Abstract
Increased generation of reactive oxygen species (ROS) has been observed in cancer, degenerative diseases, and other pathological conditions. ROS can stimulate cell proliferation, promote genetic instability, and induce adaptive responses that enable cancer cells to maintain their malignant phenotypes. However, when cellular redox balance is severely disturbed, high levels of ROS may cause various damages leading to cell death. The studies of ROS effects on biological systems, their underlying mechanisms and therapeutic implications largely depend on proper experimental models. Here we review several in vitro and in vivo models for ROS research.
Introduction
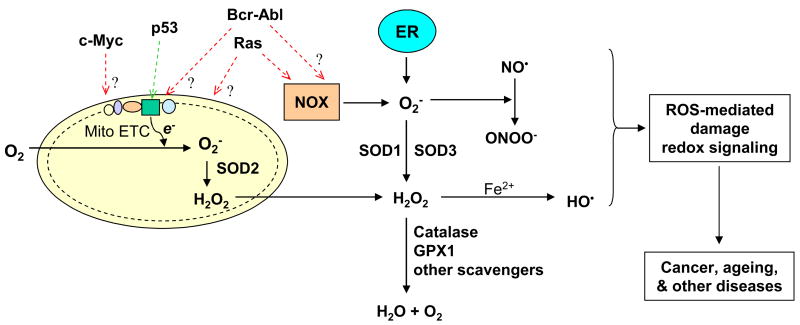
Reactive oxygen species are derived from oxygen. Reduction of molecular oxygen is a principal mechanism for generation of superoxide (O2−), which can then be converted to other ROS. In mammalian cells, the mitochondrial electron transport chain is a major site of cellular ROS generation, where the electrons escaping from their transport complexes react with oxygen to form O2− [1]. Superoxide can be rapidly converted to H2O2 by superoxide dismutases (SODs). In the presence of transitional metals, H2O2 can generate hydroxyl radicals. ROS are highly reactive and readily cause oxidative modifications to biomolecules. Due to a short half-life and limited diffusion distance, most ROS cause damage locally near the sites of production. However, H2O2 has a relatively long half-life and can travel long distances, which enable this molecule to function as a second messenger in signal transduction pathways and to cause damage at distant sites including nuclear DNA. Under physiological conditions, ROS are maintained at proper levels by a balance between its generation and elimination. As illustrated in Figure 1, the steady state of ROS would readily change if any step in the ROS production or scavenging is disturbed. An increase in ROS generation, a decrease in antioxidant capacity, or both will lead to oxidative stress. Based on this principle, experimental models of ROS stress have been created by disrupting a specific process in redox balance. For instance, several in vitro and in vivo models show that oncogenic signals such as Ras and Bcr-Abl promote ROS generation, contributing to oxidative stress in cancer cells. Suppression of ROS elimination by knocking out SODs also causes oxidative stress, leading to an increase in cancer risk.
Figure 1. ROS metabolism and major molecules that affect redox balance.
Major sites of cellular ROS generation include the mitochondrial electron transport chain (Mito ETC), the endoplasmic reticulum (ER) system, and the NAD(P)H oxidase (NOX) complex. Several major enzymes catalyzing ROS generation or elimination that have been used to create experimental models are indicated in blue. Oncogenic molecules (Ras, Bcr-Abl, c-Myc) and the tumor suppressor p53 have significant effects on ROS generation, with potential target sites indicated by dotted arrows. The question mark (?) indicates unclear mechanism. Red text indicates promotion of ROS generation; green indicates decrease of ROS production. In experimental model systems, each molecule can be knocked out, mutated, or overexpressed in vitro or in vivo. GPX1, glutathione peroxidase 1; HO•, hydroxyl radical; NO•, nitric oxide; ONOO-, peroxy nitrite, SOD, superoxide dismutase.
In vitro models
Ras models for ROS study
Ras functions as a guanine nucleotide triphosphatase (GTPase) in a large network of intracellular signaling pathways. Constitutive Ras activation due to overexpression or mutations is common in human cancers. Although sharing high degree of sequence homology, the three different Ras genes (H-Ras, N-Ras, and K-Ras) and their mutations are tissue- and tumor-type specific. Numerous in vitro and in vivo models have been established to enable the investigation of the roles of Ras in promoting malignant transformation. An in vitro experimental model system in which NIH3T3 fibroblasts were stably transformed with the constitutively active H-RasV12 revealed that Ras activation led to elevated generation of superoxide through a pathway involving flavoprotein and Rac1 of the NOX enzyme complex [2]. Interestingly, although oncogenic Ras can transform certain immortal cells to tumorigenic cells accompanied by increased ROS generation, forced activation of Ras alone seems to provoke premature senescence in primary cells from the Eμ-N-Ras transgenic mouse [3]. Inactivation of the tumor suppressor p53 or p16INK4a and a loss of histone methyltransferase Suv39h1 may circumvent the senescence provoked by Ras, leading to cancer development [4,5]. These observations suggest that the increase in ROS triggered by Ras, induces cellular senescence, and requires additional deregulations to achieve malignant transformation.
A genetically defined in vitro model using human ovarian epithelial cells for Ras-mediated transformation was created by Liu et al [6]. This model was established by sequentially introducing hTERT and H-Rasv12 genes into human ovarian surface epithelial cell lines that had previously been transfected with SV40 T/t antigens to abrogate the p53 and Rb regulatory pathways. While hTERT conferred cell immortalization, subsequent H-Rasv12 expression enabled cells to grow on soft-agar and form tumors in immunocompromised nude mice. Tumors derived from the Ras-transformed cell lines resembled ovarian cancer histopathology [6]. Proteomic analysis revealed that the notable groups of altered proteins were enzymes involved in cellular redox metabolism [7]. Using this model system, Trachootham et al showed that the Ras-transformed cells exhibited a significant increase in ROS levels compared with the hTERT immortalized parental cells, and were highly sensitive to anticancer agents that cause further oxidative stress [8]. The in vitro models of Ras-mediated transformation provide valuable tools for investigating the roles of ROS in cancer development and therapeutic implications.
Cell models of Bcr-Abl-mediated transformation and ROS stress
The chimeric Bcr-Abl tyrosine kinase is oncogenic and responsible for chronic myelogenous leukemia (CML). In cell model systems, introduction of Bcr-Abl into primary hematopoietic cells abrogates their growth factor dependence, leading to malignant transformation [9]. Several hematopoietic cell lines including BaF3, 32Dcl3, and MO7e have been stably transfected with Bcr-Abl to investigate the relationship between oncogenic signaling and ROS generation and their roles in the leukemogenesis [10]. Increased intracellular ROS and decreased protein-tyrosine phosphatases (PTPases) were observed in Bcr-Abl transformed cells. Treatment of the Bcr-Abl expressing cells with antioxidants such as N-acetyl cysteine (NAC) or pyrrolidine dithiocarbamate (PDTC) decreased cellular ROS and suppressed tyrosine phosphorylation. This insinuates ROS as a downstream mediator of Bcr-Abl and capable of regulating redox-sensitive enzyme activities such as protein kinases and protein phosphatases. A stable cell line (TonB210) harboring a tetracycline-inducible Bcr-Abl expression vector has been constructed using the murine hematopoietic BaF3 cell line [11]. Upon Bcr-Abl expression by tetracycline induction, H2O2 levels increased in a time-dependent manner [8]. Similarly, transformation of the myeloid cell line 32Dcl3 and Draa-40 by Bcr-Abl caused elevated ROS, oxidative DNA damage and chromosomal fragmentation. Inhibition of Bcr-Abl kinase by imatinib or elimination of ROS by PDTC or 5,5-dimethy-1-pyrroline-N-oxide (DMPO) reduced DNA damage [12]. Studies using these in vitro models suggest that Bcr-Abl promotes ROS generation leading to genomic instability and chromosome abnormalities. The mechanism by which Bcr-Abl induces ROS stress is till unclear, and the above cell models provide useful tools for mechanistic studies.
c-Myc models for ROS study
Myc is a helix-loop-helix leucine zipper transcription factor that regulates the expression of many genes involved in cell proliferation, metabolism, and apoptosis [13]. Overexpression of c-Myc has been implicated in diverse human cancers. Studies using the in vitro tamoxifen-controlled system have revealed an important relationship between ROS and c-Myc expression. In this model, c-Myc expression is regulated by estrogen or tamoxifen in the culture medium. A MYCER chimeric gene was created by fusion of the hormone-binding domain of the estrogen receptor to the 3′ end of Myc; the resulting MYCER was introduced into the immortal Rat1A or normal human fibroblast cells through a pBABE retroviral vector. Only in the presence of estrogen or tamoxifen can the translated Myc protein be activated and translocated to the nuclei. Overexpression of c-Myc in Rat1A cells evoked genomic instability and tumorigenicity upon subcutaneous injection into BALA/c nude mice [14]. Although introduction of MYCER into normal human fibroblasts did not cause tumor formation, genomic destabilization and significant increase in H2O2 were detected after c-Myc activation [14,15]. Upon treatment of c-Myc activated cells with NAC, a significant decrease in ROS and reduction of DNA damage were observed [15]. A major source of ROS generation in cells over-expressing c-Myc is from the mitochondria, but the role of ROS in causing DNA breaks remains to be defined [16].
Cell models of p53 for ROS study
The HCT116 p53+/+ and p53−/− cells are perhaps the most widely used in vitro model system for the study of p53 functions [17]. Recent studies using these cell lines, various gene transfections, and siRNA approaches have revealed important roles of p53 in glucose metabolism and redox regulation. For instance, p53 decreased ROS levels by inhibiting glycolysis and promoting NADPH generation from the pentose phosphate pathway (PPP) through transcriptional activation of TIGAR (TP53-induced glycolysis and apoptosis regulator) and by optimizing mitochondrial respiration through up-regulating SCO2 (synthesis of cytochrome c oxidase 2) transcription [18,19]. TIGAR is a novel isoform of phosphor-fructokinase (PFK2) that contains the FBPase domain but lacks the kinase domain. Hence, its overexpression lowers fructose-2, 6-bisphosphate levels, resulting in an inhibition of glycolysis. This causes a shift towards the PPP, leading to increased production of NADPH, the major reducing equivalent for generation of reduced glutathione (GSH) and other cellular antioxidants. Thus, TIGAR sets a glycolytic checkpoint to inhibit glycolysis, enhance ROS-scavenging capacity, and promote genomic stability [20]. SCO2 is a nuclear gene that encodes a copper binding protein required for the assembly of the mitochondrial cytochrome c oxidase II (CO II) subunit in complex IV of the respiratory chain. Disruption of the SCO2 gene caused respiratory dysfunction and switched energy metabolism toward glycolysis [19].
Suppression by siRNA and overexpression by transfection are reasonable in vitro models to evaluate the antioxidant function of p53. In a lung carcinogenesis model, p53 suppression by siRNA increased DNA oxidation and increased mutagenesis, which is abolished by NAC [21]. Interestingly, p53 seems to affect cellular ROS and cell fate in a complex fashion. In p53-negative H1299 cells, forced expression of wild-type p53 at a physiological level led to a decrease of ROS and an increase in expression of SESN1 and SESN2. However, as p53 accumulated over a long period (72 h), cellular ROS levels increased, accompanied by upregulated expression of BAX, PIG3, and Puma, leading to apoptosis [21].
In vivo models
SOD mouse models
SOD1-deficient and transgenic mouse models
SOD1 is the major superoxide scavenger enzyme catalyzing the conversion of O2− to H2O2 in the cytoplasm, mitochondrial intermembrane space, nucleus and lysosomes. SOD1-deficient mouse models have been generated by either deleting the entire five exons or by deleting exons 3 and 4 through homologous recombination [22,23]. Mice deficient in SOD1 developed normally up to early adulthood, but were vulnerable to motor neuron loss following injury [22]. More than 30% of the SOD−/− mice developed liver tumors by 20 months of age and greater than 70% of the mice developed tumor nodules [24]. Extensive oxidative damage, including increased protein oxidation, lipid peroxidation, DNA damage, and decreased cytosolic aconitase activity were observed in the liver cells. Furthermore, abnormal mitochondria with disorganized cristae and smaller size were seen in tumor cells developed in these mice. These observations suggest that disturbance in redox homeostasis plays an important role in liver cancer development. Both heterozygous and homozygous SOD1-knockout mice exhibited various degrees of diminished SOD activity. The ageing phenotype such as accelerated skeletal muscle atrophy, vascular dysfunction, elevated oxidative damage to lipid, protein and DNA were observed [25,26]. Lifespan of the SOD1−/− mice was reduced by 9 months compared to wild type mice [24]. The G93A mutation in SOD1 is a genetic abnormality observed in familial amyotrophic lateral sclerosis (ALS). A transgenic mouse model carrying the SOD1-G93A transgene was generated. Indeed, the mice developed ALS, exhibiting progressive motor deficits, paralysis, neurodegeneration, along with prominent mitochondrial abnormalities and increased ROS production in the spinal cord and brain [27,28]. Proteomic analysis of tissue samples identified that the SOD1 protein was carbonylated and aggregated in the spinal cord [29].
SOD2-deficient mouse models
SOD2 is a mitochondrial enzyme. SOD2-deficient mouse models were created either by replacing exon 1 and exon 2 with a PGK-HPRT sequence, or by deleting exon 3 [30,31]. SOD2−/− mice showed neonatal death with neurodegeneration, dilated cardiomyopathy, and metabolic acidosis. Increased mitochondrial superoxide and accumulation of oxidative DNA damage was also observed in the brain and heart. ROS-sensitive enzymes including succinate dehydrogenase, NADH-dehydrogenase, and aconitase were dramatically reduced in the SOD2-deficient mice [30]. Heterozygous SOD2+/− mice, which exhibited 50% reduction in SOD levels, appeared normal and was used to study the chronic effect of reduced SOD2 activity [32]. The mice exhibited mitochondrial oxidative damage and decreased mitochondrial membrane potential. In addition, significant elevation of 8-oxo-deoxyguanosine was observed in nuclear and mitochondrial DNA. Tumor incidence, particularly lymphoma and pituitary adenoma, increased 100% in old SOD2+/− mice compared with the wild-type mice. Surprisingly, the lifespan of the SOD2+/− mice was similar to the wild type mice. The SOD2 mouse models provide valuable tools to elucidate the involvement of mitochondrial superoxide generation in the development of neurodegenerative diseases and cancer.
SOD3-deficient or overexpression models
SOD3 is a secreted form of Cu/Zn-containing enzymes in the extracellular matrix. SOD3−/− mice were created by disrupting the SOD3 gene through homologous recombination [33]. The mice developed normally and remained healthy for at least 14 months. Interestingly, the lack of SOD3 did not cause a compensatory increase in SOD1 or SOD2 enzyme activity or an increase in other antioxidant enzymes. These observations suggest that under physiological conditions, the rate of superoxide formation in the extracellular space may be too low to cause a significant phenotypic change. However, the SOD3−/− mice were vulnerable to stress under a high oxygen condition [33]. In addition, a SOD3 transgenic mouse model (SOD3TG) with skin-specific over-expression was generated by fusion of mouse SOD3 cDNA with a keratinocyte-specific promoter [34]. Compared to wild-type mice, oxidative DNA damage was decreased in SOD3TG mice after topical treatment with 12-O-tetradecanoylphorbol-13-acetate (TPA). SOD3-overexpression reduced tumor formation by 50% in dimethylbenzanthracene (DMBA)-initiated and TPA-promoted skin carcinogenesis models. This experimental system allows the study of potential roles of extracellular ROS in skin tumorigenesis and the protective effect of SOD3.
Mouse models overexpressing catalase
Catalase, normally localized in the peroxisome, is a haem-based enzyme that eliminates H2O2. Transgenic mice carrying catalase targeted to the peroxisome, nucleus, or mitochondria have been created [35]. The mitochondria-targeted catalase transgene was designed by removing the peroxisomal localization signal and adding the mitochondrial localization signal. Similarly, a nuclear-targeted catalase transgene was generated. Decreased H2O2 and mtDNA damage along with a 5 month increase in life span was only observed in mice overexpressing the mitochondrial-targeted catalase. Also, a protective effect on aconitase activity was seen when the mitochondria of heart tissue were challenged with H2O2. These observations suggest that mitochondrial ROS production could have a greater influence on the ageing process. Prompt scavenging of H2O2 in mitochondria seems to have a profound effect on life span.
Mouse models with GPX1 knockout or overexpression
Glutathione peroxidase 1 (GPX1) is an abundant and ubiquitously expressed selenoprotein, and functions as a major intracellular peroxide-scavenging enzyme. It utilizes glutathione as a substrate to catalyze the reduction of H2O2 and lipid peroxides. GPX1 knockout (PGX1−/−) and GPX1-overexpressing transgenic (GPX1TG) mice have been developed to study the in vivo role of GPX1 in coping with oxidative stress [36–38]. GPX1−/− or GPX1TG mice exhibited no significant alterations in mRNA expression or activity of thioredoxin reductase and other selenoproteins. However, GPX1−/− mice showed increased susceptibility to oxidative injuries, whereas GPX1TG mice were more resistant to ROS stress. Interestingly, knockout of GPX1 rendered mice resistant to reactive nitrogen species (RNS)-associated cytotoxicity, whereas GPX1TG mice were sensitive to RNS-induced damage [39]. Thus, it seems that GPX1 may play opposite roles in coping with oxidative injuries mediated by ROS verse RNS. Unexpectedly, GPX1TG mice developed insulin resistance and obesity, possibly due to loss of the H2O2−mediated inhibition of protein tyrosine phosphatases in the insulin signal pathway [40]. Furthermore, transgenic mice overexpressing GPX or GPX plus SOD1 have increased incidence of tumorigenesis in DMBA/TPA-induced skin carcinogenesis, suggesting the importance of precise redox homeostasis [41].
p53 mouse models
A model of super Arf/p53 (s-Arf/p53) mice has recently been created to study the important roles of ROS in ageing and cancer. A large genomic DNA segment containing a murine p53 gene was microinjected into fertilized mouse oocytes to create p53 transgenic mice. The resulting offspring (s-p53 mice) carried an extra copy of p53 in addition to the two endogenous alleles [42]. The transgene seemed to be expressed and regulated in a similar fashion to its endogenous counterpart. The s-p53 mice showed significant decrease in ROS generation and DNA damage. The super-Arf (s-Arf) transgenic mice were generated using a similar method [43]. Since Arf stabilizes p53, dual transgenic Arf/p53 mice were generated by crossing s-Arf and s-p53 mice to study the combined effects of both genes [44]. S-Arf/p53 mice showed a significant decrease in basal ROS level, lipid peroxidation and oxidized proteins, and an increase in GSH level and Sesn1 and Sesn2 expression. These mice were resistant to oxidative damage by paraquat. Interestingly, the s-Arf/p53 mice exhibited a significant delay in ageing, although individually s-Arf or s-p53 had no impact on lifespan. In vitro studies using mouse embryo fibroblasts (MEF) from s-Arf/p53 mice showed that these cells were completely refractory to malignant transformation by E1A and H-RasV12. The p53 mouse models have provided important mechanistic insights into the relationship between ROS, tumor suppressors, and oncogenes.
Model comparison
A number of in vitro and in vivo models have been developed during the recent years to allow the study of ROS stress, its roles in the development of cancer and other diseases, and to explore the utilities of ROS-mediated mechanisms in disease prevention and therapeutics. These in vitro and in vivo models are compared in Table 1. The main features of the representative experimental model systems are listed in Table 2
Table 1.
Comparison of in vitro and in vivo models
| Model systems | Advantages | Disadvantages | Examples | Best use of model |
|---|---|---|---|---|
| In vitro Models | - Biochemical & molecular events well-defined. | - Cannot reliably predict cancer development in vivo. | Ras cell models | ROS in cancers |
| - ROS alterations readily measurable. | - Difficult to mimic tissue microenvironment | Bcr-Abl model | Anticancer drug testing | |
| - Amenable to further genetic modifications. | - Redox status and metabolism sensitive to culture conditions. | c-Myc model | ROS in leukemia (CML) | |
| - Suitable for mechanistic studies. | p53 cell models | Mitochondrial ROS | ||
| - Relatively inexpensive | Energy metabolism and redox regulation | |||
| - Adaptable for high-throughput drug screening. | Anticancer drug testing | |||
| In vivo models | - Resembles ROS stress and disease development in human. | - Time consuming and high costs. | SOD1−/− mice | Role of ROS in cancers, ageing & other diseases |
| - Genetic background well defined. | - Difficult to measure ROS in vivo | SOD2+/− mice | Mitochondrial ROS & cancer development | |
| - Allows the evaluation of ROS effects in complex tissue microenvironment. | - Limited flexibility for further genetic modifications | CatalaseTG mice | Role of H2O2 in vivo | |
| - Suitable for long-term follow up on biological consequences. | - Result interpretation could be complicated. |
Table 2.
Main features of experimental models for study of ROS-related pathological processes
| Models | Mechanisms | Main features | Application | References |
|---|---|---|---|---|
| Ras | NOX activation Mitochondria? | Chromosome remodeling, p53, p16 activation, ROS increase, altered redox | ROS in cancer & senescence | 2–8 |
| Bcr-Abl | NOX? Mitochondria? | Increased ROS, chromosomal fragmentation, DNA damage, decreased PTPase activity | Leukemia (CML) | 8, 10–12 |
| C-myc | Mitochondria Other? | Increased ROS, DNA damage, increased genomic instability | Various cancer | 14–16 |
| p53 | Mitochondria glycolysis, PPP | Alter redox homeostasis, SCO2, TIGAR, SESN1/2, PIG3, Puma, BAX activation | Longevity, ageing, cancer, apoptosis | 18–19,21, 42,44 |
| SOD1 | Affect O2− elimination | Abnormal mitochondria, oxidative DNA and protein damage | Cancer, ageing, neurodegeneration | 22–26 |
| SOD1G93ATG | Gain of toxic function | Protein carbonylation and aggregation, ROS increase, abnormal mitochondria | ALS | 27–29 |
| SOD2+/− | Mitochondrial ROS↑ | ROS increase, nDNA & mtDNA damage, altered mitochondria | Cancer | 32 |
| SOD2−/− | Mitochondrial ROS | Fe-S protein function loss, DNA oxidation, metabolic alteration, ROS | Role of ROS in cancer & development | 30–31 |
| SOD3TG | ↓ extracellular O2− | Decrease in oxidative DNA damage | ROS in skin cancer | 34 |
| Catalase | Lower ROS | Decreased ROS and mtDNA damage protected aconitase function | Role of mitochondrial ROS in longevity | 35 |
| GPX1 | Redox alteration | Aberrant ROS and RNS responses | Cancer, diabetes | 39–41 |
Summary and future perspectives
The redox reactions, with simple transfer of electrons affect almost all complex biological processes, and have profound effects on cell proliferation, cell fate, and various pathological processes. Maintaining proper redox homeostasis is essential for all living organisms. Any significant alteration in the ROS generation or elimination process is likely to change redox balance and have biological consequences; depending on the degree and duration of the redox stress. Complex redox regulatory mechanisms have evolved during evolution, but many of these regulatory pathways and signaling mechanisms remain to be investigated. A number of in vitro and in vivo models have been developed during the recent years to answer these important questions. The deteriorating effects of ROS have long been recognized, and the use of various antioxidants to counteract the harmful effects of ROS has traditionally been a major research area in disease prevention. In recent years, ROS stress in cancer cells and its potential therapeutic implications have emerged as a promising area of research. Because oncogenic signals, mitochondrial dysfunction, and active metabolism can cause ROS stress in cancer cells, it is hypothesized that such intrinsic oxidative stress may render the malignant cells highly dependent on antioxidant defense systems for survival and thus more vulnerable to further oxidative insults by exogenous agents that either enhance ROS generation or inhibit cellular antioxidant systems. Such cancer therapeutic strategies may be less toxic to normal cells due to their low basal ROS output, high antioxidant reserve, and intact redox regulatory mechanisms. The in vitro and in vivo models described above provide valuable experimental systems to evaluate the ROS-mediated therapeutic strategies for cancer treatment.
Acknowledgments
This work was support in part by grants CA085563, CA100428, and CA109041 from the National Cancer Institute, National Institutes of Health.
Footnotes
Contact details: Weiqin Lu, Department of Molecular Pathology, University of Texas M.D. Anderson Cancer Center. Box 0951, 1515 Holcombe Boulevard, Houston, TX 77030. Phone: 713-745-8617, E-mail: wqlu@mdanderson.org
Marcia Ogasawara, Department of Molecular Pathology, University of Texas M.D. Anderson Cancer Center. Box 0951, 1515 Holcombe Boulevard, Houston, TX 77030. Phone: 713-834-6069, E-mail: mogasawa@mdanderson.org
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Droge W. Free radicals in the physiological control of cell function. Physiol Rev. 2002;82(1):47–95. doi: 10.1152/physrev.00018.2001. [DOI] [PubMed] [Google Scholar]
- 2.Irani K, et al. Mitogenic signaling mediated by oxidants in Ras-transformed fibroblasts. Science. 1997;275(5306):1649–1652. doi: 10.1126/science.275.5306.1649. [DOI] [PubMed] [Google Scholar]
- 3.Serrano M, et al. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88(5):593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 4.Kemp CJ, et al. Reduction of p53 gene dosage does not increase initiation or promotion but enhances malignant progression of chemically induced skin tumors. Cell. 1993;74(5):813–822. doi: 10.1016/0092-8674(93)90461-x. [DOI] [PubMed] [Google Scholar]
- 5.Braig M, et al. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature. 2005;436(7051):660–665. doi: 10.1038/nature03841. [DOI] [PubMed] [Google Scholar]
- 6.Liu J, et al. A genetically defined model for human ovarian cancer. Cancer Res. 2004;64(5):1655–1663. doi: 10.1158/0008-5472.can-03-3380. [DOI] [PubMed] [Google Scholar]
- 7.Young TW, et al. Activation of antioxidant pathways in ras-mediated oncogenic transformation of human surface ovarian epithelial cells revealed by functional proteomics and mass spectrometry. Cancer Res. 2004;64(13):4577–4584. doi: 10.1158/0008-5472.CAN-04-0222. [DOI] [PubMed] [Google Scholar]
- 8.Trachootham D, et al. Selective killing of oncogenically transformed cells through a ROS-mediated mechanism by beta-phenylethyl isothiocyanate. Cancer Cell. 2006;10(3):241–252. doi: 10.1016/j.ccr.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 9.Daley GQ, Baltimore D. Transformation of an interleukin 3-dependent hematopoietic cell line by the chronic myelogenous leukemia-specific P210bcr/abl protein. Proc Natl Acad Sci U S A. 1988;85(23):9312–9316. doi: 10.1073/pnas.85.23.9312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sattler M, et al. The BCR/ABL tyrosine kinase induces production of reactive oxygen species in hematopoietic cells. J Biol Chem. 2000;275(32):24273–24278. doi: 10.1074/jbc.M002094200. [DOI] [PubMed] [Google Scholar]
- 11.Klucher KM, et al. Secondary mutation maintains the transformed state in BaF3 cells with inducible BCR/ABL expression. Blood. 1998;91(10):3927–3934. [PubMed] [Google Scholar]
- 12.Nowicki MO, et al. BCR/ABL oncogenic kinase promotes unfaithful repair of the reactive oxygen species-dependent DNA double-strand breaks. Blood. 2004;104(12):3746–3753. doi: 10.1182/blood-2004-05-1941. [DOI] [PubMed] [Google Scholar]
- 13.Dang CV. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol. 1999;19(1):1–11. doi: 10.1128/mcb.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Felsher DW, Bishop JM. Transient excess of MYC activity can elicit genomic instability and tumorigenesis. Proc Natl Acad Sci U S A. 1999;96(7):3940–3944. doi: 10.1073/pnas.96.7.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vafa O, et al. c-Myc can induce DNA damage, increase reactive oxygen species, and mitigate p53 function: a mechanism for oncogene-induced genetic instability. Mol Cell. 2002;9(5):1031–1044. doi: 10.1016/s1097-2765(02)00520-8. [DOI] [PubMed] [Google Scholar]
- 16.Ray S, et al. MYC can induce DNA breaks in vivo and in vitro independent of reactive oxygen species. Cancer Res. 2006;66(13):6598–6605. doi: 10.1158/0008-5472.CAN-05-3115. [DOI] [PubMed] [Google Scholar]
- 17.Bunz F, et al. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282(5393):1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 18.Bensaad K, et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126(1):107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 19.Matoba S, et al. p53 regulates mitochondrial respiration. Science. 2006;312(5780):1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- 20.Green DR, Chipuk JE. p53 and metabolism: Inside the TIGAR. Cell. 2006;126(1):30–32. doi: 10.1016/j.cell.2006.06.032. [DOI] [PubMed] [Google Scholar]
- 21.Sablina AA, et al. The antioxidant function of the p53 tumor suppressor. Nat Med. 2005;11(12):1306–1313. doi: 10.1038/nm1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reaume AG, et al. Motor neurons in Cu/Zn superoxide dismutase-deficient mice develop normally but exhibit enhanced cell death after axonal injury. Nat Genet. 1996;13(1):43–47. doi: 10.1038/ng0596-43. [DOI] [PubMed] [Google Scholar]
- 23.Huang TT, et al. Superoxide-mediated cytotoxicity in superoxide dismutase-deficient fetal fibroblasts. Arch Biochem Biophys. 1997;344(2):424–432. doi: 10.1006/abbi.1997.0237. [DOI] [PubMed] [Google Scholar]
- 24.Elchuri S, et al. CuZnSOD deficiency leads to persistent and widespread oxidative damage and hepatocarcinogenesis later in life. Oncogene. 2005;24(3):367–380. doi: 10.1038/sj.onc.1208207. [DOI] [PubMed] [Google Scholar]
- 25.Muller FL, et al. Absence of CuZn superoxide dismutase leads to elevated oxidative stress and acceleration of age-dependent skeletal muscle atrophy. Free Radic Biol Med. 2006;40(11):1993–2004. doi: 10.1016/j.freeradbiomed.2006.01.036. [DOI] [PubMed] [Google Scholar]
- 26.Didion SP, et al. Heterozygous CuZn superoxide dismutase deficiency produces a vascular phenotype with aging. Hypertension. 2006;48(6):1072–1079. doi: 10.1161/01.HYP.0000247302.20559.3a. [DOI] [PubMed] [Google Scholar]
- 27.Bogdanov MB, et al. Elevated “hydroxyl radical” generation in vivo in an animal model of amyotrophic lateral sclerosis. J Neurochem. 1998;71(3):1321–1324. doi: 10.1046/j.1471-4159.1998.71031321.x. [DOI] [PubMed] [Google Scholar]
- 28.Liu R, et al. Enhanced oxygen radical production in a transgenic mouse model of familial amyotrophic lateral sclerosis. Ann Neurol. 1998;44(5):763–770. doi: 10.1002/ana.410440510. [DOI] [PubMed] [Google Scholar]
- 29.Poon HF, et al. Redox proteomics analysis of oxidatively modified proteins in G93A-SOD1 transgenic mice--a model of familial amyotrophic lateral sclerosis. Free Radic Biol Med. 2005;39(4):453–462. doi: 10.1016/j.freeradbiomed.2005.03.030. [DOI] [PubMed] [Google Scholar]
- 30.Li Y, et al. Dilated cardiomyopathy and neonatal lethality in mutant mice lacking manganese superoxide dismutase. Nat Genet. 1995;11(4):376–381. doi: 10.1038/ng1295-376. [DOI] [PubMed] [Google Scholar]
- 31.Lebovitz RM, et al. Neurodegeneration, myocardial injury, and perinatal death in mitochondrial superoxide dismutase-deficient mice. Proc Natl Acad Sci U S A. 1996;93(18):9782–9787. doi: 10.1073/pnas.93.18.9782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Remmen H, et al. Life-long reduction in MnSOD activity results in increased DNA damage and higher incidence of cancer but does not accelerate aging. Physiol Genomics. 2003;16(1):29–37. doi: 10.1152/physiolgenomics.00122.2003. [DOI] [PubMed] [Google Scholar]
- 33.Carlsson LM, et al. Mice lacking extracellular superoxide dismutase are more sensitive to hyperoxia. Proc Natl Acad Sci U S A. 1995;92(14):6264–6268. doi: 10.1073/pnas.92.14.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim SH, et al. Overexpression of extracellular superoxide dismutase (EC-SOD) in mouse skin plays a protective role in DMBA/TPA-induced tumor formation. Oncol Res. 2005;15(7–8):333–341. doi: 10.3727/096504005776449725. [DOI] [PubMed] [Google Scholar]
- 35.Schriner SE, et al. Extension of murine life span by overexpression of catalase targeted to mitochondria. Science. 2005;308(5730):1909–1911. doi: 10.1126/science.1106653. [DOI] [PubMed] [Google Scholar]
- 36.Cheng WH, et al. Cellular glutathione peroxidase knockout mice express normal levels of selenium-dependent plasma and phospholipid hydroperoxide glutathione peroxidases in various tissues. J Nutr. 1997;127(8):1445–1450. doi: 10.1093/jn/127.8.1445. [DOI] [PubMed] [Google Scholar]
- 37.Ho YS, et al. Mice deficient in cellular glutathione peroxidase develop normally and show no increased sensitivity to hyperoxia. J Biol Chem. 1997;272(26):16644–16651. doi: 10.1074/jbc.272.26.16644. [DOI] [PubMed] [Google Scholar]
- 38.Yoshida T, et al. Transgenic mice overexpressing glutathione peroxidase are resistant to myocardial ischemia reperfusion injury. J Mol Cell Cardiol. 1996;28(8):1759–1767. doi: 10.1006/jmcc.1996.0165. [DOI] [PubMed] [Google Scholar]
- 39.Fu Y, et al. Comparative impacts of glutathione peroxidase-1 gene knockout on oxidative stress induced by reactive oxygen and nitrogen species in mouse hepatocytes. Biochem J. 2001;359(Pt 3):687–695. doi: 10.1042/0264-6021:3590687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McClung JP, et al. Development of insulin resistance and obesity in mice overexpressing cellular glutathione peroxidase. Proc Natl Acad Sci U S A. 2004;101(24):8852–8857. doi: 10.1073/pnas.0308096101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu YP, et al. Enhanced skin carcinogenesis in transgenic mice with high expression of glutathione peroxidase or both glutathione peroxidase and superoxide dismutase. Cancer Res. 1997;57(8):1468–1474. [PubMed] [Google Scholar]
- 42.Garcia-Cao I, et al. “Super p53” mice exhibit enhanced DNA damage response, are tumor resistant and age normally. Embo J. 2002;21(22):6225–6235. doi: 10.1093/emboj/cdf595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matheu A, et al. Increased gene dosage of Ink4a/Arf results in cancer resistance and normal aging. Genes Dev. 2004;18(22):2736–2746. doi: 10.1101/gad.310304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matheu A, et al. Delayed ageing through damage protection by the Arf/p53 pathway. Nature. 2007;448(7151):375–379. doi: 10.1038/nature05949. [DOI] [PubMed] [Google Scholar]