Abstract
Of the many types of biomolecules used for molecular imprinting applications, proteins are some of the most useful, yet challenging, templates to work with. One method, termed the ‘epitope approach’, involves imprinting a short peptide fragment of the protein into the polymer to promote specific adsorption of the entire protein, similar to the way an antigen binds to an antibody via the epitope. Whole lysozyme or the 16 residue lysozyme C peptide was imprinted into porous silica scaffolds using sol-gel processing. After removing template, scaffolds were exposed to lysozyme and/or RNase A, which was used as a competitor molecule of comparable size. When comparing protein- to peptide-imprinted scaffolds, similar amounts of lysozyme and RNase were bound from single protein solutions. However, while whole lysozyme-imprinted scaffolds showed about 4:1 preferential binding of lysozyme to RNase, peptide-imprinted scaffolds failed to show statistical significance, even though a slight preferential binding trend was present. These initial studies suggest there is potential for using peptide-imprinting to create specific protein-binding sites on porous inorganic surfaces, although further development of the materials is needed.
Keywords: molecular imprinting, porous silica, protein binding
Introduction
Molecularly imprinted polymers (MIPs) are used for molecular recognition because preparation is usually straight forward, they are robust, and they are chemically, mechanically, and thermally stable [1]. In general, MIPs are made with three main reaction components: cross-linking agent, functional monomer, and template molecule [2]. The template molecule is mixed with the functional monomer so that chemical bonds between the two are formed. The cross-linking agent binds to the functional monomers upon polymerization, which embeds the template molecule in a polymer matrix. Because the functional monomers bind to the template in a specific position within the polymer, removal of the molecule in will result in an imprint with preservation of both spatial and chemical features (Figure 1) [2–6]. The resulting structure of the polymer at these locations is a mirror of the template molecule that is able to preferentially rebind that molecule over competitor molecules, which do not share the same physical and chemical structure [1,2,7].
Figure 1.
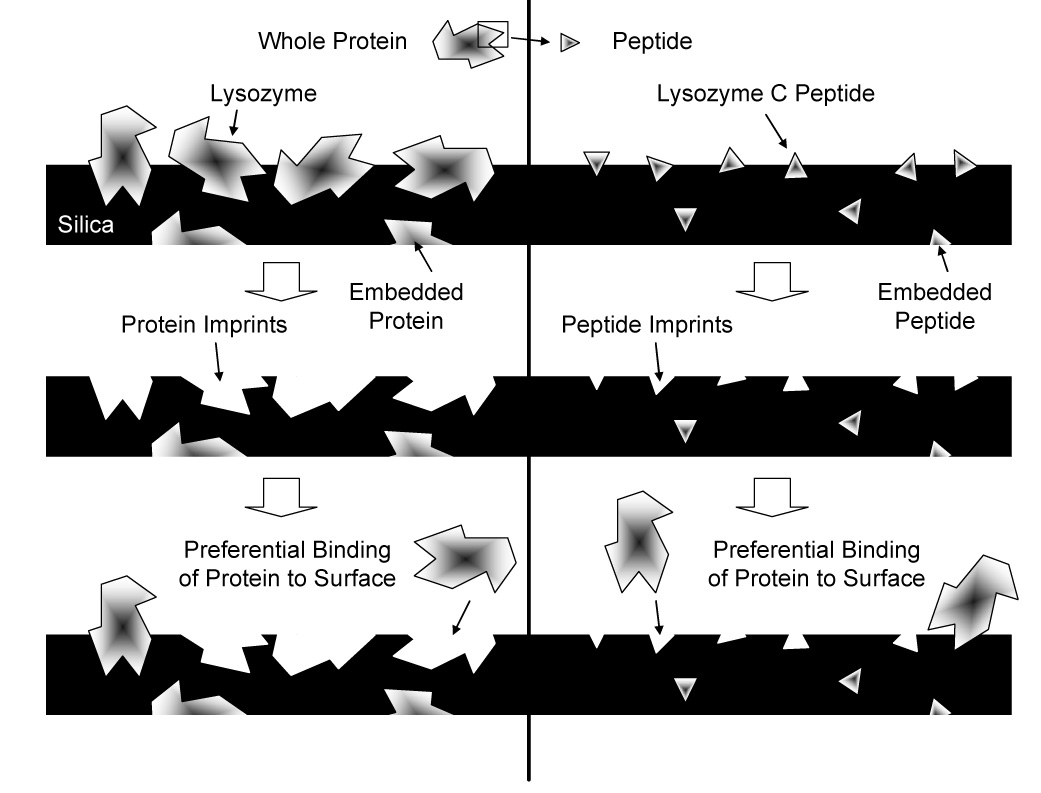
Cartoon representation of whole protein- and peptide-imprinting. On the left, the entire protein molecule creates large imprints with high surface availability. On the right, a small polypeptide sequence from the protein creates small imprints that bind the template molecule at only a specific location.
For many MIPs, small biomolecules, such as sugars, cholesterols, and certain drugs, have been used as templates because of their simplicity and stability. Recently however, proteins have become the focal point of many MIP studies for uses in medical diagnostics, drug delivery, biomaterial compatibility, and tissue engineering [5]. The chemical structure of proteins, with their numerous functional groups, allows many binding opportunities for a variety of functional monomers. While imprinting MIPs with protein is becoming a popular concept, many challenges, including the accessibility of binding sites, nonspecific binding, and flexible conformation of the molecules, remain in creating materials that selectively bind the desired molecules [2]. A newer method of molecular imprinting, called the epitope approach, is being explored [5]. The epitope approach is so named because of its similarity in concept to antigen-antibody interactions. When an antibody of the immune system binds a specific antigen, it does not need to recognize the entire molecule, but rather only a small portion of it, called the epitope. In an MIP, a peptide sequence from the surface of the protein is synthesized and imprinted into the material. When the original whole protein is exposed to the imprinted material, the region containing the peptide used as template can recognize its spatial and chemical mimic in the polymer (Figure 1) [2]. In this manner, only a portion of the entire molecule is needed for preferential binding.
The vast majority of molecular imprinting studies have used organic polymers, such as methacrylates [8–12]. Inorganic materials, however, offer advantages. For example, amorphous polysiloxane (silica) formed by sol-gel processing can provide high selectivity, high level of accessible binding sites, and faster mass transfer and binding [13].
The objective of this study was to determine if imprinting porous silica scaffolds with a 16 residue peptide (lysozyme C, 1.8 kDa) could lead to preferential binding of the whole protein (lysozyme, 14 kDa) as well as materials imprinted with the entire molecule.
Methods and Materials
Silica Scaffold Fabrication
Fabrication of the silica scaffolds began with a two step sol-gel process [14]. Tetraethoxysilane (TEOS; Fluka, Milwaukee, WI) as a cross-linking agent, 0.1M hydrochloric acid (HCl) as a catalyst, absolute ethanol as a co-solvent, and distilled water as another other co-solvent were mixed with γ-aminopropyltriethoxysilane (APS; Sigma, St. Louis, MO) as the functional monomer and sodium dodecyl sulfate (SDS) as a foaming agent. The model biomolecule lysozyme protein (chicken egg white; Sigma) or lysozyme C peptide (residues 46–61; Bachem, Torrance, CA) was added to the APS solution prior to its addition to pre-hydrolyzed TEOS. For protein imprinting, 0.1 mg was added (0.054 mg per mL of sol) to the scaffolds, and for peptide imprinting, 0.05 mg was added (0.027 mg per mL of sol). Vortex mixing the sol created bubbles that were retained in the gel and therefore gave the scaffolds a macroporous structure. As reported previously, scaffolds were about 40% porous with an average pore size of about 6 µm [14]. Blank scaffolds were made without the addition of protein. Tubes containing the samples were screw-capped and aged at 40°C for 24 hours, and then they were uncapped and dried at 40°C for 48 hours. Finally, the non-uniform top and bottom surfaces were ground off to form cylindrical silica scaffolds with dimensions of 4mm in height and 9mm in diameter.
Characterizing Scaffold Maximum Binding Capacity
In order to determine the amounts present on the surface of the scaffolds, lysozyme protein and peptide for imprinting were labeled with Alexa Fluor 350 (Molecular Probes; Eugene, OR; λex: 346nm, λem: 442nm) following the manufacturer’s instructions. The BCA Protein Assay (Pierce, Rockford, IL) with standard protein bovine serum albumin (BSA) was used to determine the concentration of protein, which was then correlated with fluorescence using a Gemini XS plate reader (Molecular Devices, Sunnyvale, CA).
Silica scaffolds loaded with labeled protein were then characterized to determine the amount of surface-accessible protein/peptide and thus how much protein could potentially rebind. Imprinted scaffolds were treated with 1 mL of 0.4 mg/mL protease (pronase E; Sigma) in 0.1 M carbonate-bicarbonate buffer, pH 8.5, for 3–24 hours to digest the protein/peptide. Measurement of fluorescence reflected the amount of protein available on the surface, which corresponded to the number of potential binding sites.
Preferential Binding
To test preferential binding of the template (lysozyme) to the scaffolds, a competitor protein having similar size but differing in chemistry was used. In these studies, ribonuclease A (RNase; MW 13.7kDa) was used to compete during rebinding of lysozyme. To distinguish between the proteins, lysozyme was labeled with Alexa 488 (Molecular Probes; λex: 495nm, λem: 519nm) and RNase with Alexa 594 (Molecular Probes; λex: 590nm, λem: 617nm). Imprinted and blank scaffolds were exposed to solutions of three protein ratios: 1) lysozyme only (1:0), 2) equal amounts of lysozyme and competitor (1:1), and 3) RNase only (0:1). Solution concentrations were based on the amounts determined as described in the previous section. For example, if 10 µg of protein/peptide was initially digested, scaffolds were exposed to 10 µg total protein in the rebinding solutions. After allowing protein to bind for 24 hr to reach steady state, scaffolds were treated with protease, and the fluorescence was measured.
Statistical Analysis
All results are plotted as mean ± standard deviation (SD). Two-way analysis of variance (ANOVA) with Bonferroni post-tests was run using the computer program Graphpad Prism v4.03 to compare effects of the type of protein and protein ratio. Student’s t-tests were used to compare results for the protein- and peptide-imprinted scaffolds. Sample size calculations were run using the program Power and Precision 2.1 to determine the number of scaffolds required for statistical power of 80%.
Results
Maximum Binding Capacity
Figure 2 shows the maximum binding capacity for both the lysozyme-imprinted and lysozyme C peptide-imprinted scaffolds. In preliminary experiments, scaffolds were digested for up to 96 hours to release surface-accessible protein/peptide, but amounts reached a plateau after about 24 hours. Nearly 68 µg of protein were released from the protein-imprinted scaffolds, while about 21 µg were released from the peptide-imprinted materials. Approximately 68% of template protein and about 41% of template peptide loaded into the sol was surface-accessible. The mass of peptide was useful for determining the number of molecules present and hence the number of potential binding sites available.
Figure 2.
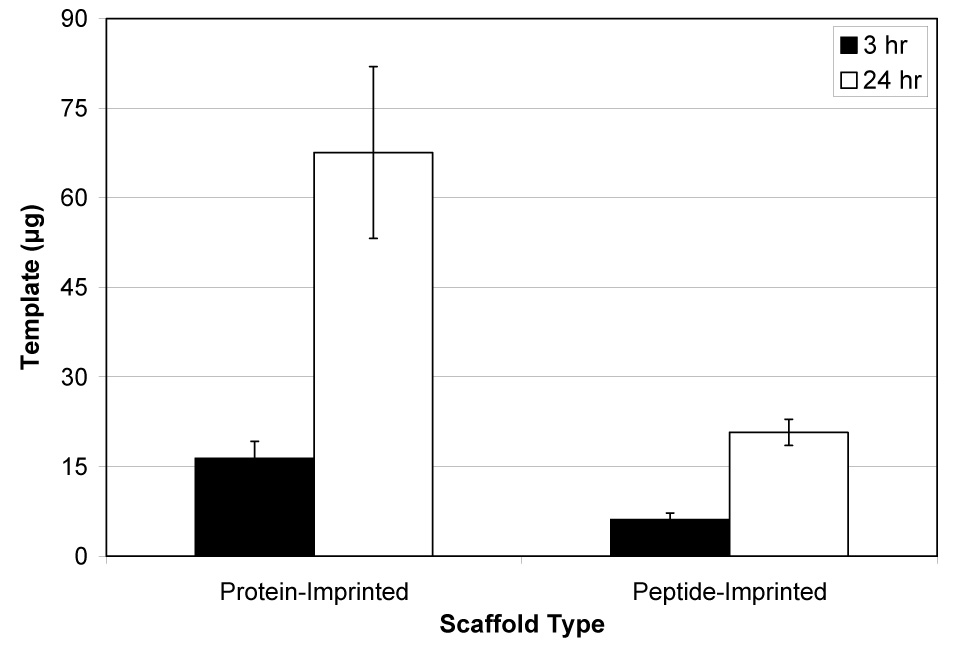
Amount of protein released from lysozyme protein-imprinted and lysozyme C peptide-imprinted silica scaffolds via digestion with protease (pronase E) for 3 or 24 hr (mean ± SD). More template was released from protein-imprinted scaffolds (p<0.001).
Preferential Binding
Figure 3 shows the difference between the amounts of proteins bound to the lysozyme-imprinted and blank scaffolds. Although binding of both proteins was detected, scaffolds bound over five times more lysozyme than RNase under noncompetitive conditions (ratios of 1:0, lysozyme only, and 0:1, RNase only) (p<0.001). When equal numbers of template and competitor protein molecules (1:1 ratio) were added, four times more lysozyme than RNase preferentially bound to the silica (p<0.01). Table 1 summarizes the amounts and percentages of lysozyme and RNase that bound to the imprinted scaffolds. Up to 8% of the available amount of lysozyme bound without competition, and about 5% bound in the presence on an equal amount of RNase. As little as 1.2–1.3% of the available RNase bound, regardless of competition.
Figure 3.
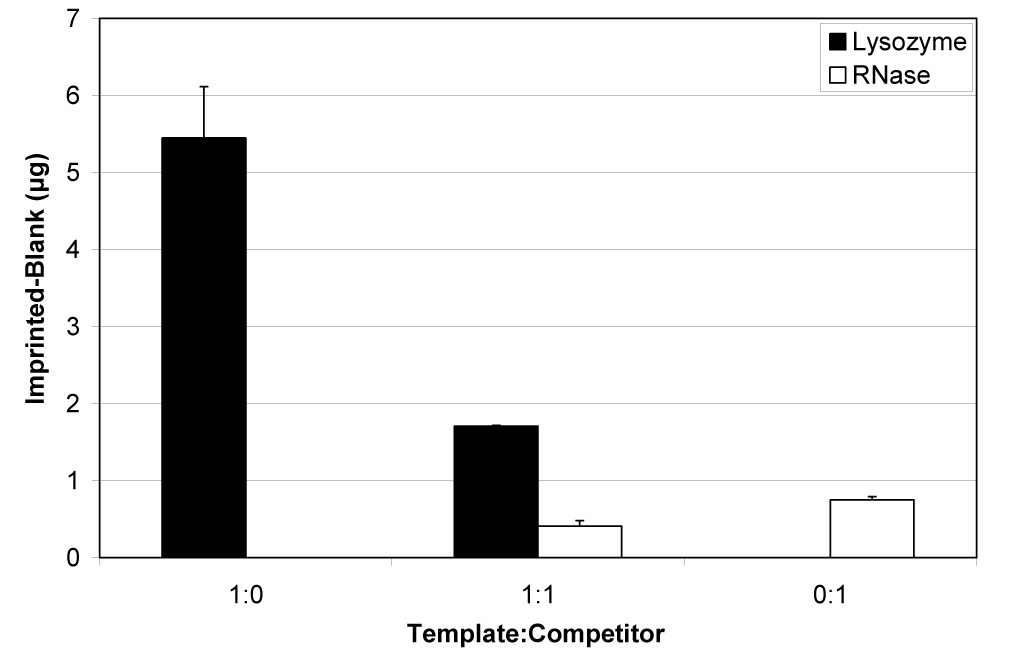
Differential binding of lysozyme and RNase A to protein-imprinted silica scaffolds under noncompetitive and competitive conditions (mean ± SD). Non-specific binding to blank/non-imprinted scaffolds was subtracted from the amounts bound to scaffolds imprinted with protein to determine preferential binding. More lysozyme than RNase bound to scaffolds from both noncompetitive (p<0.001) and competitive (p<0.01) solutions.
Table 1.
Summary of protein binding to scaffolds imprinted with either lysozyme protein or lysozyme C peptide.
| Lysozyme | RNase A | |||||
|---|---|---|---|---|---|---|
| Ratio | 1:0 | 1:1 | 0:1 | 1:0 | 1:1 | 0:1 |
| Protein-imprinted Scaffolds | ||||||
| Difference between Imprinted and Blank Scaffolds) | ||||||
| Amount Rebound (µg) | 5.45 ± 0.66 | 1.71 ± 0.01 | 0 | 0 | 0.41 ± 0.07 | 0.75 ± 0.04 |
| Percent Rebound (%) | 8.13 ± 0.99 | 5.10 ± 0.03 | 0 | 0 | 1.32 ± 0.23 | 1.21 ± 0.07 |
| Peptide-Imprinted Scaffolds | ||||||
| Difference between Imprinted and Blank Scaffolds) | ||||||
| Amount Rebound (µg) | 5.15 ± 6.88 | 0.55 ± 2.95 | 0 | 0 | 0.45 ± 0.79 | 0.67 ± 1.98 |
| Percent Rebound (%) | 8.59 ± 11.54 | 1.85 ± 9.47 | 0 | 0 | 0.83 ± 1.39 | 2.31 ± 6.87 |
Figure 4 shows the difference in amounts of lysozyme and RNase bound to peptide-imprinted silica scaffolds. As with protein-imprinted scaffolds, both lysozyme and RNase bound to the materials under noncompetitive conditions, although about five times more lysozyme was retained (p<0.05). During competition, however, comparable amounts of both proteins bound. Because of the large errors associated with the peptide-imprinted scaffolds, the competitive difference was not statistically significant. Table 1 summarizes the amounts and percentages of lysozyme and RNase that bound to the peptide-imprinted scaffolds. Whereas under noncompetitive conditions, protein- and peptide-imprinted materials behaved similarly, peptide-imprinted materials were less effective when competitor molecules were present, with less than 2% of available protein retained on the scaffolds.
Figure 4.
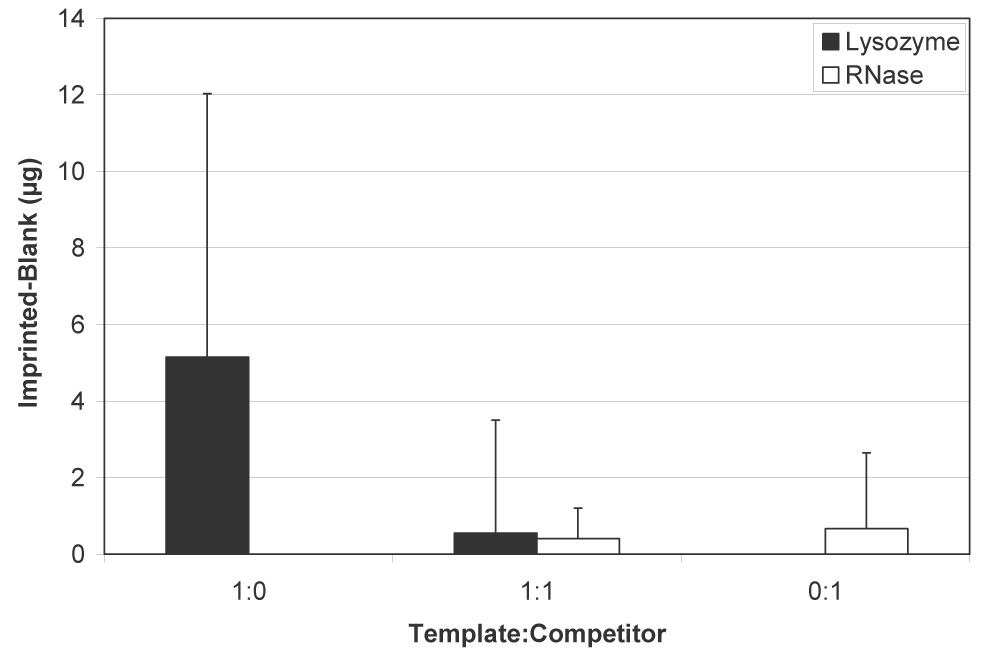
Differential binding of lysozyme and RNase A to peptide-imprinted silica scaffolds under noncompetitive and competitive conditions (mean ± SD). Non-specific binding to blank/non-imprinted scaffolds was subtracted from the amounts bound to scaffolds imprinted with peptide to determine preferential binding. Although more lysozyme than RNase bound to scaffolds from noncompetitive solutions (p<0.05), no difference was observed during competition.
Discussion
Due to their effectiveness in specific recognition of biomolecules, molecularly imprinted polymers are being developed for many chemical and medical applications, including solid phase extraction and chromatography, biosensors, and signaling polymers [15]. MIPs also have potential for use in tissue engineering, a discipline that brings together biology and materials science. For example, substrates that selectively bind particular proteins in a physiological environment facilitate desired cell and tissue responses, which are critically dependent on the type and nature of adsorbed biomolecules [16].
Because of its potential for greater stability, silica was used for the present studies [14]. During polycondensation to form the polysiloxane network, amino groups on the functional monomer APS are able to interact with the template (protein or peptide). TEOS molecules then cross-linked the monomers, locking in the position of the template in silica. The sol-gel processing used to fabricate scaffolds was simple, but to make macroporous (foamed) scaffolds eventually intended for cell ingrowth in tissue engineering applications, samples were made one at a time, which led to variability between scaffolds, as reflected in some of the results presented.
Several challenges have been identified concerning imprinting proteins in molecularly imprinted polymers, including poor accessibility of the binding sites, low binding capacity, and large non-specific binding [2], all of which were observed in the present study. In addition, some loss of protein/peptide was inevitable during fabrication of the scaffolds, such as by adsorption to tubes and grinding to make the shapes uniform. Improvements for fabricating effective scaffolds will lie with both the protein that is being imprinted and the material that constitutes the scaffold. The properties of a protein, both physical and chemical, make the fabrication process difficult. The conformation of a protein is quite flexible, and while an imprint of the protein is made in one position, it may be difficult for another molecule of the same type to perfectly fit that original shape [2]. In addition, a protein’s large molecular size, fragility, and complexity create difficulties in imprinting [17].
Maximum binding capacity experiments were useful in that they determined the amount of protein or peptide available at the surface of the scaffolds, and therefore they presented an upper limit to the amount of protein that could rebind. Due to the porous nature of the scaffolds, release of digested protein/peptide was logarithmic and reached a plateau close to 24 hours (data not shown), with molecules exposed on the outer portion of the scaffold released quickly, while those within the pores took longer to diffuse outward. This 24 hr time point was subsequently used to ensure complete digestion of bound protein/peptide.
Differences in behavior of protein- and peptide-imprinted scaffolds depend on the size of the molecules as well as their interactions with the surrounding matrix. Because of the smaller size of the peptide, which was only 12% of the molecular weight of lysozyme, more molecules could be expected to become embedded in the silica beyond the reach of the protease, so that less would be digested from the surface. In the present studies, even though a greater mass of protein was imprinted compared to peptide, the number of moles of peptide added to the sol was larger. The nearly four-fold greater molar content of peptide was used in an unsuccessful attempt to compensate for embedding. The larger, entire protein molecule, despite the smaller quantity of molecules, was more exposed and therefore was vulnerable to protease digestion.
Results from the lysozyme-imprinted scaffold experiments showed good preferential binding, which confirmed our previous findings [14]. The template protein bound significantly more to the silica scaffolds than did the competitor under both noncompetitive (1:0 and 0:1) and competitive (1:1) conditions. Experiments showed that successful preferential binding can be achieved by using whole proteins for imprints to rebind the template molecule. The peptide-imprinted scaffolds, however, did not show the same behavior. Whereas there was a significant difference in the binding of lysozyme to peptide-imprinted materials compared to RNase binding from noncompetitive solutions, the difference was not significant when equal amounts of lysozyme and RNase were present. Lack of a statistically significant effect for peptide-imprinted scaffolds can be largely attributed to great variability with these samples. Sample size calculations indicated that, whereas the experiments for protein-imprinted scaffolds were sufficiently powered, thousands of samples would be needed to detect a difference in binding of lysozyme and RNase to peptide-imprinted scaffolds with 80% power. This result strikingly underscores the differences between the two types of scaffolds. Considering that, with exception of the biomolecules, the two types of scaffolds were fabricated the same way, differences in behavior can be largely attributed to use of peptide as template.
Although Rachkov and Minoura reported some success using the “epitope approach” [5], their peptide template was approximately 44% of the desired molecule, whereas the peptide was only about 12% in the present study. Imprinting with the smaller peptide molecules greatly increased the likelihood of completely embedding them and thereby decreasing surface accessibility. This explains the lower percentage of peptide on the silica surfaces compared to those imprinted with whole lysozyme protein. Furthermore, using a small fragment of the desired protein substantially restricts the orientations in which the whole molecules can bind. As shown in Figure 1, for peptide-imprinted scaffolds, only those protein molecules with the “epitope” directed toward the surface can bind. Furthermore, only those imprint sites resulting from template molecules appropriately oriented in the condensing polysiloxane gel will be able to bind protein molecules. In contrast, imprinting with the whole protein will result in sites able to bind protein in a multitude of orientations of both the template and binding molecules.
Conclusion
Sol-gel methods were used to prepare porous silica scaffolds molecularly imprinted with protein and peptide. Protein-imprinted silica preferentially bound about five times more of the desired template molecule under noncompetitive conditions and four times more in the presence of competitor. For peptide-imprinted scaffolds, similar relative binding was observed without competition, but much less preference was seen in the presence of competitor molecules. While initial studies suggest there is potential for further development of the “epitope approach” to improve selectivity, current results show that imprinting with the entire protein molecule is most effective.
Acknowledgement
This work was supported by the National Institutes of Health (AR048700 and EB02958).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wei S, Jakusch M, Mizaikoff B. Capturing molecules with templated materials--Analysis and rational design of molecularly imprinted polymers. Anal Chim Acta. 2006;578:50–58. doi: 10.1016/j.aca.2006.06.077. [DOI] [PubMed] [Google Scholar]
- 2.Bossi A, Bonini F, Turner APF, Piletsky SA. Molecularly imprinted polymers for the recognition of proteins: The state of the art. Biosens Bioelectron. 2007;22:1131–1137. doi: 10.1016/j.bios.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 3.Sellergren B, Allender CJ. Molecularly imprinted polymers: A bridge to advanced drug delivery. Adv Drug Deliv Rev. 2005;57:1733–1741. doi: 10.1016/j.addr.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 4.Garcia-Calzon JA, Diaz-Garcia ME. Characterization of binding sites in molecularly imprinted polymers. Sensor Actuat B-Chem. 2007;123:1180–1194. [Google Scholar]
- 5.Rachkov A, Minoura N. Towards molecularly imprinted polymers selective to peptides and proteins. The epitope approach. Biochim Biophys Acta-Protein Struct M. 2001;1544:255–266. doi: 10.1016/s0167-4838(00)00226-0. [DOI] [PubMed] [Google Scholar]
- 6.Spivak DA. Optimization, evaluation, and characterization of molecularly imprinted polymers. Adv Drug Deliv Rev. 2005;57:1779–1794. doi: 10.1016/j.addr.2005.07.012. [DOI] [PubMed] [Google Scholar]
- 7.Nilsson J, Spegel P, Nilsson S. Molecularly imprinted polymer formats for capillary electrochromatography. J Chromatog B. 2004;804:3–12. doi: 10.1016/j.jchromb.2003.12.036. [DOI] [PubMed] [Google Scholar]
- 8.Ou SH, Wu MC, Chou TC, Liu CC. Polyacrylamide gels with electrostatic functional groups for the molecular imprinting of lysozyme. Anal Chim Acta. 2004;504:163–166. [Google Scholar]
- 9.Lv Y, Lin Z, Feng W, Zhou X, Tan T. Selective recognition and large enrichment of dimethoate from tea leaves by molecularly imprinted polymers. Biochem Eng J. 2007 [Google Scholar]
- 10.Oral E, Peppas NA. Dynamic studies of molecular imprinting polymerizations. Polymer. 2004;45:6163–6173. [Google Scholar]
- 11.Buchmeiser MR. New synthetic ways for the preparation of high-performance liquid chromatography supports. J Chromatog A. 2001;918:233–266. doi: 10.1016/s0021-9673(00)00129-1. [DOI] [PubMed] [Google Scholar]
- 12.Sellergren B. Noncovalent molecular imprinting: antibody-like molecular recognition in polymeric network materials. Trends Anal Chem. 1997;16:310–320. [Google Scholar]
- 13.Jiang X, Tian W, Zhao C, Zhang H, Liu M. A novel sol-gel-material prepared by a surface imprinting technique for the selective solid-phase extraction of bisphenol A. Talanta. 2007;72:119–125. doi: 10.1016/j.talanta.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 14.Lee K, Itharaju RR, Puleo DA. Protein-imprinted polysiloxane scaffolds. Acta Biomater. 2007;3:515–522. doi: 10.1016/j.actbio.2007.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Komiyama M, Takeuchi T, Mukawa T, Asanuma H. Molecular Imprinting: From Fundamentals to Applications. Weinheim: Wiley-VCH; 2003. [Google Scholar]
- 16.Sawyer AA, Hennessy KM, Bellis SL. The effect of adsorbed serum proteins, RGD and proteoglycan-binding peptides on the adhesion of mesenchymal stem cells to hydroxyapatite. Biomaterials. 2007;28:383–392. doi: 10.1016/j.biomaterials.2006.08.031. [DOI] [PubMed] [Google Scholar]
- 17.Odabasi M, Say R, Denizli A. Molecular imprinted particles for lysozyme purification. Mat Sci Eng C. 2007;27:90–99. [Google Scholar]


