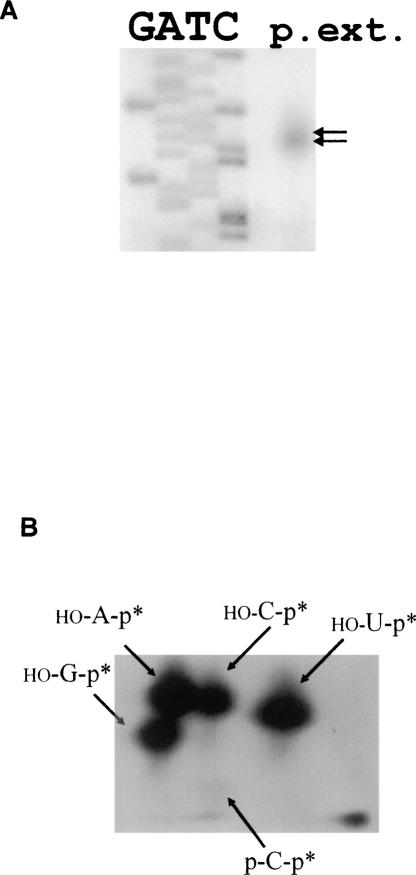
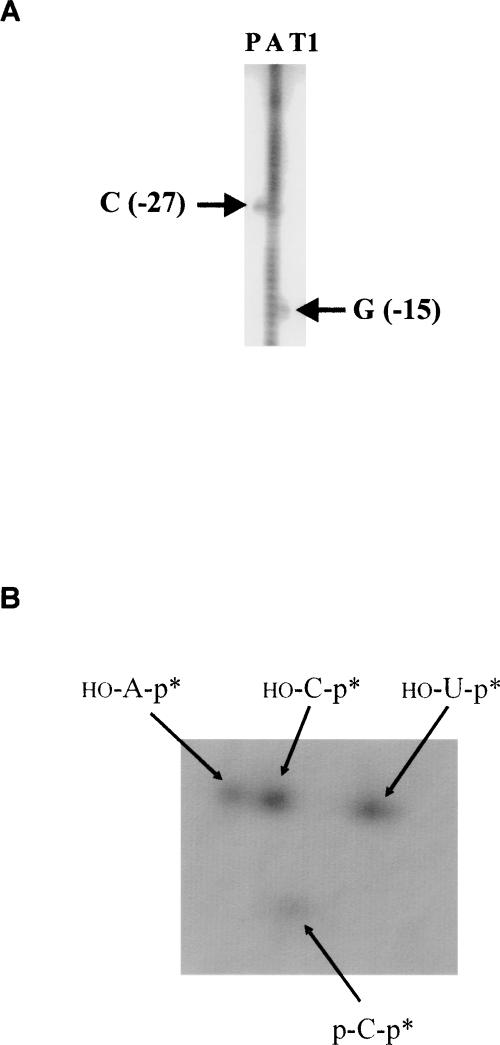
The sites of the cleavages were determined by primer extension for the cleavage that produces the larger product (band I in Fig. 1A), and by comparison against size ladders for the cleavage that produces the smaller product as described in band II in Figure 1A. The cleavages occur at C (−138) and C (−27), respectively. To verify that the 3′ products carry 5′ phosphate ends, a hallmark feature of RNase P cleavage, the purified products were digested by RNase T2 and run on thin layer chromatography plates. The results confirm that the purified products carried a phosphate at their 5′ end, and that the cleavage occurs upstream of C in both cases (Figs. 2, 3). The cleavage at C (−138) is 20% less efficient than that at C (−27). Only the cleavage at C (−138) occurs when the assays are performed in the presence of the RNase P RNA subunit (P RNA) alone (Fig. 1).
FIGURE 2.
Mapping of the cleavage at C (−138) of pbuE adenine riboswitch in vitro. (A) The larger product of the RNase P cleavage was purified from gel and mapped by primer extension (p. ext. in the figure) as described in Materials and Methods. (B) Autoradiogram of two-dimensional, thin-layer chromatography of the products of digestion by RNase T2 of the [32P]ATP internally labeled 3′ terminal fragment generated by the cleavage of B. subtilis RNase P holoenzyme (see Materials and Methods). Asterisks indicate a radioactive phosphate.
FIGURE 3.
Mapping of the cleavage at C (−27) of pbuE adenine riboswitch in vitro. (A) 3′ end terminal product labeled at the 3′ end (lane P) was loaded next to an alkali ladder (lane A) and a partial T1 digest of the riboswitch (lane T1) on an 8% PAGE gel. Based on the position of G (−15) and the alkali ladder, the cleavage was estimated to occur at C (−27). (B) Autoradiogram of two-dimensional, thin-layer chromatography of the products of digestion by RNase T2 of the [32P]CTP internally labeled 3′ terminal fragment generated by the cleavage at C (−27) (see Materials and Methods). The OH-Gp products are not visible on the autoradiogram because none of the Gs are preceded by a C and, as a result, the Gs are not radiolabeled. Asterisks indicate a radioactive phosphate.
Efficiency of the cleavage at C (−138) depends on the length of the substrate
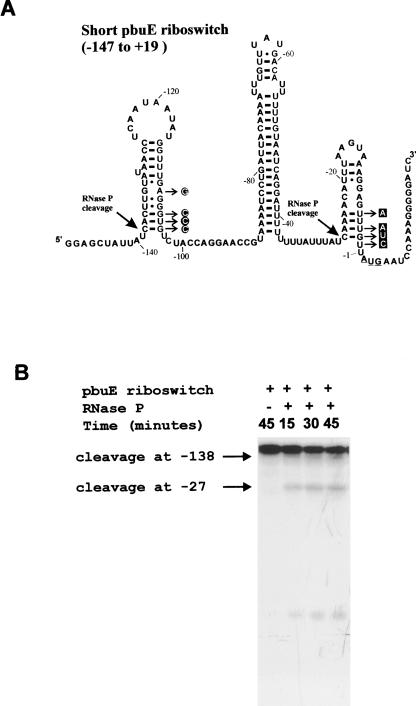
To identify the essential elements for these cleavage sites, a new construct was designed by PCR. This latter construct starts at the position −147, 16 nt shorter than the previous construct on the 5′ extremity, and ends at position +19, 12 nt longer on the 3′ end (Fig. 4A). RNase P assays on this shorter substrate (−147 to +19) showed that cleavages occur at the same positions, C (−138) and C (−27), indicating that no essential elements are present within the modified regions (Fig. 4B). However, the efficiency of the cleavage at position (−138) decreased twofold approximately compared to the one observed with the longer substrate (−165 to +4) and became a minor product of the reaction when less RNase P holoenzyme was used (Fig. 5B).
FIGURE 4.
Secondary structure model and RNase P assays of the short pbuE riboswitch (−147 to +19). (A) Secondary structure model of the short pbuE riboswitch, which starts at position (−147). The point mutations of the C (−138) cleavage mutant are encircled and indicated by arrows, whereas the point mutations at the C (−27) cleavage site are boxed. The translation start site AUG is underlined. (B) Internally labeled short adenine riboswitch, prepared as described in Materials and Methods, was incubated at 37°C in BP buffer (0.2 M NH4Cl, 50 mM MgCl2, 50 mM Tris-HCl pH 8) for 15, 30, and 45 min, in the presence of 0.5 nM of RNase P holoenzyme. The first lane served as a negative control and did not contain any RNase P. The products on the gel were quantified using a FujiX phosphorimager and ImageGauge software. The ratios of the counts recorded for both C (−27) and C (−138) cleavage products over the counts of the initial substrate were calculated, taking into consideration the number of radiolabeled Gs in each molecule. The ratio of products from cleavage at C (−138) is half that of the cleavage at C (−27).
FIGURE 5.
Effect of point mutations on RNase P cleavage. (A) Assays performed on the construct with point mutations at positions −104, −105, −106, and −109 shown in Figure 4. The short form of adenine riboswitch was used as a positive control. Internally labeled substrates were prepared as described in Materials and Methods, incubated at 37°C in BP buffer (0.2 M NH4Cl, 50 mM MgCl2, 50 mM Tris-HCl pH 8) for 60 min, in the presence and absence of 0.5 nM of B. subtilis RNase P holoenzyme, and the products were run on a 7% denaturing polyacrylamide gel. The C (−138) cleavage product is absent in the mutant substrate lane. (B) Assays performed on the construct with point mutations at positions −2, −3, −4, and −6 as shown in Figure 4. The assays were performed as in panel A, with two modifications. The incubation time was reduced to 30 min and only 0.25 nM of B. subtilis RNase P holoenzyme were used. The products on the gel were quantified using a FujiX phosphorimager and ImageGauge software. The ratios of the counts measured for the C (−27) cleavage products over the counts of the initial substrate were calculated for both the wild-type and C (−27) mutant pbuE adenine riboswitch. A drastic reduction in the mutated riboswitch (>80%) was observed compared to the wild-type riboswitch. Arrows in both parts of this figure indicate the site of cleavage products (or their absence).
Transient structures in the 5′ UTR of pbuE are recognized by RNase P
RNase P cleavage sites do not coincide with any predicted single-stranded-to-helix junction in the published secondary structure model of the adenine riboswitch (Mandal and Breaker 2004). To look for transient helical structures responsible for the cleavages, as was the case for the coenzyme B12 riboswitch (Altman et al. 2005), all possible helices longer than four base pairs that could form at the 3′ of the cleavage sites were drawn and their thermodynamic stability determined using mfold software (Zuker 2003). For the cleavage at C (−138), only one model was retained (see Fig. 1B). Our model does not match the one proposed previously by Mandal and Breaker at the “off-state,” which includes the terminator stem and a P2 helix (Mandal and Breaker 2004). The free energy of our putative conformation including the whole common sequence between the short and long riboswitch constructs (positions −147 to +3) is −37 kcal/mol. The free energy of the same sequence with the conformation formed by the P2 helix is −38 kcal/mol. Although our proposed conformation is not the most favored one energetically in the mfold program, the difference between this model and that of Mandal and Breaker (2004) is small, and a transient conformation could easily form in solution or during transcription. To validate our model, we introduced four site-specific mutations simultaneously that completely disrupted the putative helix at positions −104, −105, −106, and −109 as described in Figure 4A. These mutations inhibited the cleavage at C (−138) completely (Fig. 5A).
To verify the structures shown in Figures 1B and 4A, a construct was made that reversed the mutations in Figure 4A. Between positions −138 and −133, nucleotides were changed so the structure shown in Figure 4A now contained an intact double helix with four new G-C pairs. This structure was a good substrate for B. subtilis RNase P and was cleaved at least at 25% of the rate of the wild-type substrate in two experiments (data not shown).
The cleavage at C (−27) occurs in a region that had not been folded previously. Using the same approach as for the cleavage at C (−138), two putative helices were identified as possible candidates between positions −38 and +4. The free energy of the two putative conformations, including the whole common sequence between the short and long riboswitch constructs (−147 to +3), is identical (−34 kcal/mol). Two constructs with different point mutations that disrupt the putative helices were designed, and RNase P assays were performed on each of the mutants. The first construct was designed with two point mutations at positions +2 and +3 and did not show any significant reduction on cleavage by RNase P (not shown). The second construct with point mutations simultaneously at positions −2, −3, −4, and −6 caused a drastic decrease in RNase P cleavage (>80%) as shown in Figure 5B. The results of these assays suggest that the conformation shown in Figure 4A is mainly responsible for the cleavage at position C (−27).
Adenine has no effect on RNase P cleavage in vitro
To test whether the presence of adenine would affect the cleavage by RNase P, assays were performed in the presence of concentrations of adenine ranging between 20 nM and 10 μM. After 30 min at 37°C in the presence of 0.25 nM of RNase P holoenzyme, no significant effect on any of the cleavages was observed. This result is in agreement with those of Lemay et al. (2006): Full-length riboswitch does not bind adenine because the adenine-binding domain fails to fold due to conformational competition by the terminator stem.
RNase P cleavage in vivo increases the total observed enzymatic activity of a reporter protein
To validate that RNase P cleaves the adenine riboswitch in vivo, we transformed B. subtilis SSB318 with pDG1661, in which the 5′ UTR region of pbuE was fused to the lacZ reporter gene (Mandal and Breaker 2004). In SSB318, the original promoter of rnpB gene has been exchanged for Pspac promoter that is inducible by IPTG (Wegscheid et al. 2006). As a consequence, in the absence of IPTG the transcription of rnpB, which codes for P RNA, is considerably reduced. β-Galactosidase activity was assayed in the presence and the absence of IPTG. A genetic control, pDG1661, that contains the lacZ gene without the pbuE 5′ UTR sequence was used to compensate for background effects on the reporter protein enzymatic activity because of the decreased RNase P activity. The control strain was transformed and assayed in parallel with the experimental strain and the corresponding results were used to normalize our data. A 40% reduction of β-galactosidase activity occurred in the cells in which RNase P activity was reduced (Table 1).
TABLE 1.
Results of β-galactosidase assays in B. subtilis SSB318
DISCUSSION
pbuE adenine riboswitch is a substrate for RNase P in vitro
RNase P from B. subtilis cleaves in vitro the pbuE adenine riboswitch, but not the xpt-pbuX guanine riboswitch. The adenine riboswitch is cleaved at two different sites, C (−138) and C (−27). A relatively low efficiency of cleavage has been described for several substrates of RNase P in operon mRNAs in bacteria (Li and Altman 2004) in comparison with the cleavage of precursor tRNA. These low-activity cleavages could be important in regulatory mechanisms that create a polar effect, or they may be cleavages of misfolded RNAs during the transcription process. In fact, the physical folding of the adenine aptamer has been studied on molecules at different stages of transcription of the adenine riboswitch (Greenleaf et al. 2008). The structures studied are similar to the ones we show in Figure 1B but are not precisely identical. However, the results clearly indicate that partial transcription of the pbuE riboswitch shows that these molecules are physical realities that have some bearing on aptamer function. In fact, the appearance of transient structures for the adenine riboswitch substrate is apparent also from the mutagenesis studies. It is important to note that maintaining the intact double helix by reverse mutagenesis near the 5′ end of the adenine riboswitch restored the ability of this structure to act as a good substrate for RNase P.
The efficiency of cleavage of the wild-type adenine riboswitch at position C (−138) drops by half for the short substrate (−147 to +19) compared to the longer substrate (−165 to +6). Two hypotheses are plausible to explain this observation: (1) A longer 5′ sequence preceding the cleavage site may provide a better anchor for RNase P holoenzyme binding, or (2) the upstream sequence impacts the structure of the aptamer domain of the riboswitch and favors the conformation recognized by RNase P.
It has been shown previously that during evolution experiments in vitro that the RNase P protein subunit increases the versatility of the holoenzyme (Liu and Altman 1994) by facilitating recognition of nonprecursor tRNA sustrates. Obviously, studies of the expression of the proteins involved and detailed mechanisms of enzyme actions in vivo on these substrates should be accomplished before rigorous explanations of physiological function are given.
Since RNase P RNA cleaves only at C (−138), it is not apparent what specific role this has in function in vivo where the protein subunit is also necessary for activity of the enzyme.
RNase P cleavage of pbuE mRNA in vivo
RNase P cleavage induces a 40% increase in the expression of the lacZ reporter gene in vivo. These results are in agreement with microarray analysis performed on RNA extracted from SSB318 B. subtilis strains grown in the presence and the absence of IPTG. The pbuE mRNA is 30% higher in the presence of normal RNase P activity compared to that under decreased RNase P activity (R. Hartmann and C. Condon, unpubl.). No difference was seen for the xpt mRNA. Previously, RNase P cleavage has been reported to stabilize the his operon from Salmonella typhimurium (Alifano et al. 1996). Our observations suggest that RNase P cleavage might perform a similar function for pbuE mRNA, but the nature of physiological function must still be explored, as stated above in the Discussion.
MATERIALS AND METHODS
Strains and genomic DNA
B. subtilis 1A40 was kindly provided by Dr. Ronald R. Breaker (Yale University). To extract genomic DNA, cells were grown in 1 mL LB media at 37°C overnight. After 1 min centrifugation at 14,000 rpm, the pellet was dissolved in TE buffer (10 mM tris-HCl, pH 7.5, 1 mM EDTA) and treated with lysozyme, followed by phenol/chloroform extraction and ethanol precipitation. B. subtilis SSB318 (generously supplied by Dr. Roland Hartmann, University of Marburg, Germany) was grown in LB media supplemented with lincomycin (12.5 μg/mL), erythromycin (0.5 μg/mL), and IPTG (1 mM) (Wegscheid et al. 2006).
Substrates
Oligonucleotides were synthesized by the Keck Facility at Yale University. Radiochemicals were bought from GE Healthcare. Precursor tRNATyr (pTyr) was used as a control in RNase P assays (Guerrier-Takada et al. 1989).
The guanine riboswitch was amplified by PCR from B. subtilis genomic DNA using the following oligos: 5′-TAATACGACTCACTATAGGGAATTTTTATTTTCAGCCTATG-3′ and 5′-GGATCCTTCTTCCTCTATTTTCCGTTT-3′.
The full-length adenine riboswitch and short constructs comprised positions (−165 to +6) and (−147 to +9), respectively, relative to the translational start site of pbuE, and were amplified by PCR from B. subtilis genomic DNA using the following oligos: 5′-TAATACGACTCACTATAGGAAGCGAATCAATTAAATAGC-3′ and 5′-CTGGGGATCCATAACAAACTCC-3′ for the full-length adenine riboswitch and 5′-GGATCCCCCTTTGGATTCATAAC-3′ and 5′-TAATACGACTCACTATAGGAGCTATTATCACTGTATAACCTC-3′ for the short adenine riboswitch construct. The oligomers were designed to introduce BamHI sites at the 3′ end of the products.
Internally labeled substrates (32P-a-GTP) were transcribed in vitro from plasmids linearized with BamHI using T7 RNA polymerase (Promega) at 37°C for 18 h, in the buffer provided by the manufacturer. Reactions were stopped by adding 20 mL of 10 M urea/10% phenol dye and then purified on a preparative 7% polyacrylamide denaturing gel.
5′-end-labeled substrates were prepared by transcription from corresponding plasmids linearized with BamHI as described by Seif et al. (2006). The product was dephosphorylated using alkaline phosphatase (Roche) in a buffer provided by the manufacturer, extracted by phenol/chloform and ethanol precipitated. Dephosphorylated substrate (10 pmol) was labeled by 32P-γ-ATP using T4 polynucleotide kinase (New England Biolabs) at 37°C in the buffer provided by the manufacturer. Reactions were stopped by adding 20 μL of 10 M urea/10% phenol dye and then purified on a preparative 7% polyacrylamide denaturing gel.
Cloning and mutagenesis
Cloning of the PCR products in pUC19 and mutagenesis were done as described by Seif et al. (2006) using the following oligos: For the mutants of cleavage at the helix that contains C (−27) (Fig. 3), 5′-ATATCTATGAATCCAAAGGGGGATCC-3′, 5′-CTCCTTTACTTAAATGTTTTGA-3′. For the mutants of cleavage at the helix that contains C (−138) (Fig. 4), 5′-GGCCCTCTACCAGGAACCGTAAAATC-3′, 5′-CCAAACCATATTATTGAGGTTATAC-3′. The reverse mutants that restored the helix were made using the same oligonucleotides with the nucleotides changed to make the four new G-C pairs.
RNase P assays
Reactions were carried out in 10 μL at 37°C in 1× BP buffer (0.2 M NH4Cl, 50 mM MgCl2, 50 mM Tris pH 8) with RNasin (0.1 units/mL; Promega). The choice of salt concentration and pH were based on the analysis of ion conditions required by B. subtilis RNase P holoenzyme as described by Gardiner et al. (1985). RNase P holoenzyme (0.5 nM) of B. subtilis was used for all assays on riboswitch substrates unless otherwise indicated and 0.1 nM for tRNA precursor. RNase P reaction assays were stopped by the addition of 10 μL of 10 M urea/10% phenol dye and then analyzed on a 7% PAGE denaturing gel.
Mapping of the cleavage sites
Primer extension was performed using SuperScript II reverse transcriptase (Invitrogen) as proposed by the manufacturer using the 5′-end radiolabeled oligonucleotide: 5′-GGATCCTTCTTCCTCTATTTTCCGTTT-3′. The products were precipitated in 95% ethanol, and the pellet was dissolved in 10 mL of RNase free water and 10 mL of formamide-loading buffer. The same oligonucleotide was phosphorylated and used for the sequencing reaction, which served as a ladder.
The 5′-end-labeled substrate was cleaved by RNase P, and the products were extracted from a denaturing preparative 7% polyacrylamide gel. The products were loaded next to RNA ladders as described in the text. The alkali ladder was prepared by incubating five tubes containing 5′-end-labeled substrate at 95°C in 50 mM Na2CO3 (pH 9.2) and 1 mM EDTA. At each minute, one tube was removed and its content was ethanol precipitated. Finally, the contents of the tubes were combined dissolved in 10 mL of 10 M urea/10% phenol dye solution (0.125 mg/mL bromophenol blue, 0.125 mg/mL xylene cyanol) and loaded on a 7% polyacrylamide sequencing gel. To prepare a ladder of substrate partially digested with RNase T1 (Calbiochem), the 5′-end-labeled substrate was incubated on ice in 1× PA buffer (50 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 100 mM NH4Cl), then 0.05 units of RNase T1 were added, and the reaction was rapidly stopped by phenol after 15 sec. The products were ethanol precipitated, dissolved in 10 mL of 10 M urea/10% phenol dye solution and loaded on 7% polyacrylamide sequencing gel. For the complete digestion by RNase T2 (Sigma-Aldrich), the substrates were internally labeled and cleaved by RNase P, and the products were extracted from a preparative gel. The purified products were completely digested by RNase T2, and the mix was separated by two-dimensional thin-layer chromatography as described by Guerrier-Takada et al. (1988).
Assays for β-galactosidase activity
The pDG1661 construct that contained pbuE-LacZ was kindly provided by Dr. Ronald Breaker (Yale University) (Mandal and Breaker 2004). The construct was sequenced to confirm its integrity, and then transformed into B. subtilis SSB318, and then β-galactosidase activity was assayed as described by Mandal et al. (2003). The results were retained only for the assays in which the ratio of absorbance at 600 nm for noninduced over IPTG induced cells was lower then 0.6.
SUPPLEMENTAL DATA
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Dr. Roland Hartmann (University of Marburg, Germany) for B. subtilis SSB318 and the plasmid encoding B. subtilis RNase P protein. We thank Dr. Ciaran Condon (CNRS, Institut de Biologie Physico-Chimique, Paris, France) and Dr. Roland Hartmann (University of Marburg, Germany) for sharing the microarray data of B. subtilis SSB318 strain. We also thank Dr. Ronald R. Breaker (Yale University) for the generous gift of B. subtilis 1A40 and plasmids containing the adenine riboswitch. We further acknowledge Dr. Cecilia Guerrier-Takada, Donna Wesolowski, and all S.A. laboratory members for helpful discussions. Le Fonds Québécois de Recherche sur la Nature et les Technologies and Yale University supported E.S.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.833408.
REFERENCES
- Alifano P., Fani R., Lio P., Lazcano A., Bazzicalupo M., Carlomagno M.S., Bruni C.B., Rivellini F., Piscitelli C., Arraiano C.M., et al. Histidine biosynthetic pathway and genes: Structure, regulation, and evolution. Microbiol. Rev. 1996;60:44–69. doi: 10.1128/mr.60.1.44-69.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman S. A View of RNase P. Mol. Biosyst. 2007;3:1–5. doi: 10.1039/b707850c. [DOI] [PubMed] [Google Scholar]
- Altman S., Wesolowski D., Guerrier-Takada C., Li Y. RNase P cleaves transient structures in some riboswitches. Proc. Natl. Acad. Sci. 2005;102:11284–11289. doi: 10.1073/pnas.0505271102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothwell A.L., Garber R.L., Altman S. Nucleotide sequence and in vitro processing of a precursor molecule to Escherichia coli 4.5 S RNA. J. Biol. Chem. 1976;251:7709–7716. [PubMed] [Google Scholar]
- Christiansen L.C., Schou S., Nygaard P., Saxild H.H. Xanthine metabolism in Bacillus subtilis: Characterization of the xpt-pbuX operon and evidence for purine- and nitrogen-controlled expression of genes involved in xanthine salvage and catabolism. J. Bacteriol. 1997;179:2540–2550. doi: 10.1128/jb.179.8.2540-2550.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forti F., Sabbattini P., Sironi G., Zangrossi S., Deho G., Ghisotti D. Immunity determinant of phage-plasmid P4 is a short processed RNA. J. Mol. Biol. 1995;249:869–878. doi: 10.1006/jmbi.1995.0344. [DOI] [PubMed] [Google Scholar]
- Gardiner K.J., Marsh T.L., Pace N.R. Ion dependence of the Bacillus subtilis RNase P reaction. J. Biol. Chem. 1985;260:5415–5419. [PubMed] [Google Scholar]
- Greenleaf W.J., Frieda K.L., Foster D.A.N., Woodside M.T., Block S.M. Direct observation of hierarchical folding in single riboswitch aptamers. Science. 2008;319:630–633. doi: 10.1126/science.1151298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrier-Takada C., Altman S. Catalytic activity of an RNA molecule prepared by transcription in vitro. Science. 1984;223:285–286. doi: 10.1126/science.6199841. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C., van Belkum A., Pleij C.W., Altman S. Novel reactions of RNase P with a tRNA-like structure in turnip yellow mosaic virus RNA. Cell. 1988;53:267–272. doi: 10.1016/0092-8674(88)90388-1. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C., Lumelsky N., Altman S. Specific interactions in RNA enzyme-substrate complexes. Science. 1989;246:1578–1584. doi: 10.1126/science.2480641. [DOI] [PubMed] [Google Scholar]
- Johansen L.E., Nygaard P., Lassen C., Agerso Y., Saxild H.H. Definition of a second Bacillus subtilis pur regulon comprising the pur and xpt-pbuX operons plus pbuG, nupG (yxjA), and pbuE (ydhL) J. Bacteriol. 2003;185:5200–5209. doi: 10.1128/JB.185.17.5200-5209.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komine Y., Kitabatake M., Yokogawa T., Nishikawa K., Inokuchi H. A tRNA-like structure is present in 10Sa RNA, a small stable RNA from Escherichia coli . Proc. Natl. Acad. Sci. 1994;91:9223–9227. doi: 10.1073/pnas.91.20.9223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemay J.F., Penedo J.C., Tremblay R., Lilley D.M., Lafontaine D.A. Folding of the adenine riboswitch. Chem. Biol. 2006;13:857–868. doi: 10.1016/j.chembiol.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Li Y., Altman S. A specific endoribonuclease, RNase P, affects gene expression in polycistronic mRNAs. Proc. Natl. Acad. Sci. 2003;100:13213–13218. doi: 10.1073/pnas.2235589100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Altman S. Polarity effects in the lactose operon of Escherichia coli . J. Mol. Biol. 2004;339:31–39. doi: 10.1016/j.jmb.2004.03.041. [DOI] [PubMed] [Google Scholar]
- Liu F., Altman S. Differential evolution of substrates for an RNA enzyme in the presence and absence of its protein cofactor. Cell. 1994;77:1093–1100. doi: 10.1016/0092-8674(94)90448-0. [DOI] [PubMed] [Google Scholar]
- Mandal M., Breaker R.R. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nat. Struct. Mol. Biol. 2004;11:29–35. doi: 10.1038/nsmb710. [DOI] [PubMed] [Google Scholar]
- Mandal M., Boese B., Barrick J.E., Winkler W.C., Breaker R.R. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell. 2003;113:577–586. doi: 10.1016/s0092-8674(03)00391-x. [DOI] [PubMed] [Google Scholar]
- Seif E., Cadieux A., Lang B.F. Hybrid E. coli–mitochondrial ribonuclease P RNAs are catalytically active. RNA. 2006;12:1661–1670. doi: 10.1261/rna.52106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegscheid B., Condon C., Hartmann R.K. Type A and B RNase P RNAs are interchangeable in vivo despite substantial biophysical differences. EMBO Rep. 2006;7:411–417. doi: 10.1038/sj.embor.7400641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. J. Mol. Biol. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]