Abstract
Many prokaryotes form the amide aminoacyl-tRNAs, glutaminyl-tRNA and asparaginyl-tRNA, by tRNA-dependent amidation of the mischarged tRNA species, glutamyl-tRNAGln or aspartyl-tRNAAsn. Archaea employ two such amidotransferases, GatCAB and GatDE, while bacteria possess only one, GatCAB. The Methanothermobacter thermautotrophicus GatDE is slightly more efficient using Asn as an amide donor than Gln (kcat/KM of 5.4 s−1/mM and 1.2 s−1/mM, respectively). Unlike the bacterial GatCAB enzymes studied to date, the M. thermautotrophicus GatCAB uses Asn almost as well as Gln as an amide donor kcat/KM of 5.7 s−1/mM and 16.7 s−1/mM, respectively). In contrast to the initial characterization of the M. thermautotrophicus GatCAB as being able to form Asn-tRNAAsn and Gln-tRNAGln, our data demonstrate that while the enzyme is able to transamidate Asp-tRNAAsn (kcat/KM of 125 s−1/mM) it is unable to transamidate M. thermautotrophicus Glu-tRNAGln. However, M. thermautotrophicus GatCAB is capable of transamidating Glu-tRNAGln from H. pylori or B. subtilis, and M. thermautotrophicus Glu-tRNAAsn. Thus, M. thermautotrophicus encodes two amidotransferases, each with its own activity, GatDE for Gln-tRNA and GatCAB for Asn-tRNA synthesis.
Keywords: tRNA-dependent amidotransferase, GatCAB, GatDE, Glu-AdT, Asp-AdT
Introduction
Most prokaryotes form Gln-tRNAGln or Asn-tRNAAsn by an indirect pathway that relies on conversion of a misacylated aminoacyl-tRNA to the cognate pair.1–3 This involves two enzymes, a non-discriminating aminoacyl-tRNA synthetase (aaRS) and a tRNA-dependent amidotransferase (AdT). All known archaea1 and most bacteria3 lack glutaminyl-tRNA synthetase (GlnRS). A non-discriminating glutamyl-tRNA synthetase (ND-GluRS) forms Glu-tRNAGln.4 Amidation of the tRNA-bound Glu by glutamyl-tRNAGln amidotransferase (Glu-AdT) leads to the correctly charged Gln-tRNAGln.5 Similarly, prokaryotes lacking an asparaginyl-tRNA synthetase (AsnRS) employ a ND-aspartyl- tRNA synthetase (ND-AspRS)6 and an aspartyl-tRNAAsn amidotransferase (Asp-AdT).7, 8 All AdT enzymes catalyze three different reactions during the conversion of their mischarged tRNA substrates.9 (i) AdTs are kinases that phosphorylate the β-carboxyl group of the Asp moiety or the γ-carboxyl of the Glu moiety still bound to the tRNA forming an activated intermediate.10 (ii) The enzymes hydrolyze an amide donor (Gln or Asn) to liberate ammonia. (iii) AdTs use the liberated ammonia to amidate the activated intermediate to form the cognate aa-tRNA species, Asn-tRNAAsn or Gln-tRNAGln.
Two distinct AdTs are found in nature, the heterotrimeric GatCAB11 and the heterodimeric GatDE.1 The latter acts only as a Glu-AdT and is present in all known archaea.1 GatCAB occurs in both Bacteria11 and Archaea.1 To date all GatCAB enzymes have shown the ability to act as both a Glu-AdT and an Asp-AdT in vitro.1, 12–14
Since the bacterial GatCAB is a dual specificity enzyme that can function as an Asp-AdT and/or a Glu-AdT, what determines its in vivo role? The task of GatCAB is defined by the genomic context of the organism in terms of the occurrence of a ND-GluRS and/or a ND-AspRS, the enzymes responsible for Glu-tRNAGln and/or Asp-tRNAAsn synthesis. In Pseudomonas aeruginosa, which has a ND-AspRS but lacks a ND-GluRS, GatCAB serves just as an Asp-AdT,15 while in Bacillus subtilis GatCAB is used only as a Glu-AdT11 as it lacks a ND-AspRS but has a ND-GluRS.4 In Helicobacter pylori, which has both ND-aaRSs,16–18 GatCAB has a dual function as both an Asp-AdT and as a Glu-AdT.3, 19 In Archaea, given the specificity of GatDE as a Glu-AdT1 and the fact that GatCAB is present only when AsnRS is absent,1, 2 the in vivo role of GatCAB is assumed to be primarily as an Asp-AdT.
How do the AdTs discriminate their mischarged tRNA substrates (Glu-tRNAGln and/or Asp-tRNAAsn) from the cognate aa-tRNA species (Glu-tRNAGlu and Asp-tRNAAsp)? The GatDE and bacterial GatCAB enzymes recognize the D-loop and the first base-pair of the acceptor stem to accomplish this.20–22 The archaeal GatCAB rejects Asp-tRNAAsp not based on recognition of the first base-pair of the acceptor stem, but it instead relies on anti-determinants in tRNAAsp (the D-loop, variable loop and position 49).21, 23
Bioinformatics first showed that the glutaminase subunits of the archaeal AdT enzymes are of different biochemical origins.1 In GatDE, the glutaminase subunit (GatD) is derived from a type I L-asparaginase,1, 24, 25 while the comparable GatA subunit of GatCAB belongs to the amidase enzyme family.3, 11, 26 L-asparaginases are known to hydrolyze both Asn and Gln27 although the preferred substrate varies among enzymes from different organisms. For example, the Erwinia carotovora L-asparaginase is more efficient hydrolyzing Asn than Gln,28 while the Pseudomonas 7A enzyme is more active in the hydrolysis of Gln than Asn.29
The crystal structure of the Staphylococcus aureus GatCAB complexed with Gln or Asn suggested Gln as a better donor in transamidation.20 In agreement with that prediction, the bacterial GatCAB enzymes have been shown to be more active using Gln as the donor than Asn.3, 30, 31 The H. pylori enzyme was approximately 130-fold more efficient using Gln than Asn, mostly due to a difference in kcat.3 However, this enzyme is nearly as a efficient in forming Asn-tRNAAsn as Gln-tRNAGln in keeping with its function in the cell as both a Glu-AdT and an Asp-AdT.3
Here we present our characterization of GatDE and GatCAB from the archaeon Methanothermobacter thermautotrophicus with respect to their amide donor and mischarged tRNA substrate preferences for transamidation.
Results
Characterization of the two M. thermautotrophicus AdTs
The initial characterization of the M. thermautotrophicus AdTs suggested that both enzymes could use Gln or Asn as amide donors but did not clarify if either enzyme was more efficient with one or the other donor.1 With the bacterial GatCAB enzymes studied to date, Gln is a better amide donor for transamidation than Asn.3, 20, 30, 31 Since the glutaminase subunit of GatDE is an L-asparaginase (GatD),1, 24, 25 raised the possibility that the archaeal specific AdT would be able to efficiently use Asn as an amide donor. The previous results also showed that GatDE was specific for Glu-tRNAGln and that GatCAB could transamidate both Asp-tRNAAsn and Glu-tRNAGln but whether the enzyme could transamidate both substrates with similar efficiencies was not determined.1 The predicted role of the archaeal GatCAB as an Asp-AdT suggested that the enzyme would prefer Asp-tRNAAsn over Glu-tRNAGln as a substrate. We therefore set out to determine the amide donor preferences of the archaeal AdTs (Fig. 1) and whether the M. thermautotrophicus GatCAB is equally efficient in transamidation of Glu-tRNAGln and Asp-tRNAAsn (Fig. 2).
Figure 1. Activity of the M. thermautotrophicus GatDE (a) and the M. thermautotrophicus GatCAB (b) with Gln or Asn as the amide donor in the transamidation reaction.
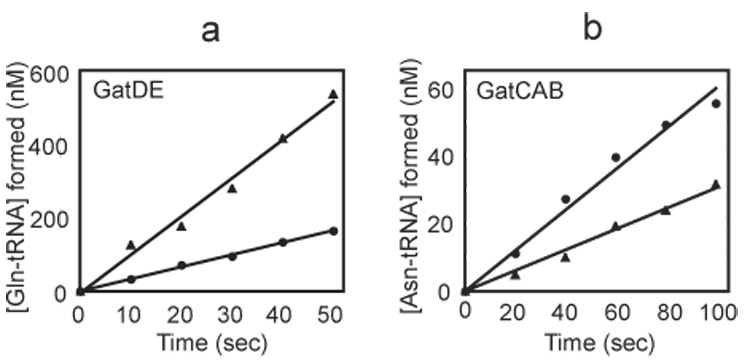
(a) Glu-tRNAGln conversion to Gln-tRNAGln by the M. thermautotrophicus GatDE (10 nM) with either Gln (4 mM) (●) or Asn (4 mM) (▲). (b) Asp-RNAAsn conversion to Asn-tRNAAsn by the M. thermautotrophicus GatCAB (5 nM) in the presence of Gln (4 mM) (●) or Asn (4 mM) (▲). In both (a) and (b), reactions were carried with 4 mM ATP and 10 µM mischarged tRNA (Glu-tRNAGln or Asp-tRNAAsn, respectively).
Figure 2. Glu-AdT (a) and Asp-AdT (b) activity of the M. thermautotrophicus GatDE and GatCAB.
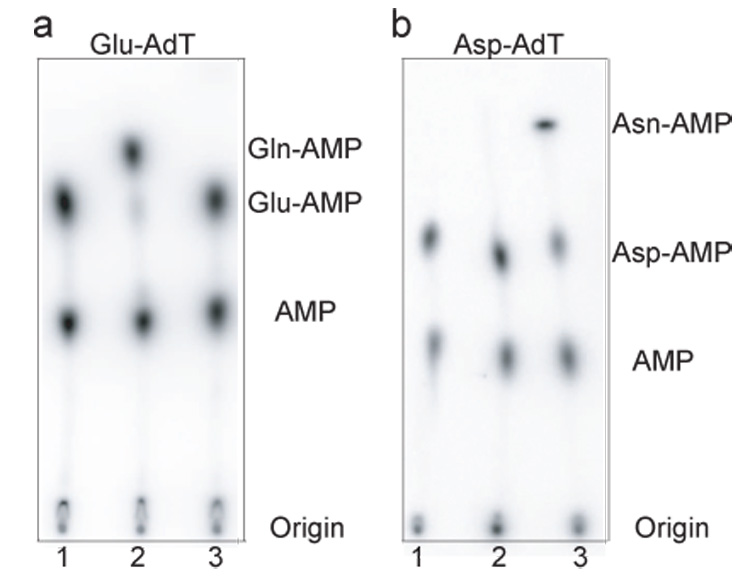
(a) Phosphorimage of a TLC separating [32P]AMP, Glu- [32P]AMP, and Gln-[32P]AMP released from M. thermautotrophicus tRNAGln by digestion with nuclease P1 following incubation with either no enzyme (lane 1), GatDE (200 nM) (lane 2) or GatCAB (200 nM) (lane 3) for 7 minutes at 37 °C in the presence of ATP (4 mM), 32P labeled M. thermautotrophicus Glu-tRNAGln (10 µM), and amide donor (4 mM). The amide donor in the GatCAB reaction was Gln and the donor in the GatDE reaction was Asn. (b) Phosphorimage of a TLC separating [32P]AMP, Asp- [32P]AMP, and Asn-[32P]AMP released from M. thermautotrophicus tRNAAsn by digestion with nuclease P1 following incubation with either no enzyme (lane 1), GatDE (200 nM) (lane 2) or GatCAB (200 nM) (lane 3) for 7 minutes at 37 °C in the presence of ATP (4 mM), 32P-labeled M. thermautotrophicus Asp-tRNAAsn (10 µM), and amide donor (4 mM). The amide donor in the GatCAB reactions was Gln and the donor in the GatDE reactions was Asn.
To test the transamidase activities of both AdTs we used a [32P]tRNA/nuclease P1 assay, which had earlier been successfully used to determine the enzymatic properties of the H. pylori GatCAB.3 This assay is sensitive, and the presence of the 32P-label in the 3’ terminal AMP of the tRNA ensures that any conversion of Glu to Gln or Asp to Asn detected is on the tRNA itself. It should be noted that the optimal growth temperature for M. thermautotrophicus is approximately 65 °C; yet these experiments were carried out at 37 °C, unless otherwise noted, in order to minimize deacylation of the aa-tRNA species. We found that at elevated temperatures our aa-tRNA substrates began to detectably deacylate over the time span of our reactions. It is speculated that in vivo complexes between ND-aaRSs and AdTs protects the aa-tRNA from deacylation.22, 32, 33
The M. thermautotrophicus GatDE utilized both amide donors (Fig. 1a) though it is approximately 5-fold more efficient with Asn than Gln as the donor for transamidating Glu-tRNAGln (Table 1). The effect is primarily due to a difference in kcat as GatDE has similar KM values for Gln and Asn (331.1 µM and 204.3 µM, respectively). Interestingly, the KM value of GatDE for Asn (204.3 µM) is between the corresponding values for the E. coli type I (3.5 mM) and type II (11.5 µM) L-asparaginases.34
Table 1.
Kinetic data for the transamidase activity of the M. thermautotrophicus GatDE*
| aSubstrate | KM (µM) | kcat (s−1) | kcat/KM (s−1/µM) (x 10−3) |
|---|---|---|---|
| Asp-tRNAAsn b | NDe | NDe | NDe |
| Glu-tRNAGln b | 1.71 ± 0.29 | 0.9 ± 0.1 | 530 ± 110 |
| ATPc | 114 ± 60 | 1.4 ± 0.2 | 12 ± 7 |
| Glnd | 331 ± 114 | 0.4 ± < 0.1 | 1.2 ± 0.5 |
| Asnd | 204 ± 48 | 1.1 ± 0.1 | 5.4 ± 1.3 |
For details of the steady-state kinetics see Materials and Methods. Experiments were done in duplicate and repeated two to four times. Standard deviations are reported.
KM was determined by varying the concentration of the substrate listed while adding the other two substrates required for transamidation in excess with 10 nM GatDE.
Asn and ATP were added in excess (4 mM and 4 mM, respectively).
10–11 µM Glu-tRNAGln and 4 mM Asn were added.
10–11 µM Glu-tRNAGln, 4 mM ATP were added.
ND stands for no-detectable activity under the conditions assayed (see text and Fig. 2).
Surprisingly, the M. thermautotrophicus GatCAB used both Gln and Asn as amide donors with similar efficiencies (Fig. 1b). The enzyme was only approximately 3-fold more efficient using Gln as the donor than Asn (Table 2). This is in contrast to the H. pylori GatCAB, which is 130-fold more efficient using Gln than Asn.3 The KM values for Gln and Asn with the M. thermautotrophicus GatCAB (6.7 µM and 11.6 µM, respectively) are slightly lower than the corresponding values with the H. pylori GatCAB (20.7 µM and 22.4 µM).3 These results suggest that the active site of the M thermautotrophicus GatA is subtly different from the S. aureus A-subunit,20 and thus allows the archaeal enzyme to use Asn nearly as well as Gln as an amide donor.
Table 2.
Kinetic data for the transamidase activity of the M. thermautotrophicus GatCAB*
| Substratea | KM (µM) | kcat (s−1) | kcat/KM (s−1/µM) (x 10−3) |
|---|---|---|---|
| Asp-tRNAAsn b | 0.78 ± 0.30 | 0.10 ± 0.01 | 125 ± 49 |
| Glu-tRNAGln b | NDe | NDe | NDe |
| ATPc | 400 ± 140 | 0.09 ± 0.01 | 0.2 ± 0.1 |
| Glnd | 6.7 ± 1.2 | 0.11 ± < 0.01 | 16.7 ± 3.0 |
| Asnd | 11.6 ± 2.6 | 0.07 ± < 0.01 | 5.7 ± 1.3 |
For details of the steady-state kinetics see Materials and Methods. Experiments were done in duplicate and repeated two to four times. Standard deviations are reported.
KM was determined by varying the concentration of the substrate listed while adding the other two substrates required for transamidation in excess with 10 nM GatCAB.
Asn and ATP were added in excess (4 mM of each).
10–11 µM Asp-tRNAAsn and 4 mM Gln were added.
10–11 µM Glu-tRNAAsn, 4 mM ATP were added.
ND stands for no-detectable activity under the conditions assayed (see text and Fig. 2).
Regarding the preferred mischarged tRNA substrate, GatDE as expected was specific for Glu-tRNAGln (Fig. 2). The KM values with the M. thermautotrophicus GatDE for Glu-tRNAGln and ATP, (1.71 µM and 114.0 µM, respectively) (Table 1) are similar to the values obtained with the H. pylori GatCAB (1.18 µM and 206.8 µM, respectively).3 The similarity in KM values is not surprising considering that the subunits (GatE and GatB) of the two AdTs that recognize Glu-tRNAGln and ATP are homologous.1 For ATP, the KM value (400 µM) with the M. thermautotrophicus GatCAB is slightly higher than that determined with the H. pylori GatCAB (206.8 µM).3 For Asp-tRNAAsn, the KM value with the M. thermautotrophicus GatCAB (0.78 µM) (Table 2) is slightly lower than the corresponding values with the Neisseria meningitidis GatCAB (1.2 µM)21 and H. pylori GatCAB (0.95 µM),3 and is slightly higher than the KD value (0.6 µM) determined for the enzyme with the mischarged substrate.23
While fully active as an Asp-AdT (Fig. 2b), under our new assay conditions purified (>95% homogeneity) M. thermautotrophicus GatCAB was unable to transamidate the homologous Glu-tRNAGln (Fig. 2a) even over an hour-long time course with 1 µM of enzyme (data not shown). We cannot rule out the possibility that the M thermautotrophicus GatCAB retains trace Glu-AdT activity with homologous Glu-tRNAGln as the aa-tRNA substrate substantially deacylated over this extended time period. However, it should be noted that no Glu-AdT activity with the M. thermautotrophicus GatCAB (1 µM) was detected within the first five minutes of the reaction during which time deacylation of the aa-tRNA was not as significant. In contrast, we were able to detect Asp-AdT activity with the M. thermautotrophicus GatCAB (5 nM) within 20 seconds (Fig. 1b).
Given that the optimal growth temperature of M. thermautotrophicus is 65 °C,35 we tested whether its GatCAB was more active at elevated temperatures (50 °C and 65 °C). As an Asp-AdT, the enzyme was approximately as active at 50 °C (apparent ktrans of 0.1 s−1) as it was at 37 °C (ktrans of 0.11 s−1, Table 3) while it was twice as active at 65 °C (apparent ktrans of 0.2 s−1). At both 50 °C and 65 °C the aa-tRNA substrate did detectably begin to deacylate over the time spans assayed (approximately 12 % at 65 °C over a time course of 100 seconds), thus the apparent Asp-AdT activity for GatCAB at these temperatures might be an underestimate of the actual activity. At both 50 °C and 65 °C, we did not detect Glu-AdT activity with the M. thermautotrophicus GatCAB (1 µM) and homologous Glu-tRNAGln (data not shown). However, trace activity may have been masked by the deacylation of the aa-tRNA (50% deacylation after approximately 9 and 6 min, respectively). The results do demonstrate however that regardless of the temperature at which M. thermautotrophicus GatCAB was assayed, at the very least, the enzyme is significantly more active as an Asp-AdT than as a Glu-AdT with homologous mischarged tRNA.
Table 3.
Transamidase activity of the M. thermautotrophicus (M.t.) GatCAB and GatDE, and the H. pylori (H.p.) GatCAB with different mischarged tRNA substrates *
| Transamidase activity (ktrans, s−1) | |||
|---|---|---|---|
| aa-tRNAa | M.t. GatCABb | M.t. GatDEc | H.p. GatCABd |
| M.t. GlutRNAGln | NDe | 0.98 ± 0.27 | 0.08 ± 0.02 |
| H.p. Glu-tRNAGln | 0.08 ± 0.02 | 0.01 ± <0.01 | 3.12 ± 0.09 |
| B.s. Glu-tRNAGln | 0.004 ± 0.003 | 0.01 ± <0.01 | 2.2 ± 0.1 |
| C.t. Asp-tRNAAsn | 0.09 ± 0.01 | NDe | 1.27 ± 0.01 |
| M.t. Asp-tRNAAsn | 0.11 ± 0.01 | NDe | 0.01 ± <0.01 |
Measurements were from three to four separate experiments. Standard deviations are reported. Reactions were carried out at 37 °C in the presence of ATP (4 mM), amide donor (4 mM), and aa-tRNA (10 µM) indicated. For reactions with the GatCAB enzymes, Gln was provided as the amide donor. In reactions with GatDE, Asn was the donor.
The aa-tRNA substrates tested were the M. thermautotrophicus (M.t.), H. pylori (H.p.), and B. subtilis (B.s.) Glu-tRNAGln, and the C. trachomatis (C.t.) and M. thermautotrophicus (M.t.) Asp-tRNAAsn.
In reactions with M.t. GatCAB, enzyme concentrations ranged from 20 nM to 1 µM.
In reactions with M.t. GatDE, enzyme concentrations ranged from 20 nM to 1 µM.
In reactions with H.p. GatCAB, enzyme concentrations ranged from 10 nM to 100 nM.
ND stands for no-detectable activity under conditions assayed.
The inability of the M. thermautotrophicus GatCAB to transamidate homologous Glu-tRNAGln is in contrast to the previous results.1 The earlier work was carried out with an enzyme preparation over-produced in Escherichia coli and partially purified (D. Tumbula-Hansen, personal communication); a similarly prepared partially purified enzyme formed Gln-tRNAGln (data not shown) using our current assay suggesting that this reaction is due to the presence of contaminating E. coli enzyme(s). It has been demonstrated that both the E. coli and the Bacillus subtilis glutamine synthetases are able to amidate Glu-tRNAGln to Gln-tRNAGln at low yield.30 The Gln-tRNAGln synthesis activity observed therefore may have been due to the presence of E. coli glutamine synthetase in the partially purified GatCAB preparations. Scenarios involving the E. coli GlnRS are less likely as the M. thermautotrophicus tRNAGln is an extremely poor substrate for the bacterial aaRS.1
Based on these results, it appears that M. thermautotrophicus encodes two AdTs, each with its own unique activity; GatDE is a Glu-AdT and only transamidates Glu-tRNAGln, while GatCAB is an Asp-AdT responsible for Asn-tRNAAsn formation.
M. thermautotrophicus GatCAB can transamidate Glu-tRNA
We then tested whether the M. thermautotrophicus GatCAB could transamidate other Glu-tRNA species. Theoretically, the primary discrimination of Asp-tRNAAsn from Glu-tRNAGln could be achieved by recognition of the Asp moiety attached to the tRNA as compared to Glu. The use of the amino acid moiety of the misacylated tRNA as an identity element had already been suggested.21, 23 Therefore we tested the ability of the M. thermautotrophicus GatCAB to use Glu-tRNAAsn as a substrate. Formation of M. thermautotrophicus tRNAAsn with homologous GluRS was possible; up to 10% of tRNAAsn was converted to Glu-tRNAAsn (data not shown). Of the two M. thermautotrophicus AdTs only GatCAB converted Glu-tRNAAsn to Gln-tRNAAsn (Fig. 3) with about 65% that of the enzyme’s Asp-AdT activity with homologous Asp-tRNAAsn as the substrate. The results reveal that M. thermautotrophicus GatCAB can amidate tRNA-bound Glu, and suggest that the AdT distinguishes Asp-tRNAAsn from Glu-tRNAGln based more on differences in the tRNA isoacceptors than discriminating Asp from Glu. However, we cannot rule out that recognition of the aminoacyl-moiety contributes to the ability of the AdT to discriminate Asp-tRNAAsn from Glu-tRNAGln. Attempts to perform the complementary experiment with Asp-tRNAGln failed because the M. thermautotrophicus AspRS was unable to acylate tRNAGln (data not shown).
Figure 3. M. thermautotrophicus GatCAB can transamidate Glu-tRNAAsn.
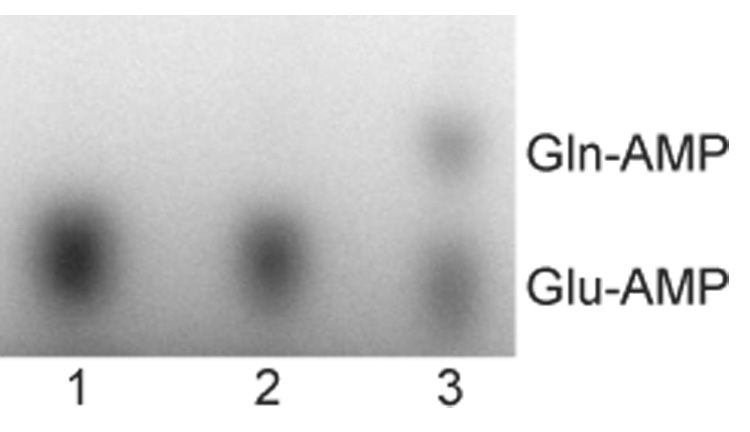
Phosphorimage of a TLC separating Glu-[32P]AMP, and Gln-[32P]AMP released from M. thermautotrophicus tRNAAsn by digestion with nuclease P1 following incubation with either no enzyme (lane 1), GatDE (200 nM) (lane 2) or GatCAB (200 nM) (lane 3), for 7 minutes at 37 °C in the presence of ATP (4 mM), 32P labeled M. thermautotrophicus Glu-tRNAAsn (1 µM), and amide donor (4 mM). The M. thermautotrophicus GluRS was only able to glutamylate up to ten percent of the M. thermautotrophicus tRNAAsn. The amide donor in the GatCAB reaction was Gln and the donor in the GatDE reaction was Asn.
The poor yield of Glu-tRNAAsn formation prevented a more quantitative examination of this unusual M. thermautotrophicus GatCAB substrate. However, we noticed that archaeal GatCAB can use bacterial Asp-tRNAAsn as a substrate approximately as well as archaeal Asp-tRNAAsn,21, 23 and bacterial GatCAB readily transamidates both bacterial Asp-tRNAAsn and Glu-tRNAGln.3, 12–14, 19, 36 Therefore, we tested whether the M. thermautotrophicus GatCAB could transamidate bacterial (H. pylori and B. subtilis) Glu-tRNAGln species (Table 3).
In contrast to M. thermautotrophicus Glu-tRNAGln, bacterial Glu-tRNAGln from H. pylori or B. subtilis were substrates for M. thermautotrophicus GatCAB (Table 3). The enzyme used the H. pylori Glu-tRNAGln as a substrate about as well as the Chlamydia trachomatis and M. thermautotrophicus Asp-tRNAAsn species (ktrans of 0.08 s−1, 0.09 s−1 and 0.11 s−1, respectively), demonstrating the M. thermautotrophicus GatCAB retains the ability to act approximately equally well as a Glu-AdT and an Asp-AdT. Interestingly, the archaeal GatCAB preferred the H. pylori Glu-tRNAGln over B. subtilis Glu-tRNAGln (ktrans of 0.08 s−1 and 0.004 s−1, respectively), suggesting that elements in the tRNA allow preferential recognition of H. pylori over B. subtilis Glu-tRNAGln by the M. thermautotrophicus GatCAB.
In keeping with the known identity elements recognized by GatDE22 and bacterial GatCAB,20, 21 the M. thermautotrophicus GatDE preferred the archaeal Glu-tRNAGln over other substrates tested (Table 3) and the H. pylori GatCAB (Table 3) favored the bacterial mischarged tRNA substrates over the archaeal ones. These two enzymes, in contrast to the archaeal GatCAB, positively recognize the first base-pair in the acceptor stem of their mischarged tRNA substrates (Fig. 4).20–22
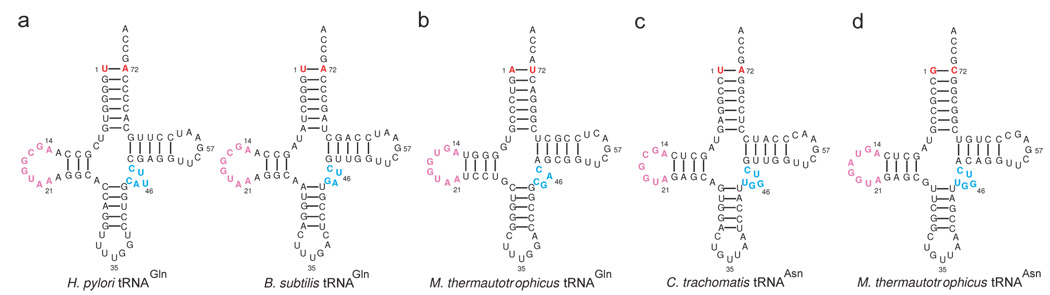
Figure 4. Known identity elements recognized by the AdTs.
Cloverleaf representations of (a) two bacterial tRNAGln isoacceptors (H. pylori and B. subtilis), (b) the M. thermautotrophicus tRNAGln, (c) the C. trachomatis tRNAAsn, and (d) the M. thermautotrophicus tRNAAsn. Known identity elements recognized by the AdTs are color-coded. The first base-pair of the acceptor stem (red) is an important recognition element for bacterial GatCAB and GatDE in their respective aa-tRNA substrates.20, 21 The D-loop (pink) is used by the archaeal and bacterial GatCAB as well as by GatDE to distinguish their mischarged aa-tRNA substrates from other aa-tRNA species.20–22 The length of the variable loop (cyan) has been demonstrated to be important for the archaeal GatCAB to distinguish Asp-tRNAAsn from Asp-tRNAAsp.21, 23
Discussion
Asn-tRNA and Gln-tRNA are formed in M. thermautotrophicus by two different enzymes
While it was established earlier that GatDE is a Glu-AdT unable to synthesize Asn-tRNAAsn,1 we have now provided in vitro evidence that M. thermautotrophicus GatCAB cannot synthesize Gln on its homologous tRNAGln and functions only as an Asp-AdT. Thus, M. thermautotrophicus probably uses two different amidotransferases for amide aminoacyl-tRNA synthesis, GatDE for Gln-tRNAGln formation and GatCAB for Asn-tRNAAsn synthesis also in vivo (Fig. 5). Given that GatCAB is only encoded in archaeal genomes that lack AsnRS,1, 2 it is plausible to assume that other archaea also employ GatCAB just as an Asp-AdT. This is in contrast to the situation in Bacteria; for instance C. trachomatis and H. pylori do not encode AsnRS and GlnRS in their genomes and employ only GatCAB to serve both as an Asp-AdT and a Glu-AdT to meet their cellular needs for both Asn-tRNAAsn and Gln-tRNAGln (Fig. 5).3, 14
Figure 5. In the archaeon, M. thermautotrophicus, there is a separate Glu-AdT, GatDE, and Asp-AdT, GatCAB, while in Bacteria like H. pylori a single AdT, GatCAB, serves both functions.
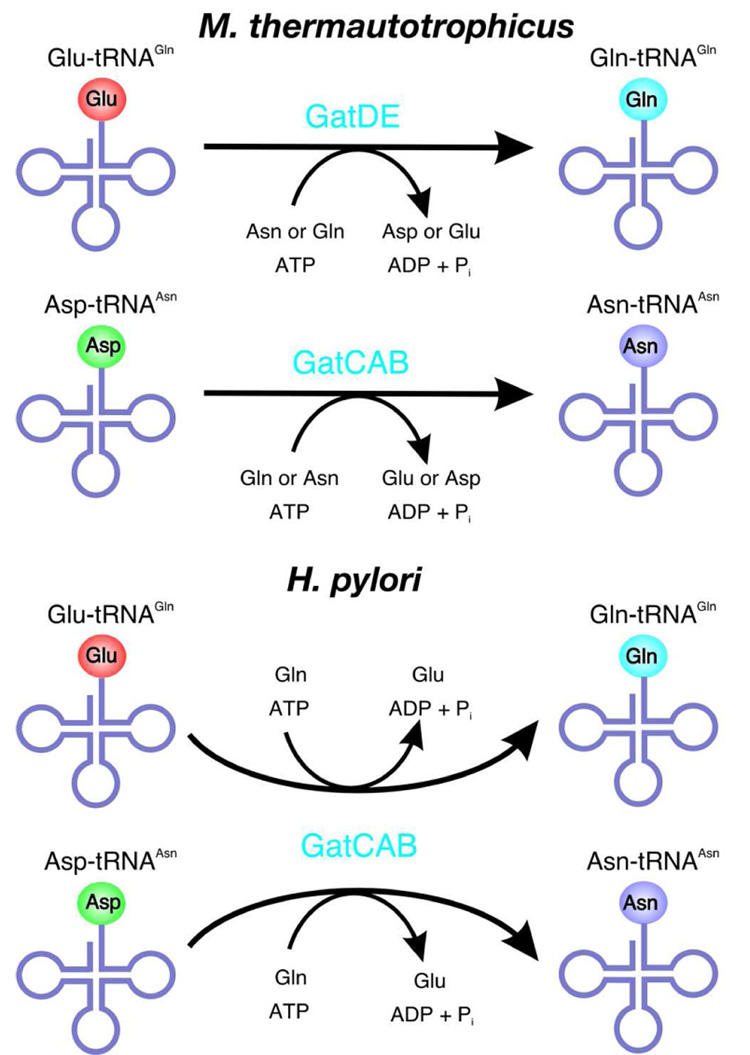
In M. thermautotrophicus, which lacks a GlnRS and an AsnRS, GatDE transamidates Glu-tRNAGln to Gln-tRNAGln while GatCAB catalyzes Asn-tRNAAsn formation from Asp-tRNAAsn. GatDE is slightly more efficient using Asn as an amide donor while the archaeal GatCAB has a slight preference for Gln over Asn. In H. pylori, which also lacks the two aaRSs, on the other hand employs just GatCAB as both a Glu-AdT and an Asp-AdT to form both amide aa-tRNA species. The H. pylori GatCAB is significantly more efficient using Gln as an amide donor than Asn.3
Given the specificity of the M. thermautotrophicus GatCAB as an Asp-AdT raises the question of how the AdT distinguishes M. thermautotrophicus Asp-tRNAAsn from M. thermautotrophicus Glu-tRNAGln. The enzyme retains the ability to transamidate Glu attached to other tRNA isoacceptors, strongly indicating the Glu bound to the tRNA is not preventing M. thermautotrophicus GatCAB from recognizing the mischarged tRNA. This is particularly highlighted by the fact that the M. thermautotrophicus GatCAB is able to transamidate H. pylori Glu-tRNAGln tRNA as well as Asp-tRNAAsn from C. trachomatis or M. thermautotrophicus. The results suggest that M. thermautotrophicus tRNAGln limits the M. thermautotrophicus GatCAB to Asn-tRNAAsn synthesis.
Evolution of substrate preferences of the archaeal AdTs
It is unkown why archaea use two AdTs while bacteria utilize only one. One thought might invoke the putative thermophilic origins of archaea.37 While in mesophiles the codon usage of Asn and Gln is approximately equal, in thermophiles the codon usage of Asn is on average twice that of Gln.38, 39 Since Gln is more labile at elevated temperatures than Asn,40 it may suggest that themophiles have different cellular demands for Asn-tRNAAsn and Gln-tRNAGln. Possibly having a separate Asp-AdT and Glu-AdT by retaining both GatCAB and GatDE may have enabled the thermophilic ancestral archaea to separate the regulation of the enzymes involved in Asn-tRNAAsn formation from those involved in Gln-tRNAGln biosynthesis to better meet the cellular needs for the two aa-tRNA species.
It is interesting to speculate whether in thermophilic organisms the relative ratios of free Asn to Gln are also elevated as compared to those in mesophiles given that Asn is more stable than Gln at higher temperatures.40 A higher relative concentration of Asn relative to Gln in thermophiles as compared to mesophiles would provide an explanation as to why the M. thermautotrophicus GatDE is slightly more efficient using Asn over Gln as an amide donor and why the M. thermautotrophicus GatCAB is only 3-fold more efficient using Gln than Asn while the H. pylori GatCAB is 130-fold more efficient using Gln over Asn.3 The H. pylori enzyme would be inefficient in a cellular environment in which the relative ratio of Asn to Gln favors Asn.3 On the other hand, the archaeal GatCAB would only be marginally less active while GatDE would be in fact more active.
Materials and Methods
General
All oligonucleotide synthesis and DNA sequencing was carried out by the Keck Foundation Biotechnology Research Laboratory at Yale University. [α-32P] ATP (10 mmol/µCi) was from Amersham Biosciences (GE Healthcare). Escherichia coli BL21(DE3) and BL21(DE3) codon plus strains were from Stratagene (La Jolla, CA). Nickel-nitrilotriacetic acid-agarose (Ni-NTA) was from QIAGEN (Chatsworth, CA). Bio-Spin 30 columns were from Bio-Rad (Hercules, CA). High purity cold L-Glu, L-Asp, L-Gln and L-Asn were from Fluka (Deisenhofen, Germany). Phenol was from American Bioanalytical (Natick, MA).
H. pylori GluRS, M. thermautotrophicus GluRS, M. thermautotrophicus GatDE, H pylori GatCAB, and C. trachomatis GluRS over-production and purifications
M. thermautotrophicus AspRS over-production and purification
The enzyme was over-produced in an E. coli BL21-CodonPlus(DE3)-RIL strain harboring the pET28a vector with M. thermautotrophicus aspS between the NdeI and BamHI restriction sites.41 Over-production was by auto-induction42 as described for the H. pylori GatCAB except the resistance was kanamycin in place of ampicillin.3 The enzyme was purified as previously described though the final purification step of passing the enzyme over a Superdex200 column was omitted.41
M. thermautotrophicus GatCAB over-production and purification
The enzyme was over-produced in an E. coli BL21-CodonPlus(DE3)-RIL strain and purified over a Ni-NTA column as previously described.41 The sample was then dialyzed in buffer Q (25 mM Hepes-KOH, pH 7.2, 5 mM 2-mercaptoethanol, 0.2 mM EDTA, and 5% glycerol). The protein was purified to > 95% homogeneity by fast protein liquid chromatography on a MonoQ column (Amersham Biosciences) that was developed with a 20–750 mM KCl gradient in buffer Q. GatCAB eluted with approximately a 135 mM concentration of KCl. The method has been used to successfully purify M. thermautotrophicus GatDE and H. pylori GatCAB.3, 24 The pure fraction of GatCAB was then dialyzed in buffer Q with 50% glycerol.
M. thermautotrophicus tRNAGln and tRNAAsn and C. trachomatis tRNAAsn production and purification
The tRNA isoacceptors were transcribed and purified as previously described.1; 43
H. pylori tRNAGln and B. subtilis tRNAGln purification
Both tRNA species were over-expressed in an E. coli XL1-Blue strain and total tRNA was purified as previously described.3, 11 The H. pylori tRNAGln was purified from total tRNA as described previously.3, 44 The B. subtilis tRNAGln was purified in a same manner3; 44 except a different 5’ biotinylated probe ( 5’- GCTGGATTCGAACCAACGCATGACGGAGTCAAAGTCC-3’) was used.
Preparation of lableled tRNA
The tRNA isoacceptors were 32P-labeled on their 3’ terminus as previously described.3
Preparation of bacterial Glu-tRNAGln and Asp-tRNAAsn
The H. pylori Glu-tRNAGln was prepared as described.3 The C. trachomatis Asp-tRNAAsn was prepared as described for the H. pylori tRNAAsn with some modification.3 Instead of 21 µM unlabled tRNAAsn, 10 µM tRNAAsn was added in the reaction and the reaction time was shortened to one hour from 14 hours. The P. aeruginosa AspRS was a gift from Jacques Lapointe (Laval University, Canada). The B. subtilis Glu-tRNAGln was prepared as described for the H. pylori tRNAGln except the C. trachomatis GluRS was used in place of the H. pylori GluRS.3
Preparation of M. thermautotrophicus Glu-tRNAGln
M. thermautotrophicus tRNAGln was glutamlyated as described previously with slight modification.22 To the reaction 600 nM of [32P]tRNAGln was added instead of the 40 nM stated.
Preparation of M. thermautotrophicus Asp-tRNAAsn
Aminoacylation of the M. thermautotrophicus tRNAAsn with Asp was carried out in 1× M. thermautotrophicus aminoacylation buffer (50 mM Hepes-KOH buffer (pH 7.2) containing 25 mM KCl, 10 mM MgCl2, and 5 mM DTT) with 24 µg/µL of pyrophosphatase (Roche), 4 mM ATP, 3 µM M. thermautotrophicus AspRS, 1 mM L-Asp, 10 µM unlabeled tRNAAsn and 600 nM 32P-labeled tRNAAsn. The reactions were incubated at 37 °C for 45 min. The samples were processed in the same manner as previously described.3
Preparation of M. thermautotrophicus Glu-tRNAAsn
Aminoacylation of the M. thermautotrophicus tRNAAsn with Glu was carried out at 37 °C for 90 min. in 1× M. thermautotrophicus aminoacylation buffer with 24 µg/µL of pyrophosphatase (Roche), 4 mM ATP, 1 µM M. thermautotrophicus GluRS, 1 mM L-Asp, 0.9 µM unlabeled tRNAAsn and 100 nM 32P-labeled tRNAAsn. The samples were processed in the same manner as previously described.3
[32P]tRNA/nuclease P1 amidotransferase assay
Glu-AdT and Asp-AdT activities of the M. thermautotrophicus GatCAB and GatDE were assayed as previously described3, 22 with some modification. Briefly, amidotransferase reaction mixtures included 1×M. thermautotrophicus AdT buffer (100 mM Hepes-KOH, pH 7.2, 12 mM MgCl2, 30 mM KCl and 5 mM DTT) and unless otherwise noted 4 mM ATP, 9 to 11 µM 32P-labeled mischarged tRNA (Asp-tRNAAsn or Glu-tRNAGln) and 4 mM amide donor (Gln or Asn) and were carried out at 37 °C (unless otherwise noted). For determination of the kinetic parameters, initial velocities were measured in duplicate while varying the concentration of one substrate and saturating with the other two. In studies with the M. thermautotrophicus GatDE, 10 nM enzyme was used and reactions were carried out over a minute. With the M. thermautotrophicus GatCAB, reactions were carried over two minutes and with 5 nM enzyme. For KM determinations of the two-amide donors, substrate concentrations varied from 3 µM to 4000 µM. For KM determinations of the mischarged tRNA substrate, concentrations varied from approximately 150 nM up to 12 µM. For KM determinations with ATP, concentrations varied from 40 µM to 4000 µM. Reaction mixes were pre-incubated at 37 °C and started by addition of enzyme. Aliquots (2 µL) of the reaction mixes were quenched, digested and processed as previously described.3, 22
Acknowledgements
We thank Patrick O’Donoghue and Sotiria Palioura for stimulating discussions and Dr. Jacques Lapointe (Laval University, Canada) for his gift of the Pseudomonas AspRS. R.L.S. is the recipient of a Kirschstein–National Research Service Award postdoctoral fellowship from the National Institute of General Medical Sciences. This work was supported by grants from the National Institute of General Medical Sciences (GM22854) and Department of Energy (DE-FG02-98ER20311).
The abbreviations used are
- aaRS
aminoacyl-tRNA synthetase
- GlnRS
glutaminyl-tRNA synthetase
- AsnRS
asparaginyl-tRNA synthetase
- ND
non-discriminating
- AdT
tRNA-dependent amidotransferase
- GluRS
glutamyl-tRNA synthetase
- Glu-AdT
glutamyl-tRNAGln amidotransferase catalyzing Gln-tRNA formation
- AspRS
aspartyl-tRNA synthetase
- Asp-AdT
aspartyl-tRNAAsn amidotransferase catalyzing Asn-tRNA formation
- NTA
Nickel-nitrilotriacetic acid-agarose
- PEI
polyethylenimine
- TLC
thin layer chromatography. The three-letter code for the amino acids is used throughout.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tumbula DL, Becker HD, Chang WZ, Söll D. Domain-specific recruitment of amide amino acids for protein synthesis. Nature. 2000;407:106–110. doi: 10.1038/35024120. [DOI] [PubMed] [Google Scholar]
- 2.Roy H, Becker HD, Reinbolt J, Kern D. When contemporary aminoacyl-tRNA synthetases invent their cognate amino acid metabolism. Proc. Natl. Acad. Sci. USA. 2003;100:9837–9842. doi: 10.1073/pnas.1632156100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sheppard K, Akochy PM, Salazar JC, Söll D. The Helicobacter pylori amidotransferase GatCAB is equally efficient in glutamine-dependent transamidation of Asp-tRNAAsn and Glu-tRNAGln. J. Biol. Chem. 2007;282:11866–118773. doi: 10.1074/jbc.M700398200. [DOI] [PubMed] [Google Scholar]
- 4.Lapointe J, Duplain L, Proulx M. A single glutamyl-tRNA synthetase aminoacylates tRNAGlu and tRNAGln in Bacillus subtilis and efficiently misacylates Escherichia coli tRNAGln1in vitro. J. Bacteriol. 1986;165:88–93. doi: 10.1128/jb.165.1.88-93.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilcox M, Nirenberg M. Transfer RNA as a cofactor coupling amino acid synthesis with that of protein. Proc. Natl. Acad. Sci. USA. 1968;61:229–236. doi: 10.1073/pnas.61.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Becker HD, Reinbolt J, Kreutzer R, Giege R, Kern D. Existence of two distinct aspartyl-tRNA synthetases in Thermus thermophilus. Structural and biochemical properties of the two enzymes. Biochemistry. 1997;36:8785–8797. doi: 10.1021/bi970392v. [DOI] [PubMed] [Google Scholar]
- 7.Curnow AW, Ibba M, Söll D. tRNA-dependent asparagine formation. Nature. 1996;382:589–590. doi: 10.1038/382589b0. [DOI] [PubMed] [Google Scholar]
- 8.Becker HD, Kern D. Thermus thermophilus: a link in evolution of the tRNA-dependent amino acid amidation pathways. Proc. Natl. Acad. Sci. USA. 1998;95:12832–12837. doi: 10.1073/pnas.95.22.12832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cathopoulis T, Chuawong P, Hendrickson TL. Novel tRNA aminoacylation mechanisms. Mol. Biosyst. 2007;3:408–418. doi: 10.1039/b618899k. [DOI] [PubMed] [Google Scholar]
- 10.Wilcox M. Gamma-phosphoryl ester of Glu-tRNAGln as an intermediate in Bacillus subtilis glutaminyl-tRNA synthesis. Cold Spring Harb. Symp. Quant. Biol. 1969;34:521–528. doi: 10.1101/sqb.1969.034.01.059. [DOI] [PubMed] [Google Scholar]
- 11.Curnow AW, Hong K, Yuan R, Kim S, Martins O, Winkler W, Henkin TM, Söll D. Glu-tRNAGln amidotransferase: a novel heterotrimeric enzyme required for correct decoding of glutamine codons during translation. Proc. Natl. Acad. Sci. USA. 1997;94:11819–11826. doi: 10.1073/pnas.94.22.11819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Curnow AW, Tumbula DL, Pelaschier JT, Min B, Söll D. Glutamyl-tRNA(Gln) amidotransferase in Deinococcus radiodurans may be confined to asparagine biosynthesis. Proc. Natl. Acad. Sci. USA. 1998;95:12838–12843. doi: 10.1073/pnas.95.22.12838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Becker HD, Min B, Jacobi C, Raczniak G, Pelaschier J, Roy H, Klein S, Kern D, Söll D. The heterotrimeric Thermus thermophilus AsptRNA(Asn) amidotransferase can also generate Gln-tRNA(Gln) FEBS Lett. 2000;476:140–144. doi: 10.1016/s0014-5793(00)01697-5. [DOI] [PubMed] [Google Scholar]
- 14.Raczniak G, Becker HD, Min B, Söll D. A single amidotransferase forms asparaginyl-tRNA and glutaminyl-tRNA in Chlamydia trachomatis. J. Biol. Chem. 2001;276:45862–45867. doi: 10.1074/jbc.M109494200. [DOI] [PubMed] [Google Scholar]
- 15.Akochy PM, Bernard D, Roy PH, Lapointe J. Direct glutaminyl-tRNA biosynthesis and indirect asparaginyl-tRNA biosynthesis in Pseudomonas aeruginosa PAO1. J. Bacteriol. 2004;186:767–776. doi: 10.1128/JB.186.3.767-776.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skouloubris S, Ribas de Pouplana L, De Reuse H, Hendrickson TL. A noncognate aminoacyl-tRNA synthetase that may resolve a missing link in protein evolution. Proc. Natl. Acad. Sci. USA. 2003;100:11297–11302. doi: 10.1073/pnas.1932482100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salazar JC, Ahel I, Orellana O, Tumbula-Hansen D, Krieger R, Daniels L, Söll D. Coevolution of an aminoacyl-tRNA synthetase with its tRNA substrates. Proc. Natl. Acad. Sci. USA. 2003;100:13863–13868. doi: 10.1073/pnas.1936123100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chuawong P, Hendrickson TL. The nondiscriminating aspartyl-tRNA synthetase from Helicobacter pylori: anticodon-binding domain mutations that impact tRNA specificity and heterologous toxicity. Biochemistry. 2006;45:8079–8087. doi: 10.1021/bi060189c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cathopoulis TJ, Chuawong P, Hendrickson TL. A thin-layer electrophoretic assay for Asp-tRNA(Asn)/Glu-tRNA(Gln) amidotransferase. Anal. Biochem. 2007;360:151–153. doi: 10.1016/j.ab.2006.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakamura A, Yao M, Chimnaronk S, Sakai N, Tanaka I. Ammonia channel couples glutaminase with transamidase reactions in GatCAB. Science. 2006;312:1954–1958. doi: 10.1126/science.1127156. [DOI] [PubMed] [Google Scholar]
- 21.Bailly M, Giannouli S, Blaise M, Stathopoulos C, Kern D, Becker HD. A single tRNA base pair mediates bacterial tRNA-dependent biosynthesis of asparagine. Nucleic Acids Res. 2006;34:6083–6094. doi: 10.1093/nar/gkl622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oshikane H, Sheppard K, Fukai S, Nakamura Y, Ishitani R, Numata T, Sherrer RL, Feng L, Schmitt E, Panvert M, Blanquet S, Mechulam Y, Söll D, Nureki O. Structural basis of RNA-dependent recruitment of glutamine to the genetic code. Science. 2006;312:1950–1954. doi: 10.1126/science.1128470. [DOI] [PubMed] [Google Scholar]
- 23.Namgoong S, Sheppard K, Sherrer RL, Söll D. Co-evolution of the archaeal tRNA-dependent amidotransferase GatCAB with tRNA(Asn) FEBS Lett. 2007;581:309–314. doi: 10.1016/j.febslet.2006.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng L, Sheppard K, Tumbula-Hansen D, Söll D. Gln-tRNAGln formation from Glu-tRNAGln requires cooperation of an asparaginase and a Glu-tRNAGln kinase. J. Biol. Chem. 2005;280:8150–8155. doi: 10.1074/jbc.M411098200. [DOI] [PubMed] [Google Scholar]
- 25.Schmitt E, Panvert M, Blanquet S, Mechulam Y. Structural basis for tRNA-dependent amidotransferase function. Structure. 2005;13:1421–1433. doi: 10.1016/j.str.2005.06.016. [DOI] [PubMed] [Google Scholar]
- 26.Harpel MR, Horiuchi KY, Luo Y, Shen L, Jiang W, Nelson DJ, Rogers KC, Decicco CP, Copeland RA. Mutagenesis and mechanism-based inhibition of Streptococcus pyogenes Glu-tRNAGln amidotransferase implicate a serine-based glutaminase site. Biochemistry. 2002;41:6398–6407. doi: 10.1021/bi012126u. [DOI] [PubMed] [Google Scholar]
- 27.Müller HJ, Boos J. Use of L-asparaginase in childhood ALL. Crit. Rev. Oncol. Hematol. 1998;28:97–113. doi: 10.1016/s1040-8428(98)00015-8. [DOI] [PubMed] [Google Scholar]
- 28.Kotzia GA, Labrou NE. Cloning, expression and characterisation of Erwinia carotovora L-asparaginase. J. Biotechnol. 2005;119:309–323. doi: 10.1016/j.jbiotec.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 29.Roberts J. Purification and properties of a highly potent antitumor glutaminase-asparaginase from Pseudomonas 7Z. J. Biol. Chem. 1976;251:2119–2123. [PubMed] [Google Scholar]
- 30.Strauch MA, Zalkin H, Aronson AI. Characterization of the glutamyl-tRNA(Gln)-to-glutaminyl-tRNA(Gln) amidotransferase reaction of Bacillus subtilis. J. Bacteriol. 1988;170:916–920. doi: 10.1128/jb.170.2.916-920.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jahn D, Kim YC, Ishino Y, Chen MW, Söll D. Purification and functional characterization of the Glu-tRNA(Gln) amidotransferase from Chlamydomonas reinhardtii. J. Biol. Chem. 1990;265:8059–8064. [PubMed] [Google Scholar]
- 32.Huot JL, Balg C, Jahn D, Moser J, Emond A, Blais SP, Chenevert R, Lapointe J. Mechanism of a GatCAB Amidotransferase: Aspartyl-tRNA Synthetase Increases Its Affinity for Asp-tRNA(Asn) and Novel Aminoacyl-tRNA Analogues Are Competitive Inhibitors. Biochemistry. 2007;46:13190–13198. doi: 10.1021/bi700602n. [DOI] [PubMed] [Google Scholar]
- 33.Bailly M, Blaise M, Lorber B, Becker HD, Kern D. The transamidosome: a dynamic ribonucleoprotein particle dedicated to prokaryotic tRNA-dependent asparagine biosynthesis. Molecular Cell. 2007;28:228–239. doi: 10.1016/j.molcel.2007.08.017. [DOI] [PubMed] [Google Scholar]
- 34.Jennings MP, Beacham IR. Analysis of the Escherichia coli gene encoding L-asparaginase II, ansB, and its regulation by cyclic AMP receptor and FNR proteins. J. Bacteriol. 1990;172:1491–1498. doi: 10.1128/jb.172.3.1491-1498.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeikus JG, Wolfe RS. Fine structure of Methanobacterium thermoautotrophicum: effect of growth temperature on morphology and ultrastructure. J. Bacteriol. 1973;113:461–467. doi: 10.1128/jb.113.1.461-467.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salazar JC, Zuniga R, Raczniak G, Becker H, Söll D, Orellana O. A dual-specific Glu-tRNA(Gln) and Asp-tRNA(Asn) amidotransferase is involved in decoding glutamine and asparagine codons in Acidithiobacillus ferrooxidans. FEBS Lett. 2001;500:129–131. doi: 10.1016/s0014-5793(01)02600-x. [DOI] [PubMed] [Google Scholar]
- 37.Woese CR. Bacterial evolution. Microbiol. Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Michelitsch MD, Weissman JS. A census of glutamine/asparaginerich regions: implications for their conserved function and the prediction of novel prions. Proc. Natl. Acad. Sci. USA. 2000;97:11910–11915. doi: 10.1073/pnas.97.22.11910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singer GA, Hickey DA. Thermophilic prokaryotes have characteristic patterns of codon usage, amino acid composition and nucleotide content. Gene. 2003;317:39–47. doi: 10.1016/s0378-1119(03)00660-7. [DOI] [PubMed] [Google Scholar]
- 40.Vickery HB, Pucher GW, Clark HE. The determination of glutamine in the presence of asparagine. Biochem. J. 1935;29:2710–2720. doi: 10.1042/bj0292710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tumbula-Hansen D, Feng L, Toogood H, Stetter KO, Söll D. Evolutionary divergence of the archaeal aspartyl-tRNA synthetases into discriminating and nondiscriminating forms. J. Biol. Chem. 2002;277:37184–37190. doi: 10.1074/jbc.M204767200. [DOI] [PubMed] [Google Scholar]
- 42.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 43.Fechter P, Rudinger J, Giege R, Theobald-Dietrich A. Ribozyme processed tRNA transcripts with unfriendly internal promoter for T7 RNA polymerase: production and activity. FEBS Lett. 1998;436:99–103. doi: 10.1016/s0014-5793(98)01096-5. [DOI] [PubMed] [Google Scholar]
- 44.Kaneko T, Suzuki T, Kapushoc ST, Rubio MA, Ghazvini J, Watanabe K, Simpson L. Wobble modification differences and subcellular localization of tRNAs in Leishmania tarentolae: implication for tRNA sorting mechanism. EMBO J. 2003;22:657–667. doi: 10.1093/emboj/cdg066. [DOI] [PMC free article] [PubMed] [Google Scholar]