Abstract
Recent studies have demonstrated a central role for the exchange protein activated by cAMP (Epac) in the inhibition of Fcγ-receptor mediated phagocytosis and bacterial killing by prostaglandin E2 (PGE2) in macrophages. However, the subcellular localization of Epac, and its primary target Rap1, has yet to be determined in primary macrophages. Therefore, we used immunofluorescent techniques and phagosome isolation to localize Epac-1 and Rap1 in alveolar macrophages. Epac-1 was predominantly expressed on punctate and tubular membranes throughout the cell body; on the plasma membrane; and co-localized with microtubule organizing centers (MTOCs). Rap1 was abundant on punctate membranes, less abundant on plasma membrane, and also found on MTOCs. Following PGE2 treatment, Epac-1, but not Rap1, accumulated on the nuclear envelope and disappeared from MTOCs. By immunofluorescent microscopy, both Epac-1 and Rap1 were seen to associate with phagosomes containing IgG-opsonized beads, but this association appeared weak, as we failed to observe such interactions in phagosomes isolated from cells at various timepoints after bead ingestion. Strikingly, however, Epac-1, but not Rap1, appeared to accumulate on maturing phagosomes, but only after PGE2 treatment (or treatment with a selective Epac-1 agonist). This association was confirmed in isolated phagosome preparations. The changes in Epac-1 localization were too slow to account for the inhibitory effects of PGE2 on phagocytosis. However, the appearance of Epac-1 on late phagosomes following PGE2 treatment might be important for suppressing H2O2 production and inhibiting the killing of intraphagosomal pathogens. The absence of Rap1 on late phagosomes suggests that the effect of Epac-1 might not require Rap1.
Introduction
The recognition and clearance of invading microorganisms is a critical function of phagocytic cells involved in innate immunity and occurs through both opsonin-dependent and -independent pathways. Phagocytes express a broad array of receptors that participate in particle recognition and internalization. Key opsonins include complement, fibronectin/vitronectin, and immunoglobulin (Ig), which bind corresponding complement, integrin, and Fcγ receptors, respectively [1]. Interestingly, the molecular mechanisms facilitating opsonin-dependent phagocytosis are different for particular opsonin/receptor pairs [1]. For example, phagocytosis of IgG-opsonized pathogens, which occurs via the Fcγ class of receptors (FcγR), involves phagocyte membrane extension around the microbe and results in the production of pro-inflammatory mediators [1]. Complement receptor mediated pathogen ingestion, on the other hand, occurs without observable membrane extension (particles “sink” into the cell) and is not generally associated with a concomitant inflammatory mediator response [1].
Regardless of the opsonin/receptor pathway involved, phagocytosis is a highly regulated process subject to both positive and negative regulation. Our laboratory has focused on the regulation of FcγR-mediated phagocytosis by lipid mediators derived from the cell membrane constituent arachidonic acid. Such metabolites, known as eicosanoids, are generated in abundance at sites of inflammation (e.g. the host-microbial interface) and influence key components of the innate immune system, namely, phagocytosis [2, 3], intracellular microbial killing [4, 5], and inflammatory mediator generation [6].
Prostaglandin (PG) E2 is an immunomodulatory eicosanoid generated by the sequential oxygenation and isomerization of arachidonic acid by cyclooxygenase and PGE2 synthase enzymes. The regulation of target cells by PGE2 occurs via signaling through 4 distinct cell membrane-associated G-protein coupled E-prostanoid (EP) receptors, termed EP1, EP2, EP3, and EP4 [7]. EP1 receptor activation provokes Gq-coupled increases in intracellular Ca2+, EP2 and EP4 receptors signal predominantly through Gs, increasing cAMP, and the EP3 receptor most commonly reduces cAMP via Gi coupling after PGE2 ligation [7]. It is through its ability to stimulate the production of intracellular cAMP (via EP2 and/or EP4) that PGE2 impairs FcγR-mediated phagocytosis [2]. And though cAMP is a well known counterregulator of FcγR phagocytosis [2, 8-10], the precise mechanisms downstream of cAMP remain to be fully defined.
Classically, cAMP signaling involves the immediate activation of protein kinase A (PKA), which phosphorylates several downstream targets, including the cAMP response element binding protein. However, PKA-independent actions of cAMP have been recognized in various experimental systems and novel targets for cAMP signaling have been described. These include cyclic nucleotide gated channels involved in the transduction of olfactory and visual signals and the guanine exchange proteins directly activated by cAMP (Epac-1 and −2) [11, 12]. Epac-1 is more generally expressed throughout cells and tissues than Epac-2 [12] and its expression has been described in diverse cell types, including cells of the innate immune system.
Recently, Aronoff et al. reported the PKA-independent, Epac-dependent suppression of FcγR-mediated phagocytosis in rat alveolar macrophages (AMs) [4]. This effect involved Epac-1, as AMs did not express Epac-2 protein [4]. Subsequent publications have confirmed an important role for Epac-1 in both FcγR-dependent and –independent phagocytic pathways [10, 13], though the mechanisms involved remain undiscovered. Epac-1 activates the small GTPases Rap1 and Rap2 [11] and Rap1 was found to associate with late endocytic/phagocytic compartments of J774.A1 macrophage-like cells [14]. Rap2 associates with secretory pathways and has not been implicated in phagocytosis [14]. It is notable that neither Rap1 overexpression nor Rap1 inhibition significantly altered FcγR-mediated phagocytosis in the J774.A1 cells [15]. Whether Epac-1-Rap1 signaling is involved in regulating FcγR-mediated phagocytosis in primary macrophages remains unclear.
Given the involvement of Epac-1 in regulating FcγR-mediated phagocytosis and bacterial killing in macrophages, and the ability of Epac-1 to activate Rap1, we hypothesized that these effector molecules would spatially and temporally associate with phagosomes during FcγR-mediated phagocytosis. We further speculated that such associations might be altered by treatment with PGE2. The following experiments were designed to test these hypotheses using two complementary approaches, immunofluorescence microscopy and dynamic phagosome isolation, to track Epac-1 and Rap1 before and during FcγR-mediated phagocytosis, in the presence or absence of PGE2.
Materials and Methods
Animals
Pathogen-free 125-150 gm female Wistar rats (Charles River Laboratories, Portage, MI) were utilized. Animals were treated according to National Institutes of Health guidelines for the use of experimental animals with the approval of the University of Michigan Committee for the Use and Care of Animals.
Reagents
RPMI-1640, and penicillin/streptomycin/amphotericin B solution were purchased from Gibco-Invitrogen (Carlsbad, CA). PGE2 from Cayman Chemical (Ann Arbor, MI) was dissolved in N2-purged DMSO and stored under N2 at -80 °C. The selective phosphodiesterase-resistant, cell permeable Epac-1 agonist 8-pCPT-2’-O-Me-cAMP was purchased from Biolog (Bremen, Germany). Dilutions were prepared immediately before use, with equivalent quantities of DMSO added to the appropriate controls. Rabbit polyclonal anti-Rap1, anti-Epac-1 and anti-α-tubulin antibodies were from Upstate (Charlottesville, VA).
Cell isolation and culture
The rat alveolar macrophage cell line NR8383 was obtained from the American Type Culture Collection (Manassas, VA). Resident rat AMs were obtained via ex vivo lung lavage as previously described [16] and resuspended in RPMI to a final concentration of 2 × 106 cells/ml. Cells were allowed to adhere to tissue-culture treated slides for 1 h (37°C, 5% CO2) followed by two washes with warm RPMI, resulting in > 99% of adherent cells identified as AMs by use of a modified Wright-Giemsa stain (Diff-Quik; American Scientific Products, McGraw Park, IL) [16]. Cells were cultured overnight in RPMI containing 10% fetal bovine serum and 1% penicillin/streptomycin/amphotericin B prior to use. The following day cells were washed 2x with warm medium to remove nonadherent cells.
Microscopic phagocytosis assay
Rat AMs (105/well) were plated and cultured overnight in glass 4-well Falcon tissue culture slides (BD, Franklin Lakes, NJ) as described above. Cells were preincubated with compounds of interest at the concentrations and times indicated in figure legends prior to the addition of phagocytic targets. Superparamagnetic, polystyrene beads (2.8 μM diameter) covalently opsonized with affinity purified sheep anti-mouse IgG (Dynabeads M-280, Dynal Biotech, Oslo, Norway) were used to study FcγR-mediated phagocytosis [17]. Beads were suspended in sterile PBS containing 0.1% bovine serum albumin and added at a ratio of 4 beads per cell for the times indicated in figure legends. Slides were then rinsed with warm PBS to remove uningested beads and processed for immunofluorescent staining as described below.
Immunofluorescent staining and fluorescence microscopy
Cells were cultured on chamber slides, fixed with 4% paraformaldehyde at room temperature for 30 min, permeabilized with 0.3% Triton X-100 in PBS, then blocked with 1% BSA in PBS containing non-immune goat serum. Rabbit anti-Epac-1 and rabbit anti-Rap1 (Abcam, Cambridge, MA) and rabbit anti-GAPDH (Santa Cruz Biotechnology, Santa Cruz, CA) antibodies were prepared in 1% BSA-PBS (1:200) and applied for 1 hour at 37 °C. Mounts were washed three times with 1% BSA-PBS and rhodamine-conjugated goat anti-rabbit secondary (1:200) was added for 1 hour at 37 °C. After washing three times, preparations were mounted using Vectashield mounting media containing 4’,6-diamidino-2-phenylindole (DAPI, Vector Laboratories). Fluorescence was visualized with a Nikon Labophot 2 microscope equipped for epifluorescence.
Phagosome isolation and immunoblot analysis
Macrophages (3 × 106/well) plated in a 6-well dish were washed twice with PBS, placed on ice for 10 min to synchronize phagocytosis, then incubated at 37°C for 10 min with 1 μM PGE2 prior to the addition of 10:1 IgG-conjugated, 3 μm, paramagnetic beads (Dynal-Invitrogen Carlsbad, California). Cells were then incubated at 37°C and phagosomes recovered at intervals to obtain early, intermediate and late phagosomes. After [18, 19], to isolate IgG-bead-containing phagosomes, macrophages were rinsed twice in PBS and scraped into 0.5 ml/dish ice-cold homogenization buffer (250 mM sucrose, 10 mM HEPES, 1 mM EDTA, pH 7.2; protease inhibitors and 1% Triton X-100). AMs were lysed during 30 min incubation on ice and the beads/phagosomes were isolated from cellular debris using a magnet (Qiagen, Valencia, CA) and washed twice in 0.5 ml of homogenization buffer without Triton X-100. Phagosomal proteins were removed from the beads by sonication (2 min with cooling on ice) followed by boiling for 3 min. The beads were collected with the magnet, then counted with a hemacytometer, and the solubilized material from an equal number of phagosomes from each experimental condition was used as a source of phagosomal proteins for subsequent electrophoresis and immunoblot analysis. Plasma membrane contamination was determined by quantitating the integral plasma membrane protein CD45 (data not shown). For immunoblot analysis, equal volumes of proteins were mixed with loading buffer (50 mM Tris-HCl (pH 6.8), 2% SDS, 100 mM DTT, 10% glycerol, and 0.1% bromophenol blue), boiled, applied to 8% SDS-polyacrylamide gels and subjected to electrophoresis. The separated proteins were transferred to nitrocellulose membranes in Trans-blot SD-Semidry Transfer Cells (Bio-Rad; 15 min at 15 mV). After transfer, the membranes were incubated in TBST buffer (150 mM NaCl, 20 mM Tris, 0.01% Tween 20 (pH 7.4)) containing 5% fat-free dry milk. The blot was probed with primary antibodies (as described above, or with antibody against the late phagosome marker flotillin (BD Biosciences, Franklin Lakes, NJ)), washed three times with TBST, and incubated with 1/5000 dilutions of peroxidase-conjugated monoclonal anti-rabbit IgG for 1 h at room temperature. The immunocomplexed peroxidase-labeled Abs were visualized by an ECL chemiluminescence kit following the manufacturer’s instruction (Amersham Biosciences). Densitometry was performed using ImageJ software.
Results
To determine the localization of Epac-1 in AMs before and during FcγR-mediated phagocytosis, rat AMs were adhered to glass chamber slides and incubated without or with IgG opsonized beads for 60 min. Immunofluorescent staining for Epac-1 in AMs without beads revealed strong membrane staining around the nucleus and throughout the cell body (Fig. 1A). Lighter staining for Epac-1 was also indicated at the plasma membrane at the margins of adherent, spread AMs. As Epac-1 has been reported to move during cell cycling [20], we considered the possibility that Epac-1 might move during phagocytosis. In AMs incubated with IgG opsonized beads, the fluorescent distribution of Epac-1 remained brightest on perinuclear membranes and on membranes throughout the cell body (Fig. 1B, C). Beads captured at the cell margin were outlined by Epac-1-positive plasma membrane (Fig 1B). IgG-opsonized beads that were not associated with cells did not fluoresce when excited with blue light (not shown), used to excite FITC-linked Epac-1 here.
Fig. 1. Subcellular localization of Epac-1 and Rap1 in AMs following phagocytosis of opsonized beads.
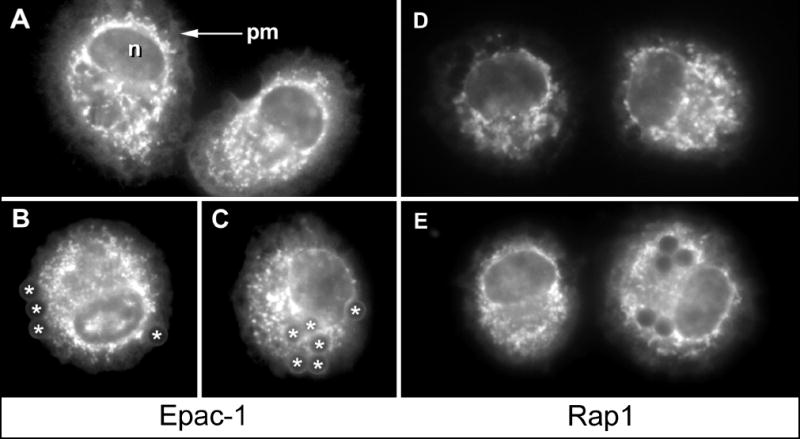
Rat AMs were incubated without (A,D) or with (B, C, E) opsonized beads for 60 min, then fixed and stained for Epac-1 (A-C) or Rap1 (D, E). Image E shows a cell that contains no beads (left) next to a cell that has engulfed five beads (right). n, nucleus; pm, plasma membrane; *, phagocytosed bead. Results are from one experiment and are representative of three.
Adherent AMs incubated without beads stained positively for Rap1 in a punctate pattern throughout the cell (Fig. 1D). Fluorescence at the plasma membrane was weak, as was staining at the nuclear envelope. Rap1 localization can be altered by phorbol esters [21], by growth factors [22] or during neutrophil degranulation [23], raising the possibility that Rap1 localization might change during FcγR-mediated phagocytosis. In phagocytosing AMs, Rap1 remained distributed in a punctate fashion throughout the cell, with light staining at the plasma membrane and nuclear envelope (Fig. 1E). Closer inspection of phagosomes showed that antibody against Epac-1 positively stained the membranes of phagosomes surrounding captured beads (Fig. 2 A, C, D). Although other brightly fluorescing membranes appeared to be associated with the phagosomes (Fig. 2C), it was not clear whether these bright membranes had actively moved to the phagosome or if the phagosome moved from the cell surface toward these membranes. Rap1-positive membranes around phagocytosed particles were also observed (Fig. 2B, E), but this staining was consistently weaker than that for Epac-1. No significant change in the bright punctate pattern of Rap1 was evident following phagocytosis and punctate regions did not appear to significantly associate with phagosomes. For both Epac-1 and Rap1, then, plasma membrane-associated protein was observed to surround the captured target in the early phagosome.
Fig. 2. Detail of subcellular localization of Epac-1 or Rap1 near phagocytic cup.
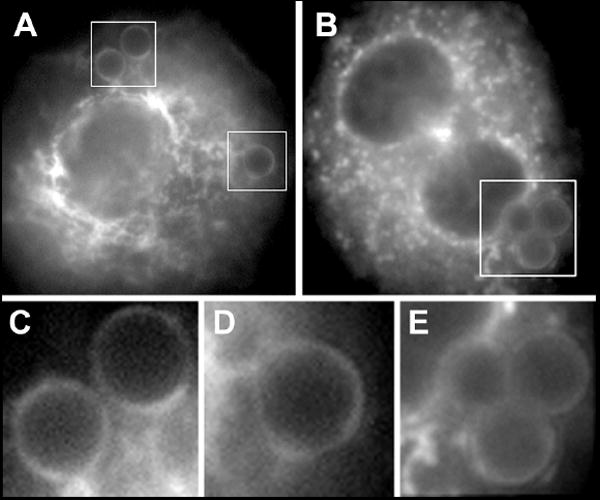
Following phagocytosis of opsonized beads for 60 min, AMs were fixed and stained for Epac-1 (A, C, D) or Rap1 (B, E). Images in C and D are details from A, and E is a detail of the indicated area in image B.
The rat AM cell line NR8383 was utilized to further investigate the localization of Epac-1 and Rap1 in phagocytosing macrophages. As in primary AMs, Epac-1 was most abundant on membranes in the cell body of resting NR8383 cells and less abundant on plasma membrane (Fig. 3A). These membranes were more tubular than those observed in primary AMs. Opsonized beads, captured within 15 min by NR8383 cells, were outlined by positive Epac-1 staining (Fig. 3B). As seen in AMs at 60 min, brighter Epac-1 membranes could be found near the lighter Epac-1 staining that surrounded the beads (Fig. 3B, inset). With longer incubation times, internalized beads were surrounded by membranes that were strongly positive for Epac-1 (Fig. 3C). In NR8383 cells, Rap1 was largely diffuse with some punctate staining (Fig. 3D); no Rap1 was associated with internalized phagosomes. For comparison, the soluble protein GAPDH appeared soluble and diffusely distributed throughout the cells (Fig. 3D). Importantly, staining for GAPDH was evident around peripheral beads (arrow, Fig. 3D), indicating that soluble proteins will also be found to be associated with these beads.
Fig. 3. Subcellular localization of Epac-1, Rap1 and GAPDH in the AM cell line NR8383.
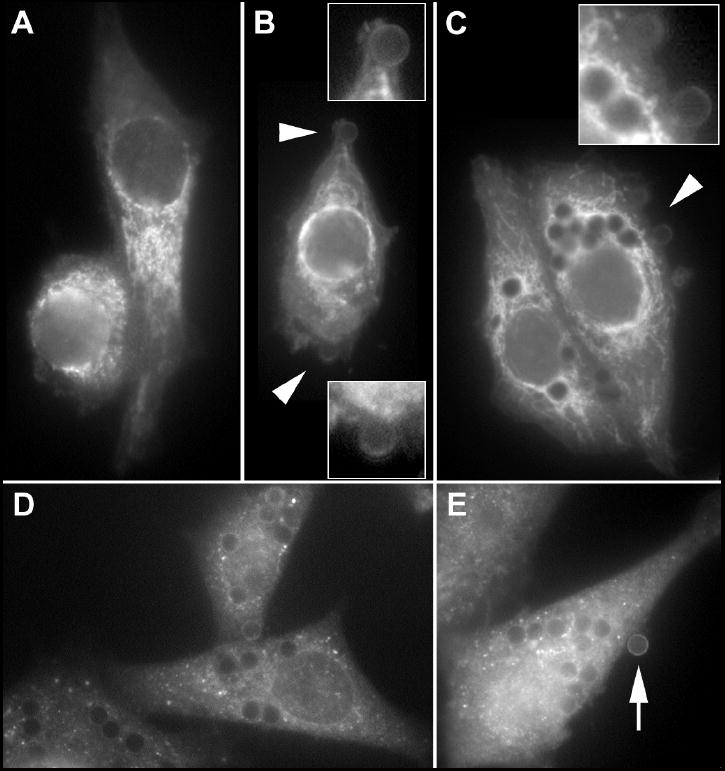
Cells were incubated without beads (A) or with opsonized beads for 15 (B) or 60 (C-E) min. Insets in (B) and (C) are details of the regions indicated by arrowheads. Arrow in (E) indicates peripherally bound bead with positive staining for GAPDH.
As noted above, Epac-1 is known to be involved in the inhibition of FcγR-mediated phagocytosis by cAMP-stimulating compounds such as PGE2 [4, 10]. Treatment of AMs with 1 μM PGE2 induced a time-dependent redistribution of Epac-1 to the nuclear envelope, with changes apparent within 30 min and more pronounced by 60 min (Fig. 4, left). Also, Epac-1 remained prominently associated with perinuclear membranes as macrophages spread in response to PGE2. In contrast, PGE2 had no discernible effect on the distribution of Rap1 in AMs (Fig. 4, right). Thus, in addition to being distributed at different sites, with Epac-1 on tubular membranes and Rap1 on punctate membranes, only Epac-1 responds to PGE2 by accumulating on the nuclear envelope and perinuclear membranes.
Fig. 4. Subcellular localization of Epac-1 and Rap1 in AMs treated with PGE2.
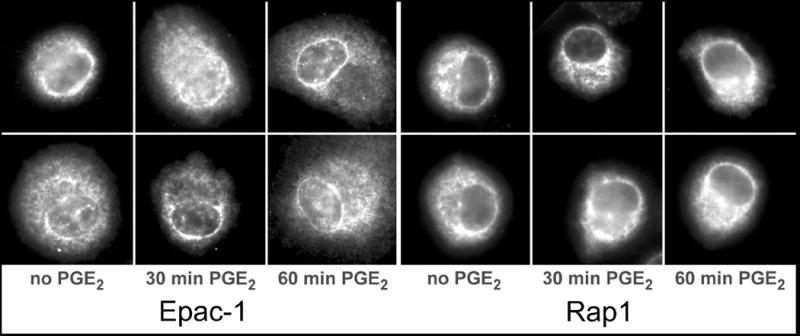
Cells were treated with 1 μM PGE2 for 0, 30 or 60 min, then fixed and stained for Epac-1 (left) or Rap1 (right) as described in Materials and Methods. Images are from one experiment and are representative of three independent experiments.
To determine the effect of PGE2 pretreatment on Epac-1 and Rap1 localization during phagocytosis, AMs were incubated with IgG opsonized beads following the addition of PGE2 Again, Epac-1 was commonly found on the nuclear envelope of PGE2-pretreated AMs, even after phagocytosis (Fig. 5A, B). Peripheral phagosomes that formed on PGE2-pretreated AMs were clearly delimited by Epac-1-containing membrane, as was seen in AMs not receiving PGE2 (Figs. 1, 2). Although the majority of phagocytosed beads appear to be restricted to the periphery of PGE2-pretreated AMs (Fig. 5), rather than moving deeper into the lysosomal field as seen in untreated AMs (Figs 1-3), this was an inconsistent finding. The distribution of Rap1 in PGE2-pretreated AMs paralleled that seen in AMs unexposed to PGE2, with abundant labeling of punctate membrane structures and weaker staining of plasma membrane, in the absence or presence of phagocytosis (Fig. 5). Thus, activation of FcγR did not substantially change the distribution of Epac-1 and Rap1 in PGE2 treated AMs, as assessed by immunofluorescent staining.
Fig. 5. Localization of Epac-1 and Rap1 following phagocytosis of opsonized beads in AMs pretreated with PGE2.
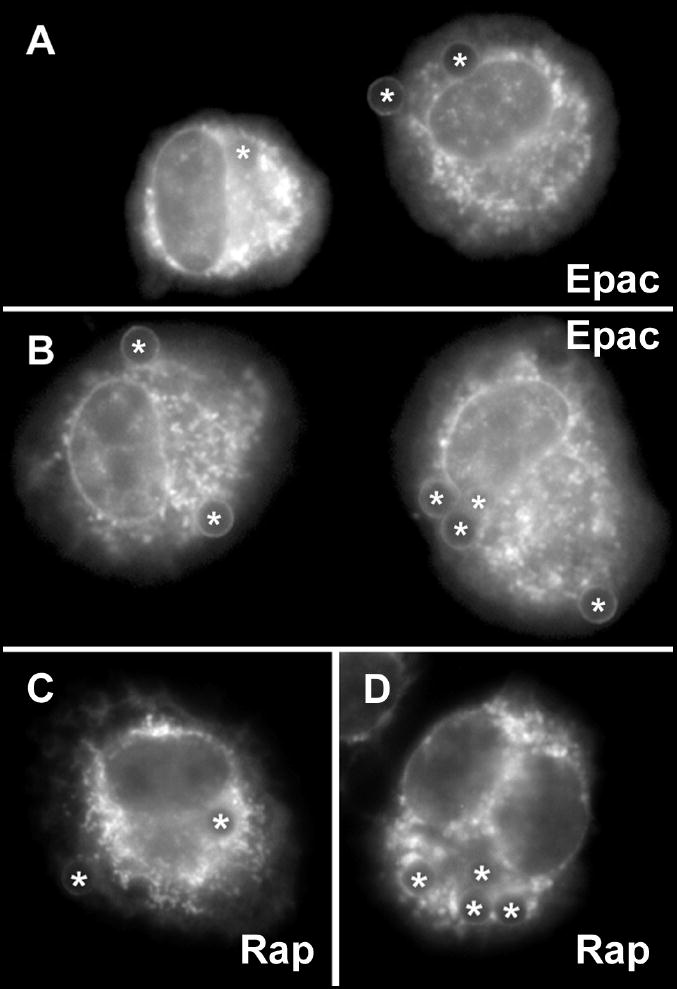
Cells were pretreated with 1 μM PGE2 for 15 min and then incubated with opsonized beads for 60 min. Two fields from independent experiments are presented for Epac-1 (A, B) and Rap1 (C, D)). Asterisks (*) indicate phagosomes containing beads.
To more clearly determine if PGE2 altered the association of Epac-1 or Rap1 with intracellular phagosomes, we isolated phagosomes using magnetic, IgG-opsonized polystyrene microbeads at various times. Flotillin was used as a marker of phagosome maturation, as it has been identified on late phagosomes [24]. As shown by immunoblot, flotillin levels increased over time in phagosomes from untreated AMs (Fig. 6). Consistent with our previously published results [2], pretreatment of AMs with 1 μM PGE2 for 5 min produced a significant decrease in the number of captured microbeads/phagosomes (data not shown). However, when equal numbers of untreated and PGE2-treated microbead/phagosomes were analyzed, flotillin increased similarly in both conditions, suggesting that PGE2 treatment did not alter the rate of phagosome maturation. However, Epac-1 was not detected on phagosomes of untreated AMs, but was readily detected on late phagosomes of PGE2 treated cells (Fig. 6). We observed a similar increase in Epac-1 association with late phagosomes when cells were treated with the selective Epac-1 agonist 8-pCPT-2’-O-Me-cAMP (1 mM) as we observed with PGE2 (data not shown). We were unable to detect significant quantities of Rap1 using this technique, suggesting that the association between Rap1 and the phagosomal membrane is loose and/or transient.
Fig. 6. Trafficking of Epac-1 to the phagosome following PGE2 treatment.
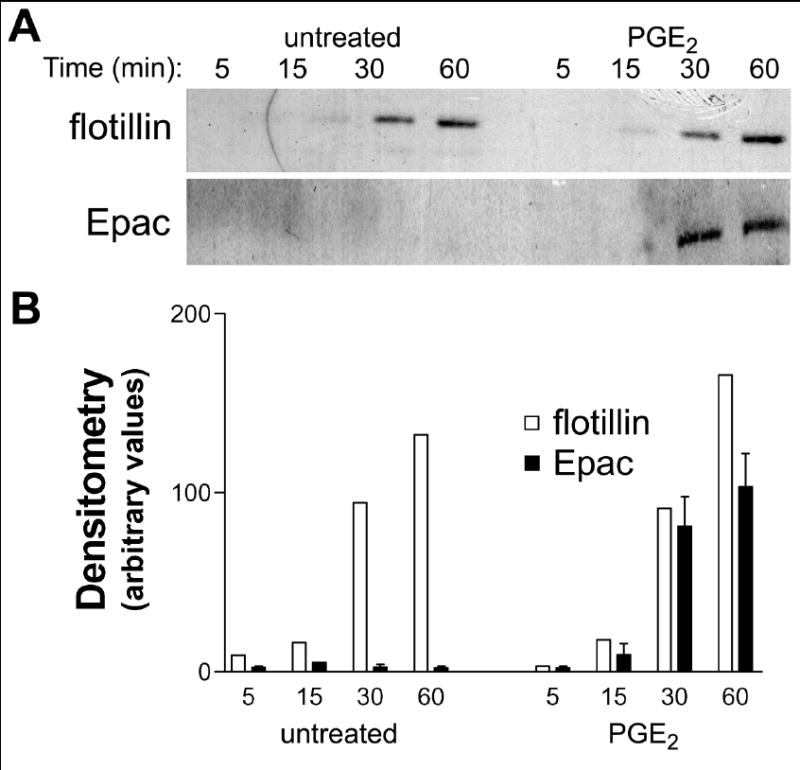
Rat AMs were pretreated for 5 min with vehicle (DMSO) or PGE2 (1 uM), followed by challenge with IgG-opsonized magnetic beads. Phagosomal membranes were harvested at the times noted and (A) assayed by immunoblot for the late phagosomal marker flotillin and for Epac-1. Densitometry (B) presents mean (+SE) data from independent experiments.
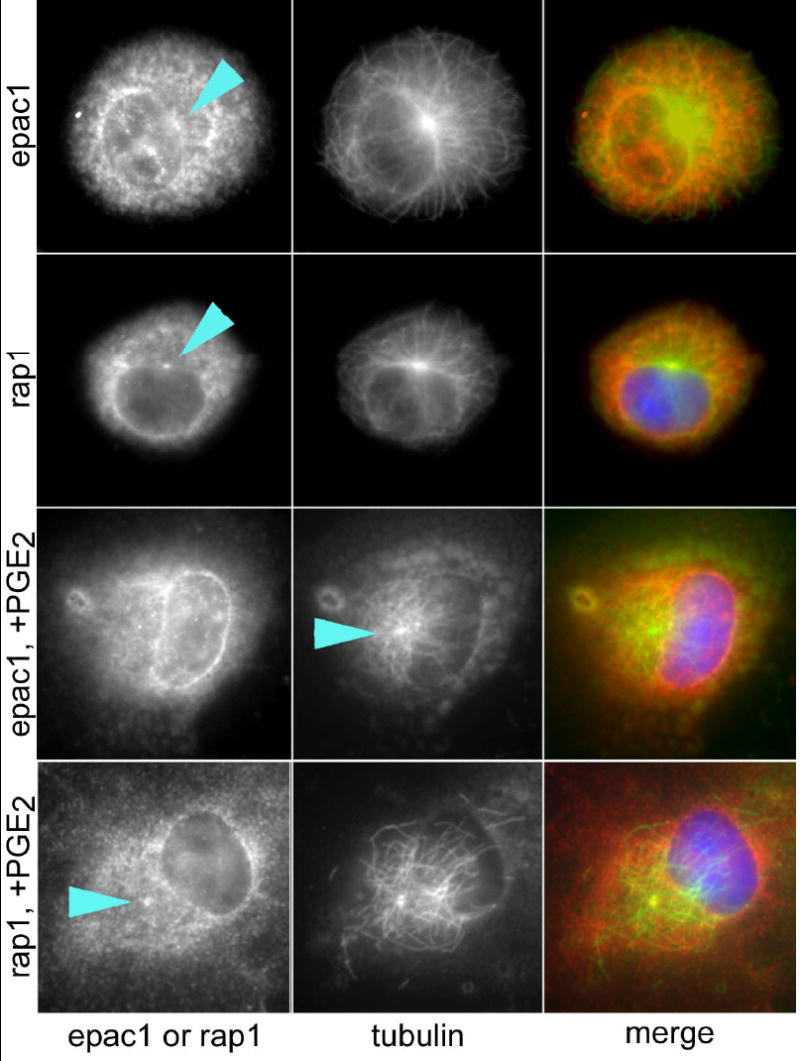
Finally, Epac-1 has also been found to be associated with microtubules or centrosomes [20]. Dual staining of untreated AMs for these proteins and α-tubulin indicated co-localization of both Epac-1 and Rap1 at microtubule organizing centers (MTOC; Fig. 7). Pretreatment of AMs with PGE2 (1 μM) for 60 min resulted in loss of Epac-1 staining at the MTOC, whereas Rap1 remained at the MTOC.
Fig. 7. Trafficking of Epac-1 to the phagosome following PGE2 treatment.
Rat AMs were pretreated for 5 min with vehicle (DMSO) or PGE2 (1 μM), followed by challenge with IgG-opsonized magnetic beads. Phagosomal membranes were harvested at the times noted and (A) assayed by immunoblot for the late phagosomal marker flotillin and for Epac-1. Densitometry (B) presents mean (±SE) data from independent experiments.
Co-localization of Epac-1 and Rap1 with MTOCs. AMs were either untreated or treated for 60 min with 1 μM PGE2 before fixing and staining for Epac-1 or Rap-1 and α-tubulin. Individual stains (left, center) are provided in black and white for optimum presentation; merged images (right) use false coloring to who Epac-1 or Rap1 as red and α-tubulin as green. The positioning of the MTOC is indicated by an arrowhead. The Epac-1 + PGE2 image is identical to that shown in Fig. 5. Results are from one experiment and are representative of three independent experiments.
Discussion
Although it is recognized that cAMP signaling, as is triggered by PGE2 in macrophages, inhibits FcγR-mediated phagocytosis, the critical role of Epac has only recently been demonstrated. In this study, we sought to determine the subcellular localization of Epac and one of its major downstream targets, Rap1, before and after PGE2 treatment. We find that both proteins are distributed on a variety of subcellular sites, but that only Epac-1 redistributes in response to PGE2.
Epac-1 localization
Epac-1 is known to have a Dishevelled, Egl-10, Pleckstrin (DEP) domain, which is involved in membrane localization [25]. In fibroblast-like COS-7 cells, Epac-1 was found to be membrane-associated with a punctate distribution, with many of these co-localizing with a mitochondrial marker, as well as some localization to nuclear membranes [20]. There is also evidence that, in COS-7 cells, Epac-1 associates with the mitotic spindle and centrosomes during metaphase, that elevation of intracellular cAMP levels directs Epac-1 to nuclear membranes and that reduction of cAMP favors cytoskeleton association of Epac-1 [20].
To our knowledge, the spatial localization of endogenous Epac-1 has not previously been studied in phagocytic cells of the innate immune system. In resting AMs, Epac-1 is most abundantly distributed on punctate membranes throughout the cell body, including in the perinuclear region, with additional lower levels evident on the plasma membrane. A similar distribution for Epac-1 was observed in the NR8383 macrophage cell line, although there were additional tubular structures that stained strongly for Epac-1. These distributions were reminiscent of the tubular and spherical lysosomes that have been described in macrophages [26, 27]. In addition to these sites, Epac-1 staining also overlapped with α-tubulin staining at the MTOC, but not with microtubules away from the MTOC.
Following cell treatment with PGE2, Epac-1 redistributed to the nuclear envelope and accumulated on late phagosomes. Both processes were relatively slow, requiring 30-60 min to produce changes that were observable by the methods used here. It is notable that Epac-1 and Rap1 appear to be associated with phagosomes from cells not exposed to PGE2 when examined by immunofluorescence microscopy (Fig. 2), however we failed to observe this association when phagosomes were isolated from untreated cells and analyzed by Western blot (Fig. 6). We speculate that the association between Epac-1 and phagosomes seen by microscopy in non-PGE2 treated cells is not a tight association, and the Epac-1 is therefore lost during the physical isolation of bead-containing phagosomes. Under the influence of PGE2, however, the Epac-1-phagosome association might become tighter and is not disrupted by phagosome isolation. This finding is further supported by our observing the soluble protein GAPDH in association with bead-containing phagosomes when observed by immunofluorescent staining (Fig. 3D) but not following phagosome isolation (not shown). The immunofluorescence data suggest that cytosolic proteins might non-specifically (and perhaps loosely) associate with phagosomes during the internalization of IgG-opsonized targets. This is in contrast to the finding that Epac-1 becomes specifically (and perhaps tightly) associated with phagosomes during Fc receptor mediated phagocytosis when cells are exposed to PGE2 (or 8-pCPT-2’-O-Me-cAMP).
Although Epac-1 redistribution in response to PGE2 may have begun earlier than 30 min, these changes may be too slow to account for the inhibitory effects of Epac-1 activation on phagocytosis. On the other hand, the striking recruitment of Epac-1 to maturing phagosomes following PGE2 treatment places this signaling protein at a site that could be important for its known capacity to inhibit both H2O2 production and bacterial killing [4]. Also, although Epac-1 was typically co-localized with MTOCs in untreated AMs, it did not co-localize with MTOCs following PGE2 treatment. We find no evidence that Epac-1 associates with microtubules away from the MTOC in AMs. The loss of Epac-1 from MTOCs following PGE2 treatment could possible reduce the ability for Epac-1 to regulate microtubule function and this might play a role in altering phagosome internalization.
Rap1 localization
Rap1 has been localized to a wide variety of membranes in different cell types. These include the golgi apparatus in rat-1 fibroblasts [28] and COS-7 cells [29], on secretory granules in parotid gland [30], on zymogen granules in pancreatic acinar cells [31], on specific granules in human neutrophils [21, 23] on endosomes of fibroblasts and T cells [32] and COS-7 cells [29], in late endosomes in C2 (muscle) cells [33] as well as fibroblasts [14], on phagosomes in J774 macrophages [14] and in plasma membrane of fibroblasts and T cells [32]. Furthermore, Rap1 reportedly moves to the nucleus of cancer cells in response to growth factors [22] and to the plasma membrane of neutrophils following degranulation [21, 23].
In this study, we localized Rap1 in primary AMs. Rap1 was, like Epac-1, most abundant on punctate membranes throughout the cell, with additional staining evident on the plasma membrane. Although positive staining for Rap1 on late phagosomes was apparent by immunofluorescent staining, Rap1 was not detected by immunoblot analysis of isolated, mature phagosomes. Rap1 was also found, by immunofluorescent staining, to co-localize with MTOCs. In our hands, Rap1 did not appear to move, either to the nucleus or to the plasma membrane, under any of the conditions examined here. Perhaps most surprisingly, we found little evidence that Rap1 accumulated on maturing phagosomes, as Epac-1 did. This raises the possibility that Epac-1 signals through pathways that are independent of Rap1. In addition, it is possible that the inhibition of killing of phagocytosed bacteria in AMs treated with PGE2, which is known to involve Epac-1 activation [4], does not require Rap1 activation. There is some precedent for Rap-independent Epac-1 action [34], though alternative targets for Epac-1 have not been convincingly defined.
In conclusion, we sought to determine the subcellular distribution of Epac-1 and its primary target, Rap1, in primary phagocytes, since activation of Epac-1 by PGE2 significantly inhibits FcγR-mediated phagocytosis in these cells. We found that both enzymes were abundant on intracellular membranes that resembled the tubular and spherical lysosomes of macrophages, with lesser amounts also on plasma membrane and MTOCs. Treatment with PGE2 led to a redistribution of Epac-1, but not Rap1, to the nuclear envelope and late phagosomes. The separation of effects on Epac-1 and Rap1 redistribution suggests that some effects of Epac-1, particularly in killing of phagocytosed bacteria, might be independent of Rap1. Earlier signaling events through Epac-1 that result in impaired phagocytosis remain to be elucidated.
Acknowledgments
Generous support for this study was provided by the National Institutes of Health grants AI43955 (TGB), HL078727 (DMA), and HL058897 (MPG), and the American Lung Association grant RG-8909-N (DMA).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Underhill DM, Ozinsky A. Phagocytosis of microbes: complexity in action. Annu Rev Immunol. 2002;20:825–852. doi: 10.1146/annurev.immunol.20.103001.114744. [DOI] [PubMed] [Google Scholar]
- 2.Aronoff DM, Canetti C, Peters-Golden M. Prostaglandin E2 inhibits alveolar macrophage phagocytosis through an E-prostanoid 2 receptor-mediated increase in intracellular cyclic AMP. J Immunol. 2004;173:559–565. doi: 10.4049/jimmunol.173.1.559. [DOI] [PubMed] [Google Scholar]
- 3.Mancuso P, Standiford TJ, Marshall T, Peters-Golden M. 5-Lipoxygenase reaction products modulate alveolar macrophage phagocytosis of Klebsiella pneumoniae. Infect Immun. 1998;66:5140–5146. doi: 10.1128/iai.66.11.5140-5146.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aronoff DM, Canetti C, Serezani CH, Luo M, Peters-Golden M. Cutting Edge: Macrophage Inhibition by Cyclic AMP (cAMP): Differential Roles of Protein Kinase A and Exchange Protein Directly Activated by cAMP-1. J Immunol. 2005;174:595–599. doi: 10.4049/jimmunol.174.2.595. [DOI] [PubMed] [Google Scholar]
- 5.Serezani CH, Aronoff DM, Jancar S, Mancuso P, Peters-Golden M. Leukotrienes enhance the bactericidal activity of alveolar macrophages against Klebsiella pneumoniae through the activation of NADPH oxidase. Blood. 2005;106:1067–1075. doi: 10.1182/blood-2004-08-3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aronoff DM, Carstens JK, Chen GH, Toews GB, Peters-Golden M. Differences between macrophages and dendritic cells in the cyclic AMP dependent regulation of lipopolysaccharide-induced cytokine and chemokine synthesis. J Interferon Cytokine Res. 2006;26:827–833. doi: 10.1089/jir.2006.26.827. [DOI] [PubMed] [Google Scholar]
- 7.Breyer RM, Bagdassarian CK, Myers SA, Breyer MD. Prostanoid receptors: subtypes and signaling. Annu Rev Pharmacol Toxicol. 2001;41:661–690. doi: 10.1146/annurev.pharmtox.41.1.661. [DOI] [PubMed] [Google Scholar]
- 8.Nambu M, Morita M, Watanabe H, Uenoyama Y, Kim KM, Tanaka M, Iwai Y, Kimata H, Mayumi M, Mikawa H. Regulation of Fc gamma receptor expression and phagocytosis of a human monoblast cell line U937. Participation of cAMP and protein kinase C in the effects of IFN-gamma and phorbol ester. J Immunol. 1989;143:4158–4165. [PubMed] [Google Scholar]
- 9.Thomas CA, Weinberger OK, Ziegler BL, Greenberg S, Schieren I, Silverstein SC, El Khoury J. Human immunodeficiency virus-1 env impairs Fc receptor-mediated phagocytosis via a cyclic adenosine monophosphate-dependent mechanism. Blood. 1997;90:3760–3765. [PubMed] [Google Scholar]
- 10.Bryn T, Mahic M, Enserink JM, Schwede F, Aandahl EM, Tasken K. The cyclic AMP-Epac1-Rap1 pathway is dissociated from regulation of effector functions in monocytes but acquires immunoregulatory function in mature macrophages. J Immunol. 2006;176:7361–7370. doi: 10.4049/jimmunol.176.12.7361. [DOI] [PubMed] [Google Scholar]
- 11.de Rooij J, Zwartkruis FJ, Verheijen MH, Cool RH, Nijman SM, Wittinghofer A, Bos JL. Epac is a Rap1 guanine-nucleotide-exchange factor directly activated by cyclic AMP. Nature. 1998;396:474–477. doi: 10.1038/24884. [DOI] [PubMed] [Google Scholar]
- 12.Kawasaki H, Springett GM, Mochizuki N, Toki S, Nakaya M, Matsuda M, Housman DE, Graybiel AM. A family of cAMP-binding proteins that directly activate Rap1. Science. 1998;282:2275–2279. doi: 10.1126/science.282.5397.2275. [DOI] [PubMed] [Google Scholar]
- 13.Makranz C, Cohen G, Reichert F, Kodama T, Rotshenker S. cAMP cascade (PKA, Epac, adenylyl cyclase, Gi, and phosphodiesterases) regulates myelin phagocytosis mediated by complement receptor-3 and scavenger receptor-AI/II in microglia and macrophages. Glia. 2006;53:441–448. doi: 10.1002/glia.20303. [DOI] [PubMed] [Google Scholar]
- 14.Pizon V, Desjardins M, Bucci C, Parton RG, Zerial M. Association of Rap1a and Rap1b proteins with late endocytic/phagocytic compartments and Rap2a with the Golgi complex. J Cell Sci. 1994;107(Pt 6):1661–1670. doi: 10.1242/jcs.107.6.1661. [DOI] [PubMed] [Google Scholar]
- 15.Caron E, Self AJ, Hall A. The GTPase Rap1 controls functional activation of macrophage integrin alphaMbeta2 by LPS and other inflammatory mediators. Curr Biol. 2000;10:974–978. doi: 10.1016/s0960-9822(00)00641-2. [DOI] [PubMed] [Google Scholar]
- 16.Peters-Golden M, Thebert P. Inhibition by methylprednisolone of zymosan-induced leukotriene synthesis in alveolar macrophages. Am Rev Respir Dis. 1987;135:1020–1026. doi: 10.1164/arrd.1987.135.5.1020. [DOI] [PubMed] [Google Scholar]
- 17.Allen LA, Aderem A. Molecular definition of distinct cytoskeletal structures involved in complement- and Fc receptor-mediated phagocytosis in macrophages. J Exp Med. 1996;184:627–637. doi: 10.1084/jem.184.2.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lutz DA, Chen XM, McLaughlin BJ. Isolation of the phagocytic compartment from macrophages using a paramagnetic, particulate ligand. Anal Biochem. 1993;214:205–211. doi: 10.1006/abio.1993.1478. [DOI] [PubMed] [Google Scholar]
- 19.Morrissette NS, Gold ES, Guo J, Hamerman JA, Ozinsky A, Bedian V, Aderem AA. Isolation and characterization of monoclonal antibodies directed against novel components of macrophage phagosomes. J Cell Sci. 1999;112(Pt 24):4705–4713. doi: 10.1242/jcs.112.24.4705. [DOI] [PubMed] [Google Scholar]
- 20.Qiao J, Mei FC, Popov VL, Vergara LA, Cheng X. Cell cycle-dependent subcellular localization of exchange factor directly activated by cAMP. J Biol Chem. 2002;277:26581–26586. doi: 10.1074/jbc.M203571200. [DOI] [PubMed] [Google Scholar]
- 21.Quinn MT, Mullen ML, Jesaitis AJ, Linner JG. Subcellular distribution of the Rap1A protein in human neutrophils: colocalization and cotranslocation with cytochrome b559. Blood. 1992;79:1563–1573. [PubMed] [Google Scholar]
- 22.Mitra RS, Zhang Z, Henson BS, Kurnit DM, Carey TE, D’Silva NJ. Rap1A and rap1B ras-family proteins are prominently expressed in the nucleus of squamous carcinomas: nuclear translocation of GTP-bound active form. Oncogene. 2003;22:6243–6256. doi: 10.1038/sj.onc.1206534. [DOI] [PubMed] [Google Scholar]
- 23.Maridonneau-Parini I, de Gunzburg J. Association of rap1 and rap2 proteins with the specific granules of human neutrophils. Translocation to the plasma membrane during cell activation. J Biol Chem. 1992;267:6396–6402. [PubMed] [Google Scholar]
- 24.Dermine JF, Duclos S, Garin J, St-Louis F, Rea S, Parton RG, Desjardins M. Flotillin-1-enriched lipid raft domains accumulate on maturing phagosomes. J Biol Chem. 2001;276:18507–18512. doi: 10.1074/jbc.M101113200. [DOI] [PubMed] [Google Scholar]
- 25.de Rooij J, Rehmann H, van Triest M, Cool RH, Wittinghofer A, Bos JL. Mechanism of regulation of the Epac family of cAMP-dependent RapGEFs. J Biol Chem. 2000;275:20829–20836. doi: 10.1074/jbc.M001113200. [DOI] [PubMed] [Google Scholar]
- 26.Swanson J, Bushnell A, Silverstein SC. Tubular lysosome morphology and distribution within macrophages depend on the integrity of cytoplasmic microtubules. Proc Natl Acad Sci U S A. 1987;84:1921–1925. doi: 10.1073/pnas.84.7.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsang AW, Oestergaard K, Myers JT, Swanson JA. Altered membrane trafficking in activated bone marrow-derived macrophages. J Leukoc Biol. 2000;68:487–494. [PubMed] [Google Scholar]
- 28.Beranger F, Goud B, Tavitian A, de Gunzburg J. Association of the Ras-antagonistic Rap1/Krev-1 proteins with the Golgi complex. Proc Natl Acad Sci U S A. 1991;88:1606–1610. doi: 10.1073/pnas.88.5.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nomura K, Kanemura H, Satoh T, Kataoka T. Identification of a novel domain of Ras and Rap1 that directs their differential subcellular localizations. J Biol Chem. 2004;279:22664–22673. doi: 10.1074/jbc.M314169200. [DOI] [PubMed] [Google Scholar]
- 30.D’Silva NJ, DiJulio DH, Belton CM, Jacobson KL, Watson EL. Immunolocalization of rap1 in the rat parotid gland: detection on secretory granule membranes. J Histochem Cytochem. 1997;45:965–973. doi: 10.1177/002215549704500706. [DOI] [PubMed] [Google Scholar]
- 31.Chen X, Walker AK, Strahler JR, Simon ES, Tomanicek-Volk SL, Nelson BB, Hurley MC, Ernst SA, Williams JA, Andrews PC. Organellar proteomics: analysis of pancreatic zymogen granule membranes. Mol Cell Proteomics. 2006;5:306–312. doi: 10.1074/mcp.M500172-MCP200. [DOI] [PubMed] [Google Scholar]
- 32.Bivona TG, Wiener HH, Ahearn IM, Silletti J, Chiu VK, Philips MR. Rap1 up-regulation and activation on plasma membrane regulates T cell adhesion. J Cell Biol. 2004;164:461–470. doi: 10.1083/jcb.200311093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pizon V, Cifuentes-Diaz C, Mege RM, Baldacci G, Rieger F. Expression and localization of RAP1 proteins during myogenic differentiation. European journal of cell biology. 1996;69:224–235. [PubMed] [Google Scholar]
- 34.Hochbaum D, Tanos T, Ribeiro-Neto F, Altschuler D, Coso OA. Activation of JNK by Epac is independent of its activity as a Rap guanine nucleotide exchanger. J Biol Chem. 2003;278:33738–33746. doi: 10.1074/jbc.M305208200. [DOI] [PubMed] [Google Scholar]


