Summary
Major insulin gene transcription factors, such as PDX-1 or NeuroD1, have equally important roles in pancreatic development and the differentiation of pancreatic endocrine cells. Previously, we identified and cloned another critical insulin gene transcription factor MafA (RIPE3b1) and reported that other Maf factors were expressed in pancreatic endocrine cells. Maf factors are important regulators of cellular differentiation; to understand their role in differentiation of pancreatic endocrine cells, we analyzed the expression pattern of large Maf factors in the pancreas of embryonic and adult mice. Ectopically expressed large Maf factors, MafA, MafB or cMaf, induced expression from insulin and glucagon reporter constructs, demonstrating a redundancy in their function. Yet in adult pancreas, cMaf was expressed in both α- and β-cells, and MafA and MafB showed selective expression in the β- and α-cells, respectively. Interestingly, during embryonic development a significant proportion of MafB-expressing cells also expressed insulin. In embryos MafB is expressed before MafA, and our results suggest that the differentiation of β-cells proceeds through a MafB+ MafA− Ins+ intermediate cell to MafB− MafA+ Ins+ cells. Furthermore, the MafB to MafA transition follows induction of PDX-1 expression(Pdx-1high) in MafB+ Ins+ cells. We suggest that MafB may have a dual role in regulating embryonic differentiation of both β- and α-cells while MafA may regulate replication/survival and function of β-cells after birth. Thus, this redundancy in the function and expression of the large Maf factors may explain the normal islet morphology observed in the MafA knockout mice at birth.
Keywords: MafA, MafB, Maf factors, insulin gene transcription factor, pancreatic development, endocrine differentiation, pancreatic islets
Introduction
Transcription factors binding to three conserved insulin enhancer elements, A3 (Ohlsson et al., 1991; Boam and Docherty, 1989), RIPE3b/C1-A2 (Sharma and Stein, 1994; Shieh and Tsai, 1991) and E1 (Crowe and Tsai, 1989; Karlsson et al., 1987) are major regulators of insulin gene expression. The loss of PDX-1, the A3 binding factor, results in mice that are apancreatic (Jonsson et al., 1994; Offield et al., 1996), whereas mice homozygous null for NeuroD1, which binds to the E1 element, have a striking reduction in the number of insulin-producing β-cells (Naya et al., 1997). These observations underscore the importance of insulin gene transcription factors in the development and differentiation of pancreatic endocrine cells. We have identified and cloned MafA, a member of the large Maf family of basic leucine zipper (bZIP) transcription factors, as the β-cell specific factor binding to the RIPE3b/C1-A2 element (Maf Response element or MARE) (Olbrot et al., 2002). RIPE3b1/MafA is a glucose-responsive factor expressed in insulin-producing cells (Sharma and Stein, 1994; Olbrot et al., 2002; Matsuoka et al., 2003; Kataoka et al., 2002).
The Maf family of transcription factors are important regulators of differentiation and regulate processes such as hematopoiesis, lens differentiation and segmentation of hind-brain (Blank and Andrews 1997; Reza and Yasuda 2004; Ogino and Yasuda, 1998; Ogata et al., 2004; Reza et al., 2002; Kim et al., 1999; Kawauchi et al., 1999; Ring et al., 2000). In the lens, Maf factors regulate expression of the major lens-specific protein crystallins (Ogino and Yasuda, 1998). Expression of avian MafA/L-Maf in neuroectodermal cells triggered lens differentiation program (Ogino and Yasuda, 1998), while the absence of cMaf prevented lens formation and the expression of crystallin genes (Kawauchi et al., 1999; Kim et al., 1999). Hence, it was surprising that the loss of MafA did not affect embryonic pancreatic development (Zhang et al., 2005a). However, after birth, the loss of MafA resulted in reduced proportion of β-cells in islets and impaired glucose tolerance, demonstrating the role of MafA in regulating β-cell replication/survival and function. In addition to MafA, pancreatic endocrine cells express other large-Maf family members (Olbrot et al., 2002; Matsuoka et al., 2003; Kataoka et al., 2004). Since all Maf family members recognize the consensus MARE element, cells expressing multiple Maf family members will have competition to regulate MARE-dependent gene expression. Thus, to understand the role of Maf factors in the differentiation of pancreatic endocrine cells, it is essential to determine the temporal and spatial expression patterns of these factors.
In this study we demonstrate that in adult mice MafA and MafB are preferentially expressed in pancreatic β- and α-cells, respectively. Similarly, at E15.5, MafA and MafB expression was observed in the insulin- and glucagon-expressing cells. However, MafB was also expressed in nearly 90% of insulin-producing cells. The ability of MafB+ cells to express insulin was associated with Nkx6.1 expression, while MafB+ Nkx6.1− cells expressed glucagon. Our results demonstrate a gradual shift in expression from MafB+ insulin+ to MafA+ insulin+ cells during fetal development, which followed the enhanced PDX-1 expression in insulin+ cells. We suggest that this switch from MafB+ to MafA+ cells is not essential for generation of insulin+ cells but may play a role in β-cell replication/survival and function. These results provide an explanation for the lack of effect on the embryonic development of pancreatic islets in MafA knockout mice.
Material and methods
Construction of expression vectors
Expression vectors were constructed by conventional molecular biology techniques. The insulin luciferase reporter constructs -238 WT LUC and its derivatives mutant 110-09m LUC have been described previously (Harrington and Sharma, 2001). The glucagon luciferase reporter construct GLU LUC (Lee et al., 1992) was obtained from Dr. Dan Drucker (Toronto, Canada). The glucagon promoter from GLU LUC was cloned into multiple cloning site (MCS) of pEGFP (Clontech, Palo Alto, CA) to generate GLU GFP. The expression vectors pcDNAMafA and ΔN-hMafA have been described previously (Olbrot et al., 2002). Mouse full-length MafB cDNA clone in pCMV-Sport6 was purchased from ATCC (Manassas, VA). cMaf cDNA from pCIcMaf (Dr. I-C Ho, BWH, Boston) was cloned into MCS of pcDNA3.1 (Clontech, Palo Alto, CA) to generate pcDNA cMaf.
Mice and Immunostaining
The day of vaginal-plug discovery was designated as E0.5 of C57BL/6 mice. Animals were anesthetized with intraperitoneal injection of Pentobarbitol (50 mg/kg) and dissected. All animal protocols were approved by the Animal Care Committee of the Joslin Diabetes Center. Pancreata were excised, fixed in 4% paraformaldehyde, embedded in paraffin, and immunostained as described previously (Laybutt et al., 2002). Antigen retrieval consisted of heating de-paraffinized tissue sections three times for 5 min each with 0.01 M citrate buffer in a microwave oven, followed by 20 min cool-down period before proceeding to the next step. The primary antibodies were guinea pig anti-insulin (Linco); guinea pig anti-glucagon (Linco); monoclonal anti-synaptophysin (Chemicon); mouse monoclonal anti-Ki-67 (BD Pharmingen); rabbit anti-Nkx6.1 (provided by P. Serup); goat anti-PDX-1 (provided by C. Wright); rabbit anti-MafB (Bethyl); rabbit anti-cMaf (Bethyl); rabbit anti-Pax6 (Covance). Our rabbit anti-MafA antibody was raised against the 329 to 347 carboxy-terminal amino acids of mouse MafA (ProSci, Poway, CA). The specificity of this antibody was confirmed using its blocking peptide in immunohistochemistry and western blotting; the pre-immune sera gave no staining at the same concentrations used in these experiments. Secondary antibodies for immunofluorescence were FITC- or Texas red-conjugated anti-rabbit, anti-mouse or anti-guinea pig IgG (Jackson ImmunoResearch). For amplification, biotinylated anti-rabbit, anti-mouse or anti-goat IgG antibodies (1:400, Jackson Immunoresearch) were used followed by streptavidin-conjugated Texas red (1:400, Jackson Immunoresearch) or streptavidin-conjugated Alexa fluor 488 (1:400, Molecular Probes). Nuclear counterstaining was achieved by DAPI mounting medium (Vector). The immunostaining shown in each figure were derived from several replicates of at least 5 sections from 3 different mice/embryos. At least 3 different pancreases were counted for quantitative results. Confocal images were taken on Zeiss LSM410 (Zeiss, Thornwood, NY); quantitations were performed using NIH Image J.
Cell culture and Immunocytochemistry
HeLa cells were cultured in the Dulbecco's modified Eagle's medium supplemented with 10% (w/v) FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin at 37°C under 5% CO2 Cells were transfected with the various expression plasmids and 1 μg of GLU GFP reporter constructs using Lipofectamine (Invitrogen, CA). 48 hours after transfection cells were fixed with 4% (w/v) paraformaldehyde for 15 min, permeabilized with 0.25% (w/v) Triton X-100 in PBS for 15 min and immunostained as described previously (Nishimura et al., 2002). After blocking with PBS containing 10% (w/v) BSA for 30 min, cells were incubated overnight with the primary antibody in PBS containing 3% (w/v) BSA, washed with PBS and incubated with secondary antibody in PBS containing 3% (w/v) BSA for 1 hr, followed by washing with PBS and mounting.
In situ hybridization
Fresh frozen slides were fixed in 10% Buffered formalin (Fisher) for 20 min at -20 C. cMaf sense and antisense probes were diluted to 1ng/ul and incubate over-night in humidity chamber at 65°C. Next wash slides for 4 X 30 min in 1 X Sodium Chloride-Sodium Citrate (SSC) (Sigma) and then for 1X 30 min in Maleaic Acid Buffer w/tween (MABT). Add primary Alkaline-Phosphatase-DIG-antibody (Roche, 1:1000) incubate over-night at 4°C. Staining was developed using BM Purple (Roche) for up to 48 hours (4°C during over-night incubation and RT during daytime). Image of stained section were taken on Olympus BH2 light microscope. Immediately following in situ hybridization, slides were washed twice with PBS and incubated with guinea pig-anti-insulin (1:200) and rabbit-anti-glucagon (1:1000) antibodies for 2 hrs at RT. Secondary antibodies donkey-anti- guinea pig-Texas Red and donkey-anti-rabbit-FITC (Vector Labs, 1:200) were used for 1 hr at RT, Confocal images were taken on Zeiss LSM410 (Zeiss, Thornwood, NY).
Luciferase Assays
HeLa cells were transfected with the indicated amount of reporter constructs of -238 WT LUC, 110-09m LUC or GLU LUC and with 1 μg of pSVβ-gal plasmid (Promega, Madison, WI). Whole cell extracts were prepared and luciferase activity was measured as previously described (Nishimura et al., 2005).
RT-PCR
Total RNA was extracted from MIN-6 or α-TC 1.6 cells and reverse-transcribed to cDNA, which was amplified by PCR with appropriate oligonucleotide primers as previously described (Olbrot et al., 2002). The results were confirmed from at least three independent samples. The following oligonucleotides were used for primers: cMaf3'ppT,5'CTTGTGAGTTTTGGCCTTATGATG3'; cMaf3'ppB, 5'GCTCTGGGGTTGTACTTTTTCTGT3'; MafA3'ppT, 5'TTTCCTCGGCAGCGTCCACTTGTA3'; MafA3'ppB, 5'GGGGGTTCCTCCGGGTTTTCTAAT3', MafB3'ppT, 5'CTGCGCCCCTAGCCCTGGACTC3'; MafB3'ppB, 5'GGCGGCCCTGGCACTCACAAA3'.
Western Blot
40 μg of nuclear extracts from indicated cell lines were run on 10% SDS-PAGE and transferred to PVDF membranes, which were subjected to Western blotting with indicated antibodies and visualized by enhanced chemiluminescence kit (Amersham Biosciences).
Results and Discussion
Differential expression of large-Maf factors in pancreatic endocrine cells
Previously we reported that in addition to the MafA, other large-Maf factors, MafB and cMaf, were expressed in pancreatic endocrine cells (Olbrot et al., 2002). In a detailed study, Matsuoka and colleagues (2003) reported that in islets MafA was expressed in β-cells, MafB in more α- than β-cells, and cMaf at extremely low levels. Surprisingly, Kataoka and colleagues were unable to demonstrate MafB expression in α-cells but showed the expression of cMaf in these cells (Kataoka et al., 2004). Hence, we reexamined the expression profile of large-Maf factors in pancreatic endocrine cells. RT-PCR was performed on the RNA isolated from mouse islets, the β-cell line MIN-6 and the α-cell line α-TC1.6. Since the large-Maf factors share sequence homology in the coding region, PCR primers were designed in the unique 3’ untranslated regions. MafA and MafB expression was mostly restricted to β and α-cell lines, respectively, while cMaf was expressed in both cell lines (Figure. 1A). To determine the relative expression of these factors, real-time PCR reactions were performed with the same cDNAs used in Figure 1A. MafA was expressed at 250-fold higher in β- than in α-cells, while MafB expression was 450-fold higher in α-cells. Consistent with the results in Figure 1A, cMaf expression was similar in both cell lines. This differential expression of MafA and MafB in pancreatic α- and β-cells is consistent with the results of Matsuoka et.al. (Matsuoka et al., 2003). We determined the expression of large-Maf proteins in hormone-producing (αTC1.6 and MIN6) and non- hormone producing (HeLa) cell lines. Protein bands corresponding to MafA were detected only in the extract from the β-cell line, while cMaf protein was expressed in both α- and β-cell lines. Anti-MafA antibody recognized multiple protein bands (most likely phospho-MafA isoforms) and pre-incubation of the antibody with specific peptide prevented this recognition (Kondo et. al. unpublished observation). Anti-MafB antibody does not work optimally in the Western blot (or the levels of MafB protein in αTC1.6 are very low), yet results from two independent samples convincingly showed expression of MafB in the α-cells (Figure 1B). Band intensities corresponding to these large-Maf factors were significantly different in each cell line. Since the affinity of antibodies for these factors may be different, these intensities may not reflect true expression levels of these proteins. Even so, the Western blot analyses are consistent with the RNA expression pattern (Figure. 1A, B).
Figure 1. Large-Maf factors are differentially expressed in pancreatic α- and β-cells.
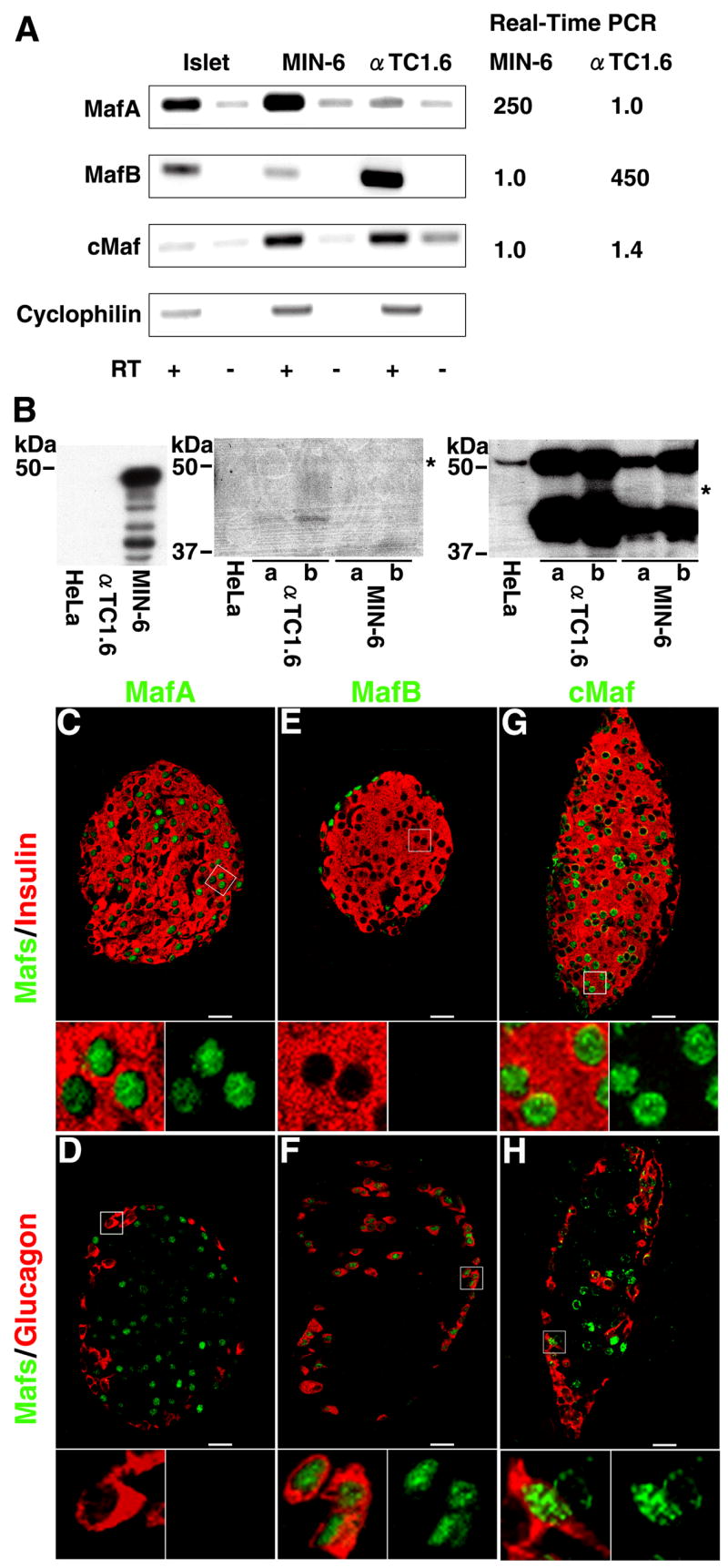
A, mRNA expression levels of different Maf factors: cDNAs prepared in the absence or the presence of reverse transcriptase (RT) from total RNA isolated from mouse islets, MIN-6 cells and α-TC1.6 cells, were used for PCR with the indicated primers. Cyclophilin was used as a control. A representative gel from at least three independent experiments is shown. The same cDNAs were used for Real Time quantitative PCR with primers and probes corresponding to the three large-Maf factors. The relative expression levels were quantitated using ΔΔCT method with 18S rRNA as a control. B, Protein expression levels of different Maf factors: Nuclear extract from MIN-6 cells, α-TC1.6 cells and HeLa cell lines was used to detect specific Maf factors by Western blot analyses as described in the Methods. Two independent samples (a and b) each from α TC1.6 and MIN-6 cell lines were used to confirm the expression of MafB and cMaf. Asterisks denote non-specific bands. C, Double immunohistochemical (IHC) detection of MafA, MafB or cMaf (green) with either insulin or glucagon (red) in the adult mouse pancreas. Note selective expression of MafA and MafB in insulin+ and glucagon+ cells respectively, while expression of cMaf can be detected in both cell types. Insets, higher magnification of demarcated area. Magnification bar = 20 μm.
In adult pancreas immunostaining for MafA was restricted to the pancreatic β-cells (Figure. 1C, D). Similarly, MafB expression was highly restricted to α-cell (Figure. 1E, 1F), and only a few rare β-cells in the adult pancreas immunostained for MafB. cMaf was expressed in both α- and β-cells (Figure. 1G, 1H). This data differs from the reported MafB expression in nearly a third (37%) of adult β-cells (Matsuoka et al., 2003). Differences in the mouse strains and/or antibodies may account for this discrepancy. Immunostaining results are consistent with those from RT-PCR and Western blot analyses, and together they confirm differential expression of MafA and MafB in pancreatic β-and α-cells.
Large-Maf factors activate expression from insulin and glucagon reporter constructs
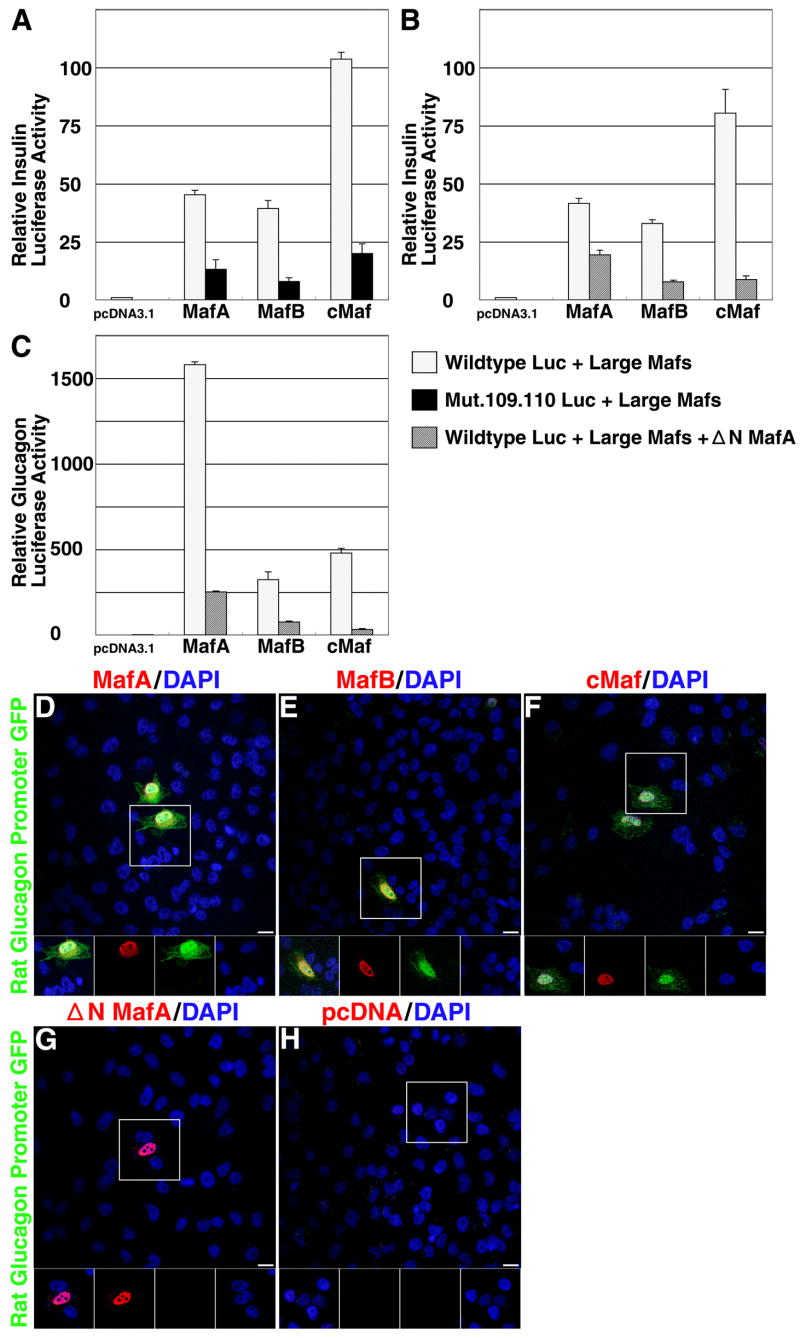
Since large-Maf family members are expressed in pancreatic β- and α-cells, we examined their effect on regulating insulin and glucagon gene expression. Rat insulin II promoter: luciferase reporter constructs (-238 WTLUC) with or without mutations in the MARE element (Harrington and Sharma, 2001; Olbrot et al., 2002) were transfected into HeLa cells with MafA, MafB or cMaf expression plasmids, or pcDNA3.1 as a control. All three large Maf factors activated insulin gene expression from the wild type construct and not from the construct with the mutated MARE element (Figure. 2A). To determine if these factors regulate expression by forming heterodimers, cells were transfected with -238 WTLUC plasmid and large-Maf expression plasmids with or without the NMafA plasmid that lacks the N-terminal activation domain of MafA (Olbrot et al., 2002). Activation of insulin gene expression mediated by these factors was inhibited by NMafA (Figure. 2B). As observed for insulin gene expression, MafA, MafB and cMaf activated glucagon gene expression, and this activation was inhibited by NMafA (Figure. 2C). To assess the ability of Maf factors to activate gene expression at a single cell level, a glucagon: GFP reporter plasmid was transfected into HeLa cells with MafA, MafB, cMaf, NMafA expression plasmids or pcDNA3.1. Cells singly transfected with MafA, MafB or cMaf induced expression from glucagon reporter construct (as observed by co-expression of Maf factors and GFP) (Figure. 2D-F) but not in those cells transfected with NMafA or pcDNA3.1 (Figure. 2G, H). These results are consistent with the published reports (Planque et al., 2001; Kataoka et al., 2004; Zhao et al., 2005), and they demonstrate that MafA, MafB or cMaf can activate expression from not only insulin but also glucagon promoter:reporter constructs.
Figure 2. MafA, MafB and cMaf can activate insulin and glucagon expression.
A, Luciferase reporter plasmids (-238 WT LUC or -122.121m LUC) were transfected into HeLa cells with the indicated expression plasmids (pcDNA3.1, MafA, MafB or cMaf). Luciferase activity was determined 48 h after transfection. Results are presented relative to the activity of -238 WT luciferase construct transfected with pcDNA3.1 ± SE (n = 4). B, Luciferase reporter plasmid (-238 WT LUC) and indicated expression plasmid were transfected into HeLa cells with or without NMafA plasmid that lacks N-terminal activation domain. Results are presented as in (a) (n=3). C, Glucagon Luciferase reporter plasmid [-2.2 Glucagon LUC, (Lee et al., 1992)] and indicated expression plasmid were transfected into HeLa cells with or without NMafA plasmid. Results are presented as in (a) (n=3). Rat glucagon: GFP reporter plasmid was transfected into HeLa cells with (D) MafA, (E) MafB, (F) cMaf (G) ΔNMafA expression plasmids or (H) pcDNA3.1. 48 h after transfection, HeLa cells were immunostained to detect GFP (green), nuclei (DAPI blue) and Maf factors (Red), using anti- MafA (D and G), MafB (E), cMaf (F), and large-Maf (H) antibodies. Each figure shows combined three-color image, while the insets show signals from individual color channels. Magnification bar = 20 μm.
Expression of MafB precedes MafA expression during pancreatic development
Large-Maf factors have highly conserved C-terminal DNA binding and protein dimerization domains (Blank and Andrews, 1997). However, homology in the N-terminal activation domain is less conserved, suggesting possible differences in their activation potentials and/or downstream targets. Thus, the expression of different large-Maf factors in the endocrine cells may differentially affect their function. Additionally, embryonic expression of these large-Maf factors may have significant impact on the differentiation of endocrine cell types. The anti-cMaf antibody that in adult pancreatic sections convincingly demonstrated cMaf expression in both α - and β-cells (Figure 1G,-H), was found unsuitable for detecting embryonic expression of this factor. Hence, to detect expression of cMaf during embryonic development, in situ hybridization was performed on the frozen pancreatic sections from E17.5 embryos; these sections were immunostained to detect insulin and glucagon expressing cells (Figure 3). Our results demonstrate that as in pancreatic islets from the adult mice, cMaf was expressed in both insulin and glucagon producing cells. Unlike cMaf, MafA is exclusively expressed in β-cells and the expression of MafB is specific to α-cells in the adult pancreas. Thus, we hypothesized that MafA is important for the differentiation of β-cells while MafB has a similar role in the α-cells. Therefore, we focused our study on examination of spatial and temporal expression patterns of MafA and MafB during embryonic development to understand their roles in the pancreas.
Figure 3. During embryonic development cMaf is expressed in both insulin+ and glucagon+ cells.
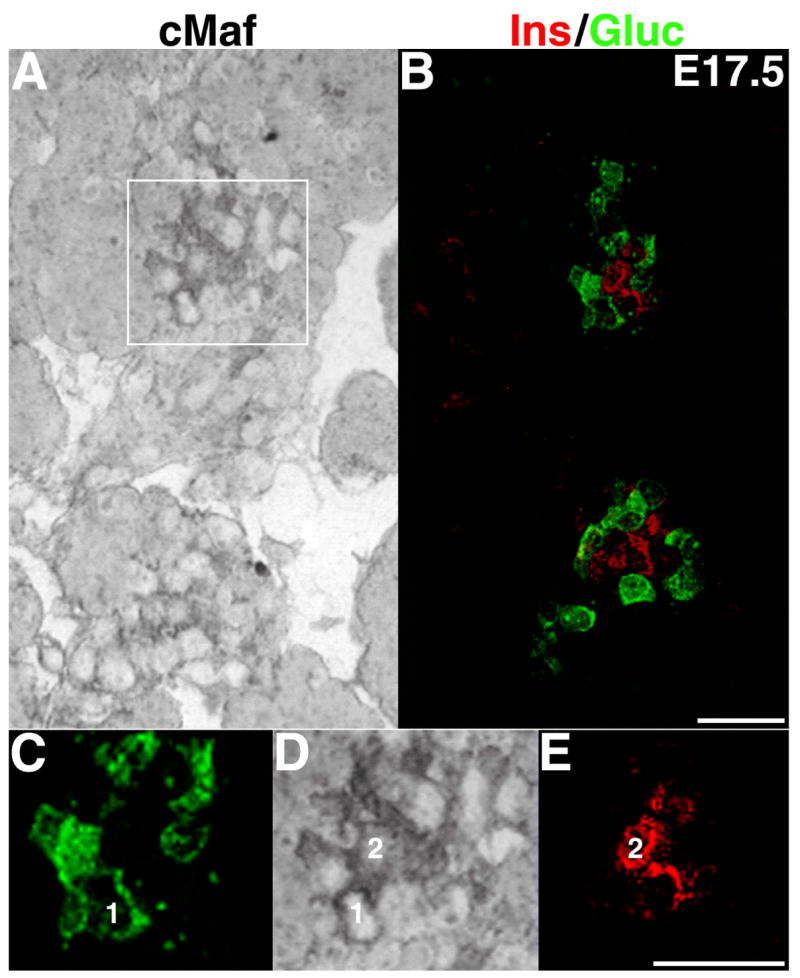
A, A light microscope image of E17.5 pancreas following in situ hybridization shows cMaf expression (dark staining). B, Confocal image of the same section stained for insulin (red) and glucagon (green). C-E, are higher magnification images of inset shown in A and the corresponding region from B. One insulin- and one glucagon-expressing cell are numbered in the higher magnification image to clearly show that cMaf is expressed in both cell types. Magnification bar = 20 μm.
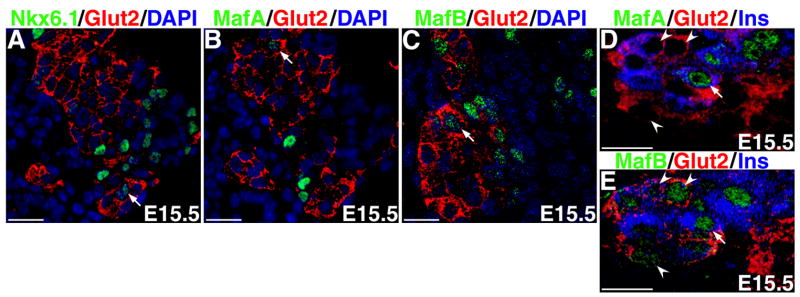
Immunostained sections of pancreas from E10.5-P14 were examined for co-expression of MafA with insulin, and MafB with glucagon. Glucagon+ cells can be seen soon after pancreatic specification, and by E10.5 glucagon+ cells are seen in the pancreatic epithelium (Figure 4C). Interestingly, cells in these early glucagon+ clusters express MafB, but occasional MafB+ cells near these clusters did not express glucagon (Figure 4C, F). These data suggest that embryonically MafB is expressed in pancreatic cell types other than glucagon or that its expression precedes that of glucagon. In contrast to glucagon+ cells that are easily detected by E10.5, insulin+ cells are rare, and their number increases only after the secondary transition (around E13.5). At E10.5 and 11.5 neither insulin+ nor MafA+ cells were found, but the rare insulin+ cells at E12.5 and E13.5 expressed MafA (Figure 4G, I). These results are similar to the report that MafA+ cells can be detected only from E13.5 (Matsuoka et al., 2004). The apparent discrepancy of our finding a few insulin+ MafA+ cells at E12.5 and Matsuoka et.al. not (2004), could be due simply to the limited number of insulin+ cells observed in the E12.5 pancreas. By E15.5, there is a significant increase in the number of insulin+ and glucagon+ cells (Figure 4J, K). All of the MafA+ cells at this stage co-expressed insulin, but a significant proportion of insulin+ cell did not express MafA. Interestingly, at E15.5 nearly all (98%) glucagon+ cells expressed MafB, but a significant number of MafB+ cells did not express glucagon. MafB is selectively expressed in the α-cells of the adult pancreatic islets; we determined that MafB expression gradually becomes restricted to the α-cells after birth, and by two weeks few MafB+ glucagon− cells are seen (Figure 4L, M, N).
Figure 4. MafA and MafB expression pattern during embryonic development.
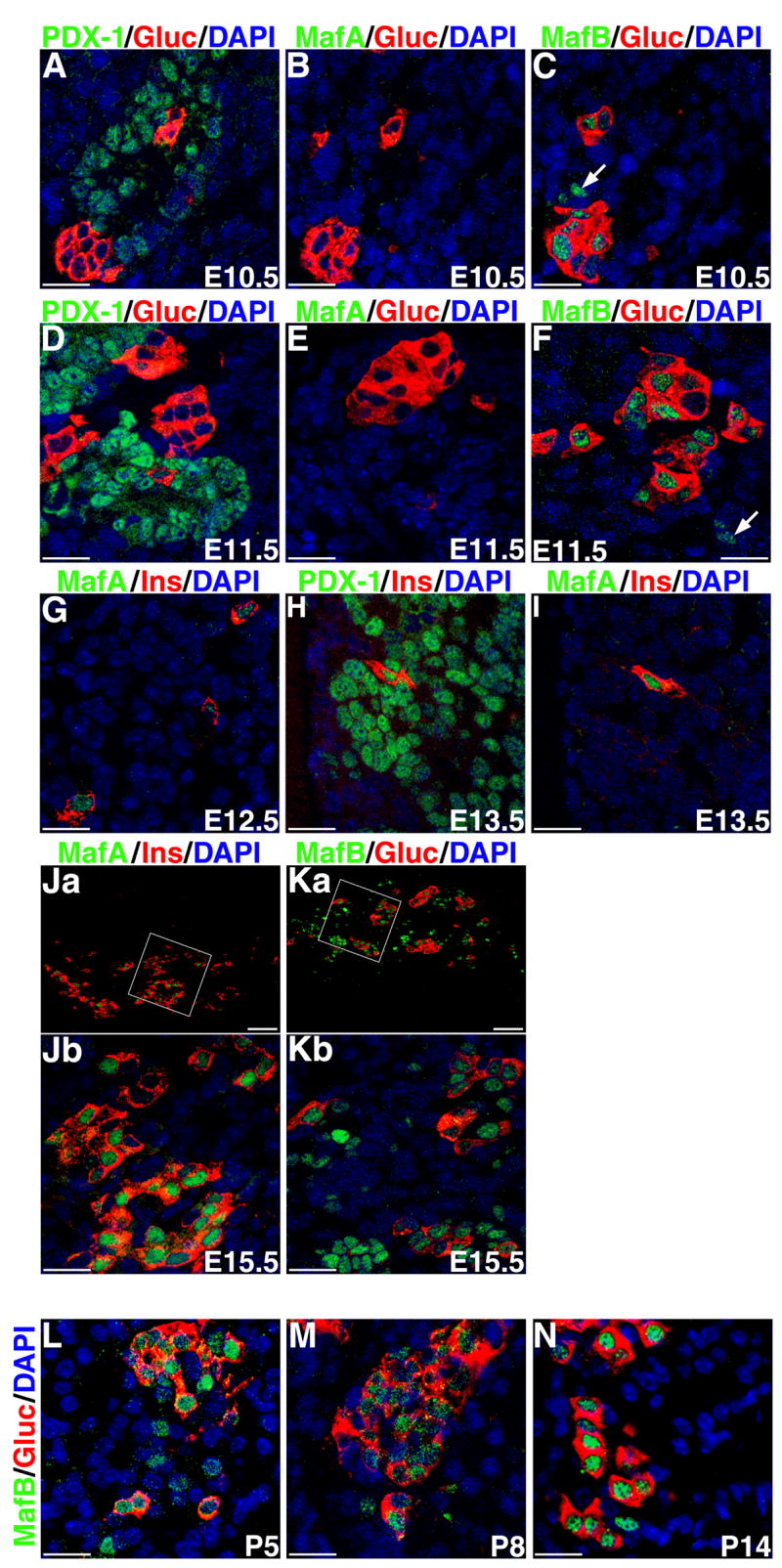
Confocal images of immunostained pancreatic sections from indicated embryonic (E10.5– E15.5) and post-natal days 5, 8, and 14. Sections were stained with indicated antibodies that recognize transcription factors PDX-1, MafA or MafB (green) or hormones insulin or glucagon (red) and nuclei (DAPI blue). Results show that MafB is expressed earlier (E10.5) than MafA (E12.5) in developing pancreas. MafA expression is restricted to insulin+ cells. At the earlier time points (E10.5–11.5), the majority of MafB+ cells express glucagon, and only occasional glucagon− MafB+ cells are seen (arrows in C and F), but around E15.5, there is a significant increase in the number of glucagon− MafB+ cells. After birth, expression of MafB again becomes restricted to glucagon+ cells. Magnification bar = 20 μm for all images except Ja and Ka, where magnification bar = 50μm.
MafB is expressed in the insulin-producing cells during development
The increased number of MafB+ glucagon− cells around secondary transition suggests an important role of this factor in regulating differentiation of other pancreatic cell-type(s). To determine if the E15.5 MafB+ glucagon− cells were endocrine cells, pancreatic sections were immunostained to detect MafB and synaptophysin expression. All MafB+ cells were synaptophysin+, but not all synaptophysin+ cells expressed MafB (Figure 5A), suggesting that MafB is expressed in only some endocrine cells at this stage of development. Since the number of insulin+ cells increases at the secondary transition, we examined if the MafB+ Glucagon− cells expressed insulin and the MafA+ cells expressed glucagon. We observed that MafA+ cells did not express glucagon, while many MafB+ cells were insulin+(Figure 5B, C). This was in direct contrast with the highly selective α-cell expression of MafB in adult islets. Quantitation showed that all the 137 MafA+ cells co-expressed insulin, while nearly 60% of MafB+ cells were insulin+ (Figure 5F). Interestingly, only half (54%) of the insulin+ cells were MafA+ whereas nearly 90% of insulin+ cells expressed MafB. These observations demonstrate that at E15.5 some insulin+ cells express MafB, a significant proportion express both MafA and MafB, and a minor population only express MafA.
Figure 5. MafB is expressed in insulin producing cells during embryonic development.
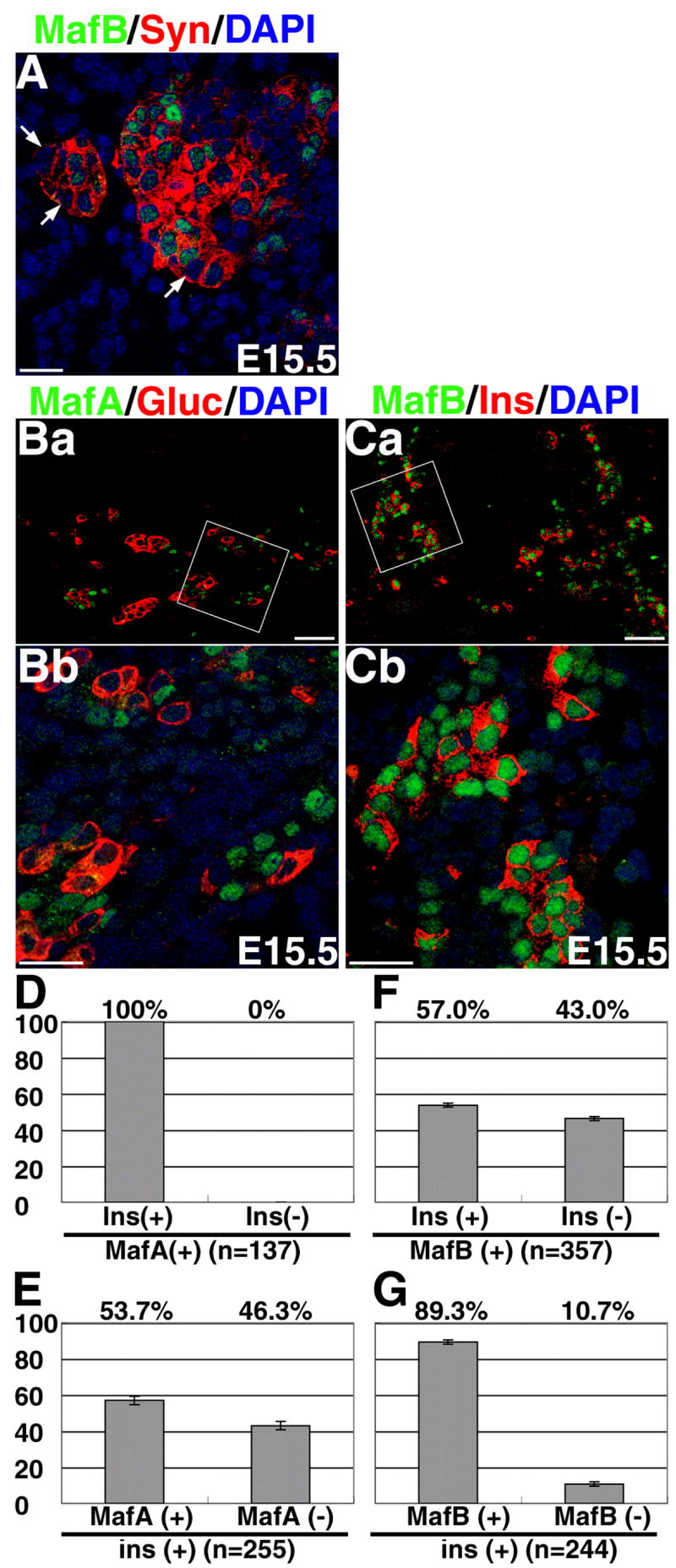
A, Confocal images of E15.5 pancreas stained for MafB (green), Synaptophysin (red) and nuclei (DAPI, Blue). All MafB+ cells are Synaptophysin+, but not all Synaptophysin+ cells are MafB+ (arrow). Images B and C show E15.5 pancreas stained for MafA (green), glucagon (red), and nuclei (DAPI, blue), or MafB (green) insulin (red) and nuclei (DAPI, blue), respectively. Images Bb and Cb show areas marked in Ba and Ca at higher magnification. Quantitation is shown as percentage of MafA+(D) or MafB+(F) cells expressing insulin or the percentage of insulin+ cells expressing MafA (E) or MafB (G). Results are from at least five independent sections from three different pancreases. A significant proportion of MafB+ cells express insulin. MafA is expressed only in the insulin+ cells, but several insulin+ cells do not express MafA. Magnification bar = (Ba, Ca): 50 μm, (A, Bb, Cb): 20 μm.
To determine MafA and MafB co-expression with insulin, we stained thin (3μm) consecutive sections of E15.5 pancreas with appropriate antibodies (Figure 6A, B). Insulin staining in consecutive sections identified cells that were present in both sections, thus permitting determination of MafB and MafA expression in the same cell. This analysis provides novel information regarding the differentiation of insulin+ cells during embryonic development: 1) all insulin+ cells expressed at least MafA or MafB; 2) in insulin+ cells, we can identify all possible combinations (only MafB, only MafA or both) of large-Maf factor expression (Figure 6A and B, and insets). The observed proportion of these three cell-types was consistent with the results in Figure 4. MafB is expressed earlier than the MafA (Figures 4 and 5) and becomes restricted to α-cells during the post-natal period (Figure 4), and adult insulin+ cells express MafA (Figure 1). Furthermore, we found that during different stages of embryonic development less than 1% of insulin+ cells (E13.5 none, E15.5 and E18.5 0.6%) are apoptotic as judged by condensed nuclei visualized with propidium iodide (data not shown); these data suggest that the majority of insulin+ MafB+ cells at E15.5 survive and give rise to the insulin+ cells found after birth. We propose a model in which in any given insulin+ cell MafB expression precedes that of MafA, with a gradual inhibition in MafB expression in the MafA+ MafB+ insulin+ cells, resulting in the generation of insulin+ MafA+ cells. A future lineage tracing study should provide definitive proof for such switch. However, this model raises several important questions, including whether MafB turns on the expression of insulin and glucagon, that is, is MafB upstream of these hormones? What regulates the choice of MafB+ cells to express either insulin or glucagon? What turns on MafA expression in MafB+ insulin+ cells? Finally, why do insulin+ cells transition from MafB+ to a MafA+ cell-type when both factors can bind to the insulin MARE and activate insulin gene expression?
Figure 6. Expression of MafA and MafB in the insulin+ cells.
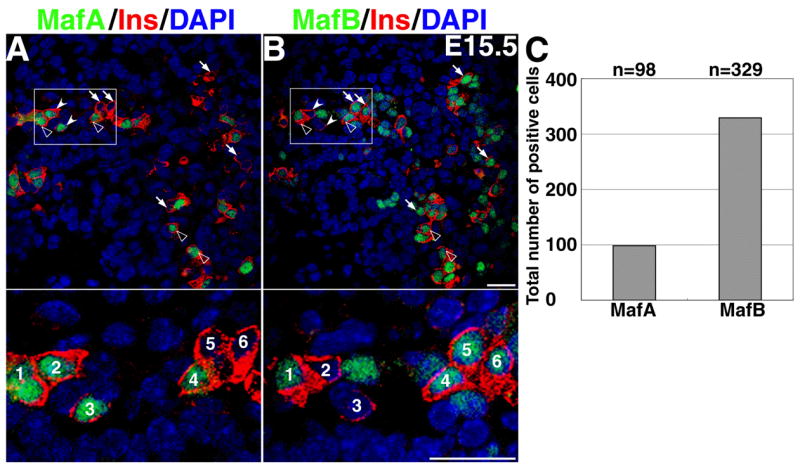
Two consecutive sections from E15.5 mouse pancreas stained with the indicated antibodies to detect insulin (red), nuclei (blue) and either MafA (A) or MafB (B). One can identify all possible combinations of MafA and MafB expression in the insulin+ cells. Cells indicated with arrows are MafB+ insulin+ but MafA−, cells indicated with the open arrow-heads express both MafA and MafB, while cells indicated by filled arrow-head demonstrate the presence of MafA+ insulin+ MafB− cells. Lower images are higher magnification of the boxed areas in A and B; cells are numbered to identify the same cells in both sections. Total number of MafA and MafB expressing nuclei at E15.5 from five adjacent pairs of sections from three different pancreases (C) shows over 3 fold higher proportion of MafB+ cells at this stage (p=0.01). Magnification bar = 20 μm.
Differential expression of Nkx6.1 in MafB+ cells defines the cells that express insulin or glucagon
The generation and analysis of MafB knockout mice will eventually address whether MafB is upstream or downstream of insulin and glucagon gene expression. However, our results suggest that MafB is an important differentiation factor for both α - and β-cells. A significant proportion of insulin+ cells expressed both MafA and MafB, but glucagon+ cells expressed only MafB, so we decided to test whether the expression of Nkx6.1 correlated with the ability of MafB+ cells to express insulin. The transcription factor Nkx6.1 is critical for the differentiation of β-cells (Sander et al., 2000), and is required for the expression of MafA (Matsuoka et al., 2004). In consecutive sections of E15.5 pancreas significantly more cells expressed Nkx6.1 than MafB+ (Figure 7A, B). As reported previously (Sander et al., 2000), at this stage, a reasonable proportion of Nkx6.1+ cells are insulin− (Figure 7B). We observed that at E15.5, the majority of insulin+ cells also expressed Nkx6.1and MafB (arrow Figure 7A, B and insets) but that MafB+ glucagon+ cells did not express NKX6.1 (closed triangle, Figure. 7C, D). Importantly, cells that were Nkx6.1+ MafB+ were glucagon− (arrows Figure. 7C, D). Thus, the ability of MafB-expressing cells to expresses glucagon or insulin depends on the co-expression of Nkx6.1.
Figure 7. MafB+ and insulin+ cells express NKX6.1 but MafB+ glucagon+ cells are NKX6.1−.
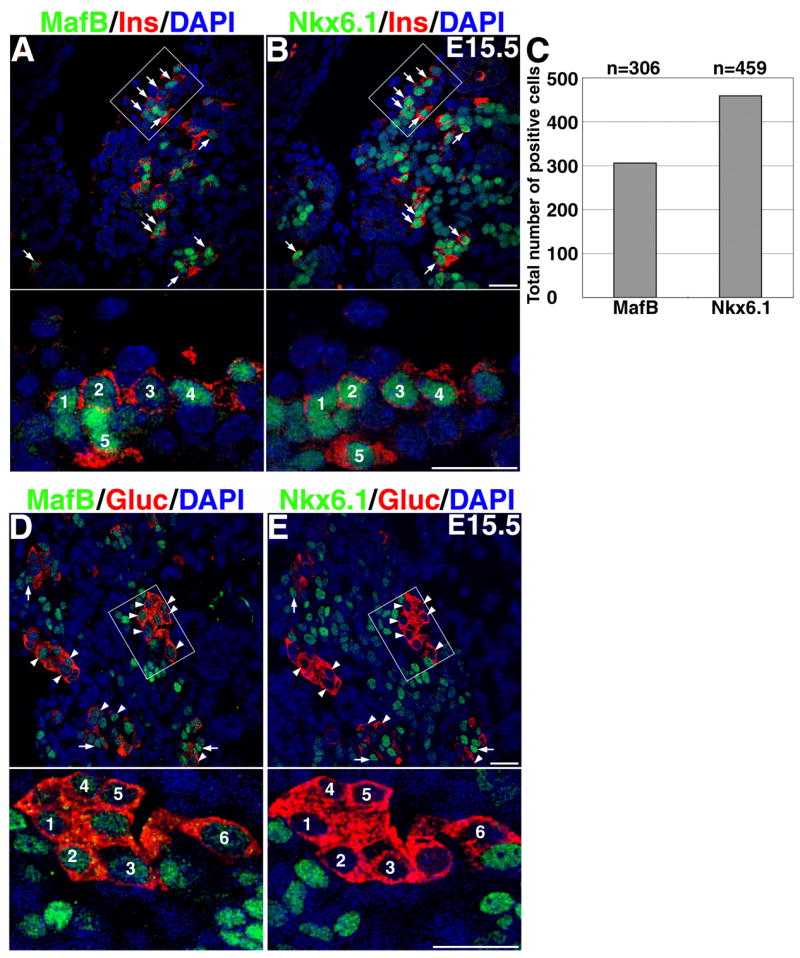
Consecutive sections A and B shows cells expressing insulin (red), nuclei (blue) and either MafB or Nkx6.1(green). Arrows denote cells that are present in both sections and express both Nkx6.1 and MafB. All of the insulin+ cells are Nkx6.1+, but not all Nkx6.1+ cells were insulin+. Higher magnification image of areas marked in D and E are shown at the bottom. Total number of nuclei expressing the indicated factor at E15.5 from five adjacent pairs of sections from three different pancreases(C). Significantly more (50% p=0.001) Nkx6.1+ cells are seen at E15.5 than MafB+ cells. D and E represent consecutive sections stained to detect glucagon (red), nuclei (blue) and MafB or Nkx6.1 (green). The arrowheads denote glucagon+ MafB+ cells that are Nk6.1−, while arrows indicate MafB+ Nkx6.1+ cells that are glucagon−. Magnification bar = 20 μm.
Nkx6.1 is upstream of MafA during pancreatic development and is important for the differentiation of insulin+ cells. However, Nkx6.1 is not likely to be upstream of MafB since glucagon+ MafB+ cells do not express Nkx6.1 [glucagon+ Nkx6.1+ cells are extremely rare (Henseleit et al., 2005)]. Even so, we cannot rule out the possibility that Nkx6.1 is upstream of MafB in cells that will turn on insulin expression. Since a few insulin+ cells can be detected in Nkx6.1 knockout mice (Henseleit et al., 2005; Matsuoka et al., 2004), Nkx6.1 may not be essential for the formation of insulin+ cells. This finding suggests two other possibilities for differentiation of insulin+ cells: 1) MafB can initiate differentiation of insulin-expressing cells and Nkx6.1 is required for the maintenance of insulin expression and induction of MafA; 2) Nkx6.1 and MafB together trigger the differentiation of insulin+ cells (we did observe a few rare MafB+ Nkx6.1+ cells that were insulin−). Demonstration of MafB expression in the remaining insulin+ cells in Nkx6.1 knockout mice would support the first possibility. The absence of insulin+ cells, even in the presence of Nkx6.1+ cells, in MafB knockout mice would also be consistent with the first possibility. However, reduced numbers of insulin+ cells in MafB knockout mice would support the second. The definitive answer for the importance of MafB and Nkx6.1 in the differentiation of insulin+ cells will only come from the analyses of these knockout mice.
Role of MafA in β-cell replication and maturation
Our results suggest a switch in the expression from MafB to MafA in insulin+ cells, raising questions such as, what regulates the switch in the transcription factor expression, and is it necessary? The MafA knockout mouse study (Zhang et al., 2005a) would suggest that the switch to MafA is required for the survival/proliferation and function of β-cells. The loss of MafA resulted in normal pancreatic morphology at birth, but after birth a reduction in the proportion of β-cells with resulting impaired glucose tolerance and diabetes (Zhang et al., 2005a).
Terminally differentiated β-cells have glucose-induced insulin secretion whereas fetal and neonatal islets are not yet glucose-responsive. The MafA knockout study showed Glut2 expression reduced in the knockout animals. Since more fetal and neonatal insulin+ cells express MafB than in adult mice, the transition from MafB to MafA expression may initiate the glucose-responsiveness of insulin+ cells by the selective induction of glucose transporter Glut2. In E15.5 pancreas (Figure 8), Glut2 is expressed in some MafA+, MafB+ or Nkx6.1+ cells. Some of these MafB+ Glut2+ or MafA+Glut2+ cells also express insulin. This suggests that the switch from MafB to MafA may not be needed for the maturation of β-cells, as judged by the induction of Glut2 expression. The observed expression of Glut2 in MafB+ or MafA+ cells may reflect differentiation of these cells from Glut2+ precursors (Pang et al., 1994). We find that at E15.5, more MafB+ than MafA+ cells express Glut2, which is consistent with our hypothesis that during differentiation of insulin+ cells, MafB is induced first, followed by the expression of MafA. This would suggest that the Glut2 expression at E15.5 is a marker of precursor cells rather than the maturation of β-cells. The lack of a correlation between MafA and Glut2 expression in insulin+ cells (Figure 8D) is not consistent with the observed reduction in the Glut2 expression in MafA null mice (Zhang et al., 2005a). However, as reported in other animal models (Thorens et al., 1990), reduction in the Glut2 expression could result from the hyperglycemia in these mice. Thus, our results demonstrate that the switch from MafB to MafA at E15.5 is not required for the induction of Glut2 expression but that MafA expression may turn-on additional genes critical for the function of adult β-cells.
Figure 8. MafB to MafA transition is not essential for Glut2 expression.
Triple IHC detection of A) Nkx6.1, B) MafA or C) MafB expression (green) with Glut2 (red) and nuclei (DAPI blue) shows that at E15.5 only some of the transcription factor expressing cells are Glut2+(arrow). Staining in E shows that some MafB+ insulin+ cells are Glut2+MafA− (arrowhead), thus demonstrating that MafA expression in the insulin+ cells is not essential for the expression of Glut2 at this stage of development. Magnification bar = 20 μm.
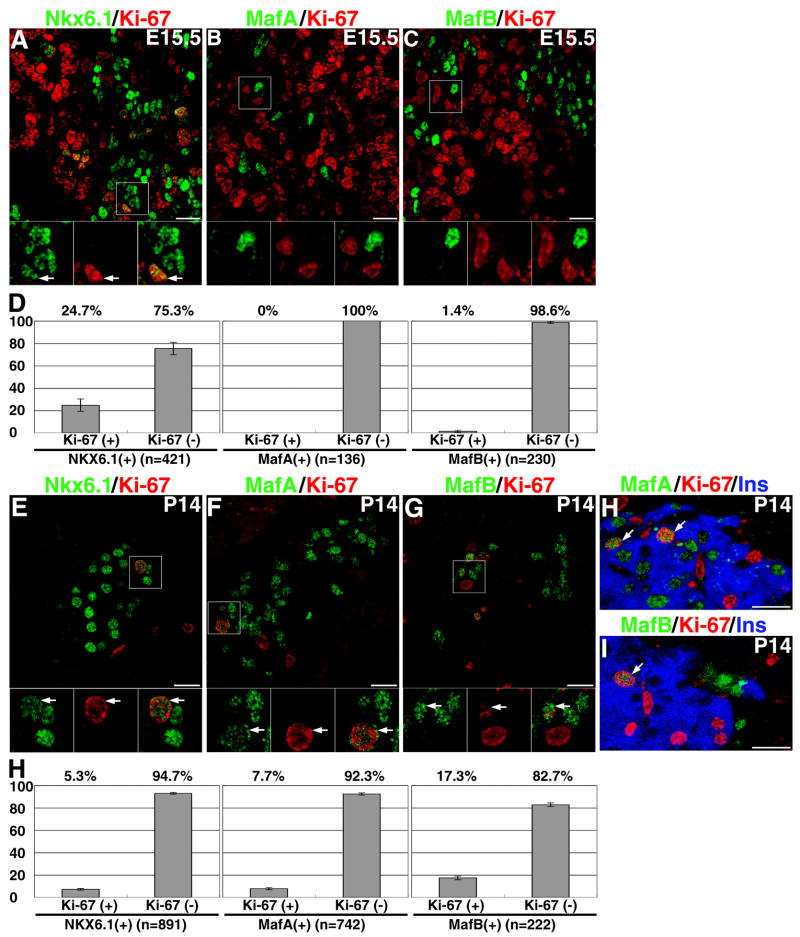
The loss of MafA results in a reduced proportion of β-cells after birth, suggesting a possible role of the transition from MafB to MafA in the replication/survival of β-cells. To test for a role of these factors in β-cell replication, the co-expression of cell cycle marker Ki67 with MafA and MafB factors was examined in E15.5 and P14 pancreas. Consistent with previous observations that newly differentiated endocrine cells do not proliferate, we found in E15.5 mice that nearly 25% of the Nkx6.1+ cells are in the cell cycle (Ki67+) but none of the MafA+ cells, and only a rare MafB+ cell expressed Ki67 (1.3%, 3 out of 230 MafB+ nuclei) (Figure 9A-D). However, from shortly before birth up to two weeks after birth, the replication rate of endocrine cells is significant (Figure 9E-H). At P14, the proportion of Ki67+ Nkx6.1+ cells was reduced significantly (5.3%) and was comparable to the proportion of Ki67+MafA+ cells (7.7%), possibly reflecting the restriction of Nkx6.1 to the β-cells. Nearly 17% of MafB+ cells expressed Ki67, showing that MafB expression does not impede replication. It is important to note that the increased replication rate of the MafA+ and MafB+ cells after birth accompanies the change from co-expression to single expression of these factors. Even though the majority of MafB+ cells at P14 are α-cell (Figure 4L), we can still detect a few Ki67+ MafB+ insulin+ cells. Thus, our results suggest that the presence of MafB in the insulin+ cell does not prevent their replication. However, the paucity of MafB+glucagon− cells at this stage and the fact that some MafB+ insulin+ cells may already express MafA preclude our being able to conclude whether the expression of MafB in the insulin+ cells impairs but does not prevent β-cell replication.
Figure 9. MafB expression does not inhibit replication of insulin+ cells.
Mouse pancreatic sections at E15.5 were stained to detect (A) NKX6.1, (B) MafA, or (C) MafB (green) with Ki67 (red) and DAPI (blue). Insets show higher magnification of demarcated areas. Arrows indicate cells expressing both Ki67 and the indicated transcription factor. D) Quantitation of Ki67+ cells showed that nearly 25% of Nkx6.1 cells are in cell cycle, while MafA and MafB expressing cells do not express Ki67 at this stage. Expression of NKX6.1 (E), MafA (F) or MafB (G) (green) in Ki67 (red) positive cells in pancreas at P14. H and I show Ki67 (red), insulin (blue) and either MafA (H) or MafB (I) in green. J. Quantitation of at least five independent sections from three different mice for each transcription factor shows similar levels of Ki67+, Nkx6.1+ and MafA+ cells, while a higher proportion of MafB+ cells express Ki67. I shows insulin+ MafB+ cell expressing Ki67 at P14 in the pancreatic islet. Magnification bar = 20 μm.
PDX-1 expression precedes induction of MafA in MafB+ Insulin+ cells
Transcription factor Nkx6.1 is upstream of MafA (Matsuoka et al., 2004), but at E15.5 not all Nkx6.1+ insulin+ cells express MafA with a significant proportion being MafB+ (Figure 7 and data not shown), suggesting that signals in addition to NKx6.1 are needed to trigger MafA expression. Transcription factors PDX-1, HB9 and Nkx2.2 are widely expressed in the early pancreatic epithelium. With time their expression gradually becomes restricted while others, such as Pax4, Pax6 and NeuroD1, are expressed in the endocrine progenitors, following Ngn3 expression. Recent studies demonstrate that the transcription factors Nkx2.2 and Pax4 work in parallel to enable β-cell differentiation (Prado et al., 2004; Wang et al., 2004). Nkx2.2, its downstream target Pax6, and Pax4 enhance expression of PDX-1, HB9 and Nkx6.1, which are required for the maturation of β-cells. Interestingly, in a recent review, Sosa-Pineda reports that loss of Pax4 inhibits MafA expression (Sosa-Pineda, 2004), suggesting a possible link between MafA expression and β-cell maturation. Unfortunately, there are no good antibodies to monitor Pax4 expression during pancreatic development and hence we cannot determine the role of this factor in MafB to MafA switch. A recent lineage-tracing study demonstrates a role for Pax6 in the expression of terminal differentiation markers such as insulin and Glut2 (Ashery-Padan et al., 2004). Since the MafB to MafA switch may regulate terminal differentiation, we analyzed the role of transcription factors PDX-1 and Pax6 in inducing MafA expression.
During lens development in the chicken, Pax6 is required for the expression of MafA/L-Maf, thus placing Pax6 upstream of MafA (Reza et al., 2002; Reza and Yasuda, 2004). To determine if Pax6 is upstream of MafA expression in the insulin+ cells, consecutive sections were stained for MafA-insulin and Pax6-insulin. Pax6 is expressed in all endocrine cells, so at E15.5 all insulin+ and several insulin− cells expressed it (Figure 10B). MafA is expressed only in some insulin+ cells (Figure 10A), so several Pax6+ insulin+ cells were MafA+ (indicated by arrows) while some were MafA− (shown in inset). Pax6 is expressed in all insulin+ cells, so we can conclude that Pax6 is upstream of MafA during β-cell differentiation but cannot determine if Pax6 is required for switching the expression from MafB to MafA.
Figure 10. Pax6 expression precedes MafA expression.
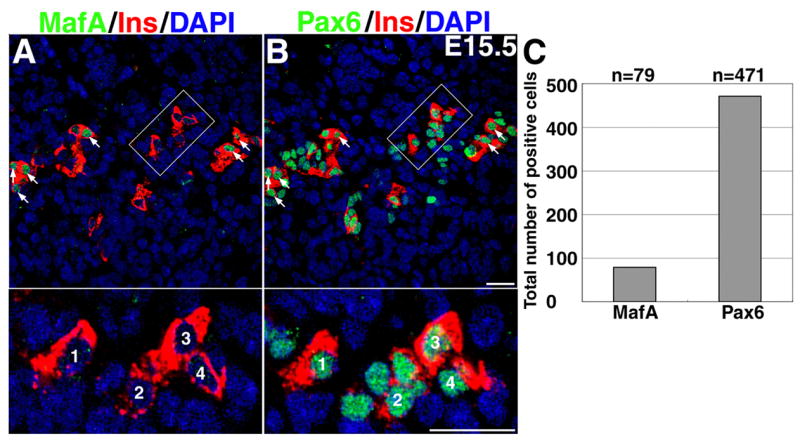
Consecutive sections from E15.5 pancreas stained to detect MafA (A) and Pax6 (B) show that all insulin+ cells express Pax6 but not MafA. Arrows in A indicate MafA+ cells. Several insulin+ Pax6+ cells that are MafA− are shown at the higher magnification. Total number of nuclei expressing the indicated factor at E15.5 from five adjacent pairs of sections from three different pancreases(C). Nearly 6-fold more nuclei were Pax6+ than MafA+ at this stage of development (p=0.004).
To assess the role of PDX-1 in triggering MafA expression in MafB+ cells, consecutive sections from E15.5 pancreases were stained for insulin and MafA, MafB or PDX-1 (Figures 11A, B and 11 C, D). As previously reported (Oster et al., 1998; Jensen et al., 2000), PDX-1 expression can be varied at this stage with most PDX-1high cells being insulin+ while few PDX-1low cells expressed insulin+. All MafA+ cells were PDX-1high, and only a few insulin+ PDX-1high cells did not express MafA (insets Figures 11A, B). Since insulin+ cells express at least one of the two large-Maf factors, Pdx-1low insulin+ MafA− cells and PDX-1high insulin+ MafA− cells must express MafB. Figures 11C and D confirm these predictions and demonstrate that at E15.5, insulin+ MafB+ cells could express either PDX-1high or PDX-1low but that MafA is expressed only in cells with high levels of PDX-1. These data suggest that PDX-1 is enhanced prior to the expression of MafA but after MafB expression and so may play a role in turning on MafA in MafB+ cells. MafA can regulate PDX-1 expression in adult islets (Samaras et al., 2003). However, the presence of PDX-1high MafA− insulin+ cells at secondary transition stage suggests that MafA does not activate PDX-1 expression at this developmental stage. Our results demonstrating that increased expression of PDX-1 in insulin+ cells at secondary transition occurs before MafA induction suggest that the MafB to MafA switch proceeds through a PDX-1high intermediate stage. Analysis of endocrine-cell specific PDX-1 knockout mice (PDX-1PBCreERTM, (Zhang et al., 2005b)) could determine the role of PDX-1 in this switch.
Figure 11. Induction of PDX-1high in the insulin+ cells precedes the switch in expression from MafB+ to MafA+.
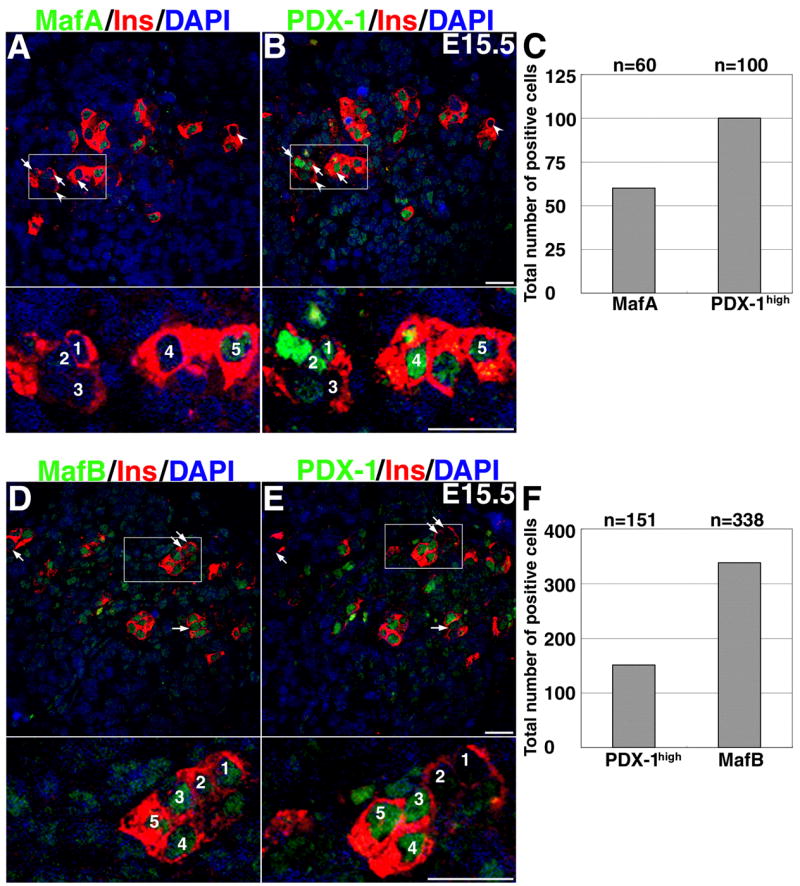
Co-expression of MafA (A) and PDX-1high (B) in the insulin+ cells were determined by staining consecutive sections. Total number of nuclei expressing the indicated factor at E15.5 from five adjacent pairs of images from three different pancreases(C, F). At E15.5, nearly 66% more nuclei were expressing high level of PDX-1high than MafA (p=0.006), yet at this stage not all insulin+ cells were PDX-1high. Arrows indicate PDX-1high cells that were MafA−, and arrowheads indicate insulin+ cells that are MafA− PDX-1low. D and E show that some insulin+ MafB+ cells were PDX-1low (arrows), while others were PDX-1high. There are nearly two-fold more MafB+ than the Pdx-1high cells at this stage (F, p=0.01). These results show that PDX-1high expression is induced after MafB but before MafA expression.
In summary, we have shown that the differentiation of β-cells (insulin+ MafA+) from pancreatic endocrine precursors goes through an intermediate stage of insulin+ MafB+ that after the induction of PDX-1 gives rise to insulin+ MafB+ MafA+ cells, which eventually become insulin+ MafA+ cells (Figure 12). Since insulin+ MafB+ cells are detected first and can be seen even after birth, our results suggest that MafA expression is not necessary for the formation of insulin+ cells during embryonic development and that MafB is sufficient for the formation of insulin+ cells. This observation provides an explanation for the normal looking pancreatic islets in MafA knockout mice at postnatal day 1(Zhang et al., 2005a). Our data provides strong evidence for a role of MafB in regulating differentiation of both α- and β-cells, with the possibility that MafB, either independently or together with Nkx6.1, triggers insulin expression. Analyses of MafB and Nkx6.1 knockout mice for the expression of MafA, MafB and insulin should further define the role of these factors in triggering insulin expression.
Figure 12. Model of Maf factor transition and the differentiation of insulin+ cells.
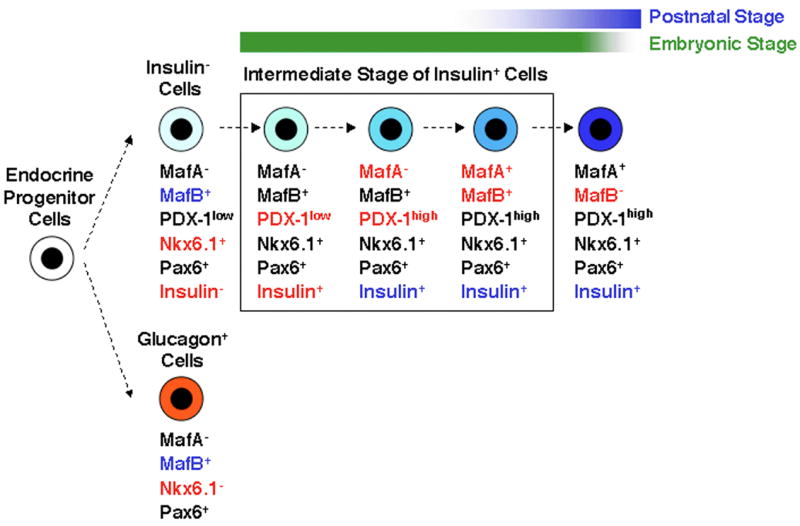
Endocrine precursors expressing both MafB and Nkx6.1 differentiate towards an insulin+ phenotype, while precursors expressing MafB alone follow a pathway to glucagon-expressing cells. In addition to MafB and Nkx6.1, initial insulin+ cells are Pax6+, PDX-1low and MafA−. Subsequently, there is induction of PDX-1high expression, which is followed by expression of MafA in the insulin+ cells. Towards the later stages of embryonic development and the early neonatal period, insulin+ MafA+ MafB+ cells can be seen and these then differentiate into insulin+ MafA+ MafB− cells.
Acknowledgments
We thank Dr I-C Ho for providing cMaf expression plasmid, Dr. Dan Drucker for glucagon:Luciferase reporter, Dr. Chris Wright for PDX-1 antibody, Drs. O. Madsen and P. Serup and NIH funded Beta Cell Biology Consortium for Nkx6.1 antibody. This study was supported in part by research grants NIH DK060127 and one from American Diabetes Association to AS, postdoctoral fellowships from Juvenile Diabetes Research Foundation (3-2005-74) to WN and from The Adler Foundation to TK, IEK is recipient of NIH institutional training grant (T32), and the Media, Animal, and Advanced Microscopy (Histology and Confocal facilities) Cores of the Joslin Diabetes Endocrinology Research Center (NIH DK-36836).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Maf factors are important for pancreatic differentiation
References
- Ashery-Padan R, Zhou X, Marquardt T, Herrera P, Toube L, Berry A, Gruss P. Conditional inactivation of Pax6 in the pancreas causes early onset of diabetes. Dev Biol. 2004;269:479–488. doi: 10.1016/j.ydbio.2004.01.040. [DOI] [PubMed] [Google Scholar]
- Blank V, Andrews NC. The Maf transcription factors: regulators of differentiation. TIBS. 1997;22:437–441. doi: 10.1016/s0968-0004(97)01105-5. [DOI] [PubMed] [Google Scholar]
- Boam DSW, Docherty K. A tissue-specific nuclear factor binds to multiple sites in the human insulin-gene enchancer. Biochem J. 1989;264:233–239. doi: 10.1042/bj2640233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe DT, Tsai M-J. Mutagenesis of the rat insulin II 5'-flanking region defines sequences important for expression in HIT cells. Mol Cell Biol. 1989;9:1784–1789. doi: 10.1128/mcb.9.4.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington RH, Sharma A. Transcription factors recognizing overlapping C1-A2 binding sites positively regulate insulin gene expression. J Biol Chem. 2001;276:104–113. doi: 10.1074/jbc.M008415200. [DOI] [PubMed] [Google Scholar]
- Henseleit KD, Nelson SB, Kuhlbrodt K, Hennings JC, Ericson J, Sander M. NKX6 transcription factor activity is required for alpha- and beta-cell development in the pancreas. Development. 2005;132:3139–3149. doi: 10.1242/dev.01875. [DOI] [PubMed] [Google Scholar]
- Jensen J, Heller RS, Funder-Nielsen T, Pedersen EE, Lindsell C, Weinmaster G, Madsen OD, Serup P. Independent development of pancreatic alpha- and beta-cells from neurogenin3-expressing precursors: a role for the notch pathway in repression of premature differentiation. Diabetes. 2000;49:163–176. doi: 10.2337/diabetes.49.2.163. [DOI] [PubMed] [Google Scholar]
- Jonsson J, Carlsson L, Edlund T, Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature. 1994;371:606–609. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- Karlsson O, Edlund T, Moss JB, Rutter WJ, Walker MD. A mutational analysis of the insulin gene transcription control region: expression in beta cells is dependent on two related sequences within the enhancer. Proc Natl Acad Sci U S A. 1987;84:8819–8823. doi: 10.1073/pnas.84.24.8819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka K, Han SI, Shioda S, Hirai M, Nishizawa M, Handa H. MafA is a glucose-regulated and pancreatic beta-cell-specific transcriptional activator for the insulin gene. J Biol Chem. 2002;277:49903–49910. doi: 10.1074/jbc.M206796200. [DOI] [PubMed] [Google Scholar]
- Kataoka K, Shioda S, Ando K, Sakagami K, Handa H, Yasuda K. Differentially expressed Maf family transcription factors, c-Maf and MafA, activate glucagon and insulin gene expression in pancreatic islet alpha- and beta-cells. J Mol Endocrinol. 2004;32:9–20. doi: 10.1677/jme.0.0320009. [DOI] [PubMed] [Google Scholar]
- Kawauchi S, Takahashi S, Nakajima O, Ogino H, Morita M, Nishizawa M, Yasuda K, Yamamoto M. Regulation of lens fiber cell differentiation by transcription factor c-Maf. J Biol Chem. 1999;274:19254–19260. doi: 10.1074/jbc.274.27.19254. [DOI] [PubMed] [Google Scholar]
- Kim JI, Li T, Ho IC, Grusby MJ, Glimcher LH. Requirement for the c-Maf transcription factor in crystallin gene regulation and lens development. Proc Natl Acad Sci U S A. 1999;96:3781–3785. doi: 10.1073/pnas.96.7.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laybutt DR, Weir GC, Kaneto H, Lebet J, Palmiter RD, Sharma A, Bonner-Weir S. Overexpression of c-Myc in beta-cells of transgenic mice causes proliferation and apoptosis, downregulation of insulin gene expression, and diabetes. Diabetes. 2002;51:1793–1804. doi: 10.2337/diabetes.51.6.1793. [DOI] [PubMed] [Google Scholar]
- Lee YC, Asa SL, Drucker DJ. Glucagon gene 5'-flanking sequences direct expression of SV40 large T antigen to the intestine producing carcinoma of the large bowel in transgenic mice. J Biol Chem. 1992;267:10705–10708. [PubMed] [Google Scholar]
- Matsuoka TA, Artner I, Henderson E, Means A, Sander M, Stein R. The MafA transcription factor appears to be responsible for tissue-specific expression of insulin. Proc Natl Acad Sci U S A. 2004;101:2930–2933. doi: 10.1073/pnas.0306233101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka TA, Zhao L, Artner I, Jarrett HW, Friedman D, Means A, Stein R. Members of the large Maf transcription family regulate insulin gene transcription in islet beta cells. Mol Cell Biol. 2003;23:6049–6062. doi: 10.1128/MCB.23.17.6049-6062.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naya FJ, Huang HP, Qiu Y, Mutoh H, DeMayo FJ, Leiter AB, Tsai MJ. Diabetes, defective pancreatic morphogenesis, and abnormal enteroendocrine differentiation in BETA2/neuroD-deficient mice. Genes Dev. 1997;11:2323–2334. doi: 10.1101/gad.11.18.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura W, Salameh T, Kondo T, Sharma A. Regulation of insulin gene expression by overlapping DNA-binding elements. Biochem J. 2005;392:181–189. doi: 10.1042/BJ20050970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura W, Yao I, Iida J, Tanaka N, Hata Y. Interaction of synaptic scaffolding molecule and Beta -catenin. J Neurosci. 2002;22:757–765. doi: 10.1523/JNEUROSCI.22-03-00757.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offield MF, Jetton TL, Labosky P, Ray M, Stein R, Magnuson M, Hogan BLM, Wright CVE. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development. 1996;122:983–985. doi: 10.1242/dev.122.3.983. [DOI] [PubMed] [Google Scholar]
- Ogata A, Shimizu T, Abe R, Shimizu H, Sakai M. Expression of c-maf and mafB genes in the skin during rat embryonic development. Acta Histochem. 2004;106:65–67. doi: 10.1016/j.acthis.2003.10.001. [DOI] [PubMed] [Google Scholar]
- Ogino H, Yasuda K. Induction of lens differentiation by activation of a bZIP transcription factor, L-Maf. Science. 1998;280:115–118. doi: 10.1126/science.280.5360.115. [DOI] [PubMed] [Google Scholar]
- Ohlsson H, Thor S, Edlund T. Novel insulin promoter- and enhancer-binding proteins that discriminate between pancreatic alpha- and beta-cells. Mol Endocrinol. 1991;5:897–904. doi: 10.1210/mend-5-7-897. [DOI] [PubMed] [Google Scholar]
- Olbrot M, Rud J, Moss LG, Sharma A. Identification of beta -cell-specific insulin gene transcription factor RIPE3b1 as mammalian MafA. Proc Natl Acad Sci U S A. 2002;99:6737–6742. doi: 10.1073/pnas.102168499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oster A, Jensen J, Serup P, Galante P, Madsen OD, Larsson LI. Rat endocrine pancreatic development in relation to two homeobox gene products (Pdx-1 and Nkx 6.1) J Histochem Cytochem. 1998;46:707–715. doi: 10.1177/002215549804600602. [DOI] [PubMed] [Google Scholar]
- Pang K, Mukonoweshuro C, Wong GG. Beta cells arise from glucose transporter type 2 (GLUT2) expresing epithelial cells of the developing rat pancreas. Proc Natl Acad Sci USA. 1994;91:9559–9563. doi: 10.1073/pnas.91.20.9559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planque N, Leconte L, Coquelle FM, Benkhelifa S, Martin P, Felder-Schmittbuhl MP, Saule S. Interaction of Maf transcription factors with Pax-6 results in synergistic activation of the glucagon promoter. J Biol Chem. 2001;276:35751–35760. doi: 10.1074/jbc.M104523200. [DOI] [PubMed] [Google Scholar]
- Prado CL, Pugh-Bernard AE, Elghazi L, Sosa-Pineda B, Sussel L. Ghrelin cells replace insulin-producing beta cells in two mouse models of pancreas development. Proc Natl Acad Sci U S A. 2004;101:2924–2929. doi: 10.1073/pnas.0308604100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reza HM, Ogino H, Yasuda K. L-Maf, a downstream target of Pax6, is essential for chick lens development. Mech Dev. 2002;116:61–73. doi: 10.1016/s0925-4773(02)00137-5. [DOI] [PubMed] [Google Scholar]
- Reza HM, Yasuda K. Roles of Maf family proteins in lens development. Dev Dyn. 2004;229:440–448. doi: 10.1002/dvdy.10467. [DOI] [PubMed] [Google Scholar]
- Ring BZ, Cordes SP, Overbeek PA, Barsh GS. Regulation of mouse lens fiber cell development and differentiation by the Maf gene. Development. 2000;127:307–317. doi: 10.1242/dev.127.2.307. [DOI] [PubMed] [Google Scholar]
- Samaras SE, Zhao L, Means A, Henderson E, Matsuoka TA, Stein R. The islet beta cell-enriched RIPE3b1/Maf transcription factor regulates pdx-1 expression. J Biol Chem. 2003;278:12263–12270. doi: 10.1074/jbc.M210801200. [DOI] [PubMed] [Google Scholar]
- Sander M, Sussel L, Conners J, Scheel D, Kalamaras J, Dela Cruz F, Schwitzgebel V, Hayes-Jordan A, German M. Homeobox gene Nkx6.1 lies downstream of Nkx2.2 in the major pathway of beta-cell formation in the pancreas. Development. 2000;127:5533–5540. doi: 10.1242/dev.127.24.5533. [DOI] [PubMed] [Google Scholar]
- Sharma A, Stein R. Glucose-induced transcription of the insulin gene is mediated by factors required for B-cell-type-specific expression. Mol Cell Biol. 1994;14:871–879. doi: 10.1128/mcb.14.2.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh S-Y, Tsai M-J. Cell-specific and ubiquitous factors are responsible for the enhancer activity of the rat insulin II gene. J Biol Chem. 1991;266:16708–16714. [PubMed] [Google Scholar]
- Sosa-Pineda B. The gene Pax4 is an essential regulator of pancreatic beta-cell development. Mol Cells. 2004;18(3):289–294. [PubMed] [Google Scholar]
- Thorens B, Weir GC, Leahy JL, Lodish HF, Bonner-Weir S. Reduced expression of the liver/beta-cell glucose transporter isoform in glucose-insensitive pancreatic beta cells of diabetic rats. Proc Natl Acad Sci USA. 1990;87:6492–6496. doi: 10.1073/pnas.87.17.6492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Elghazi L, Parker SE, Kizilocak H, Asano M, Sussel L, Sosa-Pineda B. The concerted activities of Pax4 and Nkx2.2 are essential to initiate pancreatic beta-cell differentiation. Dev Biol. 2004;266:178–189. doi: 10.1016/j.ydbio.2003.10.018. [DOI] [PubMed] [Google Scholar]
- Zhang C, Moriguchi T, Kajihara M, Esaki R, Harada A, Shimohata H, Oishi H, Hamada M, Morito N, Hasegawa K, Kudo T, Engel JD, Yamamoto M, Takahashi S. MafA is a key regulator of glucose-stimulated insulin secretion. Mol Cell Biol. 2005;25:4969–4976. doi: 10.1128/MCB.25.12.4969-4976.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Fujitani Y, Wright CV, Gannon M. Efficient recombination in pancreatic islets by a tamoxifen-inducible Cre-recombinase. Genesis. 2005;42:210–217. doi: 10.1002/gene.20137. [DOI] [PubMed] [Google Scholar]
- Zhao L, Guo M, Matsuoka TA, Hagman DK, Parazzoli SD, Poitout V, Stein R. The islet beta cell-enriched MafA activator is a key regulator of insulin gene transcription. J Biol Chem. 2005;280:11887–11894. doi: 10.1074/jbc.M409475200. [DOI] [PubMed] [Google Scholar]